Abstract
Genome-wide association studies have identified SNPs that are sensitive for tau or TDP-43 pathology in frontotemporal lobar degeneration (FTLD). Neuroimaging analyses have revealed distinct distributions of disease in FTLD patients with genetic mutations. However, genetic influences on neuroanatomical structure in sporadic FTLD have not been assessed. In this report we use novel multivariate tools, eigenanatomy and sparse canonical correlation analysis (SCCAN), to identify associations between SNPs and neuroanatomical structure in sporadic FTLD. MRI analyses revealed that rs8070723 (MAPT) was associated with grey matter variance in the temporal cortex. DTI analyses revealed that rs1768208 (MOBP), rs646776 (near SORT1) and rs5848 (PGRN) were associated with white matter variance in the midbrain and superior longitudinal fasciculus. In an independent autopsy series we observed that rs8070723 and rs1768208 conferred significant risk of tau pathology relative to TDP-43, and rs646776 conferred increased risk of TDP-43 pathology relative to tau. Identified brain regions and SNPs may help provide an in vivo screen for underlying pathology in FTLD and contribute to our understanding of sporadic FTLD.
Keywords: Frontotemporal lobar degeneration, Neuroimaging, Genetics, Biomarkers
1. Introduction
There is increasing neuroimaging evidence that genetic factors influence grey matter (GM) and white matter (WM) neuroanatomy in Alzheimer’s disease (AD) (Jahanshad et al., 2013; Shen et al., 2010). Genome-wide association (GWA) studies of autopsy-confirmed neurodegenerative disease cases have related several single nucleotide polymorphisms (SNPs) to the risk of accumulating specific types of histopathologic abnormality. In this study, we combine our knowledge of the genetic basis for frontotemporal lobar degeneration (FTLD)(DeJesus-Hernandez et al., 2011; Höglinger et al., 2011; Renton et al., 2011; Van Deerlin et al., 2010) with neuroimaging in an effort to identify novel genetic and neuroanatomic associations that may be used to improve the diagnostic accuracy of FTLD. We additionally introduce a data-driven technique for investigating genetic and neuroanatomic associations that provides a novel approach for biomarker discovery.
FTLD is a common cause of early-onset neurodegenerative dementia. About 20% of familial FTLD patients have a genetically-identified mutation (Wood et al., 2013). Autopsy studies of FTLD have demonstrated that the vast majority of patients have either tau inclusions (FTLD-tau) or a TDP-43 proteinopathy (FTLD-TDP)(Mackenzie et al., 2010). SNPs have been identified through case-control GWA or other association studies of autopsy-confirmed FTLD-tau (Höglinger et al., 2011) and FTLD-TDP (Renton et al., 2011; Van Deerlin et al., 2010), but have not been evaluated comparatively in FTLD-tau relative to FTLD-TDP. Neuroimaging studies suggest distinct neuroanatomic distributions of disease in FTLD-tau and FTLD-TDP (Seltman and Matthews, 2012; Whitwell et al., 2011b) and small sample studies have shown distinct anatomic distributions of GM and WM disease associated with genetic mutations (Rohrer et al., 2010; Whitwell et al., 2012). Diffusion tensor imaging (DTI) studies of WM achieve high sensitivity and specificity for predicting FTLD-tau or FTLD-TDP in patients with known pathology or genetic mutations and these findings were validated in a detailed neuropathologic examination (McMillan et al., 2013). This observation is also consistent with previous neuropathologic reports suggesting that FTLD-tau has relatively increased WM disease compared to FTLD-TDP (Forman et al., 2002; Geser et al., 2009).
In this study we combine SNPs with GM and WM neuroimaging to evaluate the hypothesis that genetic risk factors, or SNPs, may be reflected in differential brain morphology of sporadic FTLD-tau or FTLD-TDP. While traditional approaches to biomarker discovery typically involve retrospective studies of “gold-standard” autopsy-proven cases, we use a data-driven prospective approach that takes advantage of high-powered, multivariate statistics (see Figure 1 for a schematic diagram). Our novel approach for biomarker discovery integrates neuroimaging and genetic markers in FTLD using two approaches. “Eigenanatomy” uses dimensionality reduction to identify anatomically-constrained correlated voxels that account for the greatest variance in the entire dataset and thus minimizes multiple comparisons problems that are common in voxelwise neuroimaging studies (Avants et al., 2012; McMillan et al., 2013). We also use sparse canonical correlation analysis (SCCAN) for the multivariate integration of imaging and genetics by identifying correlations across independent matrices of data (Avants et al., 2010). We first evaluate the hypothesis that GM and WM neuroanatomic structure are related to genetic variation in sporadic FTLD patients. We then evaluate the hypothesis that SNPs associated with neuroanatomic structure confer risk for a specific histopathological subtype of FTLD pathology in a large independent, autopsy-confirmed cohort of sporadic FTLD.
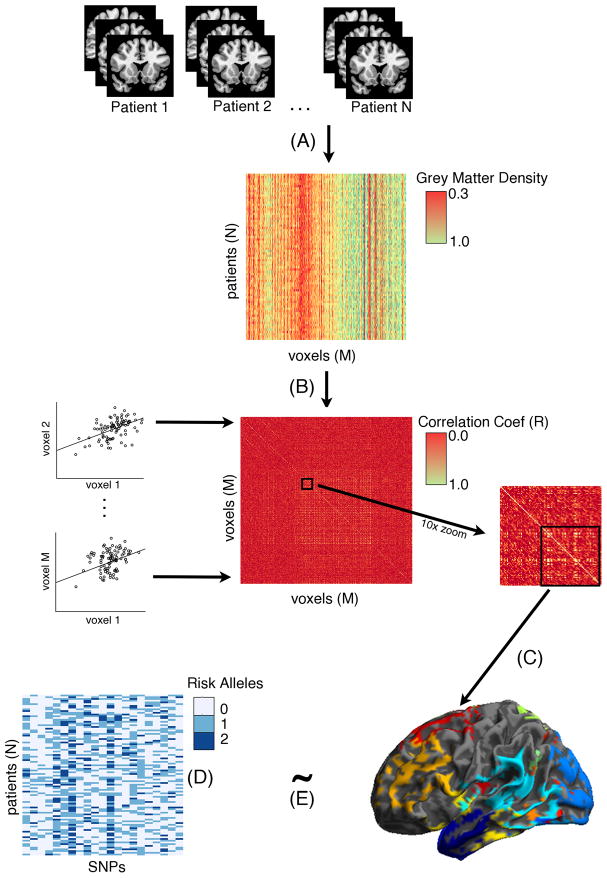
Figure 1. Schematic illustration of sparse canonical correlation analysis approach.
(A) All individual patients’ volumetric MRIs are represented in a matrix containing all voxels (M) X all patients (N). Colors in the matrix refer to grey matter density and it can be observed that there are different bands of density observed in columns; (B) Each GM voxel is correlated with each other GM voxel to generate a correlation matrix. Scatterplots illustrate correlations between voxels (Voxel1 ~ Voxel2…). For illustration purposes we displayed a 1000 × 1000 voxel matrix (center) and then a 10x zoom of a subcomponent of the matrix (right). (C) Clusters of anatomically-constrained correlated voxels are identified using Eigenanatomy, a sparse singular value decomposition method that identifies volumes of interest (VOI) that account for the most variance in the brain. (D) Represents a matrix of patients (N) X 21 SNPs used in this analysis and colors refer to 0–2 risk allele copies. (E) Eigenanatomy VOIs and the SNP matrix are analyzed using sparse canonical correlation analysis (SCCAN), in which each matrix is correlated with one another and a sparsity constraint is applied to push lower model weights to zero and thus remove from the overall model.
2. Methods
Neuroimaging Participants
92 patients were recruited from the Penn Frontotemporal Degeneration Center at the University of Pennsylvania and diagnosed with a FTLD-spectrum neurodegenerative disease by a board-certified neurologist using published criteria (see Supplementary Table 1 for clinical phenotypes). The patient cohort was comprised of 37 females and 55 males who had an overall mean age of 63.20 years (sd=8.51), mean disease duration of 4.09 years (sd=2.54), and mean education of 15.43 years (sd=2.95). All patients and their caregivers participated in an informed consent procedure approved by University of Pennsylvania Institutional Review Board.
All patients selected for this study were screened for research participation using an autopsy-validated cerebrospinal fluid (CSF) ratio of total-tau to beta-amyloid < 0.34, which has been cross-validated across two independent autopsy series and achieves 95.5% accuracy of screening FTLD and AD (Irwin et al., 2012b). To investigate “sporadic” FTLD we additionally excluded patients who had a known genetic mutation that has been associated with FTLD-TDP, including GRN (Baker et al., 2006) and C9orf72 expansions (DeJesus-Hernandez et al., 2011; Renton et al., 2011); or with an FTLD-tau associated MAPT mutation (Hutton et al., 1998). We further classified our cases using a previously published pedigree classification criteria (Wood et al., 2013): cases with a “medium” (N=9) or “low” (N=7) family history were negative for C9orf72 expansions, GRN, and MAPT and have a less than 12% chance of having a mutation detected (Wood et al., 2013); only 3 of our cases had a “high” family history and were negative when screened for 43 genetic mutations previously associated with neurodegenerative diseases; and the remaining cases were either rated as “apparent sporadic” or had too small of a family to accurately determine family history. By omitting cases with genetic mutations we also minimized overlap of cases previously reported in DTI and GM analyses of individuals with genetic or autopsy-confirmed FTLD (McMillan et al., 2013; only 3 autopsy cases from the previous report were included in the current study: 1 CBD; 1 FTLD-TDP, and 1 FTLD-ALS).
Independent Autopsy Series
We queried the Penn Brain Bank for autopsy samples that had a primary neuropathological diagnosis of a FTLD-tau, including progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), Pick’s disease (PiD), and agyrophilic grain disease (AGD), or a diagnosis of FTLD-TDP, including FTLD with TDP-43 inclusions or amyotrophic lateral sclerosis (ALS). Neuropathologic diagnoses were established according to consensus criteria (Mackenzie et al., 2010) by an expert neuropathologist (JQT) using immunohistochemistry with established monoclonal antibodies specific for pathogenic tau (mAb PHF-1) (Otvos et al., 1994) and TDP-43 (mAbs p409/410 or 171) (Lippa et al., 2009; Neumann et al., 2009). Patients who were included in the neuroimaging analysis were excluded from the independent autopsy series analysis. We further excluded cases with a secondary neuropathological diagnosis (e.g., AD, vascular disease) or a known FTLD genetic mutation: all FTLD-tau cases were screened for MAPT mutations; all FTLD-TDP patients were screened for GRN mutations and a C9orf72 expansion. This resulted in a total of 153 sporadic FTLD-spectrum patients, FTLD-tau (N=62) and FTLD-TDP (N=91; see Supplementary Table 2).
Genetic Analysis
We selected 21 SNPs from a custom-designed Pan-Neurodegenerative Disease-oriented Risk Allele panel (PANDoRA (v.1); Table 1) previously associated with FTLD-TDP or FTLD-tau in case-control GWA studies (Carrasquillo et al., 2010; Höglinger et al., 2011; Van Deerlin et al., 2010) or previously implicated in FTLD (Rademakers et al., 2008; 2005). The panel was designed using MassARRAY Assay design software in 2 multiplex reactions with 27 and 24 SNV respectively. See Supplementary Materials for detailed genotyping methods. Each SNP was coded using an additive model, where 0=homozygous for the non-risk allele, 1=heterozygous for the risk allele, and 2=homozygous for the risk allele. “Risk allele” refers to the allele previously associated with disease risk in prior case control studies.
Table 1.
SNPs previously associated with FTLD-tau or FTLD-TDP included in the neuroimaging analysis and their risk allele frequencies.
| Disorder | Ch | SNP | Gene | Risk Allele | Risk Allele Frequency |
|---|---|---|---|---|---|
|
| |||||
| FTLD-TAU | 17 | rs1052553 | MAPT | G | 18.51% (30) |
| FTLD-TAU (PSP) | 12 | rs11568563 | SLCO1A2 | G | 9.52% (16) |
| FTLD-TAU (PSP) | 6 | rs12203592 | IRF4 | T | 22.58% (35) |
| FTLD-TAU (PSP) | 1 | rs1411478 | STX6 | A | 24.11% (34) |
| FTLD-TAU (PSP) | 3 | rs1768208 | MOBP | T | 45.83% (66) |
| FTLD-TAU (PSP) | 10 | rs2142991 | BMS1 | C | 20.25% (32) |
| FTLD-TAU (PSP) | 17 | rs242557 | MAPT | A | 31.34% (36) |
| FTLD-TAU (PSP) | 6 | rs2493013 | EXOC2 | A | 8.97% (14) |
| FTLD-TAU (PSP) | 2 | rs6547705 | CD8B | G | 23.57% (37) |
| FTLD-TAU (PSP) | 1 | rs6687758 | Intergenic | G | 25.16% (39) |
| FTLD-TAU (PSP) | 4 | rs6852535 | IL2/IL21 | A | 16.00% (24) |
| FTLD-TAU (PSP) | 2 | rs7571971 | CD8B/EIF2AK3 | T | 26.45% (41) |
| FTLD-TAU (PSP) | 17 | rs8070723 | MAPT | A | 79.75% (130) |
| TAU | 2 | rs4499362 | EPC2 | T | 22.50% (36) |
| FTLD-TDP | 7 | rs1020004 | TMEM106B | C | 36.91% (55) |
| FTLD-TDP | 7 | rs1990622 | TMEM106B | A | 27.40% (40) |
| FTLD-TDP/ALS | 9 | rs2814707 | C9orf72 | T | 5.16% (8) |
| FTLD-TDP | 7 | rs3173615 | TMEM106B | C | 72.60% (106) |
| FTLD-TDP/ALS | 9 | rs3849942 | C9orf72 | T | 5.19% (8) |
| FTLD-TDP | 17 | rs5848 | GRN | T | 18.57% (26) |
| FTLD-TDP | 1 | rs646776 | SORT1/CELSR2/PSRC1 | C | 27.89% (41) |
Neuroimaging Analysis
High resolution volumetric (1mm3) MRI volumes and diffusion weighted images (DWI) were acquired and pre-processed using a previously described pipeline with ANTs software (see Supplementary Materials for details) (Avants et al., 2011; McMillan et al., 2013). To analyze GM density and FA of WM we employed Eigenanatomy (available for free download in ANTs; https://github.com/stnava/sccan) (Avants et al., 2012; McMillan et al., 2013). Eigenanatomy involves identifying volumes-of-interest (VOIs) composed of correlated voxels that maximally account for the greatest variance in the entire dataset (see Figure 1.B). By reducing the dimensionality of the data from over 1M voxels to a much smaller number of Eigenanatomy VOIs, we can perform high-powered statistics. In the current study we identified 20 VOIs for each modality (GM and FA) that in total accounted for over 95% of the variance in the dataset of each modality. To identify these VOIs, all normalized images for each modality are first transformed into a number-of-subjects (N) by number-of-voxels matrix where voxels are selected to lie within an explicit mask (see Figure 1.A). For GM and FA we used a threshold of 0.4 or greater to define our mask. Sparse singular value decomposition is then used to identify the first sparse eigenvectors from the data matrix that account for the greatest amount of variance, the second sparse eigenvectors that account for the second most amount of variance, and so on. The ANTs implementation of Eigenanatomy employs a sparseness penalty on the eigenvectors such that (1) eigenvector are both sparse (i.e. have many zero entries) and non-negative and (2) the non-zero voxels are clustered and exceed a cluster extent threshold (>100 adjacent voxels). The sparseness and non-negativity allows the eigenvectors to be interpreted as weighted averages of the original data, resembling a distributed version of a traditional region of interest (Figure 1.C). We refer to each of these distributed regions as an Eigenanatomy VOI.
To relate the identified Eigenanatomy VOIs to SNPs we performed a sparse canonical correlation analysis (SCCAN) for each neuroimaging modality (Figure 1.E), as previously reported (Avants et al., 2010). SCCAN is based on classic canonical correlation (CCA) analysis which can be used to evaluate the multivariate association between two datasets, such as genetics and Eigenanatomy VOIs. SCCAN methods, like classical CCA, compute eigenvectors that maximize the Pearson correlation between the input modalities. However, unlike classical CCA methods that can achieve very low weight values for variates that account for minimal variance, sparse methods apply a penalty in an effort to push variates that account for minimal variance to zero and therefore output a subset of significant variates that contribute to a statistical model. In our case the input modalities are comprised of one matrix of Eigenanatomy VOIs that represent a neuroimaging modality dataset and one matrix of additive SNP values that represent the genetic risk factors. In this study, we did not constrain the polarity of the SCCAN eigenvectors so that we could determine both positive and negative correlations between neuroimaging and genetics. We performed independent SCCAN analyses to relate GM to SNPs and then to relate FA to SNPs. The significance of SCCAN results was tested by permuting the imaging input matrix (FA or GM) over the SNP matrix for 1000 permutations and we report the results that survive p<0.001. The correlation value produced by the original ordering of the data is then compared to the correlations produced over a set of random permutations and thus is controlled for risk of Type I error rates in a manner similar to correction for multiple comparisons. For each neuroimaging modality we report the canonical weights for sparse correlations that like univariate regression beta weights, provide a measure of the relative contribution of each variate to the overall canonical model. To increase interpretability of these canonical weights we also plot the mean FA or GM for each significant VOI and SNP in our SCCAN models.
3. Results
Integration of DTI and SNPs
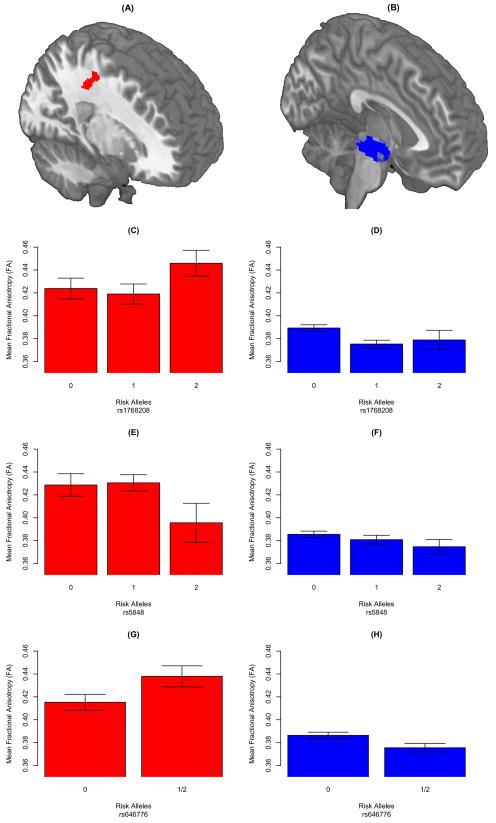
SCCAN revealed a significant association between two VOIs and three SNPs (p<0.001; canonical weights are reported following each features). VOIs were located in the midbrain (8.7E-02; Figure 2.B) and in the right superior longitudinal fasciculus (−4.7E-02; SLF; Figure 2.A). The SNPs included rs1768208 (−8.6E-02), rs646776 (−4.3E-02), and rs5848 (−4.5E-02). The canonical weights suggest that rs1768208 is the most highly associated SNP with WM and the midbrain is the most highly associated WM VOI with the three SNPs. The bar graphs in Figure 2.C–2.H summarize the direction of the associations for each SNP and WM VOI. In the midbrain, increased copy numbers of risk alleles for each SNP are associated with decreased fractional anisotropy (FA). In the SLF, increased rs5848 risk allele copies are associated with reduced FA, while fewer risk allele copies of rs1768208 and rs646776 are associated with reduced FA. A post-hoc multivariate regression revealed no significant association between SNPs and WM VOIs with demographic factors (Age + Education + Gender; F(3,85)=1.863; p=0.142). We also observed no association of clinical phenotype with SNPs and WM VOIs [F(7,84)=0.736; p=0.642].
Figure 2. White matter regions that are significantly associated with FTLD SNPs.
(A) Superior longitudinal fasisculus (SLF) and (B) the midbrain, were related to rs1768208 (MOBP), rs646776 (near SORT1), and rs5848 (GRN). Figures 2.C–2.H summarize the direction of the associations for each SNP and WM VOI: (C) fewer copies of rs178208 risk alleles are associated with reduced FA in SLF; (D) more rs178208 risk allele copies are associated with reduced FA in midbrain; (E) more rs5848 risk allele copies are associated with reduced FA in SLF; (F) more rs5848 risk allele copies are associated with reduced FA in midbrain; (G) fewer rs646776 risk allele copies are associated with reduced FA in SLF; and (H) more rs646776 risk allele copies are associated with reduced FA in midbrain. For (G) and (H) we illustrate a dominant model since only a few cases were homozygous for the risk allele.
Integration of GM Density and SNPs
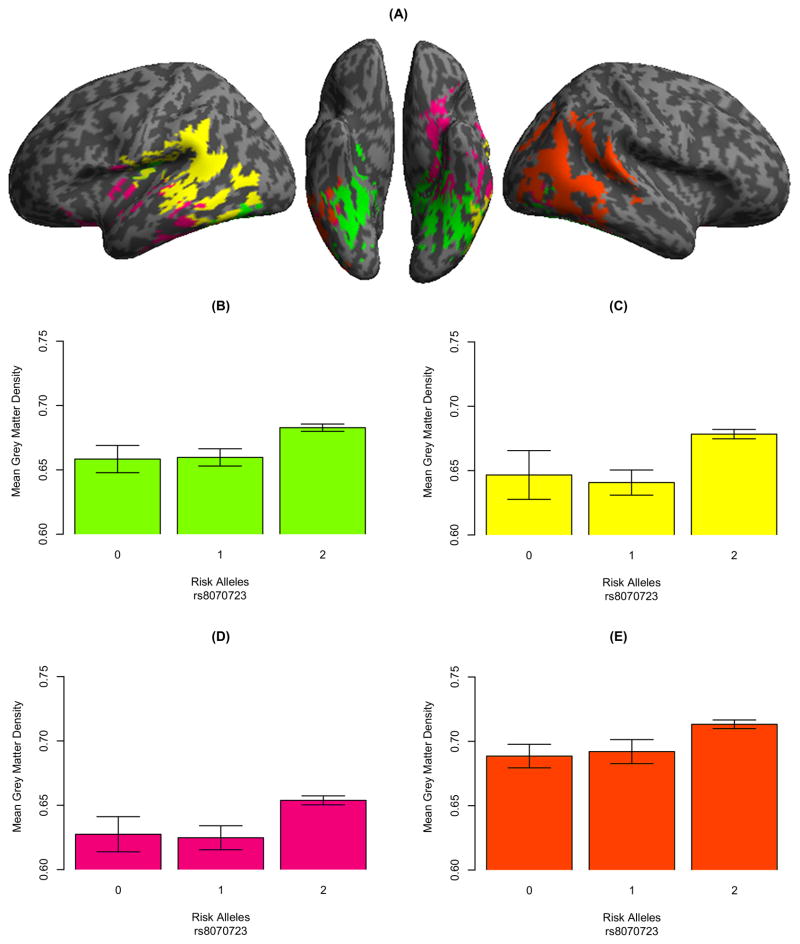
SCCAN revealed an association between a single SNP, rs8070723 (1.1E-01), and 4 GM VOIs (p<0.001): bilateral posterior ventral temporal cortex (5.3E-02; Figure 3.A; green), left temporoparietal cortex (3.2E-02; Figure 3.A; yellow), left ventral frontotemporal (1.5E-02; Figure 3.A; magenta), and right temporooccipital cortex (2.4E-02; Figure 3.A; green). The bar graphs in Figure 3 (panels B–E) illustrate that fewer rs8070723 risk allele copies are associated with decreased GM volume. A post-hoc multivariate regression did not reveal any associations between SNPs and WM with demographic factors [F(3, 85)=1.448; p=0.235] or any associations with clinical phenotype [F(7, 84)=0.516; p=0.821].
Figure 3. Grey matter regions that are significantly associated with FTLD SNPs.
(A) Four GM volumes of interest were related to rs8070723 (MAPT); Panels B–E illustrate that fewer rs8070723 risk allele copies are associated with reduced GM.
Neuropathological Confirmation of Risk Factors
We evaluated the odds ratio (OR) of each significant SNP from our neuroimaging analysis in FTLD-TDP (N=91) relative to FTLD-tau (N=62) in an independent autopsy series of sporadic FTLD cases (see Table 2 for risk allele frequencies). Carriers of rs1768208 risk alleles have a significantly increased risk of FTLD-tau relative to FTLD-TDP (OR=1.69; p=0.030). The SNP rs8070723 also confers an increased risk of FTLD-tau relative to FTLD-TDP (OR=3.36; p=0.005). Conversely, rs646776 risk allele carriers have about half the likelihood of developing FTLD-tau relative to FTLD-TDP (OR=0.50; p=0.030). rs5848 does not confer significant risk of a specific FTLD subtype (OR=1.13; p=0.627).
Table 2.
Risk allele frequencies in the independent autopsy series of SNPs identified in the neuroimaging analysis and odds ratio (95% confidence intervals) of FTLD-tau.
| SNP | FTLD-tau | FTLD-TDP | tau Odds Ratio | P-Value |
|---|---|---|---|---|
|
| ||||
| rs1768208 | 43.55% (54) | 31.32% (57) | 1.69 (1.02–2.79) | 0.030 |
| rs646776 | 14.52% (18) | 25.27% (46) | 0.50 (0.26–0.95) | 0.031 |
| rs5848 | 36.29% (45) | 33.52% (61) | 1.13 (0.68–1.87) | 0.627 |
| rs80707231 | 91.12% (113) | 75.27% (137) | 3.36 (1.62–7.56) | 0.0005 |
Note.
For all SNPs we report minor allele frequencies, except rs8070723 for which the major allele (A/A) has previously been associated with MAPT risk.
4. Discussion
SNPs have been associated with GM and WM neuroanatomic structure in AD (Jahanshad et al., 2013; Shen et al., 2010), but these associations have rarely undergone independent validation at autopsy and have not been assessed in FTLD. Since FTLD is predominantly caused by a single misfolded protein, either tau or TDP-43, it is an ideal target for pharmaceutical development (Boxer et al., 2013). This is in contrast to disease like AD that result from both tau tangles and amyloid plaque inclusions. It is therefore critical to improve our ability to diagnose FTLD using quantitative biomarkers and to better understand the underlying biological mechanisms that contribute to FTLD neurodegeneration. Our novel approach identified several genetic and imaging associations in living FTLD patients, and we determined that these associations confer risk of a specific FTLD histopathological subtype in an independent autopsy series. These findings validate the importance of specific regional WM and GM abnormalities in screening for FTLD and these novel genetic and neuroanatomic associations emphasize the influence of SNPs on neuroanatomic structure, and provide a proof-of-concept for using the current approach in ongoing biomarker discovery research.
The SNP rs8070723 associated with GM is located in the microtubule-associated protein tau (MAPT) gene. It tags the H1 haplotype which is a common variant of MAPT (Höglinger et al., 2011). The H1 haplotype has consistently been associated with 4-repeat tauopathies including PSP (Baker et al., 1999) and CBD (Di Maria et al., 2000) relative to controls, though we are unaware of a direct comparison between FTLD-tau and FTLD-TDP that evaluated the specificity of this haplotype. The current study indicates that the major risk-conferring allele (A/A) is nearly always present in FTLD-tau (>92%).
The GM associations with rs8070723 suggest that MAPT H1 (A/A) is protective of GM atrophy compared to carriers of one or more H2 (G) alleles. The direction of this association is consistent with prior reports that the MAPT H2 haplotype may be a more aggressive form of neurodegeneration. For example, H2 has been associated with reduced brain weight at autopsy (Connelly et al., 2011), lower SNAP-25 synaptic transmission (Connelly et al., 2011), and reduced glucose metabolism (Laws et al., 2007). The H2 haplotype has also been associated with increased neurofibrillary tangle burden in Alzheimer’s and Lewy body diseases (Wider et al., 2012), but to our knowledge pathological burden of MAPT haplotype has not been investigated in FTLD. The locus of the rs8070723 GM association included bilateral posterior ventral temporal, left temporoparietal, left ventral frontotemporal, and right temporooccipital cortices. Our autopsy analysis demonstrated that H1 confers increased risk of FTLD-tau, and in this comparative study H2 therefore confers increased FTLD-TDP risk; it is thus possible that this GM distribution reflects FTLD-TDP disease. Indeed, FTLD-TDP appears to be associated with a temporal distribution of disease, as observed in the semantic variant of primary progressive aphasia which is statistically biased toward TDP-43 pathology (Grossman, 2010), while FTLD-tau is more associated with a frontal distribution (Whitwell et al., 2009).
We also observed that another SNP, rs1768208, confers increased risk of developing FTLD-tau relative to FTLD-TDP. In a previous GWA study of PSP patients, rs1768208 conferred increased risk of FTLD-tau relative to case controls (Höglinger et al., 2011). This observation suggested that rs1768208 is associated with the presence of tau pathology, but the specificity of the association was not evaluated. The current study performed a comparative evaluation relative to patients with FTLD-TDP in order to determine the specificity of rs1768208 as a marker of FTLD-tau. In another study, rs1768208 was not associated with late-onset AD risk relative to case controls (Liu et al., 2013), providing additional evidence that this SNP is not a non-specific marker of any neurodegenerative disease.
The presence of more risk allele (T) copies of rs1768208 is associated with reduced FA in the midbrain. The midbrain is commonly reported in WM neuroimaging and neuropathologic studies of PSP (Dickson et al., 2010). Volumetric WM and DTI studies have revealed reduced FA in the midbrain of PSP patients relative to healthy adults (Whitwell et al., 2011a; 2011c). The observation that rs1768208 has a WM correlate in the midbrain is consistent with this SNP’s location in the region on chromosome 3 coding for myelin-associated oligodendrocyte basic protein (MOBP). MOBP is involved in protecting the integrity of the myelin sheath (Yamamoto et al., 1999) and is highly expressed in the WM of midbrain regions (Montague et al., 2006). We additionally observed that fewer copies of rs1768208 risk alleles is associated with reduced FA in the SLF. While SLF has previously been associated with FTLD-tau (McMillan et al., 2013), it is possible that the 4-repeat subtype of tau found in PSP and related to rs1768208 has distinct neural correlates relative to 3-repeat tau subtypes. However, our post hoc analyses demonstrated that clinical syndrome is not driving any of the observed neuroanatomic and genetic associations.
In our comparative autopsy analysis we observed that the SNP rs646776 conferred a twofold increased risk of FTLD-TDP relative to FTLD-tau. rs646776 is located near sortilin (SORT1) as well as CELSR2 and PSRC1, and has been associated with regulation of plasma progranulin levels: each copy of the risk allele yielded nearly a 15% decrease in proganulin (Carrasquillo et al., 2010). The current study suggests that risk allele copies for rs646776 are associated with increased FA in SLF, which is consistent with previous neuroimaging and neuropathological observations suggesting that SLF is relatively preserved in FTLD-TDP (McMillan et al., 2013).
Another SNP, rs5848, is located in the 3’ untranslated region of GRN. A previous study suggested that homozygous risk allele carriers of rs5848 have a 3.2 fold increased risk of FTLD-TDP compared to case controls (Rademakers et al., 2008). Risk allele carriers of rs5848, like rs646776, have been reported to have lower progranulin plasma levels in a clinically-defined cohort (Hsiung et al., 2011). We observed that risk allele carriers of rs5848 have reduced FA in the SLF. However, the direction of this pattern is contrary to a previous neuroimaging and neuropathological report suggesting that SLF is relatively spared in FTLD-TDP in comparison to FTLD-tau (McMillan et al., 2013). However, the role of rs5848 in FTLD-TDP has been controversial. Plasma progranulin levels were not affected in a subset of sporadic patients with presumed FTLD-TDP pathology due to FTLD-ALS or presumed FTLD-tau (e.g., CBS, PSP) (Hsiung et al., 2011), and several studies have suggested that rs5848 may not be associated with sporadic FTLD-TDP (Rollinson et al., 2011; Simón-Sánchez et al., 2009). Our comparative autopsy study also suggests that rs5848 may not specifically increase the risk for sporadic FTLD-TDP since risk allele carriers in our cohort are equally likely to have FTLD-TDP or FTLD-tau pathology.
The observation that SNPs related to FTLD-TDP have WM associations is consistent with neuropathological studies reporting microglial activation, myelin changes, and gliosis in GRN mutation carriers (Kelley et al., 2009). GRN mutations have been strongly associated with FTLD-TDP pathology (Gass et al., 2006). Also, progranulin is expressed in microglia (Ahmed et al., 2007) and in the midbrain of ALS patients (Irwin et al., 2009) who have FTLD-TDP pathology. A DTI study of WM reported that GRN mutation carriers have reduced FA relative to sporadic FTLD patients, but these analyses were restricted to a priori regions of interest in the corpus callosum (Bozzali et al., 2013). In another DTI study, FA reductions in WM of the extreme capsule, inferior frontal-occipital fasciculus and uncinate were reported in asymptomatic GRN mutation carriers relative to control non-carriers. Moreover, volumetric measures of WM and GM did not reveal any differences across carriers and non-carriers (Borroni et al., 2008). This suggests that the DTI modality used in our study may provide a more sensitive pre-clinical marker of FTLD pathology.
Since WM abnormalities in the midbrain were associated with FTLD-tau and FTLD-TDP SNPs, this region may be used to screen for the presence of any FTLD-related disease on an individual subject basis and then SNP analyses can be used to evaluate if these WM changes are associated with risk for a specific FTLD spectrum histopathologic abnormality. The midbrain may be an optimal region to evaluate presence of disease in WM since this region is hypothesized to be affected in early stages of TDP-43 (Geser et al., 2011) associated with ALS and may be an early locus of pathologically-modified tau associated with PSP or CBD (Irwin et al., 2012a).
The biomarker discovery approach proposed in this report identified neuroanatomic and genetic associations in a living cohort of FTLD patients and then validated the meaningfulness of these associations in an independent autopsy series. Rather than focusing on the neuroanatomical correlates of a single SNP, our analysis procedure additionally benefits by supporting selection of a subset of meaningful SNPs with neuroimaging correlates from a large panel of SNPs. Other approaches have included voxel-wise studies in which a GWA study is performed at each individual voxel (k=31,622) of the entire brain (Stein et al., 2010). While quantitative trait analyses such as voxel-wise measures can yield increased statistical power relative to categorical case-control GWA studies (Potkin et al., 2009), these studies require large numbers of cases which is less feasible in uncommon but informative neurodegenerative disorders like FTLD. Our data-driven VOI method, Eigenanatomy, also allows us to reduce neuroimaging comparisons to a small set of statistically determined regions that yields increased statistical power and minimizes multiple comparison problems.
In sum, we demonstrate that GM and WM neuroanatomic structure in sporadic FTLD is related to genetic risk factors, and that these SNPs confer risk specifically for FTLD-tau or FTLD-TDP histopathologic abnormalities. These observations also emphasize that SNPs modify neuroanatomic structure, and underline the practical contribution of SNPs and neuroimaging to biomarker discovery. This report also demonstrates a novel method for evaluating neuroanatomical correlates of genetic risk factors associated with neurodegenerative disease.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health: AG043503, T32-AG000255, AG017586, NS044266, AG032953, AG010124, and the Wyncote Foundation. JBT is supported by the Alfonso Martín Escudero Foundation.
Footnotes
The data contained in this manuscript has not been previously published, has not been submitted elsewhere and will not be submitted elsewhere while under consideration at Neurobiology of Aging.
All participants were treated in accordance with an Institutional Review Board protocol approved by the University of Pennsylvania and all participants and caregivers participated in an informed consent procedure.
Conflicts of Interest
All authors have no relevant conflicts of interest to disclose.
References
- Ahmed Z, Mackenzie IRA, Hutton ML, Dickson DW. Progranulin in frontotemporal lobar degeneration and neuroinflammation. J Neuroinflammation. 2007;4:7. doi: 10.1186/1742-2094-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B, Dhillon P, Kandel BM, Cook PA, McMillan CT, Grossman M, Gee JC. Eigenanatomy Improves Detection Power for Longitudinal Cortical Change. Presented at the Medical image computing and computer-assisted intervention: MICCAI; 2012. pp. 206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Cook PA, Ungar L, Gee JC, Grossman M. Dementia induces correlated reductions in white matter integrity and cortical thickness: a multivariate neuroimaging study with sparse canonical correlation analysis. Neuro Image. 2010;50:1004–1016. doi: 10.1016/j.neuroimage.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuro Image. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J, Hardy J, Lynch T, Bigio E, Hutton M. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet. 1999;8:711–715. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- Borroni B, Alberici A, Premi E, Archetti S, Garibotto V, Agosti C, Gasparotti R, Di Luca M, Perani D, Padovani A. Brain magnetic resonance imaging structural changes in a pedigree of asymptomatic progranulin mutation carriers. Rejuvenation Res. 2008;11:585–595. doi: 10.1089/rej.2007.0623. [DOI] [PubMed] [Google Scholar]
- Boxer AL, Gold M, Huey E, Gao FB, Burton EA, Chow T, Kao A, Leavitt BR, Lamb B, Grether M, Knopman D, Cairns NJ, Mackenzie IR, Mitic L, Roberson ED, Van Kammen D, Cantillon M, Zahs K, Salloway S, Morris J, Tong G, Feldman H, Fillit H, Dickinson S, Khachaturian Z, Sutherland M, Farese R, Miller BL, Cummings J. Frontotemporal degeneration, the next therapeutic frontier: molecules and animal models for frontotemporal degeneration drug development. Alzheimers Dement. 2013;9:176–188. doi: 10.1016/j.jalz.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzali M, Battistoni V, Premi E, Alberici A, Giulietti G, Archetti S, Turla M, Gasparotti R, Cercignani M, Padovani A, Borroni B. Structural Brain Signature of FTLD Driven by Granulin Mutation. J Alzheimers Dis. 2013;33:483–494. doi: 10.3233/JAD-2012-121273. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Nicholson AM, Finch N, Gibbs JR, Baker M, Rutherford NJ, Hunter TA, DeJesus-Hernandez M, Bisceglio GD, Mackenzie IR, Singleton A, Cookson MR, Crook JE, Dillman A, Hernandez D, Petersen RC, Graff-Radford NR, Younkin SG, Rademakers R. Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet. 2010;87:890–897. doi: 10.1016/j.ajhg.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly SJ, Mukaetova-Ladinska EB, Abdul-All Z, Alves da Silva J, Brayne C, Honer WG, Mann DMA. Synaptic changes in frontotemporal lobar degeneration: correlation with MAPT haplotype and APOE genotype. Neuropathol Appl Neurobiol. 2011;37:366–380. doi: 10.1111/j.1365-2990.2010.01150.x. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GYR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Maria E, Tabaton M, Vigo T, Abbruzzese G, Bellone E, Donati C, Frasson E, Marchese R, Montagna P, Munoz DG, Pramstaller PP, Zanusso G, Ajmar F, Mandich P. Corticobasal degeneration shares a common genetic background with progressive supranuclear palsy. Ann Neurol. 2000;47:374–377. doi: 10.1002/1531-8249(200003)47:3<374::aid-ana15>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- Dickson DW, Ahmed Z, Algom AA, Tsuboi Y, Josephs KA. Neuropathology of variants of progressive supranuclear palsy. Curr Opin Neurol. 2010;23:394–400. doi: 10.1097/WCO.0b013e32833be924. [DOI] [PubMed] [Google Scholar]
- Forman MS, Zhukareva V, Bergeron C, Chin SSM, Grossman M, Clark C, Lee VMY, Trojanowski JQ. Signature tau neuropathology in gray and white matter of corticobasal degeneration. Am J Pathol. 2002;160:2045–2053. doi: 10.1016/S0002-9440(10)61154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, Crook R, Melquist S, Kuntz K, Petersen R, Josephs K, Pickering-Brown SM, Graff-Radford N, Uitti R, Dickson D, Wszolek Z, Gonzalez J, Beach TG, Bigio E, Johnson N, Weintraub S, Mesulam M, White CL, Woodruff B, Caselli R, Hsiung GY, Feldman H, Knopman D, Hutton M, Rademakers R. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- Geser F, Martinez-Lage M, Robinson J, Uryu K, Neumann M, Brandmeir NJ, Xie SX, Kwong LK, Elman L, McCluskey L, Clark CM, Malunda J, Miller BL, Zimmerman EA, Qian J, Van Deerlin V, Grossman M, Lee VMY, Trojanowski JQ. Clinical and pathological continuum of multisystem TDP-43 proteinopathies. Arch Neurol. 2009;66:180–189. doi: 10.1001/archneurol.2008.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser F, Prvulovic D, O’Dwyer L, Hardiman O, Bede P, Bokde ALW, Trojanowski JQ, Hampel H. On the development of markers for pathological TDP-43 in amyotrophic lateral sclerosis with and without dementia. Prog Neurobiol. 2011;95:649–662. doi: 10.1016/j.pneurobio.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman M. Primary progressive aphasia: clinicopathological correlations. Nat Rev Neurol. 2010;6:88–97. doi: 10.1038/nrneurol.2009.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglinger GU, Melhem NM, Dickson DW, Sleiman PMA, Wang LS, Klei L, Rademakers R, de Silva R, Litvan I, Riley DE, van Swieten JC, Heutink P, Wszolek ZK, Uitti RJ, Vandrovcova J, Hurtig HI, Gross RG, Maetzler W, Goldwurm S, Tolosa E, Borroni B, Pastor P, Cantwell LB, Han MR, Dillman A, van der Brug MP, Gibbs JR, Cookson MR, Hernandez DG, Singleton AB, Farrer MJ, Yu C-E, Golbe LI, Revesz T, Hardy J, Lees AJ, Devlin B, Hakonarson H, Müller U, Schellenberg GD PSP Genetics Study Group. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung GYR, Fok A, Feldman HH, Rademakers R, Mackenzie IRA. rs5848 polymorphism and serum progranulin level. Journal of the neurological sciences. 2011;300:28–32. doi: 10.1016/j.jns.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Irwin D, Lippa CF, Rosso A. Progranulin (PGRN) expression in ALS: an immunohistochemical study. Journal of the neurological sciences. 2009;276:9–13. doi: 10.1016/j.jns.2008.08.024. [DOI] [PubMed] [Google Scholar]
- Irwin DJ, Cohen TJ, Grossman M, Arnold SE, Xie SX, Lee VMY, Trojanowski JQ. Acetylated tau, a novel pathological signature in Alzheimer’s disease and other tauopathies. Brain. 2012a;135:807–818. doi: 10.1093/brain/aws013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DJ, McMillan CT, Toledo JB, Arnold SE, Shaw LM, Wang LS, Van Deerlin V, Lee VMY, Trojanowski JQ, Grossman M. Comparison of cerebrospinal fluid levels of tau and Aβ 1–42 in Alzheimer disease and frontotemporal degeneration using 2 analytical platforms. Arch Neurol. 2012b;69:1018–1025. doi: 10.1001/archneurol.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, Jack CR, Saykin AJ, Green RC, Weiner MW, Medland SE, Montgomery GW, Hansell NK, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM Alzheimer’s Disease Neuroimaging Initiative. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proceedings of the National Academy of Sciences. 2013;110:4768–4773. doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley BJ, Haidar W, Boeve BF, Baker M, Graff-Radford NR, Krefft T, Frank AR, Jack CR, Shiung M, Knopman DS, Josephs KA, Parashos SA, Rademakers R, Hutton M, Pickering-Brown S, Adamson J, Kuntz KM, Dickson DW, Parisi JE, Smith GE, Ivnik RJ, Petersen RC. Prominent phenotypic variability associated with mutations in Progranulin. Neurobiol Aging. 2009;30:739–751. doi: 10.1016/j.neurobiolaging.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws SM, Perneczky R, Drzezga A, Diehl-Schmid J, Ibach B, Bäuml J, Eisele T, Förstl H, Kurz A, Riemenschneider M. Association of the tau haplotype H2 with age at onset and functional alterations of glucose utilization in frontotemporal dementia. Am J Psychiatry. 2007;164:1577–1584. doi: 10.1176/appi.ajp.2007.06091456. [DOI] [PubMed] [Google Scholar]
- Lippa CF, Rosso AL, Stutzbach LD, Neumann M, Lee VMY, Trojanowski JQ. Transactive response DNA-binding protein 43 burden in familial Alzheimer disease and Down syndrome. Arch Neurol. 2009;66:1483–1488. doi: 10.1001/archneurol.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu QY, Yu JT, Miao D, Ma XY, Wang HF, Wang W, Tan L. An exploratory study on STX6, MOBP, MAPT, and EIF2AK3 and late-onset Alzheimer’s disease. Neurobiol Aging. 2013;34:1519, e13–7. doi: 10.1016/j.neurobiolaging.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, Ince PG, Kamphorst W, Revesz T, Rozemuller AJM, Kumar-Singh S, Akiyama H, Baborie A, Spina S, Dickson DW, Trojanowski JQ, Mann DMA. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119:1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan CT, Irwin DJ, Avants B, Powers J, Cook P, Toledo JB, Wood EM, Van Deerlin VM, Lee CM-Y, Trojanowski JQ, Grossman M. White Matter Imaging Helps Dissociate Tau from TDP-43 in Frontotemporal Lobar Degeneration. Journal of Neurology, Neurosurgery, and Psychiatry. 2013 doi: 10.1136/jnnp-2012-304418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague P, McCallion AS, Davies RW, Griffiths IR. Myelin-associated oligodendrocytic basic protein: a family of abundant CNS myelin proteins in search of a function. Dev Neurosci. 2006;28:479–487. doi: 10.1159/000095110. [DOI] [PubMed] [Google Scholar]
- Neumann M, Kwong LK, Lee EB, Kremmer E, Flatley A, Xu Y, Forman MS, Troost D, Kretzschmar HA, Trojanowski JQ, Lee VMY. Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 2009;117:137–149. doi: 10.1007/s00401-008-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos L, Feiner L, Lang E, Szendrei GI, Goedert M, Lee VM. Monoclonal antibody PHF-1 recognizes tau protein phosphorylated at serine residues 396 and 404. J Neurosci Res. 1994;39:669–673. doi: 10.1002/jnr.490390607. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Torri F, Keator DB, Macciardi F. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cognitive Neuropsychiatry. 2009;14:391–418. doi: 10.1080/13546800903059829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, Finch N, Rutherford NJ, Crook RJ, Josephs KA, Boeve BF, Knopman DS, Petersen RC, Parisi JE, Caselli RJ, Wszolek ZK, Uitti RJ, Feldman H, Hutton ML, Mackenzie IR, Graff-Radford NR, Dickson DW. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Melquist S, Cruts M, Theuns J, Del-Favero J, Poorkaj P, Baker M, Sleegers K, Crook R, De Pooter T, Bel Kacem S, Adamson J, Van den Bossche D, Van den Broeck M, Gass J, Corsmit E, De Rijk P, Thomas N, Engelborghs S, Heckman M, Litvan I, Crook J, De Deyn PP, Dickson D, Schellenberg GD, Van Broeckhoven C, Hutton ML. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum Mol Genet. 2005;14:3281–3292. doi: 10.1093/hmg/ddi361. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ ITALSGEN Consortium. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JD, Ridgway GR, Modat M, Ourselin S, Mead S, Fox NC, Rossor MN, Warren JD. Distinct profiles of brain atrophy in frontotemporal lobar degeneration caused by progranulin and tau mutations. NeuroImage. 2010;53:1070–1076. doi: 10.1016/j.neuroimage.2009.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollinson S, Rohrer JD, van der Zee J, Sleegers K, Mead S, Engelborghs S, Collinge J, De Deyn PP, Mann DMA, Van Broeckhoven C, Pickering-Brown SM. No association of PGRN 3′UTR rs5848 in frontotemporal lobar degeneration. Neurobiol Aging. 2011;32:754–755. doi: 10.1016/j.neurobiolaging.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Seltman RE, Matthews BR. Frontotemporal lobar degeneration: epidemiology, pathology, diagnosis and management. CNS Drugs. 2012;26:841–870. doi: 10.2165/11640070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack CR, Weiner MW, Saykin AJ Alzheimer’s Disease Neuroimaging Initiative. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. NeuroImage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Sánchez J, Seelaar H, Bochdanovits Z, Deeg DJH, van Swieten JC, Heutink P. Variation at GRN 3′-UTR rs5848 is not associated with a risk of frontotemporal lobar degeneration in Dutch population. PLoS ONE. 2009;4:e7494. doi: 10.1371/journal.pone.0007494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, Saykin AJ, Shen L, Foroud T, Pankratz N, Huentelman MJ, Craig DW, Gerber JD, Allen AN, Corneveaux JJ, Dechairo BM, Potkin SG, Weiner MW, Thompson P Alzheimer’s Disease Neuroimaging Initiative. Voxelwise genome-wide association study (vGWAS) NeuroImage. 2010;53:1160–1174. doi: 10.1016/j.neuroimage.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Sleiman PMA, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, Arnold SE, Mann DMA, Pickering-Brown SM, Seelaar H, Heutink P, van Swieten JC, Murrell JR, Ghetti B, Spina S, Grafman J, Hodges J, Spillantini MG, Gilman S, Lieberman AP, Kaye JA, Woltjer RL, Bigio EH, Mesulam M, Al-Sarraj S, Troakes C, Rosenberg RN, White CL, Ferrer I, Lladó A, Neumann M, Kretzschmar HA, Hulette CM, Welsh-Bohmer KA, Miller BL, Alzualde A, Lopez de Munain A, McKee AC, Gearing M, Levey AI, Lah JJ, Hardy J, Rohrer JD, Lashley T, Mackenzie IRA, Feldman HH, Hamilton RL, Dekosky ST, van der Zee J, Kumar-Singh S, Van Broeckhoven C, Mayeux R, Vonsattel JPG, Troncoso JC, Kril JJ, Kwok JBJ, Halliday GM, Bird TD, Ince PG, Shaw PJ, Cairns NJ, Morris JC, McLean CA, DeCarli C, Ellis WG, Freeman SH, Frosch MP, Growdon JH, Perl DP, Sano M, Bennett DA, Schneider JA, Beach TG, Reiman EM, Woodruff BK, Cummings J, Vinters HV, Miller CA, Chui HC, Alafuzoff I, Hartikainen P, Seilhean D, Galasko D, Masliah E, Cotman CW, Tuñón MT, Martínez MCC, Munoz DG, Carroll SL, Marson D, Riederer PF, Bogdanovic N, Schellenberg GD, Hakonarson H, Trojanowski JQ, Lee VMY. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Avula R, Master A, Vemuri P, Senjem ML, Jones DT, Jack CR, Josephs KA. Disrupted thalamocortical connectivity in PSP: a resting-state fMRI, DTI, and VBM study. Parkinsonism Relat Disord. 2011a;17:599–605. doi: 10.1016/j.parkreldis.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR, Parisi JE, Knopman DS, Boeve BF, Petersen RC, Dickson DW, Josephs KA. Imaging signatures of molecular pathology in behavioral variant frontotemporal dementia. J Mol Neurosci. 2011b;45:372–378. doi: 10.1007/s12031-011-9533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Jack CR, Senjem ML, Parisi JE, Boeve BF, Knopman DS, Dickson DW, Petersen RC, Josephs KA. MRI correlates of protein deposition and disease severity in postmortem frontotemporal lobar degeneration. Neurodegener Dis. 2009;6:106–117. doi: 10.1159/000209507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Master AV, Avula R, Kantarci K, Eggers SD, Edmonson HA, Jack CR, Josephs KA. Clinical correlates of white matter tract degeneration in progressive supranuclear palsy. Arch Neurol. 2011c;68:753–760. doi: 10.1001/archneurol.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Weigand SD, Boeve BF, Senjem ML, Gunter JL, DeJesus-Hernandez M, Rutherford NJ, Baker M, Knopman DS, Wszolek ZK, Parisi JE, Dickson DW, Petersen RC, Rademakers R, Jack CR, Josephs KA. Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain. 2012;135:794–806. doi: 10.1093/brain/aws001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wider C, Ross OA, Nishioka K, Heckman MG, Vilariño-Güell C, Jasinska-Myga B, Erketin-Taner N, Rademakers R, Graff-Radford NR, Mash DC, Papapetropoulos S, Duara R, Uchikado H, Wszolek ZK, Farrer MJ, Dickson DW. An evaluation of the impact of MAPT, SNCA and APOE on the burden of Alzheimer’s and Lewy body pathology. Journal of Neurology, Neurosurgery, and Psychiatry. 2012;83:424–429. doi: 10.1136/jnnp-2011-301413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood EM, Falcone D, Suh E, Irwin DJ, Chen-Plotkin AS, Lee EB, Xie SX, Van Deerlin VM, Grossman M. Development and Validation of Pedigree Classification Criteria for Frontotemporal Lobar Degeneration. JAMA Neurology. 2013 doi: 10.1001/jamaneurol.2013.3956. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Yoshikawa H, Nagano S, Kondoh G, Sadahiro S, Gotow T, Yanagihara T, Sakoda S. Myelin-associated oligodendrocytic basic protein is essential for normal arrangement of the radial component in central nervous system myelin. Eur J Neurosci. 1999;11:847–855. doi: 10.1046/j.1460-9568.1999.00490.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.