Abstract
The rodent genus Peromyscus is the most numerous and species rich mammalian group in North America. The naturally occurring diversity within this genus allows opportunities to investigate the genetic basis of adaptation, monogamy, behavioral and physiological phenotypes, growth control, genomic imprinting, and disease processes. Increased genomic resources including a high quality genetic map are needed to capitalize on these opportunities. We produced interspecific hybrids between the prairie deer mouse (Peromyscus maniculatus bairdii) and the oldfield mouse (Peromyscus polionotus) and scored meiotic recombination events in backcross progeny. A genetic map was contructed by genotyping of backcross progeny at 185 gene-based and 155 microsatellite markers representing all autosomes and the X chromosome. Comparison of the constructed genetic map with the molecular maps of Mus and Rattus and consideration of previous results from interspecific reciprocal whole chromosome painting allowed most linkage groups to be unambiguously assigned to specific Peromyscus chromosomes. Based on genomic comparisons, this Peromyscus genetic map covers approximately 83% of the Rattus genome and 79% of the Mus genome. This map supports previous results that the Peromyscus genome is more similar to Rattus than Mus. For example, coverage of the 20 Rattus autosomes and the X chromosome is accomplished with only 28 segments of the Peromyscus map, but coverage of the 19 Mus autosomes and the X chromosome requires 40 chromosomal segments of the Peromyscus map. Furthermore, a single Peromyscus linkage group corresponds to about 91% of the rat and only 76% of the mouse X chromosomes.
Introduction
Members of the genus Peromyscus constitute the most numerous and species rich group of mammals in North America. The best known members of the genus are the deer mouse (P. maniculatus) and the white-footed mouse (P. leucopus). Peromyscus species have an appearance superficially similar to old world mice (e.g., species of the genera Mus or Apodemus), but represent a distantly related lineage. Mus and Rattus diverged approximately 10-12 million years ago, while their most recent common ancestor with the deer mouse was ca. 25-40 million years ago (Steppan et al. 2004).
Basic research of Peromyscus biology covers a variety of disciplines. Peromyscus occur in a wide range of habitats including sea-level wetlands, beaches, forests, deserts, and elevations up to 14,000 feet. Thus, these animals are ideal for studies on the genetic basis of adaptation to different environmental conditions, such as the genetic basis of coat color (Hoekstra et al. 2006; Linnen et al. 2009) and adaptation to life at high altitudes (Storz et al. 2011; Storz et al. 2010; Storz et al. 2009). Their ubiquitous presence in North America also means that Peromyscus are found at many sites contaminated with toxic chemicals and may be useful as biomarkers of contamination. Peromyscus have been employed in numerous studies on the responses to chemicals, such as PCBs (Voltura and French 2007) and Aroclor 1254 (Wu et al. 1999) as well as being used to demonstrate transgenerational effects of BPA exposure (Jasarevic et al. 2011).
Peromyscus species carry several pathogens important to public health. For example, P. maniculatus acts as the primary carrier of Sin Nombre virus responsible for hantaviral pulmonary syndrome (Hjelle et al. 1995; Nichol et al. 1993). P. leucopus plays a role in the transmission of Lyme disease, both harboring the spirochaete Borrelia burgdorferi and acting as the host species for the larval stage of the tick which transmits the bacteria to humans (Anderson et al. 1987; Magnarelli et al. 1988) .
Peromyscus has also been used to study behavioral and environmental effects on physiology and endocrinology (Bester-Meredith and Marler 2003; Martin et al. 2008a; Pyter et al. 2006; Trainor et al. 2007; Trainor et al. 2010) as well as immunology (Martin et al. 2007; Martin et al. 2008b; Pyter et al. 2005). Other phenotypic differences could inform our understanding of repetitive movements common in obsessive-compulsive disorder (Korff et al. 2008; Tanimura et al. 2010), while still others could address the relationship between blood glucose regulation and stress (Oriel et al. 2008). Peromyscus is also a potentially useful model for studying the genetic bass of monogamy and polygamy (Foltz 1981; Ribble 1991; Turner et al. 2010). Perhaps most useful of all is the ability of some species of Peromyscus to hybridize in the laboratory. This ability has allowed development of unique mammalian models of hybrid dysgenesis (including severe developmental defects) and epigenetic misregulation (Duselis and Vrana 2010; Vrana et al. 1998; Vrana et al. 2013)
Despite the wide range of habitat types in which Peromyscus occur, members of the genus are easily reared under standard Mus husbandry conditions (Dewey and Dawson 2001), thus making it possible to determine the genetic basis of naturally occurring variants through the use of controlled crosses in the laboratory. Two species are of particular importance in genetic analysis: the prairie deer mouse, P. maniculatus bairdii, and the oldfield mouse, P. polionotus. Hybrids produced by mating P. maniculatus females with P. polionotus males are viable and fertile, thus allowing genetic analysis of important phenotypic differences between the species and natural variants within these species. These species differ markedly in many characteristics where variation is not found in other well-established mammalian models such as Mus and Rattus. Although extensive genetic and genomic tools are available for Mus and Rattus, Peromyscus genomic resources are still being developed to enhance the utility of Peromyscus as an experimental system. Approximately 79,000 P. maniculatus expressed sequence tags (ESTs) from various tissues have deposited in GenBank. Approximately 2900 ESTs from testes and placenta have been more thoroughly analyzed with many assigned to Mus or Rattus chromosomes or a biological function (Glenn et al. 2008). Additional transcriptome sequencing has been conducted (e.g., Genbank accessions GH457086.1 - GH545901.1), but the summaries have not yet been published. Whole genome sequencing of P. maniculatus, P. polionotus, P. leucopus and P. californicus has been completed, and assembly of the genomes is underway (https://www.hgsc.bcm.edu/content/Peromyscus-genome-project).
Even with the nascent genomic resources, the utility of Peromyscus as a model for modern genetic research will be significantly enhanced by a linkage map with the position of markers on chromosomes derived from traditional genetic mapping methods. A linkage map of Peromyscus will facilitate genome assembly and increase researchers’ ability to identify genes associated with phenotypic variation and/or involved in important biological processes.
Two nascent genetic maps of Peromyscus have already been constructed, however, there is little useful overlap between the two. Nearly 80 genes have been mapped and assigned to several linkage groups (Ramsdell et al. 2008) in which comparative linkage analysis revealed greater synteny and gene order conservation between Peromyscus and Rattus than between Peromyscus and Mus. A second genetic map was produced by (Steiner et al. 2007) using ca. 120 markers, mostly microsatellites. This map focused on the location of genes controlling adaptive coat color in the light-pigmented beach mice, P. polionotus leucocephalus. Although useful in their own right, the small amount of overlap in genetic markers used in the two studies allows little ability to merge the two genetic linkage maps into one with greater marker density.
Here we report on the development of a marker-dense genetic map suitable for a great diversity of studies. By increasing the genomic resources available for this genus, the unique and significant phenotypic differences found within Peromyscus may finally make discovering the genetic bases of these phenotypes tractable. A genetic map, therefore, is essential for further development of these animals as a model system and to exploit their large potential as a mammalian small animal research model.
Materials and methods
Animals used in this study were from the P. maniculatus (BW) and P. polionotus (PO) stocks maintained at the Peromyscus Genetic Stock Center, University of South Carolina (http://stkctr.biol.sc.edu). Animal care is provided by the University of South Carolina Animal Care Facility which is fully accredited by American Laboratory Animal Care Committee. The research described here was approved by the University of South Carolina IACUC.
Development of backcross panels for genetic mapping
The mapping of autosomal genes was done using a P. maniculatus × (P. maniculatus × P. polionotus) F1 backcross panel. Twenty-two progeny from each of four different backcross families were used for analysis (i.e. for a total of 88 animals). Mapping of X-linked loci was done using a panel of offspring from a backcross of (P. maniculatus × P. polionotus) F1 × P. polionotus. This panel consisted of 145 animals from ten different matings (families).
DNA from liver tissue of all parents and backcross progeny was isolated using standard SDS/proteinase K and phenol/chloroform methods (Maniatis et al. 1982) or via the Qiagen DNeasy Blood & Tissue Kit. DNA concentration and quality were assessed by using ratio of absorbance at 260 and 280 nm.
Development of gene based PCR marker assays
Primers were developed from P. maniculatus EST sequences (Glenn et al. 2008) as previously described (Ramsdell et al. 2008). Selected genes were also targeted by designing primers based on conserved mammalian sequences as previously described (Loschiavo et al. 2007; Ramsdell et al. 2008)
PCR methods
Qiagen HotStarTaq and HotStarTaqPlus kits were used for amplification of specific DNA sequences for individual genes. Conditions were as suggested by the supplier with some optimization for different genes. The PCR cycling conditions for HotStarTaqPlus (HSP) were 95°C for 5 min followed by 35 cycles of: 95°C for 30s, annealing for 30s at temperatures between 48-65°C depending on the gene, 72°C for 30s per 0.5kb, 72°C for 10 min, 4°C hold. HotStarTaq (HS) used 14.5 min for the initial denaturation at 95°C. The same basic conditions were used for Touchdown PCR and were as follows: (1) TD65HSP used 20 cycles with annealing at 65°C for 30s minus 0.5 °C/cycle followed by 20 cycles with annealing at 55°C; (2) TD60HSP and TD55HSP used starting annealing temperatures of 60°C and 55°, respectively, and ramped down over 20 cycles by 10°C followed by 20 cycles at the lower temperature. HotStarTaq was also used in touchdown protocols with just the longer initial denaturation temperature and are referred to as TD65HS, TD60HS or TD55HS. Amplification of some DNA sequences required additional MgCl2 or Qiagen Q-solution. Some loci were amplified using Invitrogen Taq Polymerase with the suggested methods; this was the method used when only annealing temperature is given in Appendix 1. Amplification products were digested with the appropriate restriction endonuclease and digestion products were resolved on 2% agarose or 7.5 polyacrylamide gels, depending on the size of fragments to be resolved.
Microsatellite genotyping
Primers and amplification methods of microsatellites were as previously described (Weber et al. 2010). Genotyping of some loci was done by analyzing amplification products on agarose or polyacrylamide gels of appropriate concentration for the size of the amplification product.
Screening of BAC library
The Peromyscus maniculatus rufinus BAC library CHORI-233 was obtained from the BACPAC Resources of the Children Hospital Oakland Research Institute (http://bacpac.chori.org/library.php?id=280). Specific primers (see Table 1) were used to produce DNA fragments for the genes Pacrg, Hba, and Mpo. The fragments were gel purified, radiolabeled by random priming (Manufacturer’s instructions) and used all together to probe the 5 filters representing the library. Clones identified by hybridization were obtained from BACPAC resources, and the BACs were purified and rescreened by Southern blotting methods using the individual probes.
Table 1.
Synteny Conservation between the Peromyscus Genetic Map and Rattus and Mus Genomes.
| Rattus | Mus | |||||
|---|---|---|---|---|---|---|
| Segments | Total Length1 |
Number | Average Length |
Total Length |
Number | Average Length |
| Syntenic-12 | 727.2 | 11 | 66.1 | 698.1 | 18 | 38.8 |
| Syntenic-23 | 980.1 | 13 | 75.4 | 687.5 | 16 | 43 |
| Syntenic-34 | 540.4 | 4 | 135.1 | 687.9 | 6 | 114.7 |
| Total | 2247.7 | 2073.5 | ||||
| Genome Size | 2719 | 2639 | ||||
| Coverage (%) | 82.6 | 78.5 | ||||
Length is in Mb
Syntenic-1 is the length of all segments of the molecular maps of Rattus or Mus having genes in the same order as the Peromyscus genetic map.
Syntenic-2 is the molecular length of segments in the Rattus or Mus genome having synteny conservation with the Peromyscus genetic map but gene order is altered by one inversion and/or insertion.
Syntenic-3 is the length of syntenic segments conserved but with gene order differing from the Peromyscus genetic map by two or more inversions and/or insertions.
Fluorescence in situ hybridization
Confluent cell cultures of Peromyscus maniculatus fibroblasts were treated with colcemid, harvested by centrifugation, resuspended in hypotonic solution and washed with 3:1 methanol:acetic acid. Chromosomes were spread on dry, acetone cleaned slides per standard protocols, air-dried overnight, and dehydrated in an ethanol series.
Chromosome paint FISH was performed as previously described (Mlynarski et al. 2008). BAC DNA probes were labeled with fluorescein-12-dUTP (green), Texas Red-5-dUTP (red) or Diethylaminocoumarin-5-dUTP (aqua) by nick-translation per manufacturer’s protocol (Roche 11745808910). 50 ng of labeled BAC DNA with 1 μg of Cot1 in Hybrisol VII (Qbiogene) was hybridized to denatured slides under a sealed coverslip at 37° C in a humid chamber for 20 hours. Slides were washed at 72° C in 0.4XSSC and subsequently counterstained with a 1:3 dilution of DAPI in Vectashield (Vector Laboratories). Images were captured on an Olympus AX70 fluorescence microscope equipped with appropriate filters and a CCD camera. Images were analyzed with Genus (Applied Imaging). Chromosome identification was made by examining inverted DAPI banding of probe hybridizations and confirmed by FISH (for all but BAC 402) using flow-sorted chromosome paints.
Construction of genetic linkage maps
Linkage maps were created using JoinMap 4 (Kyazma B.V., Wageningen, Netherlands). The mapping population included 86 individuals in four families genotyped at 340 polymorphic loci resulting from a backcross (described above). Linkage groups were determined by a LOD score of 4.0. Regression maps were created from the linkage groups using Kosambi’s function. Identical methods were used to map X-linked genes in the 145 backcross progeny derived from the (P. maniculatus × P.polionotus)F1 × P. polionotus cross.
Assignment of linkage groups to specific chromosomes
The linkage groups were assigned to specific Peromyscus chromosomes using the results of reciprocal hybridization of isolated whole Mus and Rattus chromosomes with Peromyscus chromosome spreads with visualization by FISH (Mlynarski et al. 2008). Comparisons of the Peromyscus genetic map with the Mus and Rattus molecular maps (Sinha and Meller 2007) also aided in assignment. The standardized Peromyscus cytogenetic nomenclature (Greenbaum et al. 1994) was used for identification of Peromyscus chromosomes.
Results
Development of gene-based markers
The genes for which methods have been developed for genotyping of allelic variants are listed in Appendix 1. Also given are the primers used for amplification, the PCR conditions for production of the DNA amplification product, and the restriction enzyme used to differentiate BW and PO allelic products. Few BW and PO initial amplification products differ in size and those are listed in the table along with the method of resolution by either agarose or acrylamide gel electrophoresis.
Peromyscus genetic map covering all autosomes and the X-Chromosome
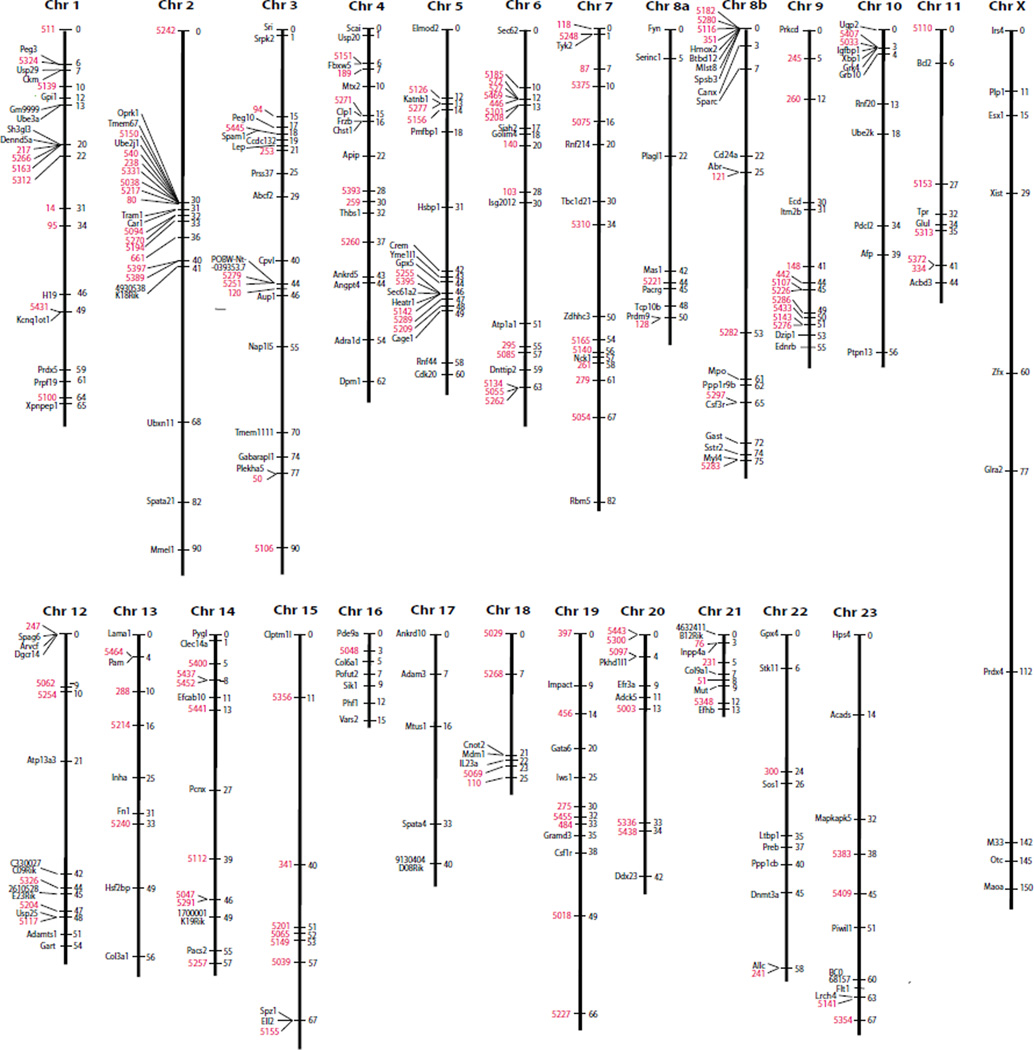
A genetic map of the Peromyscus genome based upon recombination frequency between markers among the backcross progeny is shown in Fig. 1. The genetic map shows the locations of 185 gene and 155 microsatellite loci for a total of 340 markers.
Figure 1. Peromyscus genetic linkage map.
Type I genetic markers are shown in black and microsatellite loci are indicated in red. The linkage groups are associated with the Peromyscus chromosomes according to the standardized nomenclature of the karyotype. Two linkage groups are assigned to chromosome 8.
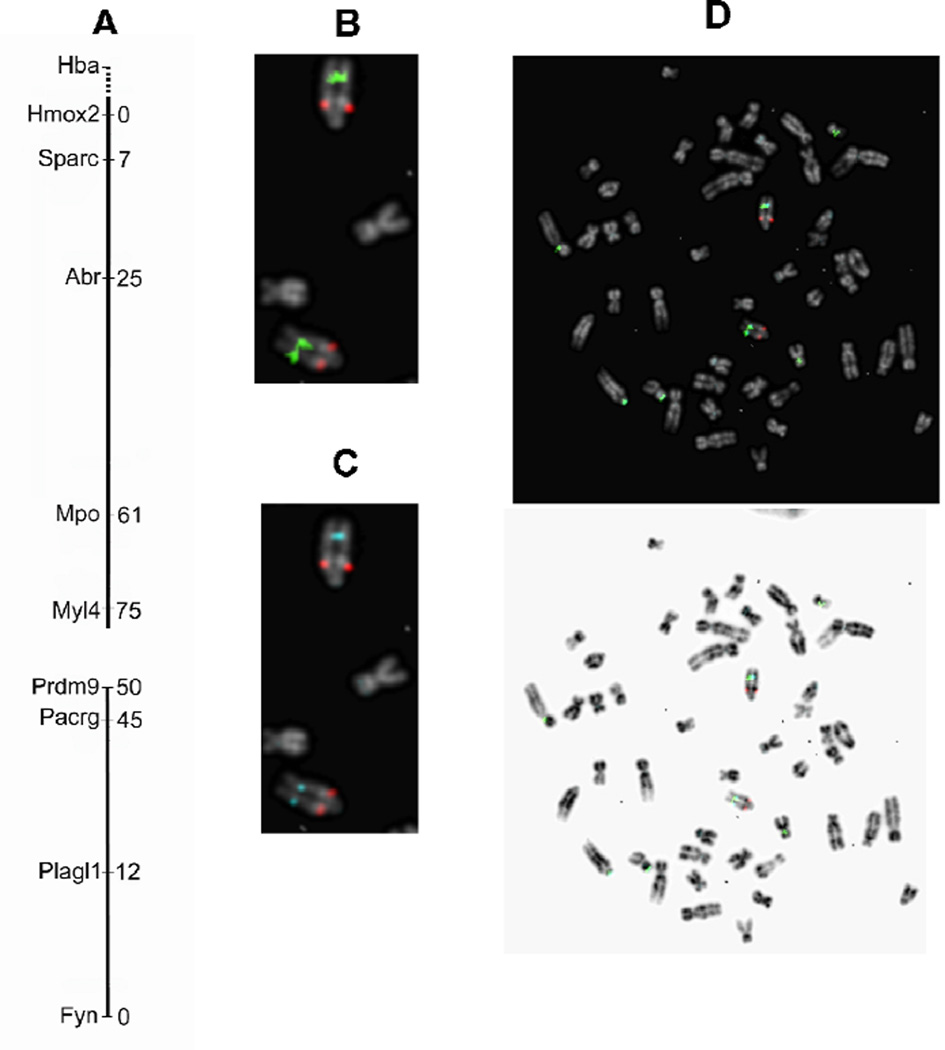
Initially, one more autosomal linkage group was detected than there are autosomes in Peromyscus. Two linkage groups are associated with Peromyscus chromosome 8. Confirmation of this association was obtained from FISH analysis using labeled BAC chromosomes harboring specific genes on the two linkage groups (Fig. 2). The P. maniculatus CHORI-233 BAC clones which harbor the Hba, Pacrg and Mpo genes are 501B24, 396H17 and 312P13, respectively. Hba was not placed on the genetic map using this backcross panel data although it was very loosely associated with Canx on chromosome 8b. Thus, Hba was used as a marker possibly located near the end of the linkage group assigned to chromosome 8a. The previous linkage group constructed using the female as the hybrid animal did show linkage between Hba and Canx on chromosome 8 (Ramsdell et al. 2006).
Figure 2. Linkage groups 8a and 8b are both located on Peromyscus Chromosome 8.
(A). The two linkage groups are aligned according to their location on Chromosome 8. BAC clones containing different genes were fluorescently labeled as described in Materials and Methods. (B). FISH to Peromyscus metaphase chromosomes with BACs harboring the Hba (red) and the Pacrg (green) genes. (C). Results of hybridization with BACs harboring the Hba (red) and Mpo (aqua) genes. (D). Merged hybridization results showing that the Pacrg and Mpo genes are close together while Hba is more proximal to the centromere of Chromosome 8.
The orientation of the two linkage groups on chromosome 8 as suggested by FISH analysis is shown in Fig. 2A. The FISH analysis using the Hba-harboring BAC clone (labeled red), and the Pacrg-harboring BAC clone (labeled green) is shown in Fig. 2B. As can be seen, the Hba signal is located proximal to the centromere whereas the Pacrg gene is located more distally. The location of Hba and Mpo (aqua labeled) determined by FISH is shown in Fig. 2C, and Mpo and Pacrg are very close together on the chromosome. This proximity is confirmed in Fig.2D where both green and aqua are visualized and their position overlaps. From these data, we infer that the two linkage groups are in the order shown in Fig. 2A.
Comparative analysis of linkage relationships between Peromyscus, Rattus and Mus
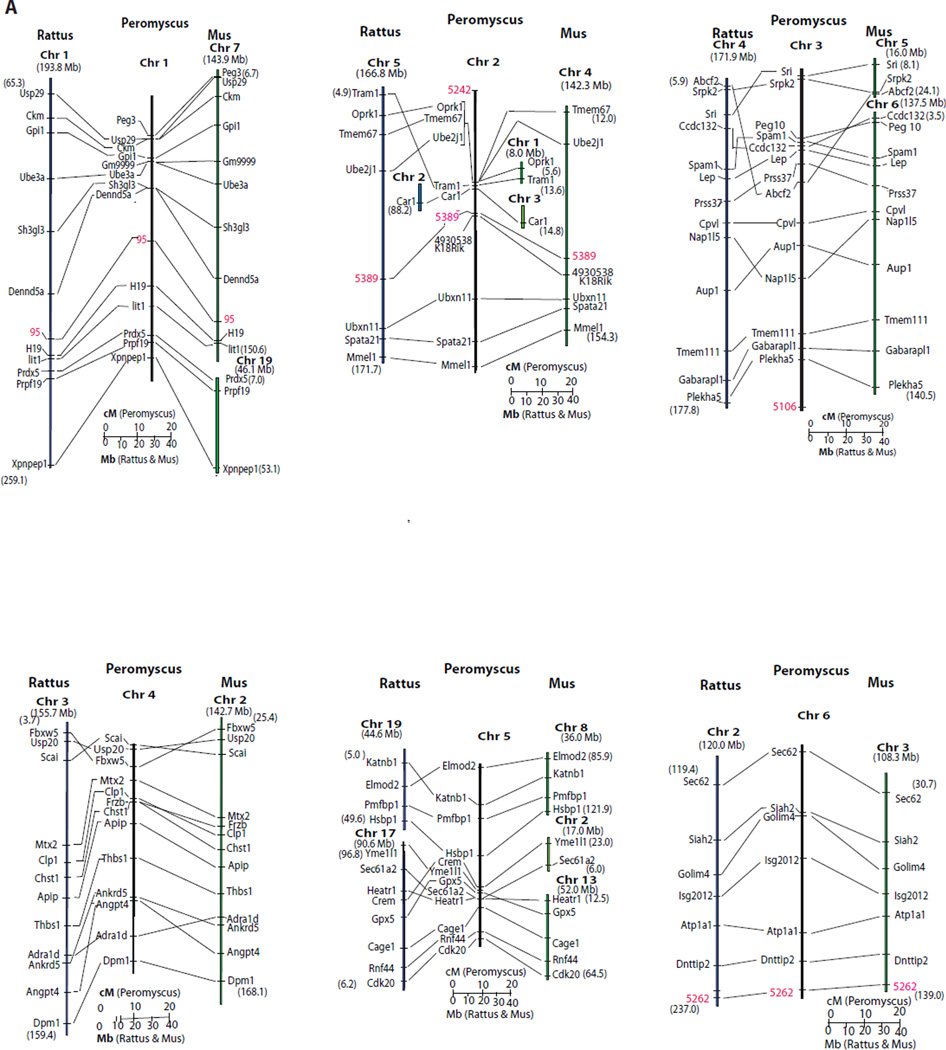
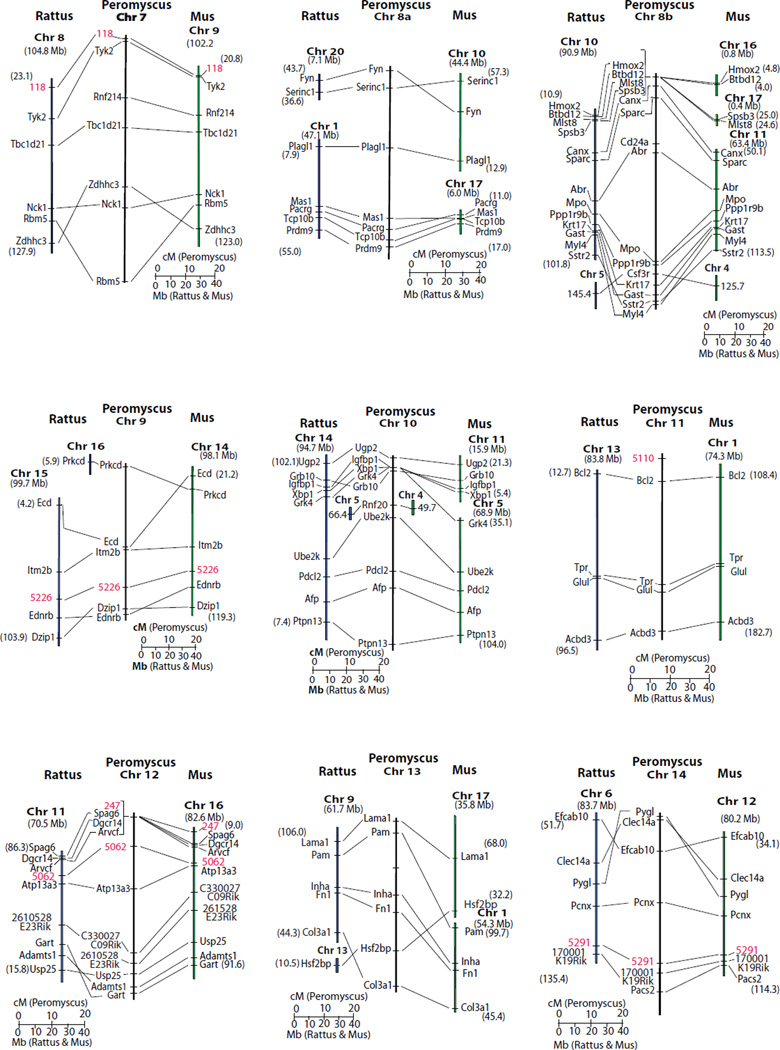
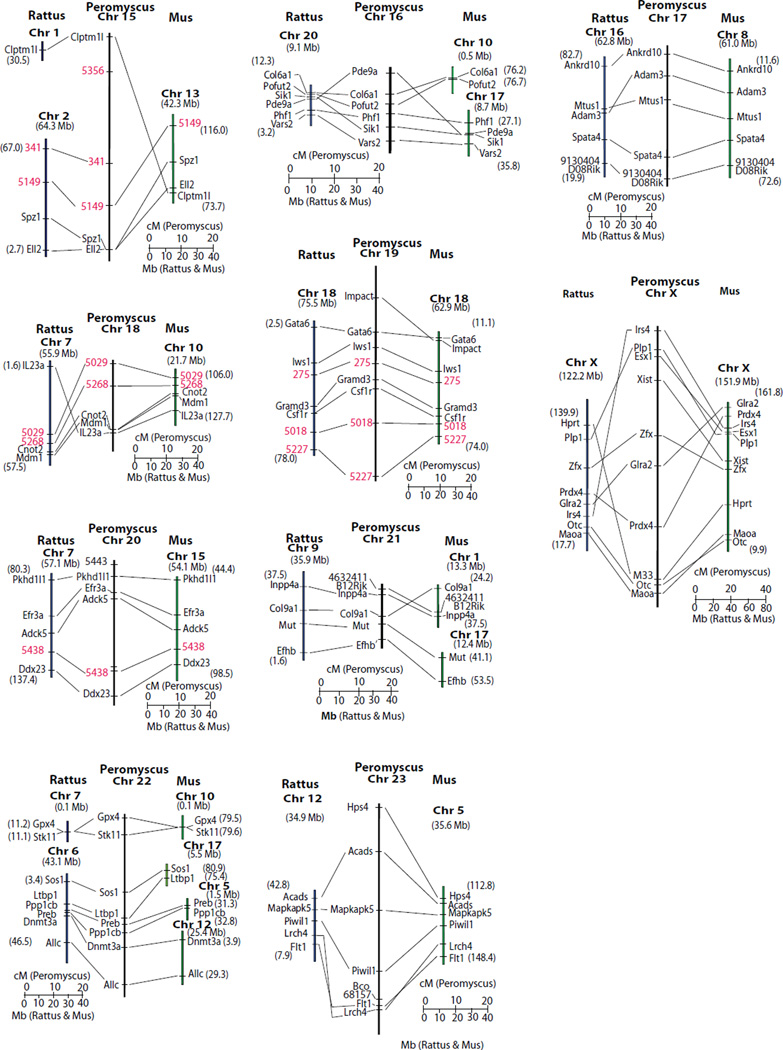
The Peromyscus genetic linkage map is compared to the Rattus and Mus molecular maps in Fig. 3. In a few instances, the order of genes not resolved by recombination events is inferred based upon the molecular order in the Rattus and Mus genomes (i.e. areas of Peromyscus chromosomes 1, 2, 8b, 10 and 12). Given these assumptions, synteny, as well as gene order, is conserved between chromosome 1 and much of Rattus Chr 1 and a significant portion of Mus Chr 7. Synteny, and gene order, is conserved between Peromyscus chromosomes 6, 11 and 20 and both Rattus and Mus corresponding genomic regions. Other regions of shared synteny are extensive but have single or multiple inversion and/or insertion differences between the Peromyscus map and the other two species.
Figure 3. Comparative relationships of the genetic map of Peromyscus with the molecular map of Rattus and Mus.
For Rattus and Mus segments the numbers in parentheses indicate the molecular position at the ends of the chromosomal segment. The Rattus and Mus chromosomes are indicated in bold. Molecular locations of Mus loci come from Genome Reference Consortium Mouse Build 38 (GRCm38) and Rattus gene positions are from RGSC Genome Assembly v3.4. Fig. 3A. Peromyscus chromosomes 1-6 compared to Mus and Rattus. Fig. 3B. Peromyscus chromosomes 7-14 compared to Mus and Rattus. Fig. 3C. Peromyscus chromosomes 14-23 and the X compared to Mus and Rattus. The M33 locus (Vrana et al. 2000) on the Peromyscus X chromosome is not orthologous to Hprt but its sequence is found very near this gene in both Mus and Rattus.
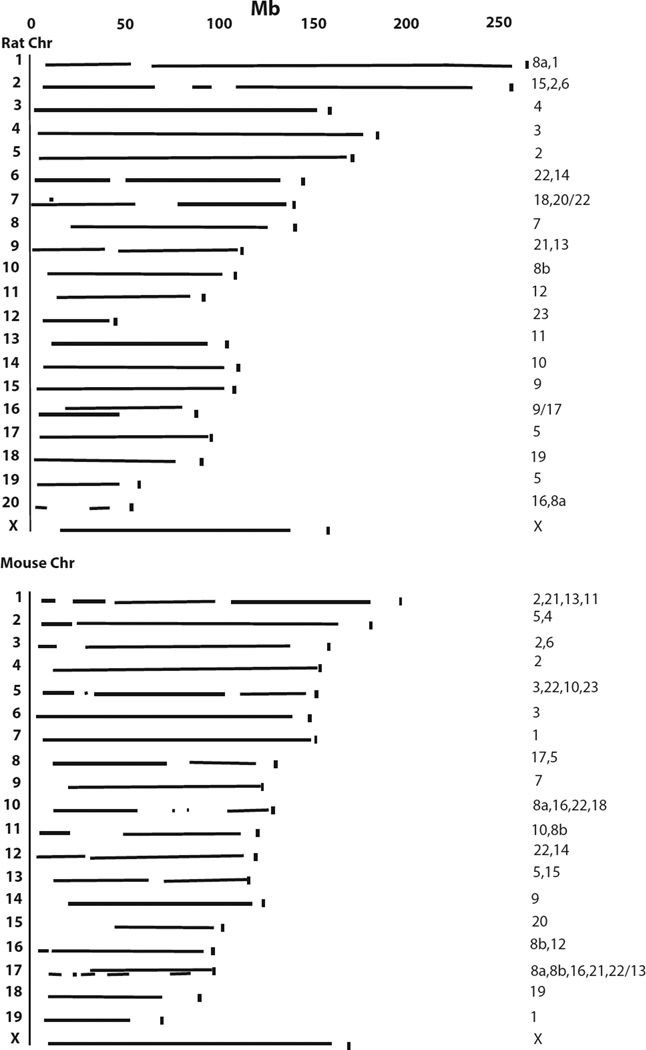
While the Peromyscus map contains regions of low marker density, the map size suggests it covers much of the genome based on comparisons to Rattus and Mus genomes (Fig.4). In addition, fewer and longer Peromyscus genetic map segments are involved in the coverage of the Rattus genome than the Mus genome.
Figure 4. Coverage of the Peromysus genetic map revealed by comparison to the Rattustus and Mus genomes.
The length of the corresponding Peromyscus mapped segments containing homologous Mus or Rattus chromosomes are indicated. The vertical mark at the end of each chromosome indicates the full length of the Rattus or Mus chromosome. The numbers at the end of the linkage segments are the Peromyscus chromosomal linkage group segments in order corresponding to the Rattus or Mus chromosomes. In cases of overlap of corresponding Peromyscus linkage segments (Rattus 7, 16 and Mus 17) the number(s) before the slash represent segment(s) on the bottom and those after the slash represent the segment(s) on the top.
A summary of those portions of the Peromyscus genetic map sharing synteny with Mus or Rattus is given in Table 1. Longer regions of shared synteny, including shared gene order, occur between Peromyscus and Rattus than occur with Mus (Table 1). Longer syntenic segments without complete gene co-linearity are also conserved between Peromyscus and Rattus than between Peromyscus and Mus. When compared to the Rattus and Mus molecular maps, the current Peromyscus genetic map has regions of synteny conservation and total size representing approximately 83% of the Rattus and 79% of the Mus genomes.
Assignment of linkage groups to Peromyscus chromosomes
The Peromyscus genetic map as linkage groups conditionally assigned to chromosomes, and orientation of the linkage group was possible on some Peromyscus chromosomes. The linkage groups on Peromyscus chromosomes 1, 5 and 10 are aligned with the short arm of the chromosome at the top of the illustrated linkage group based on correlation between the comparative map and hybridization to Mus isolated whole chromosomes. Peromyscus Chr 2 has a gap in hybridization to Mus Chr 4 possibly due to the insertions of Mus Chr1 and Mus Chr 3 homologous, short sequences revealed in the comparative map somewhere near the centromere region. This suggests this linkage group also is aligned with the short arm of Peromyscus Chr 2 at the top.
The complexity of Peromyscus Chr 8 is revealed by both comparative mapping and hybridization analysis. Hybridization of Mus whole chromosome probes correlates with the comparative mapping, strongly confirming the chromosome assignment of the linkage group. Only hybridization to Mus Chr 4 was not revealed previously (although this is based on a single marker). The longest regions of the rat genome containing orthologous genes mapped to Peromyscus chromosome 8 are on chromosomes 1 and 10; both these rat chromosomes also have sequences hybridizing to Peromyscus chromosome 8.
Linkage groups assigned to Chr 21 and Chr 22 are tentative. While Chr 21 has sequences homologous to Mus Chr 17, hybridization of Rattus Chr 9 was not detected to this chromosome. A similar situation holds for Peromyscus Chr 22, with Mus Chr 10 hybridizing but not the other chromosomes containing orthologous genes. Further, Rattus Chr 6 contains orthologous genes to Peromyscus Chr 22 but does not show hybridization. These inconsistencies are likely the result of the small size of these chromosomes.
All other Peromyscus chromosomes are assigned a single linkage group consistent with their hybridization to single Rattus and Mus chromosome regions that contain orthologous genes. Orientation of the linkage group on these chromosomes could not be determined. All X-linked genes identified in Peromyscus are also X-linked in both Mus and Rattus. Extensive inversion differences exist between the species, however.
Discussion
This medium density genetic map represents a major advance in Peromyscus laboratory genetics. The gene-based markers mapped here will provide important and useful landmarks in the genome, while the microsatellites will aid in fine-mapping of phenotypic traits.
The Peromyscus genetic map covers approximately 83% of the Rattus and 79 % of the Mus genomes. The entire Peromyscus genetic map presented here including the X chromosome is 1499 cM in length compared to 1445 cM for Mus (Cox et al. 2009) and 1503 cM for Rattus (Brown et al. 1998). Approximately 20 percent of the Peromyscus genome is likely not included in the current genetic map. Thus, identification and assignment of additional markers to the map would appear to make the Peromyscus linkage map larger than that of Mus or Rattus. A recent linkage map constructed for the prairie vole (Microtus ochrogaster) suggested to cover about 90% of the genome was 1707 cM in length, based on recombination analysis in both sexes (McGraw et al. 2011). Thus, the Peromyscus genetic map may be closer in size to that of the prairie vole.
There is confidence in the identity of synteny and homologous relationships between the Peromyscus linkage map and the genome of other species including assignment of linkage groups to chromosomes. This is primarily due to supporting FISH studies and the number of gene-based markers used in map construction. The FISH studies (Mlynarski et al. 2008) using both isolated chromosomes from Mus and Rattus as probes on Peromyscus chromosome spreads as well as the reciprocal were further supported by synteny relationships between Mus and Rattus at the molecular level (Sinha and Meller 2007).
However, minor differences are observed between results with the reciprocal chromosomal hybridization studies and older ones using only isolated chromosomes of a single species for hybridization analysis on Peromyscus chromosome spreads. For example, previous FISH analysis using only Mus chromosome paint probes indicated that Mus Chr 3 hybridized to two regions of Peromyscus Chr 3 (Dawson et al. 1999). However, the use of both Mus and Rattus chromosome probes suggests Mus Chr 3 and Rattus Chr 2 correspond to Peromyscus Chr 6 (Mlynarski et al. 2008) and large regions of synteny between Mus Chr 3 and Rattus Chr 2 exists at the molecular level (Sinha and Meller 2007) supporting this conclusion. The Peromyscus linkage group assigned to Chr 6 is syntenic with a long region of Rattus Chr 2 and Mus Chr 3.
Previous FISH analysis with three Mus paint probes that suggested hybridization of Mus Chr 7 and Chr 9 to Peromyscus Chr 1 and Chr 7, respectively (Dawson et al. 1999), are in complete agreement with the results using both Mus and Rattus chromosome probes (Mlynarski et al. 2008). However, most of Mus Chr 3 was presumed to hybridize to two regions of Peromyscus Chr 3 (Dawson et al. 1999), but the cross-species whole chromosome FISH using both Rattus and Mus isolated chromosomes as probes suggested Mus Chr 3 hybridizes entirely to Peromyscus Chr 6 as does Rattus Chr 2 (Mlynarski et al. 2008).
The use of both Mus and Rattus whole chromosome probes on Peromyscus chromosome spreads provides additional evidence of the homology relationships between Peromyscus Chr 6, Rattus Chr 2 and Mus Chr 3. Moreover, Peromyscus Chr 3 hybridizes entirely with Rattus Chr 4 and Mus Chr 6 to a considerable extent with only the proximal region of the long arm of Peromyscus Chr 3 hybridizing to Mus Chr 5. These results are supported by the synteny relationships between Mus and Rattus analyzed at the sequence level (Sinha and Meller 2007). Peromyscus chromosomes 3 and 6 are of similar overall size with similar centromere position and somewhat similar banding pattern (Greenbaum et al. 1994) possibly accounting for this discrepancy.
Despite attempts to unify chromosome 8 via additional markers predicted to map to this region, no linkage was obtained. One explanation for the anomalous chromosome 8 results is that this region contains a recombination hotspot, possibly induced by the inter-specific hybridization. Interestingly, the Prdm9 locus is located within this region. Prdm9 encodes a histone methyltransferase which has been implicated in determining recombination hotspots (Baudat et al. 2010) . Morever, Prdm9 has been implicated in reproductive isolation between Mus musculus musculus and Mus musculus domesticus.(Mihola et al. 2009). P. maniculatus and P. polionotus have been shown to differ in the number of zinc-finger domains encoded by this locus (Oliver et al. 2009) consistent with the hypothesis that Prdm9 plays a role in this phenomenon.
This study also offers insights into chromosomal evolution. For example, prior studies have indicated inversions in P. polionotus relative to P. maniculatus on chromosomes 5, 6 and 14 (Greenbaum and Baker 1978). Despite these inversions, there is no obvious evidence of inhibition of recombination as might be predicted. However, each of these chromosomes has substantial regions with a paucity of markers. Thus, increasing marker density may yet reveal regions of inhibited recombination. The nascent genome sequences of both BW and PO will aid in such efforts.
This combination of the genetic map and genomes will allow immediate genetic characterization and gene discovery of natural variants and mutants within the Peromyscus species complex, such as those maintained at the Peromyscus Genetic Stock Center (PGSC, http://stkctr.biol.sc.edu/). That is, rigorous positional cloning approaches are now feasible.
Future resource development will include greatly increasing the density of this map through traditional and newer genome wide approaches such as RAD-seq (Davey et al. 2011) which can be used for gene discovery and facilitating increased density of the genetic map. This approach has recently been used to identify regions in the genome controlling the complex behavior of nest-building in Peromyscus (Weber et al. 2013). The results suggested that at least three genetic regions on different linkage groups control this behavior which is consistent with the previous suggestion based on genetic analysis that two or more loci control this phenotype (Dawson et al. 1988). Other differences between these two species that might be similarly investigated include differences in repetitive behavior (Shorter et al., submitted), mating system, cholesterol levels (Wiedmeyer et al., submitted) as well as the aforementioned differences in genomic imprinting/growth control and glucose homeostasis.
Acknowledgements
This work was supported primarily by NIH GM069601 (MJD, TG), and also by NIH P40 OD010961 (MRF,GS) and NSF 0444165 (MRF, GS). We thank the Colony Manager of the Peromyscus Genetic Stock Center, Janet Crossland, for invaluable help in breeding and record keeping of the animals. We thank M. Peters, T. Tuberville, S. Lance, K. Jones, A. McKee for assistance in genotyping of microsatellite loci.
Appendix 1. Conditions for genotyping markers
| Gene | Forward Primer | Reverse Primer | PCR Conditiona | ENZYMEb |
|---|---|---|---|---|
| 1700001K19Rik | TGAAGGCATCTGGAGAGAG | TCTTCTGAGTCGCCATCATA | TD65HSP+2.0mM | HpyCH4IV |
| 2610528E23Rik | AGGTGTATCACACGCTTCTC | CAGGTCACTGACAGCATTTA | TD55HSP+0.5XQ | BsmAI |
| 4632411B12Rik | GAAAGATCAGAACCATCACT | GACTTAGGAGAACAGCTTGA | TD55HSP | HpyCH4III |
| 4930538K18Rik | AAAACACAGGAAGCGTAGG | ATGGCAATAGGGACGATGAC | TD65HSP | HphI |
| 5430411K18Rik | GTGGCTGGAACAGGTGTAC | GTCGTTCTGCTCCATCTGA | TD55HSP | MnlI |
| 9130404D08Rik | CCTGATCTGGGTCTGAGT | ATTCCAAAGCTCTGAGAAGT | TD55HSP | HpaII |
| Abcf2 | CTGCTCAGTGACACCAAACT | AACCCAATCCGTGTAAGATC | TD65HS+0.5XQ | 2%AG |
| Abr | CAAGAGAGGTCAGGAATG | GGACTGGAAACGGTACTT | TD55HSP+0.5XQ | AvaII |
| Acads | GAGCTGGCTGCCTCTGAG | GCTGGTGCCCTCGTAGAT | TD65HSP+0.5XQ | PvuII |
| Acbd3 | TGAACTCTTGCTGGGAACCAGGAT | TGCTGAGCTTCATTCAGTTGGAGC | 60HS+1XQ | SspI |
| Adam3 | ACTGGACATCGTATTTATCT | GGGTCCTAATTATTTGTGTA | TD65HSP | Ssp1 |
| Adamts1 | AAGAAACCAAAGCATTACAT | AAAGATTCTGCCCAGACTCT | TD55HSP+0.5XQ | MboI |
| Adck5 | GGTATGAGGAGGTGATGTCT | ATCGGCTCATAATCAAACTC | TD65HS+0.5XQ | MspI |
| Adra1d | CAACTATTTCATCGTGAACCTGG | TACACGCGGCAGTACATGAC | 54HS | BtgI |
| Afp | CAAGAAGACATCCCAACTAT | GACGTGTTTTTGCAGTTCT | TD65HSP+2.0mM | MspI |
| Allc | TTACTTCTCAGGGAGCTATG | CGCTAATGACTCCAAACTC | TD55HSP | AluI |
| Angpt4 | GCGACATGGACAATGATAAC | AGTGTTGATGGACTGGATAG | TD65HSP+2.0mM | MwoI |
| Ankrd10 | CTCCCAAAGATTCTGAGTCT | TACAGTCTGTTTCAGGTGAG | TD55HSP | NlaIV |
| Ankrd5 | ACAGAAGAAAAGAAAGTACA | GCAACTGCTATGACTTAGAG | TD65HSP+2.0mM | HaeIII |
| Apip | CATGACAGCAGCCTTAGAG | AAAAAAAAGCCAGTGTACT | TD55HSP+4mM | StyI |
| Arvcf | GACAATGAGGTCCGTGAG | ATGGGTCAGCGTCTGTAAG | TD65HSP | AvaI |
| Atp13a3 | ATCTACCTTTTTGACAAC | ACAGACCGATGACACTTA | TD55HSP | EcoRI |
| Atp1a1 | TTGCCTCCATTGTGACTG | CACTCTCAGCCTGTTCATAG | TD65HSP+2mM | BsaJI |
| Aup1 | GGAGCGGCTCTTTGACTC | GGTGCTACAGGTGGTGAG | TD65HSP+0.5XQ | Eco0109I |
| BC068157 | GGAGAGAGAAAGGATTTA | AGTCACAGCCCTTCTCTA | TD55HSP+2.0mM | ScrFI |
| Bcl2 | CACCCCTTCATCCAAGAATG | TACCCTGTTCTCCCTTG | HS58+1XQ | RsaI,12%AC |
| Btbd12 | CTGATGAAGTGGTCGAAGTT | CCGCTCCAGATTCAAGT | TD65HS | MboI |
| C330027C09Rik | AGTTCTTTGACAGCGATACT | GATGGTGAAGGATCAGATTA | TD65HSP | HindIII |
| Cage1 | TGGGGATATCTCAAGTCT | AGAATAACAAGTACCGAAGA | TD55HSP+4.0mM | MseI |
| Canx | TGTGGAATGCAGATGGGTGCTAGA | TGTTTGCTTCAAGGAGAACCCTGC | 60HS | BfaI |
| Car1 | CCTGGTCTTGATCTTGAACT | TATTTTGGCTCTCTGACTCA | TD65HS+0.5XQ | SfcI |
| Ccdc132 | GACAGTTAAAAACAGAGAAC | GGGATCTCATATCAATAAGA | TD55HS+1XQ | Cell IEXTRa |
| Cd24a | ATGCAAATCAATTCCATAAC | GAGAGCAGAATTACGTTGA | TD55HSP+0.5XQ | TaqI |
| Cdk20 | GCCTTTGAGTTCATGCTGTC | CAGGCCAAAGTCAGCTATCT | TD65HSP+2.0mM | NlaIII |
| Chst1 | CCATCACAGGACCAGTTA | ACAGCCTTCCAGGAACATT | TD55HSP+4mM | MboI |
| Ckm | AACACCCACAACAAGTTCAAGCTG | GTCTGGATGACATCGTCYASAGTG | TD55HSP+2.0mM | BstUI |
| Clec14a | ACTGGAAGCAAATAAATAAC | ACATTGGAACAGGAAGATAC | TD55HS+1XQ | NlaIII |
| Clp1 | TCCGAGGCTGCTTCTATC | GGTAGCACAGTGGAAGACAT | TD55HSP | AvaII |
| Clptml1 | GAATGTGATGGTGGATGACT | TATGGGGAGGTAGTGTACTG | TD65HS+0.5XQ | PstI |
| Cnot2 | TTATGTGGTCAGAAAATACA | CACAAAAGACTCAACACAAT | TD55HSP | DdeI |
| Col3a1 | GGACCAGGAAGTGATGGGAA | ACTTTCTCCTTGACTTCCCT | 60HS | 12%AC |
| Col6a1 | ATTGACATCCTCTTCGTGCT | CATCTGGTTGTGGCTGTACT | TD65HSP+0.5XQ | BstEII |
| Col9a1 | ATCAGGATTGGCCAAGATGA | GGAATCCTGAAGTCTACATT | 53HS+1XQ | RsaI |
| Cpvl | GGGCCTTATGTTATCACAAG | TAAAGCATGGAAAGGGTAGT | TD55HSP | HpaII |
| Crem | TTGCAGAGACAGATGATTCT | GAATNCCAGGCNCATCAGAG | TD65HSP+2.0mM | RsaI |
| Csf1r | GGGACAGCACGAGAATATAG | GTAGCAGCAGTATTCAGTGA | TD55HSP+2.0mM | DdeI |
| Csf3r | CTTTGCCACCAACATCTGG | GCTGGAAGGCTGATGTGAAG | TD55HSP | AvaII |
| Ddx23 | GCCGAACAAGACATCTCAAC | GTTCGCCTCTTTTCCATTAG | TD55HSP | MwoI |
| Dennd5a | TGTGAAGCACTTCCATAAAC | CACTCTCCACAAAGCAACAG | TD55HSP+2.0mM | AvaI |
| Dgcr14 | GTCGGGAGGGGATGTAGA | GAGGANAGCTGGCGGAAC | TD55HSP+0.5XQ | HinPI |
| Dnmt3a | TACAACAAGCAGCCCATGTACC | TCTCCTCTGCACCAAGAAGG | 59HS | BstU I |
| Dnttip2 | AAANTCAGGTGTAATGACAG | CATAGACCCTGGACTAAGTA | TD55HSP | AvaII |
| Dpm1 | TACCGAAAAGAAGTTCTAC | TGAAATGAACACGGTAAGT | TD55HSP | EarI |
| Dzip1 | GGGGATAAACCAGTTACT | TCCCGAGGGTTCAC | TD55HSP | 2%AG |
| Ecd | GGTGATGAAAGAGCACTAGAC | TGGTATCAGAAACAGGAAGTA | TD65HSP+1XQ | DraI |
| Ednrb | ATGACGCCACCCACTAAGAC | GATGATGCCTAGCACGAACA | 62; 2.0mM | Msp I |
| Efcab10 | AGTTATTCTATGTCCATCAT | AAAAAACCGAGAGAGTAT | TD55HSP | AvaII |
| Efhb | GCACTCAAAAGGTTCTGTTA | TGGAGCTTGAAATGAGACTA | TD55HSP | RsaI |
| Efr3a | ACAGGAAAAGGAAAAGAGAC | CCAATATTTGAGCAAGTCTA | TD55HS+1XQ | MnlI |
| Ell2 | AATCCGCCTCAGACTGTA | AGCGCAACCTTCTTTAACTC | TD65HSP+2.0mM | BglII;HindIII |
| Elmod2 | GTGAACTGGACAGATGTGTA | CCTGGTAGAGCTGCTTATAG | TD65HSP | RsaI |
| Esx1 | GTTTGGTTTCAGAACAGAAGG | GATATTTCCAAATAACCTCCACA | 52HS | MboI |
| Fbxw5 | CGGGGATGAGTTCTTAT | TGTAGCTCCAGTTGTATG | TD55HSP+0.5XQ | RsaI |
| Flt1 | CACAAAGATCCCAAAGAGA | AATGAAATGCTGGAGCTGTA | TD60HSP | EarI |
| Fn1 | AGAAGAGAGAGCCCCTGATTG | TGAAGATTGGGGTGTGGAAG | 54HSP | NcoI |
| Frzb | CTATGAAGAyGAGGAACGTTCCAG | CACTTCTCAGCTATAGAGCCTTC | 52; 2.0mM | MnlI |
| Fyn | TCTTTTTAGTTGAATCAGGT | CAGGTGGAGAGAGGTTACAG | TD55HSP | MnlI |
| Gabarapl1 | GAGCTGTAGGGGACCATAC | GGACGCTCTTCATTTGTAGA | TD55HSP | EarI |
| Gart | CAAGTAATAGCTAAAGGACA | GCTTTAGGACTCCAGGTATT | TD55HSP | HpyCH4V |
| Gast | CGCCTGCTCTGAAGCTTCTTG | GGCCGAAGTCCATCCATCC | 58HS | MseI,BsmAI |
| Gata6 | ATGGGTGTAAGAAGCATGTC | GTGGCAGAAACTGTGACAAT | TD55HSP+0.5XQ | TaqaI |
| Glra2 | TCAGGAAAATGCTGCCTCGT | ACACACCATCTCCATAAATACTGTAG | 59 | HpyCH4IV |
| Glul | AAGCAAGCGGCACCAGTACC | CACGTGCGGATGAGAGCTTCT | TD55HSP | NheI |
| Golim4 | TCCTGGTACTGTTCATCAAC | ATATAAACCCAGCAGATGAC | TD65HS+0.5XQ | HhaI |
| Gpi1 | GTGTGGACCACCAGACAGG | TTCCGTATGGGGTGCTGGGTC | TD55HSP+2.0mM | AciI |
| Gpx4 | GAATTCTCAGCCAAGGACAT | TTTTCATCCATTTCCACAGT | TD55HSP+0.5XQ | BsaJI |
| Gpx5 | CTGGAAATTAGGTACGTATC | AGAGCCAGGAGAAAATATAG | TD55HSP | StuI |
| Gramd3 | GGACATTTCTGAGACGTGAC | AAACCACCTCGGACAGA | TD55HSP+2.0mM | HpyCH4V |
| Grb10 | CATGCCAAATGAGAGTAA | GCAGGCACACATACAG | TD55HSP+0.5xQ | 2%AG |
| Grk4 | AGACCTAAAGCCAGAGAATA | GCTAGACCAAGATCTGAAAT | TD55HSP+2.0mM | SspI |
| H19 | GCACTAAGTCGTTTGCACTGG | CTGTTTCCAGACTAGGCGAG | 52 | AccI |
| Hba | CCCACCACCAAGACCTACTT | CGGTATTTGGAGGTCAGCAC | TD65HS | MspI |
| Heatr1 | GGGATTGATGGTCAGATCTA | CATCTAATGTTCTGGGGTAT | TD55HSP+1XQ | MseI |
| Hmox2 | GAAACATTAAGAAGGAGCTA | CTCATTCTGCCCTACATAGT | TD55HS | HinPI |
| Hps4 | ACCCCGATTTTCCTGAGT | CAGGGAGGGTTCTGAGAT | TD60HSP+0.5XQ | MboII |
| Hsbp1 | GTGACGGTTCCTAGACAT | GGACATGATCTGAAACTT | TD65HSP+0.5XQ | BfaI |
| Hsf2bp | GTATGCCTTCATGCCTTG | CCTGGTACTCAAGCACAC | TD65HSP | HaeIII |
| Hsp90b1 | GAATGACATCAAACCAATAT | TCCATATTCATCAAACAGAC | TD55HSP+2.5mM | EcoRI |
| Igfbp1 | TTTTACCTGCCAAACTGCAAC | GGTAGACGCACCAGCAGAG | TD55HSP | PvuII |
| Il23a | AGCCAGATCTGAGAAGCAGG | CTGCTCCRTGGGCAAAGACC | HS66 | MspI |
| Impact (#1) | TGGGTGAAAATATGGGCATT | AGGGCATGGACCACAGATAG | 49; 2.0mM | BfaI |
| Impact (#2) | CTGGTATGGAGGGATTCTGC | GCCCATATTTTCACCCAGTC | 49; 2.0mM | BssSI |
| Inha | TCAGCTCAGCTGTGGTTCC | CTCGTACTTGAAAGAGTAACCTCCA | TD55HSP | BssHII |
| Inpp4a | CAGCAGCTTCACAAACTG | CGTTGACGAGAGCTTGAC | TD65HSP+0.5XQ | DdeI |
| Irs4 | TAGGCTTGCTCCCGCTGGA | RAGCCCCGGCCATTTCCTGAGC | 64HS | Bme1580I |
| Isg20l2 | TTTCATCCCAAGTCTCTCAC | GCTAATTTTCTGGAGGATTC | TD65HSP | PstI |
| Itm2b | AGAAACCTGTTGGAGCTACT | GCTCATGGAGGAGGTAAG | TD65HSP+2.0mM | BamHI |
| Iws1 | TTGACAGTGGCGTGAT | ATTAATTTCCCTGCCATATC | TD55HSP | MboI |
| Katnb1 | AGGATGCCATGTCACAGATA | TTGCAGGAGTTTCTCAATCT | TD55HSP+0.5XQ | MspI |
| Lit1 | ATGTTGGAGGGAGGGGTATC | CCTTTCACAGGGGTCACCTA | TD55HSP+0.5xQ | HpyCH4III |
| Krt17 | GGAAGCCGACATCAATGG | TTCTTCAGGTAGGCCAGCTC | 52HS | MboI |
| Lama1 | GAGGGGTTTGCATCA | CCACAGGGTCACAGTTAC | HS59-64 | 2%AG |
| Lep | GACCTTAGCCCTGAATGCTG | TGCTTTGCTTCATATCCATCC | 52 | BsrBI |
| Lrch4 | ACCGAAGTCCAGTGCTAC | ACAACGGACTCTGGAGAG | TD65HSP | BccI |
| Ltbp1 | TAACACAGAGGGCTCTTACA | CCAGAGGACGGCTACA | TD55HSP | 2%AG |
| M33 | GCTCCCGTGTCATTTCTTTCAC | AGACAAGAGCAGTCATTCTGTCACC | 52 | length |
| Maoa | ATCAAGTGCATGGTGTATTACA | ATCTTCAATCATCATGCA | 52 | BstNI |
| Mapkapk5 | AGGAAACTGGACACTGTTAG | AAGAACGGTTTGCACTGA | TD55HSP | BsrI |
| Mas1 | GTGCACTGGGTCATYATGAGCAT | GCATTTCATCTTTGAAAGCCCTGGT | 62HSP | AvaII |
| Mdm1 | AAGTCCAAGGCAGACAGA | TTGTCTCAGCTTCGGAGTAG | TD65HSP | HinfI |
| Mlst8 | GGGCGGTAGCAGATGTAG | AGGCCCCACTGACAGTAAT | TD65HS | HphI |
| Mmel1 | CTTCAGAACATGGACAAATC | CGGTCTTTGACATCGGAATC | TD65HS+0.5XQ | HindIII |
| Mpo | CCACACCCTCATCCAACC | GTCCCCGGATCTCATCC | TD55HSP+2.0mM | PvuII |
| Mtus1 | GTAGATTCTCAAGCGACTTC | GCCCATGAAACCACTAA | TD65HSP | HaeIII |
| Mtx2 | TCTCTGAGAAAGGGCTTGA | GATGGGGGAACAAGACTCT | TD55HSP | BglII |
| Mut | TACTGTGGAAGAAAGCAATA | AGGGTTGTCTGAATCATAAC | TD55HS+1XQ | RsaI |
| Myl4 | AAGGAGCAGGGCACCTATG | GATGCAGCCATTGGCATC | 60HS | NheI |
| Nap1l5 | GCAGGATCTCTCTGGACCAGCTC | CCGTGTAGCTCCTGGATCTTGGC | 62 | BtsI |
| Nck1 | AGTATCACTAAAAGCACAAG | TNTCATACAGTACCAAACAG | TD55HSP/4.0mM | MboI |
| Oprk1 | CCTGGCTTTGGCRGATGCTTTRGT | TTCCCTGACTTTGGTGCCTCCAAG | 60HS | RsaI |
| Otc | ATGCTATGGCTATCAGCAGATCTGAAA | AGACAATCTTGTTCGAGTACTTCT | 55 | Fnu4HI |
| Pacrg | GAGGAAGAGCTGTACAAGTC | GACTACAGCCAGCAGAAGAG | TD65HSP+0.5XQ | Fnu4HI |
| Pacs2 | CACAATTCCAACCTTCACA | GGTTCATTCCCTTTGTTG | TD55HSP | AvaII |
| Pam | CATTCAGATTCCGAAAAATAGT | GAATCGGAGGAGGAGGAGTACT | TD55HSP+0.5XQ | HinFI |
| Pcnx | AGATTCTGGAAGGGATTAAC | TGACAGTCACGTATTTATCA | TD55HS+1XQ | MboI |
| Pdcl2 | TAAGAGATTTTGGCATTC | TCATCTTCTCATACGGTTTA | TD55HS+2.0mM | NlaIII |
| Pde9a | CTTCCAGTACCTGCTCTC | GGCTCTGTCTCTTGTGTC | TD65HSP+2.0mM | pFLMI |
| Peg3 | CTGGGTCAGCCATTCGATG | GCTCTGCAGCCTCCACTTC | TD55HSP+2.0mM | MspI |
| Peg10 | TGGTCAGTAGACGCATGAGC | CGCATGAAGCGCCTGGATCTTGGC | 59+2.0mM | HpyCH4III |
| Phf1 | GCTCCTGTGTTGTGTCTGTC | AGTCCATACGGCAGAGAGAG | TD65HSP | PstI |
| Piwil1 | GCATCGACTGTTACCATGAC | TCCACGGTCCTGAAAGAC | TD55HSP+0.5XQ | HpaI |
| Pkhd1l1 | CAATTCCATTTAGCAACTGT | CTGAAGGCTCTCGATTCT | TD65HSP+2.0mM | MnlI |
| Plagl1 | GTTCAACCGCAAGGACCAC | TCTTGGTGTGACGAGTGAGG | 54HSP+2.0mM | DdeI |
| Plekha5 | CCATCCCAGGATAATTGTAT | GGTCGCTCCTTCTTTTCTTC | TD55HSP | BfaI |
| Plp1 | GGTCCCAGAGAGAATTAGGCTAGA | GTACACAGGCGCAGCAGAGCA | 58HS | MseI |
| Pmfbp1 | AGGATGAAAAAGCTCAACAT | CAGCATCTTGAGGTCACTCT | TD55HSP | BsrI |
| Pofut2 | GGAGTCCACCTGAGAAGA | AAAAAAACCTGGCATGAG | TD55HSP+2.0mM | RsaI |
| Ppp1cb | GGAACAGATTCGGAGAA | AGCTCGACAAATCAAGTCTA | TD55HSP | PmlI |
| Ppp1r9b(#1) | GTCCGAGARAGAAGCAGAC | CGGAAGAAACTTCCTGTACTG | 62; 2.0mM | BclI |
| Ppp1r9b(#2) | GGA CCC CCT GGT CAAACG | GAC CCA ACC CTC TCCCGY GA | 62; 2.0mM | MseI |
| Prdm9 | CAAAGAACAAATGAGATCTGA | GTCTTYCTGTAATTGTTGAGATG | TD55HSP+0.5xQ | length |
| Prdx4 | TTYYGCGCTYGCKTGGTCATGGA | TCTTSGCTTTGCTTAGRTGCAG | 52HS | Fnu4HI |
| Prdx5 | GGATGATTCTTTGGTGTCTC | GGCAGACACAAATCAAGACT | TD65HS+0.5XQ | HpyCH4IV |
| Preb | ACCACTGCTACCTGAGTCTT | CTCTGTCGCCATCTACATAG | TD65HS+0.5XQ | DdeI |
| Prkcd | GGATGTGCAAAGAGAATATA | ATCCTGATGTTTCCTGTCAT | TD65HSP+2.0mM | DdeI |
| Prpf19 | GTCTCACCAAGGAAGTCACT | AGCCTGTGGTTTCAGAGTAG | TD65HS+0.5XQ | BsmAI |
| Prss37 | GAAATCTCAAAAGCAGAATC | CAGTCTTTGTCAGTCATCAC | TD55HS+1XQ | MboII |
| Ptpn13 | TTTTTCTTCTCTTTCCTCTG | AGTCCTCAAAGAGCAAATCT | TD65HSP+2.0mM | SmlI |
| Pygl | AAGAAAACCTTCGCCTATAC | ATCTTTGCTACCCCATTCAC | TD55HS+1XQ | NheI |
| GM9999 | GGC CTT CCA ATA ACCAC | CTGTGCAGTTGGGTGATCTT | TD65HSP | RsaI |
| Rbm5 | CTTCAACCTTTTTTTTTAAT | GTTGATGAAGAGGAAAACAG | TD55HSP+0.5XQ | AflII |
| Rnf20 | TTCATACTCTTTGCGGACCT | GAGGAAGTGGTTAAGGAGAC | TD65HS+0.5XQ | SacI |
| Rnf214 | AGCCTTCCTCAGATCCCTAC | AAAAGACAAAGAGCGAGGAT | TD55HSP | MboI |
| Rnf44 | ACATGATGCCACGAAGAC | GTTAGCCTTCAGCCACTT | TD65HSP+2.0mM | MseI |
| Scai | GCCTGAGTTGGTTGTTA | CAGTATTAAAGCGATGAGTA | TD55HSP | BsrI |
| Sec61a2 | TAAGACATGGATAGAAGTGT | AATTTTAGTTCTCTTGTGAA | 50HSP | Cell IEXTR |
| Sec62 | TAAAAGCCTGTTGCAGTAGT | GGTCATCGGGTTGATTAT | TD55HSP+2.0mM | MseI |
| Serinc1 | TGTTCTACCTTCTCCTCTCT | AAGAAAGCTCCAACAATAAT | TD65HSP+2.0mM | TaqI |
| Sh3gl3 | CGCGTTCACAGGCTTTAACT | TAGAGGCAGCATTGGACTAT | TD55HSP+O.5xQ | Cac8I |
| Siah2 | AATTTCTACATGCTGTCAGT | ATGTGTAAACAGGTCATCAC | TD55HSP+2.0mM | AvaI |
| Sik1 | GTTCCGCATCCCCTTCTT | CAAGGGCCTGCATGATAC | TD55HSP+0.5XQ | PstI |
| Sos1 | ATTCCAGAGCCTGAGCCAACAGA | CTCTTACTGTTCCAATAAATTCYTCCA | 62HS | Msp1 |
| Spag6 | AAGCAGAGATTTTCCCAGTT | CCTCCTGCGTTAACAATCAG | TD65+2.0mM | HphI |
| Spam1 | CCCAATTTTAAAGTCTCTTC | GCCTGTCACAATATTTTACA | TD55HSP | SfaNI |
| Sparc | CCCCAATGTTTAAAATGTTTGG | TTTCGTGTGTCTGTTACTTCCCT | TD55HSP+2.0mM | HhaI |
| Spata21 | CTCCAAGACCCGAGAGAC | GATCAGCACAGGATGGTATG | TD55HSP+2.0mM | MslI |
| Spata4 | TCTGTGAAATTGGGGTAGT | TTAAAAGTCAAGCTGGATAAC | TD55HSP+2.0mM | DdeI |
| Spsb3 | CAAGTGCTGCATAACAAG | ACCCACTGACATCATGTA | TD65HSP | BsmAI |
| Spz1 | TAGGTCGTCTGAGATGTCAC | AAGGGGNCAGTGATCCNATT | TD65HS+0.5XQ | Cell IEXTR |
| Sri | TGGAGACTTGTCGCCTTAT | TTCCACTCCTATCGCTGTC | TD55HSP | EarI |
| Srpk2 | GCATTCTGGGGAAGACTA | ATTGAAGAATTCCCGAGAA | TD55HS+2.5mM | Cell IEXTR |
| Sstr2 | CTCACCATCATCTGTCTTTGCTAC | TGGAGGTCTCCATTGAGGAG | TD55HSP | HhaI |
| Stk11 | GTGCTGTACAATGAGGAGAA | CTCAAGCAGAACTCCATCAC | TD55HS+1XQ | Cac8I |
| Tbc1d21 | CATGGGAACTTCACAGAGAC | CAGGGGATCTTTGTCATATA | TD55HSP | StyI |
| Tcp10b | AAAAATTCTACCCGGATGGC | GATGGTTTTGTCTCCGTTCC | 62hSP+0.2mM | 2%AG |
| Thbs1 | CTTTCACTTTGGGGCTTAGA | TCTCTATTCCAATGGCAATG | TD55HSP | NlaIII |
| Tmem111 | CTCCTGTCATCTGCTCCT | ATCCTGGTACTTCCTCAA | TD55HSP+0.5XQ | 2%AG |
| Tmem67 | CGCCTGTTCCATTATATC | GCTGTAAAACACCACCTTCT | TD55HSP | HpaI |
| Tpr | AGATCAAATGGAAAAAGAAA | TCCACTCCTTTTCTATCTCA | TD55HSP+2.0mM | RsaI |
| Tram1 | AATGGAGCGGTCACATC | TTCTGATTGCCAGTTGA | TD55HS+0.5XQ | BglII |
| Tyk2 | CAGAGGCAAGTGAATCTC | AGAGTCTTCCAGCGATTC | TD65HSP+0.5XQ | Fnu4HI |
| Ube2j1 | TAATGGACGATTTGAAGTAG | GCTCCCTCTCCTTTAGTT | TD55HSP+0.5XQ | 2%AG |
| Ube2k | TAAGAGATTTTGGCATTC | TCATCTTCTCATACGGTTTA | TD65HS | AleI |
| Ube3a | GCATAGTCCTCGGTCTG | ATCATACATCATTGGGTTAC | TD55HSP+2.0mM | BsaHI |
| Ubxn11 | GTGGAAAGGTGCTGAAGATC | GGAGATCATAGTGGAGACAC | TD65HS+0.5XQ | StyI |
| Ugp2 | TAACTCTGGCTTGCTCGACACCTT | TCCACCAGTCTCAGTTTGCCTTCA | 60HS+2.0mM | AvaII |
| Usp20 | TCTCGATGGGTTGGTTGT | CATGGTAGCCGCTGTG | TD55HSP | DdeI |
| Usp25 | CCTCTCCAGACTCAAACATC | TTCCTTATCTGTGCTTATCA | TD55HSP+4.0mM | 2%AG |
| Usp29 | TAGCTTAGTGCTTTGGTGTTGC | AGGGAGTTGACGTCAACGGAGT | 52HSP | HphI |
| Vars2 | GGATGCACATCATGAAGA | CTACAGCCCGCAGTATGT | TD65HSP | StyI |
| Xbp1 | TGGCCTTGTGATTGAGAACCAGGA | ACAGCGTCAGAATCCATGGGAAGA | TD65HSP | HindIII |
| Xist | CTCCCCCTTTAAGAGGGAACTTGA | TACCCGGAAGTGACCACAAT | 55 | BstNI |
| Xpnpep1 | TGGATTCGATGGCTCTG | ATGCTCTTCCGTGATGAT | TD55HSP | BfaI |
| Yme1l1 | ANATAACACAAGGAGCATTT | ATCAGAATTCGATGAGATGT | TD55HSP | MboII |
| Zdhhc3 | AACAACTGTGTCGGCNANAA | CAAGGAAATGAGAGCTATGT | TD55HSP | AvaI |
| Zfx | GCTCCGCAAGCTCTCAGGAAGT | AAGGAAAGGAGCATAAGTGATC | 52 | HaeII |
mM, concentration of magnesium chloride
CEL EXTR, Pimpkin M, Caretti E, Canutescu A, Yeung JB, Cohn H, Chen Y, Oleykowski C, Bellacosa, and Yeung AR. 2007. Recombinant nucleases CEL I from celery and SP I from spinach for mutation detection. BMC Biotechnology 7, 29.
AG, agarose; AC polyacrylamide
Contributor Information
Jane Kenney-Hunt, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA.
Adrienne Lewandowski, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA.
Travis C. Glenn, Savannah River Ecology Laboratory, University of Georgia, Aiken, SC 29801, USA
Julie L. Glenn, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA Savannah River Ecology Laboratory, University of Georgia, Aiken, SC 29801, USA.
Olga V. Tsyusko, Savannah River Ecology Laboratory, University of Georgia, Aiken, SC 29801, USA
Rachel J. O’Neill, Department of Molecular and Cell Biology, University of Connecticut, Storrs, CT 06269-2131, USA
Judy Brown, Department of Molecular and Cell Biology, University of Connecticut, Storrs, CT 06269-2131, USA.
Clifton M. Ramsdell, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA
Quang Nguyen, Department of Biological Chemistry, University of California, Irvine CA 92799, USA.
Tony Phan, Department of Biological Chemistry, University of California, Irvine CA 92799, USA.
Kimberly S. Shorter, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA
Michael J. Dewey, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA
Gabor Szalai, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA.
Paul B. Vrana, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA
Michael R. Felder, Department of Biological Sciences and Peromyscus Genetic Stock Center, University of South Carolina, Columbia, SC 29208, USA
References
- Anderson JF, Johnson RC, Magnarelli LA, Hyde FW, Andreadis TG. New infectious spirochete isolated from short-tailed shrews and white-footed mice. J. Clin. Microbiol. 1987;25:1490–1494. doi: 10.1128/jcm.25.8.1490-1494.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester-Meredith JK, Marler CA. The Association Between Male Offspring Aggression and Paternal and Maternal Behavior of Peromyscus Mice. Ethology. 2003;109:797–808. [Google Scholar]
- Brown DM, Matise TC, Koike G, Simon JS, Winer ES, Zangen S, McLaughlin MG, Shiozawa M, Atkinson OS, Hudson JR, Jr, Chakravarti A, Lander ES, Jacob HJ. An integrated genetic linkage map of the laboratory rat. Mamm Genome. 1998;9:521–530. doi: 10.1007/s003359900812. [DOI] [PubMed] [Google Scholar]
- Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, Wergedal JE, Bult C, Paigen B, Flint J, Tsaih SW, Churchill GA, Broman KW. A new standard genetic map for the laboratory mouse. Genetics. 2009;182:1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Reviews Cancer. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Dawson WD, Lake CE, Schumpert SS. Inheritance of burrow building in Peromyscus. Behavior Genetics. 1988;18:371–382. doi: 10.1007/BF01260937. [DOI] [PubMed] [Google Scholar]
- Dawson WD, Young SR, Wang Z, Liu LW, Greenbaum IF, Davis LM, Hall BK. Mus and Peromyscus chromosome homology established by FISH with three mouse paint probes. Mamm Genome. 1999;10:730–733. doi: 10.1007/s003359901080. [DOI] [PubMed] [Google Scholar]
- Duselis AR, Vrana PB. Aberrant Growth and Pattern Formation in Peromyscus Hybrid Placental Development. Biology of Reproduction. 2010;83:988–996. doi: 10.1095/biolreprod.110.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn JL, Chen CF, Lewandowski A, Cheng CH, Ramsdell CM, Bullard-Dillard R, Chen J, Dewey MJ, Glenn TC. Expressed sequence tags from Peromyscus testis and placenta tissue: analysis, annotation, and utility for mapping. BMC Genomics. 2008;9:300. doi: 10.1186/1471-2164-9-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum IF, Baker RJ. Determination of the primitive karyotype for Peromyscus. J. Mammalogy. 1978;59:820–834. [PubMed] [Google Scholar]
- Greenbaum IF, Gunn SJ, Smith SA, McAllister BF, Hale DW, Baker RJ, Engstrom MD, Hamilton MJ, Modi WS, Robbins LW, Rogers DS, Ward OG, WD D, Elder FFB, Lee MR, Pathak S, Stangle FBJ. Cytogenetic nomenclature of deer mice, Peromyscus (Rogentia): revision and review of the standardized karyotype. Cytogenet Cell Genet. 1994;66:181–195. doi: 10.1159/000133696. [DOI] [PubMed] [Google Scholar]
- Hjelle G, Krolikowski J, Torrez-Martinez N, Chavez-Giles F, Vanner C, Laposata E. Phylogenetically distinct hantavirus implicated in a case of hantavirus pulmonary syndrome in the northeastern United States. J. Med. Virol. 1995;46:21–27. doi: 10.1002/jmv.1890460106. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- Jasarevic E, Sieli PT, Twellman EE, Welsh THJ, Schachtman TR, Roberts RM, Geary DC, Rosenfeld CS. Disruption of adult expression of sexually selected traits by developmental exposure to bisphenol A. Proc Natl Acad Sci U S A. 2011;108:11715–11720. doi: 10.1073/pnas.1107958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korff S, Stein DJ, Harvey BH. Stereotypic behaviour in the deer mouse: pharmacological validation and relevance for obsessive compulsive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:348–355. doi: 10.1016/j.pnpbp.2007.08.032. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–1098. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschiavo M, Nguyen QK, Duselis AR, Vrana PB. Mapping and Identification of Candidate Loci Responsible for Peromyscus Hybrid Overgrowth. Mamm Genome. 2007;18:75–85. doi: 10.1007/s00335-006-0083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnarelli LA, Anderson JF, Hyland KE, Fish D, Mcaninch JB. Serologic analyses of Peromyscus leucopus, a rodent reservoir for Borrelia burgdorferi, in Northeastern United States. J. of Clinical Microbiology. 1988;26:1138–1141. doi: 10.1128/jcm.26.6.1138-1141.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Martin LB, Navara KJ, Weil ZM, Nelson RJ. Immunological memory is compromised by food restriction in deer mice Peromyscus maniculatus. American Journal of Physiology-Regulatory Integrative and Comparative Physiology. 2007;292:R316–R320. doi: 10.1152/ajpregu.00386.2006. [DOI] [PubMed] [Google Scholar]
- Martin LB, Weil ZM, Nelson RJ. Fever and sickness behaviour vary among congeneric rodents. Functional Ecology. 2008a;22:68–77. [Google Scholar]
- Martin LBn, Navara KJ, Bailey MT, Hutch CR, Powell ND, Sheridan JF, Nelson RJ. Food restriction compromises immune memory in deer mice (Peromyscus maniculatus) by reducing spleen-derived antibody-producing B cell numbers. Physiol Biochem Zool. 2008b;81:366–372. doi: 10.1086/587090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Davis JK, Young LJ, Thomas JW. A genetic linkage map and comparative mapping of the prairie vole (Microtus ochrogaster) genome. BMC Genet. 2011;12:60. doi: 10.1186/1471-2156-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vicek C, Schimenti JC, Foreijt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Mlynarski EE, Obergfell CJ, Rens W, O'Brien PC, Ramsdell CM, Dewey MJ, O'Neill MJ, O'Neill RJ. Peromyscus maniculatus--Mus musculus chromosome homology map derived from reciprocal cross species chromosome painting. Cytogenetic and Genome Research. 2008;121:288–292. doi: 10.1159/000138900. [DOI] [PubMed] [Google Scholar]
- Nichol ST, Spiropoulou CF, Morzunov SP, FRollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness (see comments) science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- Oliver PI, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriel RC, Wiley CD, Dewey MJ, Vrana PB. Adaptive Genetic Variation, Stress & Glucose Regulation. Disease Models and Mechanisms. 2008;1:255–263. doi: 10.1242/dmm.000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyter LM, Neigh GN, Nelson RJ. Social environment modulates photoperiodic immune and reproductive responses in adult male white-footed mice (Peromyscus leucopus) American Journal of Physiology-Regulatory Integrative and Comparative Physiology. 2005;288:R891–R896. doi: 10.1152/ajpregu.00680.2004. [DOI] [PubMed] [Google Scholar]
- Pyter LM, Trainor BC, Nelson RJ. Testosterone and photoperiod interact to affect spatial learning and memory in adult male white-footed mice (Peromyscus leucopus) European Journal of Neuroscience. 2006;23:3056–3062. doi: 10.1111/j.1460-9568.2006.04821.x. [DOI] [PubMed] [Google Scholar]
- Ramsdell CM, Lewandowski AA, Weston Glenn JL, Vrana PB, O'Neill RJ, Dewey MJ. Comparative genome mapping of the deer mouse (Peromyscus maniculatus) reveals greater similarity to rat (Rattus norvegicus) than to the lab mouse (Mus musculus) BMC Evolutionary Biology. 2008;8:65–78. doi: 10.1186/1471-2148-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdell CM, Thames EL, Weston JL, Dewey MJ. Development of a deer mouse whole-genome radiation hybrid panel and comparative mapping of Mus chromosome 11 loci. Mammalian Genome. 2006;17:37–48. doi: 10.1007/s00335-005-0051-x. [DOI] [PubMed] [Google Scholar]
- Sinha AU, Meller J. Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Biotechnology. 2007;8:9. doi: 10.1186/1471-2105-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 2007;5:1880–1889. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Natarajan C, Cheviron ZA, Hoffmann FG, Kelly JK. Altitudinal Variation at Duplicated β-Globin Genes in Deer Mice: Effects of Selection, Recombination, and Gene Conversion. Genetics. 2011 doi: 10.1534/genetics.111.134494. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Runck AM, Moriyama H, Weber RE, Fago A. Genetic differences in hemoglobin function between highland and lowland deer mice. J Exp Biol. 2010;213:2565–2574. doi: 10.1242/jeb.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, Moriyama H, Weber RE, Fago A. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci U S A. 2009;106:14450–14455. doi: 10.1073/pnas.0905224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura Y, Yang MC, Ottens AK, Lewis MH. Development and temporal organization of repetitive behavior in an animal model. Dev Psychobiol. 2010;52:813–824. doi: 10.1002/dev.20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor BC, Lin S, Finy MS, Rowland MR, Nelson RJ. Photoperiod reverses the effects of estrogens on male aggression via genomic and nongenomic pathways. Proc Natl Acad Sci U S A. 2007;104:9840–9845. doi: 10.1073/pnas.0701819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor BC, Takahashi EY, Silva AL, Crean KK, Hostetler C. Sex differences in hormonal responses to social conflict in the monogamous California mouse. Horm Behav. 2010;58:506–512. doi: 10.1016/j.yhbeh.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltura MB, French JBJ. Effects of dietary PCB exposure on reproduction in the white-footed mouse (Peromyscus leucopus) Arch Environ Contam Toxicol. 2007;52:264–269. doi: 10.1007/s00244-006-0045-z. [DOI] [PubMed] [Google Scholar]
- Vrana PB, Fossella JA, Matteson P, del Rio T, O'Neill MJ, Tilghman SM. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nature Genetics. 2000;25:120–124. doi: 10.1038/75518. [DOI] [PubMed] [Google Scholar]
- Vrana PB, Guan XJ, Ingram RS, Tilghman SM. Genomic imprinting is disrupted in interspecific Peromyscus hybrids. Nature Genetics. 1998;20:362–365. doi: 10.1038/3833. [DOI] [PubMed] [Google Scholar]
- Vrana PB, Shorter KR, Szalai G, Felder MR, Crossland JP, Veres M, Allen JE, Dewey MJ, Dawson WD. Peromyscus (Deer Mice) as Developmental Models. WIREs Developmental Biology. 2013 doi: 10.1002/wdev.132. in press. [DOI] [PubMed] [Google Scholar]
- Weber JN, Peters MB, Tsyusko OV, Linnen CR, Hagen C, Schable NA, Tuberville TD, McKee AM, Lance SL, Jones KL, Fisher HS, Dewey MJ, Hoekstra HE, Glenn TC. Five Hundred Microsatellite Loci for Peromyscus. Conserv Genet. 2010;11:1243–1246. doi: 10.1007/s10592-009-9941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JN, Peterson BK, Hoekstra HE. Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. Nature. 2013;493:402–406. doi: 10.1038/nature11816. [DOI] [PubMed] [Google Scholar]
- Wu PJ, Greeley EH, Hansen LG, Segre M. Immunological, hematological, and biochemical responses in immature white-footed mice following maternal Aroclor 1254 exposure: A possible bioindicator. Arch Environ Contam Toxicol. 1999;36:469–476. doi: 10.1007/pl00006620. [DOI] [PubMed] [Google Scholar]