Abstract
Mutations in C9ORF72, SOD1, TARDBP and FUS genes account for approximately two third of familial cases and 5% of sporadic amyotrophic lateral sclerosis (ALS) cases. We present the first case of an ALS patient carrying a de novo nonsense mutation in exon 14 of the FUS gene (c.1483c>t; p.R495X) in a young patient with an apparently familial ALS. This mutation cause a phenotype characterized by a young age at onset, a rapid course (<24 months) and a bulbar onset with early respiratory involvement with a predominant lower motor neuron disease. De novo mutations could account for a sizable number of apparently sporadic ALS patients carrying mutations of ALS-related genes.
1. Introduction
Amyotrophic lateral sclerosis (ALS) is neurodegenerative disorder of the adult life, characterized by a progressive loss of cortical, bulbar and spinal motor neurons. Approximately 5.10% of patients have a family history of disease, whereas the remaining 85–90% of cases appear to occur sporadically in the community. To date, mutations of at least 15 genes have been described to be related to familial ALS, the most common in Caucasian populations being C9ORF72 (Renton et al, 2011; DeJesus-Hernandez 2011), SOD1 (Rosen et al, 1993), TARDBP (Sreedharan et al, 2008) and FUS (Kwiatkowski et al, 2009; Vance et al, 2009), accounting for about 60% of familial cases and 5% of apparently sporadic patients (Chiò et al, 2012; van Blitterswijk et al, 2012; Kenna et al, 2013). The detection of genetic mutations in apparently sporadic ALS cases has been variously explained as reduced gene penetrance, misdiagnosis of ALS or early death in preceding generations, non-paternity, or de novo mutations (Chio et al, 2013).
Here, we present a case of an apparently familial ALS patient carrying a de novo missense mutation of the FUS gene.
2. Methods
While performing mutational screening of large series of ALS cases in Piemonte region, Italy, we detected a young onset apparently familial ALS patient carrying the p.R495X nonsense mutation (c.1483c>t) in exon 14 of FUS which causes the truncation of the final 32 amino acids of the protein from the C-terminus of FUS, abrogating a putative nuclear localization signal (Bosco et al, 2010). A first cousin of her maternal grandmother also had ALS, and was negative for this mutation. Since both her parents were still alive and not affected by ALS, we searched for the mutation in the parents.
2.1 Genetic analysis
Genomic DNA was extracted using a Biorobot MDX DSP (Qiagen Inc.). Exons 1 to 15 of FUS were sequenced as previously described (Vance et al, 2009; Lai et al, 2010, Chiò et al, 2009). In order to exclude that a SNP under the primers could lead to selective amplification of only normal allele, a second PCR and sequence was performed using a second set of primers with different binding site. PCR products were sequenced using the Big-Dye Terminator v3.1 sequencing kit (Applied Biosystem) and run on an ABIPrism 3100Avant genetic analyzer. Exon 14 was also sequenced in 368 control Italian individuals (Chiò et al, 2009; Lai et al, 2010). Quantitative fluorescence polymerase chain reaction (QF-PCR) was performed to assess paternity and maternity of the proband, with a multiplex analysis of short tandem repeats (STRs) located on five chromosomes (Devyser Resolution kit, Devyser). The electropherograms in all 5 chromosomes confirmed the paternity and the maternity of the proband.
2.2 Standard Protocol Approvals and Patient Consents
The study was approved by the Ethical committee of our institution. The patient and her family members signed a written informed consent.
3. Case history
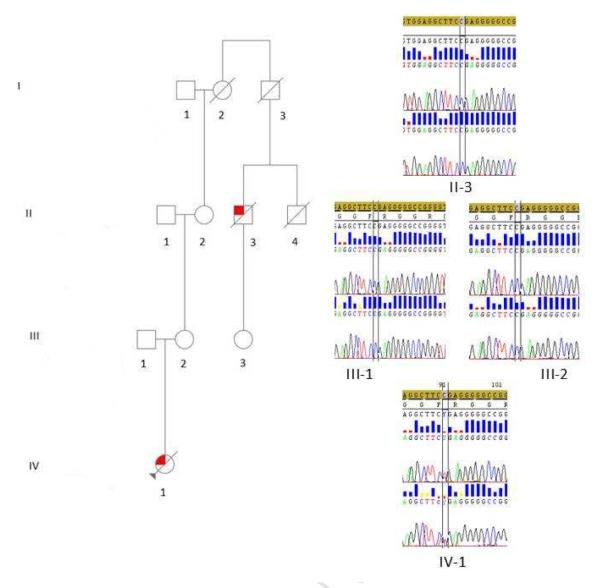
The patient's family pedigree is shown in Figure 1. The patient (III-5) was a 30 year-old woman who developed mild dysphagia and dysarthria at the age of 28 years. One year later she was referred to our ALS center because of a rapid worsening of bulbar symptoms and the onset of generalized asthenia. At neurological examination, the tongue with atrophic with fasciculation. Diffuse fasciculation were seen at upper and lower limbs, but muscle strength was normal. Deep tendon reflexes were hyperactive. Babinski and Hoffman signs were not present. She was cognitively normal. Neurophysiological examination demonstrated chronic and active denervation of tongue (genioglossus) and chronic denervation of proximal muscles of upper limbs, with normal repetitive nerve stimulation test. Cerebrospinal fluid examination was normal. Creatine kinase serum levels were raised. Head MRI showed a cortical atrophy at the precentral gyri; brain spectroscopy revealed a reduction of the NAA/Cr ratio in the motor cortex, more pronounced at left. She was diagnosed as possible ALS. In the following months, she underwent percutaneous radiological gastrostomy due to worsening of dysphagia and weight loss, and non-invasive ventilation due to a rapidly evolving respiratory failure. She refused tracheostomy and deceased from respiratory failure 24 months after the onset of symptoms.
Figure 1.
Family pedigree with chromatograms of part of exon 14 of FUS gene. Square indicates male; circle, female; slash, deceased; solid symbol, affected; and arrow, index patient. Chromatograms of part of exon 14 of FUS gene of the proband (IV-1), her parent (III-1 and III-2) and her grandmother's cousin (II-3) are shown.
She had no mutation in SMN1, SOD1, TARDBP and C9ORF72. She carried a c.1483c>t (p.R495X) truncating mutation of the FUS gene.
Her father (III-1) is 75 years of age and is healthy. Her mother (III-2) is 70 years old and healthy. Her maternal grandmother (II-2) is 91 and healthy. Her maternal great-grandmother (I-1) died at 87 from hearth failure.
A 82 year-old first cousin of the proband's maternal grandmother (II-3) developed right foot drop, subsequently progressing to proximal muscles and to the left lower limb. He was referred to our ALS clinic 6 months after the onset of symptoms. Brisk reflex with clonus at lower limbs and bilateral Babinski sign were found. Wasting at the right hand was also observed, with diffuse fasciculations at all limbs. Creatine kinase serum levels were raised. Neurophysiological examination demonstrated chronic and active denervation at upper and lower limbs. He could not undergo MRI due to claustrophobia. He was diagnosed as probable ALS. One year after the onset of symptoms he developed respiratory failure and underwent noninvasive ventilation. He died 31 months after the onset of ALS. He was cognitively normal. He had no mutation in FUS, SOD1, TARDBP and C9ORF72 genes. His brother (II-4) died at 60 due to renal failure. His daughter (III-3) is 65 years-old and healthy. His father (I-2) died at 54 from hepatic cirrhosis.
Neither the father nor the mother or the first cousin of the maternal grandmother carry the FUS mutation identified in the proband, nor any other mutation of genes related to ALS.
4. Discussion
Here we report a proven case of de novo FUS mutation presenting as a familial ALS. The parents as well the ALS affected first cousin of her maternal grandmother did not carry the mutation, and highly informative polymorphic markers confirmed paternity and maternity. This truncating mutation has been previously described both in familial (Bosco et al, 2010; Yang et al, 2010; Waibel et al, 2013) and apparently sporadic ALS patients (Kwon et al, 2012; van Blitterswijk et al, 2012; Zou et al, 2013). This particular mutation has not been detected in a large number of neurologically normal controls (Kwiatkowski et al, 2009; Vance et al, 2009; van Blitterswijk et al, 2012).
De novo mutations account for at least a part of patients affected by genetic diseases, as well known for familial adenomatous polyposis (Bisgaard et al, 1994) and Duchenne muscular dystrophy (Grimm et al, 2012). Recently, the detection of de novo mutations in patients with unaffected parents has led to the discovering of novel genes of autism (O'Roak et al, 2012; Neale et al, 2012; Sanders et al, 2012) and schizophrenia (Xu et al, 2011; Girard et al, 2011). In ALS, whole exome sequencing in young onset sporadic patients with unaffected relatives (so-called trios) represents a good resource for the discovery of new ALS genes, as recently demonstrated by a proof-of-concept study on 47 trios which identified several possible genetic mutations (Chesi et al, 2013).
The analysis of our pedigree raises some considerations. First, according to the proposed classification of familial ALS (FALS) (Byrne et al, 2011), our patient was initially classified as possible FALS, since a first cousin of her grand-mother also was diagnosed with ALS. The identification of the de novo mutation in the proband demonstrated that the two ALS cases in the same family were indeed phenocopies. The co-occurrence of two ALS cases in the same pedigree by chance is not negligible, since the risk of developing ALS in the Italian population is 1 out of 300 male and 1 out of 450 female (Chiò et al, 2009).
Second, the proband carried a truncating mutation of FUS. Three different truncating FUS mutations have been described, one in exon 15 in a French case (c.1555 C>T p.Q519X) (Belzil et al, 2011) and two in exon 14, i.e. a large heterozygous deletion of 47 nucleotides (p.G478LfsX23) in a German patient (Waibel et al, 2013) and the single nucleotide nonsense mutation found in our patients (c.1483c>t, p.R495X), which had been already described in familial and sporadic ALS patients (Table 1). Truncating mutations of FUS are characterized by an age at onset between the 2nd and 4th decade, a rapid course (usually <24 months) and a predominant bulbar phenotype. Interestingly, the p.R495X mutations has been detected both in Caucasian and in Asian patients.
Table 1.
Summary of FUS gene nonsense mutations identified in ALS patients
| Mutation | Transmission | Gender | Age at onset (years) | Onset | Duration (months) | Ancestry | Reference |
|---|---|---|---|---|---|---|---|
| p.G478LfsX23 | Familial | 1 M, 1 F | 21–26 | 2 B | 23–24 | Caucasian | Waibel et al, 2013 |
| c.1483c>t (p.R495X) | Familial | 2 M, 3 F | 14–39 | 1 B, 1 S* | 9–98 | Caucasian | Yan et al, 2010 |
| c.1483c>t (p.R495X) | Familial | 5 M, 3 F | 16–59 | 4 B, 4 S | 11–36 | Caucasian | Bosco et al, 2010 |
| c.1483c>t (p.R495X) | Sporadic | F | 29 | B | NR | Korean | Kwon et al, 2012 |
| c.1483c>t (p.R495X) | Sporadic | F | 19 | B | 24 | Caucasian | van Blitterswijk et al, 2012 |
| c.1483c>t (p.R495X) | Sporadic | M | 22 | S | >5 | Chinese | Zou et al, 2013 |
| c.1483c>t (p.R495X) | Familial | 1 M, 2 F | 31–36 | 3 B | 12–18 | Caucasian | Waibel et al, 2013 |
| c.1483c>t (p.R495X) | De novo | F | 28 | B | 24 | Caucasian | Present paper |
| c.1555 C>T (p.Q519X) | Familial | NR | NR | NR | NR | NR | Belzil et al, 2011 |
the site of onset of 3 cases is not reported. F, female; M, male; B, bulbar; S, spinal; NR, not reported.
Third, several de novo mutations involving the FUS gene have been described, including two missense mutations (c.1561C>T [p.R521C], one case, and c.1574 C>T [p. P525L], two cases) (Chiò et al, 2011; Conte et al, 2012, Zhou et al, 2013), a heterozygous 2-base pair deletion (c.1509_1510delAG [p.G504Wfs*12]) (Zhou et al, 2013), and a heterozygous splice-site mutation in FUS intron 13 (IVS13-2A>G) (Dejesus-Hernandez et al, 2010). Currently, only one de novo mutation in the SOD1 gene has been reported (Alexander et al, 2002). The relatively high frequency of de novo mutations in the FUS gene could be related to the younger age at onset usually associated with mutations in this gene compared to other ALS-related genes (Millecamps et al, 2010; Yan et al, 2010; Chiò et al, 2012), so that it is more likely that both patient's parents are still available for DNA analysis. Another possibility is that FUS gene is particularly susceptible to mutations (i.e. “mutational hotspots”), presumably because of the characteristics of the surrounding sequence (Chiò et al, 2011).
We have identified a proven de novo mutation in the FUS gene in a patient with an apparently familial ALS. This finding, together with previous identifications of other de novo mutations in ALS-related genes, indicates that at least a part of apparently sporadic patients are in fact characterized by de novo mutations that cannot be demonstrated due to the unavailability of the DNA of patients' parents. The relative frequency of proven de novo mutations in the FUS is therefore likely to be due to the young age at onset of subjects carrying these mutations.
Acknowledgements
Adriano Chiò had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. We thank the patient and her family for having collaborated to this study.
Funding/Support: This work was funded by grants of Fondazione Vialli e Mauro for ALS Research Onlus, Federazione Italiana Giuoco Calcio (FICG), Ministero della Salute (Ricerca Sanitaria Finalizzata 2007, grant RF-PIE-2007-635695, and 2010, grant RF-2010-2309849), and Joint Programme - Neurodegenerative Disease Research (Strenght Project). The research leading to these results has received funding from the European Community's Health Seventh Framework Programme (FP7/2007–2013) (grant agreement no. 259867). This work was supported in part by the Intramural Research Program of the NIH, and the National Institute on Aging (project Z01 AG000949-02). Funding organizations had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure: None reported.
Data contained in the manuscript being submitted have not been previously published, have not been submitted elsewhere and will not be submitted elsewhere while under consideration at Neurobiology of Aging.
References
- Alexander MD, Traynor BJ, Miller N, Corr B, Frost E, McQuaid S, Brett FM, Green A, Hardiman O. “True” sporadic ALS associated with a novel SOD1 mutation. Ann Neurol. 2002;52:680–3. doi: 10.1002/ana.10369. [DOI] [PubMed] [Google Scholar]
- Bäumer D, Hilton D, Paine SM, Turner MR, Lowe J, Talbot K, Ansorge O. Juvenile ALS with basophilic inclusions is a FUS proteinopathy with FUS mutations. Neurology. 2010;75:611–618. doi: 10.1212/WNL.0b013e3181ed9cde. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, St-Onge J, Daoud H, Desjarlais A, Bouchard JP, Dupré N, Camu W, Dion PA, Rouleau GA. Identification of a FUS splicing mutation in a large family with amyotrophic lateral sclerosis. J Hum Genet. 2011;56:247–249. doi: 10.1038/jhg.2010.162. [DOI] [PubMed] [Google Scholar]
- Belzil VV, Valdmanis PN, Dion PA, Daoud H, Kabashi E, Noreau A, Gauthier J, S2D team. Hince P, Desjarlais A, Bouchard JP, Lacomblez L, Salachas F, Pradat PF, Camu W, Meininger V, Dupré N, Rouleau GA. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73:1176–1179. doi: 10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard ML, Fenger K, Bülow S, Niebuhr E, Mohr J. Familial adenomatous polyposis (FAP): frequency, penetrance, and mutation rate. Hum Mutat. 1994;3:121–125. doi: 10.1002/humu.1380030206. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr., Sapp P, McKenna-Yasek D, Brown RH, Jr., Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granule. Hum Mol Genet. 2010;19:4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S, Bede P, Elamin M, Kenna K, Lynch C, McLaughlin R, Hardiman O. Proposed criteria for familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:157–159. doi: 10.3109/17482968.2010.545420. [DOI] [PubMed] [Google Scholar]
- Chesi A, Staahl BT, Jovičić A, Couthouis J, Fasolino M, Raphael AR, Yamazaki T, Elias L, Polak M, Kelly C, Williams KL, Fifita JA, Maragakis NJ, Nicholson GA, King OD, Reed R, Crabtree GR, Blair IP, Glass JD, Gitler AD. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Mora G, Calvo A, Mazzini L, Bottacchi E, Mutani R, PARALS Epidemiology of ALS in Italy. A 10-year populaton-based study. Neurology. 2009;72:725–731. doi: 10.1212/01.wnl.0000343008.26874.d1. [DOI] [PubMed] [Google Scholar]
- Chiò A, Calvo A, Moglia C, Ossola I, Brunetti M, Sbaiz L, Lai SL, Abramzon Y, Traynor BJ, Restagno G. A de novo missense mutation of the FUS gene in a “true” sporadic ALS case. Neurobiol Aging. 2011;32:553e23–553e26. doi: 10.1016/j.neurobiolaging.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Calvo A, Mazzini L, Cantello R, Mora G, Moglia C, Corrado L, D'Alfonso S, Majounie E, Renton A, Pisano F, Ossola I, Brunetti M, Traynor BJ, Restagno G, PARALS Extensive genetics of ALS: a population-based study in Italy. Neurology. 2012;79:1983–1989. doi: 10.1212/WNL.0b013e3182735d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò A, Battistini S, Calvo A, Caponnetto C, Conforti FL, Corbo M, Giannini F, Mandrioli J, Mora G, Sabatelli M, ITALSGEN Consortium. Ajmone C, Mastro E, Pain D, Mandich P, Penco S, Restagno G, Zollino M, Surbone A. Genetic counselling in ALS: facts, uncertainties and clinical suggestions. J Neurol Neurosurg Psychiatry. 2013 Jul 6; doi: 10.1136/jnnp-2013-305546. doi:10.1136/jnnp-2013-305546. [DOI] [PubMed] [Google Scholar]
- Conte A, Lattante S, Zollino M, Marangi G, Luigetti M, Del Grande A, Servidei S, Trombetta F, Sabatelli M. P525L FUS mutation is consistently associated with a sevr form of juvenile amyotrophic lateral sclerosis. Neuromus Dis. 2012;21:73–75. doi: 10.1016/j.nmd.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Dejesus-Hernandez M, Kocerha J, Finch N, Crook R, Baker M, Desaro P, Johnston A, Rutherford N, Wojtas A, Kennelly K, Wszolek ZK, Graff-Radford N, Boylan K, Rademakers R. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum Mutat. 2010;31:E1377–E1389. doi: 10.1002/humu.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GYR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geshwind DH, Wszolek Z, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademarkers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, Thibodeau P, Bachand I, Bao JY, Tong AH, Lin CH, Millet B, Jaafari N, Joober R, Dion PA, Lok S, Krebs MO, Rouleau GA. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Grimm T, Kress W, Meng G, Müller CR. Risk assessment and genetic counseling in families with Duchenne muscular dystrophy. Acta Myol. 2012;31:179–183. [PMC free article] [PubMed] [Google Scholar]
- Kenna KP, McLaughlin RL, Byrne S, Elamin M, Heverin M, Kenny EM, Cormican P, Morris DW, Donaghy CG, Bradley DG, Hardiman O. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. J Med Genet. 2013;50:776–83. doi: 10.1136/jmedgenet-2013-101795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr., Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kwon MJ, Baek W, Ki CS, Kim HY, Koh SH, Kim JW, Kim SH. Screening of the SOD1, FUS, TARDBP, ANG, and OPTN mutations in Korean patients with familial and sporadic ALS. Neurobiol Aging. 2012;33:1017.e17–1017.e23. doi: 10.1016/j.neurobiolaging.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Lai SL, Abramzon Y, Schymick JC, Stephan DA, Dunckley T, Dillman A, Cookson M, Calvo A, Battistini S, Giannini F, Caponnetto C, Mancardi GL, Spataro R, Monsurro MR, Tedeschi G, Marinou K, Sabatelli M, Conte A, Mandrioli J, Sola P, Salvi F, Bartolomei I, Lombardo F, ITALSGEN Consortium. Mora G, Restagno G, Chiò A, Traynor BJ. FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging. 2010;32:550.e1–550.e4. doi: 10.1016/j.neurobiolaging.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Boillée S, Le Ber I, Seilhean D, Teyssou E, Giraudeau M, Moigneu C, Vandenberghe N, Danel-Brunaud V, Corcia P, Pradat PF, Le Forestier N, Lacomblez L, Bruneteau G, Camu W, Brice A, Cazeneuve C, Leguern E, Meininger V, Salachas F. Phenotype difference between ALS patients with expanded repeats in C9ORF72 and patients with mutations in other ALS-related genes. J Med Genet. 2012;49:258–263. doi: 10.1136/jmedgenet-2011-100699. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser R, Jabado O, Peralta Z, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y, Lewis L, Han Y, Voight BF, Lim E, Rossin E, Kirby A, Flannick J, Fromer M, Shakir K, Fennell T, Garimella K, Banks E, Poplin R, Gabriel S, DePristo M, Wimbish JR, Boone BE, Levy SE, Betancur C, Sunyaev S, Boerwinkle E, Buxbaum JD, Cook EH, Jr, Devlin B, Gibbs RA, Roeder K, Schellenberg GD, Sutcliffe JS, Daly MJ. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, ITALSGEN Consortium. Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, Rahmani Z, Krizus A, McKenna-Yasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Van den Bergh R, Hung W-Y, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericak-Vance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH., Jr. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha RC, Choi M, Overton JD, Bjornson RD, Carriero NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Günel M, Roeder K, Geschwind DH, Devlin B, State MW. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, van Es MA, Hennekam EA, Dooijes D, van Rheenen W, Medic J, Bourque PR, Schelhaas HJ, van der Kooi AJ, de Visser M, de Bakker PI, Veldink JH, van den Berg LH. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:3776–3734. doi: 10.1093/hmg/dds199. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waibel S, Neumann M, Rosenbohm A, Birve A, Volk AE, Weishaupt JT, Meyer T, Mueller U, Andersen PM, Ludolph AC. Truncating mutations in FUS/TLS give rise to a more aggressive ALS-phenotype than missense mutations: a clinico-genetic study in Germany. Eur J Neurol. 2013;20:540–546. doi: 10.1111/ene.12031. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Deng HX, Siddique N, Fecto F, Chen W, Yang Y, Liu E, Donkervoort S, Zheng JG, Shi Y, Ahmeti KB, Brooks B, Engel WK, Siddique T. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology. 2010;75:807–814. doi: 10.1212/WNL.0b013e3181f07e0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou ZY, Cui LY, Sun Q, Li XG, Liu MS, Xu Y, Zhou Y, Yang XZ. De novo FUS gene mutations are associated with juvenile-onset sporadic amyotrophic lateral sclerosis in China. Neurobiol Aging. 2013;34:1312.e1–1312.e8. doi: 10.1016/j.neurobiolaging.2012.09.005. [DOI] [PubMed] [Google Scholar]