Abstract
Several lines of inquiry point to overlapping molecular mechanisms between late-onset Alzheimer disease (AD) and age-related macular degeneration (AMD). We evaluated summarized results from large genome-wide association studies (GWAS) for AD and AMD to test the hypothesis that AD susceptibility loci are also associated with AMD. We observed association of both disorders with genes in a region of chromosome 7 including PILRA, and ZCWPW1 (peak AMD SNP rs7792525, MAF=19%, OR=1.14, p=2.34×10−6), and with ABCA7 (peak AMD SNP rs3752228, MAF=0.054 OR=1.22, p=0.00012). Next, we evaluated association of AMD with genes in AD-related pathways identified by canonical pathway analysis of AD-associated genes. Significant associations were observed with multiple previously identified AMD risk loci and two novel genes: HGS (peak SNP rs8070488, MAF=0.23, OR=0.91, p=7.52×10−5), which plays a role in the clathrin-mediated endocytosis signaling pathway, and TNF (peak SNP rs2071590, MAF=0.34, OR=0.89, p=1.17×10−5), which is a member of the atherosclerosis signaling and the LXR/RXR activation pathways. Our results suggest that AMD and AD share genetic mechanisms.
Keywords: Alzheimer disease, Age related macular degeneration, genetic association, gene-based test, pathway analysis
1. Introduction
Age-related macular degeneration (AMD) is the most common form of severe blindness and vision loss among those over 60 years of age(Congdon, et al., 2004). The common dry form (i.e. non-neovascular) accounts for approximately 85–90% of AMD cases and the advanced acute wet form (i.e. exudative, neovascular) is responsible for the majority of persons with AMD who are legally blind. AMD pathogenesis is complex—a result of both genetic and environmental risk factors.
There are several well-established common genetic risk factors for AMD. The loci with the most robust replication across multiple populations and identified as having the strongest effect on AMD risk (odds ratio, OR, for a single risk allele > 3) are CFH and ARMS2/HTRA1(Edwards, et al., 2005,Haines, et al., 2005,Jakobsdottir, et al., 2005,Klein, et al., 2005,Rivera, et al., 2005). Candidate gene association studies identified other AMD risk genes in the complement pathway including C2/CFB(Gold, et al., 2006), C3(Maller, et al., 2007,Yates, et al., 2007), and CFI (Fagerness, et al., 2009) The APOE gene has been linked to AMD, with the ε2 and ε4 alleles associated with increased risk and decreased risk of AMD, respectively(McKay, et al., 2011). Other studies were unable to confirm these associations(Pang, et al., 2000,Schultz, et al., 2003) that are attenuated or absent when adjusted for age(Adams, et al., 2012). Many other loci of modest effect have also been identified and replicated in genome-wide association studies (GWAS) including CETP, LIPC, TIMP3, VEGFA, TNFRSF10A, COL10A1, COL8A1/FILIP1L, SLC16A8, IER3/DDR1, TGFBR1, RAD51B, ADAMTS9/MIR548A2, and B3GALTL(Chen, et al., 2010,Fritsche, et al., 2013,Yu, et al., 2011). Several of these loci, including CFH, C3, LIPC, and DDR1, have multiple risk variants that independently contribute to disease risk(Fritsche, et al., 2013,Gold, et al., 2006,Li, et al., 2006,Maller, et al., 2006).
Like AMD, late-onset Alzheimer disease (AD) is a common disorder among the elderly that has a strong but complex genetic basis including a major contribution by APOE. The ε4 allele confers increased risk of AD and the ε2 is protective(Corder, et al., 1994,Corder, et al., 1993,Farrer, et al., 1997), effect directions opposite to those reported with AMD (McKay, et al., 2011). A hypothesis-driven study demonstrated that SORL1 is genetically associated with AD(Rogaeva, et al., 2007), a finding subsequently confirmed by GWAS(Lambert, et al., 2013,Miyashita, et al., 2013). Large-scale consortium GWAS studies have successfully identified 20 other modest effect loci including PICALM, CR1, CLU, BIN1, ABCA7, CD2AP, CD33, EPHA1, MS4A4A/MS4A6E, HLA-DRB5/HLADRB1, SLC24A4/RIN3, DSG2, INFP5D, MEF2C, NME8, ZCWPW1, CELF1, FERMT2, and CASS4(Harold, et al., 2009,Hollingworth, et al., 2011,Lambert, et al., 2009,Lambert, et al., 2013,Naj, et al., 2011,Seshadri, et al., 2010).
Multiple lines of evidence indicate that AD and AMD risk may share molecular mechanisms. Analyses of a sample from a cardiovascular-health study(Baker, et al., 2009) found that early AMD was associated with low cognitive functioning. Additionally, late-stage AMD has been associated with incident AD(Klaver, et al., 1999). Both AMD and AD are characterized by abnormal extra-cellular deposits: amyloid-β (Aβ) plaques in AD and drusen in AMD. These deposits share a similar molecular composition including complement factor proteins(Dentchev, et al., 2003). Aβ has been found in drusen and AMD retinas(Dentchev, et al., 2003). Anti-Aβ substrate reduced pathological features in a mouse model of AMD(Ding, et al., 2008). In addition, drusen contain the AD-related APOE protein (Mullins, et al., 2000). AMD and AD share indicators of poor vascular health as risk factors. Smoking(Anstey, et al., 2007,Klein, et al., 1993), hypertension and/or higher blood pressure(Hyman, et al., 2000,Kennelly, et al., 2009,The Eye Disease Case-Control Study Group, 1992,van Leeuwen, et al., 2003), and atherosclerosis(Casserly and Topol, 2004,van Leeuwen, et al., 2003) are risk factors for both disorders. AMD risk is related to lower levels of high-density lipoprotein cholesterol (HDL-c) and both disorders are associated with higher serum cholesterol(Hyman, et al., 2000,Kivipelto, et al., 2001,The Eye Disease Case-Control Study Group, 1992), although not all studies replicate this(Klein, et al., 2003,Reitz, et al., 2004).
Given the evidence of etiological overlap between these two disorders, we hypothesized that findings from AD genetic studies can inform a search for novel AMD genes. This mirrors examination of AMD risk variants in complement factor genes as possible risk factors for AD(Gatta, et al., 2008,Hamilton, et al., 2007,Le Fur, et al., 2010,Proitsi, et al., 2012,Zetterberg, et al., 2008). These studies have found that complement factor genes with variants which are highly predictive of AMD play at most a modest role in the risk of AD. However, the direction of effect, the genetic models which appear most predictive, and which SNPs in the genes are associated can differ between the disorders(Gatta, et al., 2008,Proitsi, et al., 2012). In this study, we incorporated results from individual SNP, gene-based, and biological pathway analyses of AD in a design to discover additional AMD risk loci.
2. Materials and methods
2.1 Materials
Primary data for this investigation are summarized results for a common set of more than two million HapMap2 imputed SNPs from the Age-related Macular Degeneration Genetics (AMDGene) Consortium GWAS for AMD(Fritsche, et al., 2013) and the Alzheimer Disease Genetics Consortium (ADGC) for AD(Naj, et al., 2011). The ADGC sample includes 11,840 cases and 10,931 controls from 15 different studies(Naj, et al., 2011). Top-ranked findings from the AD GWAS were examined in the AMDGene GWAS reported by Fritsche et al. (2013) which contained data from more than 7,600 cases and 50,000 controls who were enrolled in 14 separate studies.
2.2 Genetic Analyses
Our first line of investigation was an analysis to identify SNPs that are associated with risk to both disorders. Because the focus of our study was on identification of new AMD risk loci, we excluded SNPs within 1 megabase (Mb) of a genome-wide significant (p<5×10−8) SNP from the AMDGene GWAS. In this approach, the AD dataset was used for discovery and the top-ranked SNPs were “replicated” in the AMD dataset. We examined association of AMD with all SNPs meeting a Benjamini–Hochberg false-discovery rate (FDR; (Benjamini and Hochberg, 1995) of < 10% in the AD dataset. These SNPs were considered to be significantly associated with AMD if the p-value exceeded a Bonferroni-adjusted significance threshold of 0.05 divided by the number of examined SNPs. Because the association between SNPs in the APOE region and AMD is well documented and the region over which linkage disequilibrium (LD) extends is very large, we excluded SNPs and genes located between positions 45.3 and 45.8 Mb on chromosome 19.
Next, we employed a genome-wide gene-based approach to identify Alzheimer genes to be tested for association with AMD. This analysis was performed by first identifying the peak SNP within 5 kb of each gene and then applying a multiple-testing correction based on the effective number of independent tests represented by the SNPs in the gene as determined by the Li and Ji method(Li and Ji, 2005) which accounts for linkage disequilibrium (LD). Determining the significance of a gene using the peak SNP adjusted with the Li and Ji approach contrasts with other gene-based methods (e.g., VEGAS(Liu, et al., 2010) which consider information from multiple SNPs in the gene region. LD between SNPs was estimated using information derived from the Caucasian controls in the MMAP AMD sample(Chen, et al., 2010,Fritsche, et al., 2013). These corrected p-values were then used to compute a FDR for each gene(Benjamini and Hochberg, 1995). For this investigation, genes were considered as possible AD loci for subsequent analyses with AMD if the gene-based test statistic exceeded a 10% FDR cutoff. These AD-implicated genes were then evaluated for association with AMD using the peak SNP approach described above. A gene was determined to be significantly associated with AMD if its gene-level corrected significance (pcorrected) was less than 0.05/k where k is the number of putative AD genes examined.
Finally, we applied a biological pathway approach to identify additional genes which are involved in processes related to AD based on canonical pathway analysis using the Ingenuity pathway analysis (IPA) software; http://www.ingenuity.com). We identified AD-related pathways in three ways. First, we investigated the list of all genes with an FDR of less than 10% in a test of association with AD (i.e., the genes examined for association with AMD as described above). In a separate analysis, we looked for pathways which were enriched for the genes in the AlzGene database (www.alzgene.org) which were significant according to a meta analysis of curated information from the literature using AlzGene’s methodology (Lars Bertram Personal Communication, June 16, 2012). The significant genes in the AlzGene and GWAS FDR10% lists are not independent because AlzGene incorporated significant results from published GWAS including the ADGC GWAS. We also conducted a separate pathway analysis of the 10% FDR genes after excluding the genes in high LD with APOE. The methodology for these analyses is similar to the gene-enrichment analyses of AD risk performed by Jones et al. (Jones, et al., 2010), however, that study examined enrichment in gene-ontology (GO)(Harris, et al., 2004) and KEGG(Kanehisa, et al., 2006) functional categories, whereas IPA utilizes the Ingenuity Knowledge Base (IKB)—a curated database of gene-gene interactions and functions summarizing the findings of over 200,000 scientific articles. The canonical pathway analysis implemented in IPA entails computation of an enrichment score based on the number of genes in the provided target list that fall into a specific canonical pathway (similar to a GO term) and calculates the probability that this pathway is enriched for the genes using Fisher’s Exact Test of association. A Benjamini– Hochberg false discovery rate (FDR) approach was used to adjust the resulting p-values for the multiplicity of INGENUITY pathways examined within each of the three gene sets (FDR). Because this method often yields many highly significant pathways and each pathway may contain hundreds of genes, we restricted our attention to the most significantly and consistently implicated pathways; that is, pathways that are most enriched for AD genes across the three analyses. Genes in each of these pathways were evaluated for association with AMD using a gene-level test as described above. Within each pathway, genes were considered significant if they had a p-value less than 0.05/k where k is the number of genes in the pathway.
Figures were generated using R (http://www.r-project.org) and LocusZoom (http://csg.sph.umich.edu/locuszoom/). Presented genomic positions are based on the hg19 (February 2009) human genome assembly. LD estimates and LD-plots were computed using Haploview (http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview) based on the white non-Hispanic subjects comprising the 1000 Genomes EUR sample. Minor allele frequency estimates were also derived from the EUR sample.
3. Results
A total of 226 SNPs in the summary results for the AD meta-analysis dataset exceeded a 10% FDR cutoff and were more than 1 Mb away from a previously established genome-wide significant AMD association. None of these SNPs survived Bonferroni correction (p<2.2×10−4) when evaluated for association with AMD. In fact, only ABCA7 SNP rs3752246, which was associated with AD (p = 5.79×10−7), was nominally associated with AMD (p = 0.038). To ensure that the lack of correspondence between the two disorders was not an artifact of overly stringent discovery criteria in the AD sample, we also examined all 485 SNPs which were associated with AD at p<10−4 and not within one Mb of known AMD loci. The most significant finding among these SNPs was rs12539172 in C7ORF71 (AD: p = 6.35×10−5; AMD: p = 0.0016) which did not exceed the multiple testing correction threshold of 0.05/485= 0.00010. Hence, loosening the criteria for SNPs associated with AD did not yield significant evidence of a shared locus with AMD.
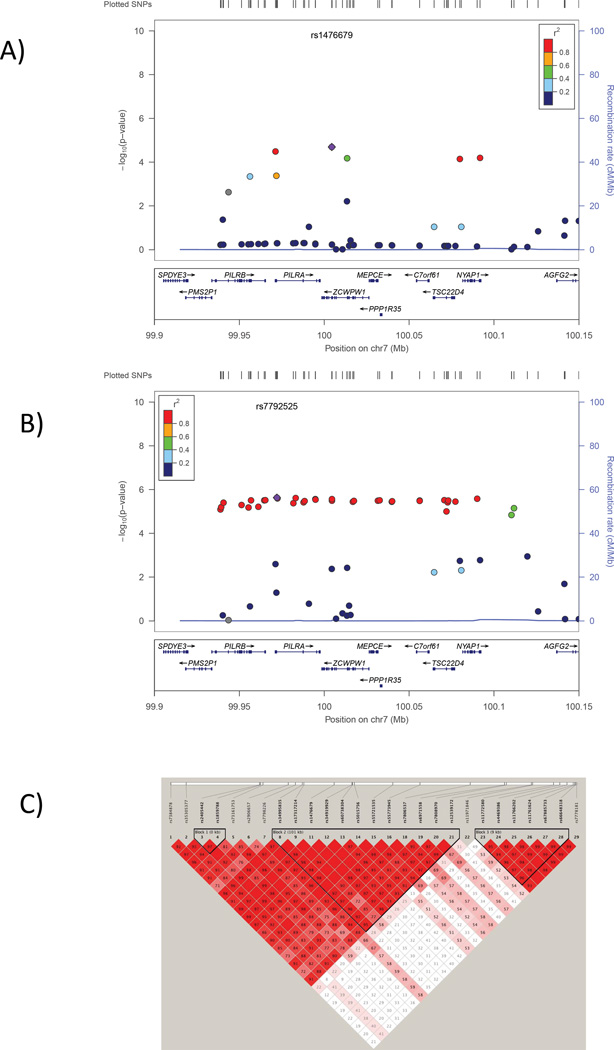
Next, we tested the hypothesis that, even though the same variants may not be shared across the two disorders, other variants in AD-related genes may be associated with AMD risk. Based on a FDR cutoff of 10%, 59 genes were identified which are potentially AD-related, including 24 genes in the APOE region and several previously established AD genes: ABCA7, BIN1, PICALM, CD2AP, SORL1, EPHA1and the MS4A–cluster genes (Supplementary Table 1). We evaluated the group of 35 genes outside of the APOE region for association with AMD. Three of these were significantly associated with AMD after correcting for the number of genes examined including PILRA, ZCWPW1, and ABCA7 (Table 1). PILRA and ZCWPW1 are adjacent genes on chromosome 7q22 whose 3’ UTR regions are separated by 773 bp. The most significant associations to AMD within 5 kb of each gene (that is, the peak SNP on which the gene-level significance is based) is rs7792525 for PILRA (p=7.02×10−6) and rs11771241 for ZCWR1P1 (p=1.61×10−5; Table 1). In fact, rs11771241 is less than 5 kb from ZCWR1P1 but within the boundaries of PILRA. Similar evidence of association was observed for other SNPs in both of these loci with AD and AMD (Figure 1). The association peak for both disorders extends across a region of high LD from approximately 99.90 to 100.15 Mb (Figure 1) encompassing several other genes including PLIRB, MEPCE, PPP1R35, C7ORF61, TSC22D4, and NYAP1. The congruence of results for AD and AMD in this region is modest at the SNP level. The most significantly associated AD SNP, rs1476679, is nominally significantly associated with AMD (p=0.0042, OR=0.93). However, the most-significantly associated AMD SNPs are not associated with AD. For example, the top AMD SNP, rs7792525, is not associated with AD (p=0.51). Thus, the functional variants for AMD and AD in this region may not be identical.
Table 1.
AD-related genes which were significantly associated with AMD at the gene level (pcorrected < 0.0014) after correcting for the 35 autosomal non-APOE region genes examined. The most significant SNP within 5 kb of each gene is shown.
| GENE | SNP | CHR | BP | MA | MAFa | OR | p | pcorrected |
|---|---|---|---|---|---|---|---|---|
| PILRA | rs7792525 | 7 | 99972122 | G | 17.9% | 1.14 | 2.34E-06 | 7.02E-06 |
| ZCWPW1 | rs11771241 | 7 | 99994785 | A | 19.4% | 1.14 | 2.68E-06 | 1.61E-05 |
| ABCA7 | rs3752228 | 19 | 1041164 | T | 3.6% | 1.22 | 0.00012 | 0.0012 |
MAF= Minor allele frequency, based on 1000 Genomes EUR population.; pcorrected=The gene-level corrected significance based on the peak SNP adjusted for effective number of tests within a gene.
Figure 1.
Association of SNPs in the chromosome 7q22 region with AD (panel A) and AMD (panel B). Significance of the association of each SNP (circles) is plotted as the negative logarithm of the p-value. Correlations (r2) between tested SNPs with the SNP most significantly associated with AD (ZCWPW1 SNP rs1476679) and AMD (PILRA SNP rs7792525) are indicated using the color scheme shown in the legend. Map positions and direction of transcription of genes in the region are shown in the bottom portion of each panel. C) Linkage disequilibrium (LD) across the same region. The measure of disequilibrium (D’) between each pair of SNPs is shown in each square. A summary of the strength of LD across the region is indicated by color such that dark red shading indicates very strong LD and white shading indicates weak or no LD. Discrete blocks of LD are demarcated with black triangles.
Pathway analysis of genes identified as significantly associated with AD by gene-based analyses (FDR<10%) revealed very significant associations with the clathrin-mediated endocytosis signaling (FDR=6.61×10−6), LXR/RXR activation (FDR=1.10×10−4) and atherosclerosis signaling (FDR=1.10×10−4) pathways (Table 2A). These pathways are not entirely distinct; approximately 40% of the genes are common to the atherosclerosis and LXR/RXR activation pathways. The same three pathways were also the most significant based on analyses of genes which were nominally significant using the AlzGene methodology (Table 2B), however their order based on significance level is reversed: LXR/RXR Activation (FDR = 6.31×10−11), atherosclerosis signaling (FDR = 6.31×10−11), and clathrin-mediated endocytosis signaling (FDR = 1.55×10−6). We repeated the pathway analysis with the top genes identified using the gene-based (FDR< 10%) approach after excluding genes in LD with APOE (Table 2C). The three “top” pathway from the previous analyses were again significantly enriched for AD genes. The clathrin-mediated endocytosis signaling pathway remained significant after FDR correction (FDR=0.0053), but the LXR/RXR activation and atherosclerosis signaling pathways did not (FDR=0.11 for each). Therefore, we examined the clathrin-mediated endocytosis signaling, LXR/RXR activation, and atherosclerosis signaling pathways genes for AMD-predisposing variants.
Table 2.
The canonical pathways most-significantly enriched for Alzheimer-related genes based on an INGENUITY analysis of the A) the 59 genes with < 10% FDR in association with AD, B) the 54 nominally significant AD loci using AlzGene’s methodology, and C) the 36 genes with 10% FDR after excluding the other APOE-region genes such as TOMM40 and PVRL2.
| A) Top Pathways from FDR 10% Alzheimer-associated Genes | ||||
|---|---|---|---|---|
| Ingenuity Canonical Pathways | p | FDR* | Ratio | Genes |
| Clathrin-mediated Endocytosis Signaling | 1.20E-7 | 6.61E-06 | 0.036 |
APOC1, APOE, CD2AP PICALM, APOC4, APOC2, CLU |
| LXR/RXR Activation | 5.89E-06 | 1.10E-04 | 0.037 |
APOC1, APOE, APOC4 APOC2, CLU |
| Atherosclerosis Signaling | 5.89E-06 | 1.10E-04 | 0.037 |
APOC1, APOE, APOC4 APOC2, CLU |
| IL-12 Signaling and Production in Macrophages |
9.55E-06 | 1.35E-04 | 0.032 |
APOC1, APOE, APOC4 APOC2, CLU |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages |
3.89E-05 | 4.37E-04 | 0.024 |
APOC1, APOE, APOC4 APOC2, CLU |
| B) Top Pathways from AlzGene Nominally Significant Genes | ||||
|---|---|---|---|---|
| Ingenuity Canonical Pathways | p | FDR* | Ratio | Genes |
| LXR/RXR Activation | 1.00E-12 | 6.31E-11 | 0.074 |
IL33, APOC1, APOE, IL1A LDLR, TF, APOC4, IL1B, TNF CLU |
| Atherosclerosis Signaling | 1.00E-12 | 6.31E-11 | 0.073 |
IL33, APOC1, IL8, APOE, IL1A APOC4, IL1B, CCR2, TNF, CLU |
| Clathrin-mediated Endocytosis Signaling | 4.07E-08 | 1.55E-06 | 0.041 | APOC1, APOE, CD2AP, LDLR, TF, PICALM, APOC4, CLU |
| Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza |
1.38E-07 | 3.89E-06 | 0.11 | IL33, IL8, IL1A, IL1B, TNF |
| Role of Cytokines in Mediating Communication between Immune Cells |
4.37E-07 | 9.77E-06 | 0.091 | IL33, IL8, IL1A, IL1B, TNF |
| C) Top Pathways from FDR 10% Alzheimer-associated Genes- Excluding genesin LD with APOE. | ||||
|---|---|---|---|---|
| Ingenuity Canonical Pathways | p | FDR* | Ratio | Genes |
| Clathrin-mediated Endocytosis Signaling | 1.17E-04 | 0.0053 | 0.020 | APOE,CD2AP,PICALM,CLU |
| Role of Tissue Factor in Cancer | 0.011 | 0.11 | 0.017 | PTK2B,HBEGF |
| RhoA Signaling | 0.012 | 0.11 | 0.017 | PTK2B,EPHA1 |
| LXR/RXR Activation | 0.012 | 0.11 | 0.015 | APOE,CLU |
| Atherosclerosis Signaling | 0.012 | 0.11 | 0.015 | APOE,CLU |
| IL-12 Signaling and Production in Macrophages |
0.015 | 0.11 | 0.013 | APOE,CLU |
Significance adjusted for multiple pathways examined using Benjamini–Hochberg false discovery rate (FDR).
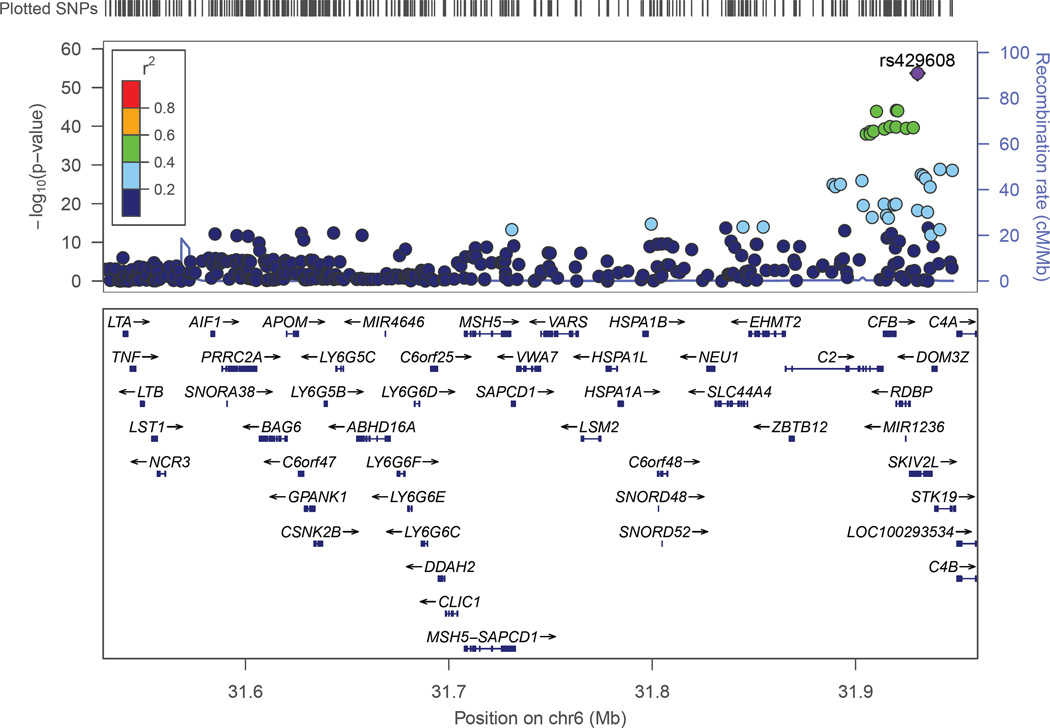
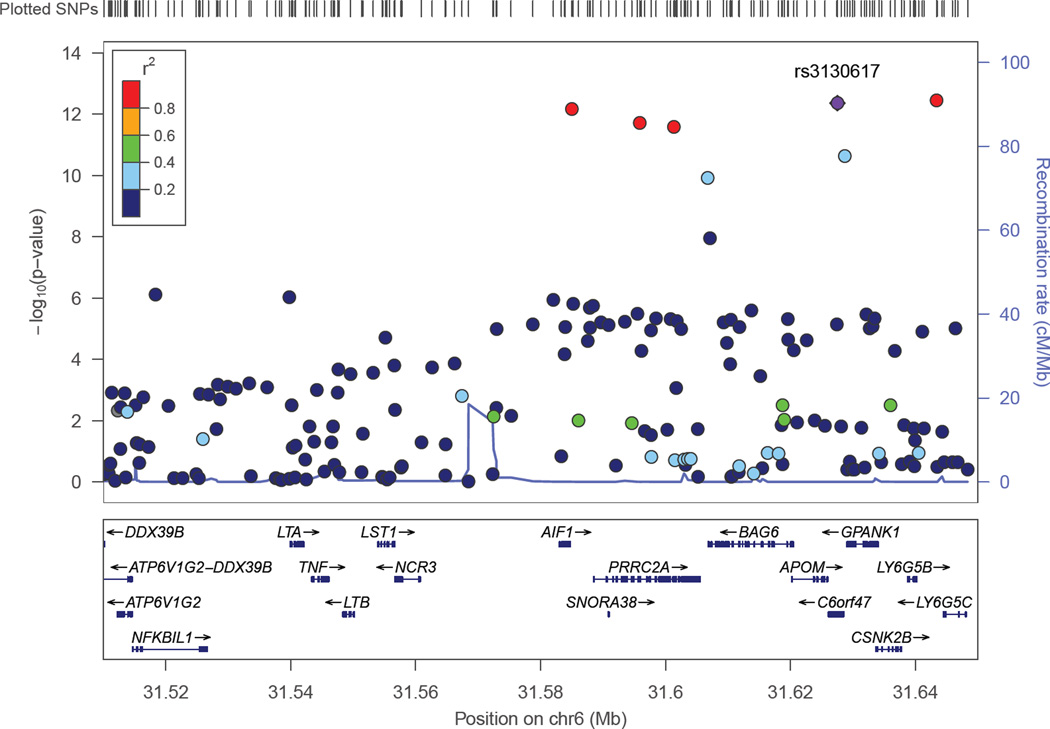
The clathrin-mediated endocytosis signaling pathway has 192 genes (excluding genes in the APOE region which were not evaluated for association with AMD). Three of these genes are significantly associated with AMD after correction for multiple testing (Table 3). The two most significant genes, APOM and CSNK2B, are in moderate LD (r2=0.275) and located in the major histocompatibility region between 250 kb and 300 kb away from the C2-CFB locus (Figure 2). These SNPs are not in LD with the peak C2-CFB SNP (rs429608, r2<0.05), however, several SNPs in the region including APOM and CSNK2B were genome-wide significant in the large AMD GWAS (Fritsche, et al., 2013). Thus, on the basis of these results alone, it is unclear whether these are veritable independent associations. The third significant gene identified from this pathway was HGS. The association peak was observed with rs8070488 (p=1.88×10−5) which is a synonymous coding variant (P595P). Association of AMD with SNPs in this region has not been reported previously. None of the HGS-region SNPs were associated with AD (p>0.25).
Table 3.
Clathrin-mediated endocytosis signaling pathway genes that are significantly associated (pcorrected<0.00026) with AMD after correcting for the 192 tests of genes outside of the APOE region.
| GENE | SNP | CHR | BP | MA | MAFa | OR | P | pcorrected |
|---|---|---|---|---|---|---|---|---|
| APOM | rs3130617 | 6 | 31627523 | C | 22.6% | 0.83 | 4.39E-13 | 4.39E-12 |
| CSNK2B | rs805262 | 6 | 31628733 | T | 46.3% | 1.20 | 2.44E-11 | 2.69E-10 |
| HGS | rs8070488 | 17 | 79663931 | C | 25.5% | 0.91 | 1.88E-05 | 7.52E-05 |
MAF= Minor allele frequency, based on 1000 Genomes EUR population.; pcorrected=The gene-level corrected significance based on the peak SNP adjusted for effective number of tests within a gene.
Figure 2.
Association of AMD with SNPs in the major histocompatibility locus region. Significance of the association of each SNP (circles) is plotted as the negative logarithm of the p-value. Correlations (r2) between tested SNPs with the SNP most significantly associated with AMD in this region (rs429608) are indicated using the color scheme shown in the legend. Map positions and direction of transcription of genes in the region are shown in the bottom portion of the figure.
Six LXR/RXR activation pathway genes were significantly associated with AMD after correction for the 132 genes examined in the pathway (Table 4). Genome-wide significant SNPs were found in three genes in the MHC region (APOM, C4A, and C4B), CETP, and C3. The TNF SNP was also significant after multiple test correction (p=1.17×10−5). TNF SNPs were not in LD with the peak C2-CFB SNP located more than 330 kb away, but were modestly correlated with APOM (r2=0.173) and CSNK2B (r2=0.083) (Figure 2). We also tested association of AMD with a different set of 132 atherosclerosis signaling genes and obtained significant results with SNPs in four genes including APOM and TNF, which were identified in the other two pathways studied, and COL10A1 which is among the previously reported AMD genes (Fritsche, et al., 2013,Yu, et al., 2011). The fourth gene, PLA2G12A (Table 5), is located near CFI, another previously established AMD risk locus (Arakawa, et al., 2011,Fagerness, et al., 2009).
Table 4.
LXR/RXR activation pathway genes that were significantly associated (pcorrected<0.00038) with AMD after correcting for the 132 tests of genes outside of the APOE region.
| GENE | SNP | CHR | BP | MA | MAFa | OR | p | Pcorrected |
|---|---|---|---|---|---|---|---|---|
| C3 | rs2230199 | 19 | 6718387 | C | 20.4% | 1.46 | 1.74E-26 | 2.95E-25 |
| C4A | rs389512 | 6 | 31947594 | C | 13.5% | 0.69 | 2.11E-29 | 8.44E-29 |
| APOM | rs3130617 | 6 | 31627523 | C | 22.6% | 0.83 | 4.39E-13 | 4.39E-12 |
| CETP | rs1864163 | 16 | 56997233 | A | 26.8% | 0.80 | 7.68E-13 | 1.23E-11 |
| C4B | rs6472 | 6 | 32007849 | C | 11.2% | 0.80 | 4.52E-09 | 1.81E-08 |
| TNF | rs2071590 | 6 | 31539768 | A | 36.5% | 0.89 | 9.76E-07 | 1.17E-05 |
MAF= Minor allele frequency, based on 1000 Genomes EUR population.; pcorrected=The gene-level corrected significance based on the peak SNP adjusted for effective number of tests within a gene.
Table 5.
Atherosclerosis signaling pathway genes that were significantly associated (pcorrected<0.00038) with AMD after correcting for the 132 tests of genes outside of the APOE region.
| GENE | SNP | CHR | BP | MA | MAFa | OR | P | pcorrected |
|---|---|---|---|---|---|---|---|---|
| APOM | rs3130617 | 6 | 31627523 | C | 22.6% | 0.83 | 4.39E-13 | 4.39E-12 |
| COL10A1 | rs3812111 | 6 | 116443735 | A | 39.2% | 0.89 | 7.19E-08 | 5.76E-07 |
| PLA2G12A | rs17586561 | 4 | 110648632 | A | 42.2% | 1.12 | 1.26E-06 | 5.02E-06 |
| TNF | rs2071590 | 6 | 31539768 | A | 36.5% | 0.89 | 9.76E-07 | 1.17E-05 |
MAF= Minor allele frequency, based on 1000 Genomes EUR population; pcorrected=The gene-level corrected significance based on the peak SNP adjusted for effective number of tests within a gene.
4. Discussion
The goal of this study was to identify novel AMD loci in a hypothesis-driven approach based on the idea that AMD and AD have shared genetic underpinnings. Although we did not observe significant overlap in genetic association at the single-SNP level, our gene-based analyses and investigation of biological pathways implicated several novel AMD loci including ABCA7, HGS, and PILRA/ZCW1P1.
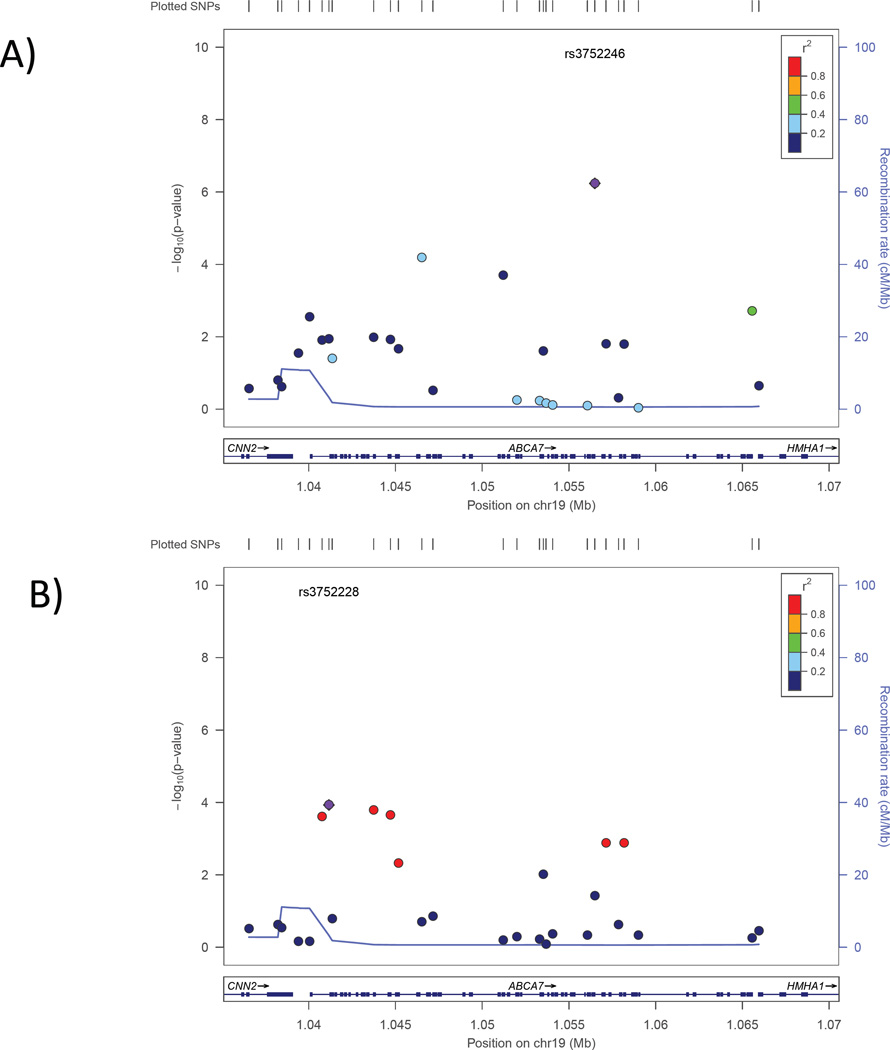
ABCA7 is an established risk gene for AD in both Caucasians(Hollingworth, et al., 2011,Naj, et al., 2011) and African Americans(Logue, et al., 2011,Reitz, et al., 2013). It is unlikely that the ABCA7 association is confounded with variants in the C3 gene—a known AMD risk locus located approximately 6 Mb away. The peak AMD ABCA7 SNP (rs3752228) is 5,356 bp away from rs3764650 and 15,328 bp away from rs3752246, which are the peak AD risk SNPs in ABCA7 identified by Hollingworth et al. (2011) and Naj et al. (2011) respectively. Rs3752228 is not in LD with either of these two AD-associated SNPs (r2<0.01). It is worth noting that rs3752228 is nominally associated with AD (p=0.011). Conversely, the AD-associated SNP rs3752246 is nominally associated with AMD (p=0.038). However, the direction of effect for both SNPs differs between AD and AMD.
The region of LD containing PILRA, ZCWPW1 and several adjacent genes (PLIRB, MEPCE, PPP1R35, C7ORF61, TSC22D4, and NYAP1) on chromosome 7q22 has also not been associated previously with AMD. Because of high LD among these genes (Figure 1C), we are not able to conclude with certainty whether the association to AMD is explained by a single variant or multiple variants in PILRA, ZCWPW1, or perhaps one of the other genes in the LD block. Recently, ZCWPW1 emerged as a new AD risk locus in the largest GWAS for this disorder to date (Lambert, et al., 2013). The zinc finger CW domain encoded by this gene is a motif of about 60 residues that functions as a histone modification reader and thus is involved in epigenetic regulation(He, et al., 2010). PILRA, the paired-immunoglobulin-like type 2 receptor, binds with herpes simplex virus-1 (HSV-1) which is neurotoxic to sensory neurons in the eye and brain(Satoh, et al., 2008). Several but not all studies have shown an increase of HSV-1 in brains of AD subjects compared to controls(Hill, et al., 2007), and particularly among subjects with the APOE ε4 allele(Itzhaki and Lin, 1998). Although there are no reports linking HSV-1 to AMD, PILRA has a role in the entry of HSV-2 into retinal pigment epithelial cells(Shukla, et al., 2009). Sequencing and molecular experiments will be necessary to determine which of these genes and variants therein are causally linked to AD and AMD.
HGS is a member of the clathrin-mediated endocytosis signaling pathway. It is a zinc-finger protein that is ubiquitously expressed in all tissues and localizes to the surface of early endosomes(Komada, et al., 1997). HGS has a role in lysosomal sorting and down-regulation of membrane receptors via the endosomal sorting complex required for transport (ESCRT) pathway (Bache, et al., 2003,Bilodeau, et al., 2002). Recently, it has been demonstrated that HGS is a regulator of endosomal cholesterol trafficking(Du, et al., 2012).
Analysis of AD-related pathways led us to previously established AMD-risk loci including C3, COL10A1, and CETP. Our investigation of AD pathway genes also revealed significant associations of AMD with genes near recognized AMD loci. PLA2G12A is located 10 kb from the 3’UTR of CFI (r2 between peak CFI and PLA2G12A SNPs is 0.34) and, thus, may not be a true AMD risk gene. APOM and CNSK2B (which harbor genome-wide significant SNPs but not considered as independent AMD loci in the AMDGene GWAS; (Fritsche, et al., 2013) and TNF span 89 kb of the MHC region on chromosome 6 (Figure 2) and, thus, may represent one association signal. Supporting the idea that there are at least two distinct AMD susceptibility loci in the MHC, we observed that the top APOM, CNSK2B, and TNF SNPs are not in LD with the genome-wide significant association peak at rs429608 in the C2-CFB region (r2<0.05). Nonetheless, further studies are needed to determine whether associations with APOM, CNSK2B, and TNF are tagging functional variants in only one of these genes or in other genes in or near C2-CFB.
Pathway analysis of known and potentially related AD genes yielded numerous gene networks, the most significant of which were the clathrin-mediated endocytosis (CME) signaling, LXR/RXR activation and atherosclerosis signaling pathways. There is a growing body of evidence linking these pathways to AD and AMD. These pathways incorporate many of the GO processes identified in a prior gene enrichment analysis of AD risk loci (Jones, et al., 2010) such as reverse cholesterol homeostasis, cholesterol transport, and cholesterol efflux. CME is one of the major mechanisms by which LDL, nutrients, and hormones are internalized into the cell. CME is also central to the processing of Ab (Wu and Yao, 2009). Few studies have investigated the possible role of endocytosis in AMD. Bando et al. (2007) found increased levels of clathrin and adaptin (two key CME proteins) in AMD donor eye tissue compared to non-AMD donor eyes, and concluded that this may be due to increased endocytosis in AMD tissue, perhaps triggered by high levels of LDL.
The relationship between AMD and atherosclerosis signaling was previously observed in the pathway analysis of AMD genes by Fritsche et al. (Fritsche, et al., 2013). The LXR/RXR activation pathway plays a role in atherosclerosis and in cholesterol homeostasis. Prior work demonstrated that lipid metabolism and lipid processing are involved in AMD (Ebrahimi and Handa, 2011,Kishan, et al., 2011). Similarities in the composition of drusen in AMD maculae, amyloid plaques in the AD brain, and atherosclerotic plaques have been noted (Mullins, et al., 2000). Two of the replicated GWAS-significant AMD loci—namely LIPC and CETP —are associated with HDL-C levels(Kathiresan, et al., 2009,Willer, et al., 2008), although the CETP risk allele is associated with higher levels of HDL-C and the LIPC risk allele is associated with lower HDL-C levels(Chen, et al., 2010). ABCA7, one of the novel AMD loci identified in this study, has a major role in the enhancement of phagocytosis, and its interaction with apolipoproteins further increases this function (Tanaka, et al., 2011).
One of the genes that emerged from the AD pathway analysis and shown to be associated with AMD is TNF. TNF is a pro-inflammatory cytokine that is involved in both initiating and limiting/terminating the inflammatory response to prevent tissue damage (Makhatadze, 1998,Marino, et al., 1997). It is expressed at low levels in the brain, but is up-regulated in AD patients (Fillit, et al., 1991). In animal studies administration of 3,6′-Dithiothaliodmide reduces TNF expression and reduces AD-related traits and behaviors (Tweedie, et al., 2012). Early studies examining the treatment of AD with the anti-TNF agent etanercept—which is approved for use in the treatment of arthritis—were encouraging (Tobinick and Gross, 2008a,Tobinick and Gross, 2008b). TNF is a strong candidate AMD gene based on the role of inflammation in AMD. The related tumor necrosis factor receptor superfamily, member 10a (TNFRSF10A) gene has been implicated in multiple AMD studies (Arakawa, et al., 2011,Fritsche, et al., 2013). Several anti-TNF agents have been or are being evaluated in conjunction with AMD. A study in monkeys found that anti TNF-a agent ESBA105 reduced the appearance of laser-induced choroidal neovascularization, indicating that it may be preventative of AMD (Lichtlen, et al., 2010). However, a pilot study using the anti-TNF agent infliximab (Remicade) found that the injections were not well tolerated (Giganti, et al., 2010).
By design, novel AMD SNPs identified by testing association with established AD risk variants implicitly have a pleiotropic effect. Solovieff et al. (Solovieff, et al., 2013) recently compared methods for establishing polygenic effects at a locus or region. Accordingly, our two-stage design is “robust” because moderate strength associations will not be missed in the discovery GWAS sample of more than 20,000 cases and controls. Several other approaches discussed in this review could not be applied in our study because they require all traits to be measured in the same individuals, or individual level data, or are effective only when the number of traits being examined is large. The PRIMe method (Huang, et al., 2011), is similar to our gene-level investigation, but PRIMe examines LD-based regions rather than gene regions for association with multiple traits. We also applied a meta-analysis approach to identify loci underpinning both AD and AMD by searching for SNPs having moderately significant signals for each trait which became genome-wide significant after combining results from the analyses of both disorders. The results of these analyses (not presented) were similar to the two-stage analysis of the individual SNPs. The only genome-wide significant SNPs from the meta-analysis were driven by known risk loci of one trait or the other rather than any joint effect.
The strengths of this study include a hypothesis-driven approach, which reduced considerably the number of tests, hence allowing an opportunity for finding AMD-risk loci that would be over-looked in GWAS, and the availability of very large AD and AMD GWAS datasets. However, our association analyses were performed using summarized results from the constituent datasets, thus precluding evaluation of more complex models to test for gene-gene interaction or effects of dominant alleles. We could also not perform conditional analyses necessary to explore confounding amount loci from the same region (e.g., loci in the MHC region). By design, the novel AMD variants we identified did not achieve genome-wide significance likely because of small effect size or low frequency of risk variants. Thus, the associations with these variants are unlikely to explain a substantial proportion of AMD heritability and will need to be confirmed in independent datasets. However, the documentation of a link between these genes and risk of AMD is useful as it helps more closely delineate the molecular mechanisms leading to AMD.
Supplementary Material
Figure 3.
Association of SNPs in the ABCA7 region with AD (panel A) and AMD (panel B). Significance of the association of each SNP (circles) is plotted as the negative logarithm of the p-value. Correlations (r2) between tested SNPs with the SNP most significantly associated with AD (rs3752246) and AMD (rs3752228) are indicated using the color scheme shown in the legend. Map positions and direction of transcription of genes in the region are shown in the bottom portion of each panel.
Figure 4.
Association of AMD with SNPs in the portion of the major histocompatibility locus encompassing TNFAPOM and CSNK2B. Significance of the association of each SNP (circles) is plotted as the negative logarithm of the p-value. Correlations (r2) between tested SNPs with the SNP most significantly associated with AMD in this region (rs3130617) are indicated using the color scheme shown in the legend. Map positions and direction of transcription of genes in the region are shown in the bottom portion of the figure.
Acknowledgements
We gratefully acknowledge the technical assistance of Grant Duclos. This work was supported by NIH grants R01-AG025259, PG30-AG13846, U01-AG032984, and R01-EY0144581, an unrestricted grant from Research to Prevent Blindness, Inc. NY, NY to the Department of Ophthalmology and Visual Sciences, University of Utah, and by a grant from the Edward and Della Thome Memorial Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors have no conflict of interest to report.
Data:
These analyses have not been published elsewhere. This manuscript is not under consideration elsewhere.
Human Subjects
The present investigation occurred with appropriate IRB oversight and human subject protections.
Author Statements
All authors have reviewed the manuscript and have approved of its contents and validate the accuracy of the data.
References
- Adams MK, Simpson JA, Richardson AJ, English DR, Aung KZ, Makeyeva GA, Guymer RH, Giles GG, Hopper J, Robman LD, Baird PN. Apolipoprotein E gene associations in age-related macular degeneration: the Melbourne Collaborative Cohort Study. Am J Epidemiol. 2012;175(6):511–518. doi: 10.1093/aje/kwr329. [DOI] [PubMed] [Google Scholar]
- Anstey KJ, von Sanden C, Salim A, O’Kearney R. Smoking as a risk factor for dementia and cognitive decline: a meta-analysis of prospective studies. Am J Epidemiol. 2007;166(4):367–378. doi: 10.1093/aje/kwm116. [DOI] [PubMed] [Google Scholar]
- Arakawa S, Takahashi A, Ashikawa K, Hosono N, Aoi T, Yasuda M, Oshima Y, Yoshida S, Enaida H, Tsuchihashi T, Mori K, Honda S, Negi A, Arakawa A, Kadonosono K, Kiyohara Y, Kamatani N, Nakamura Y, Ishibashi T, Kubo M. Genome-wide association study identifies two susceptibility loci for exudative age-related macular degeneration in the Japanese population. Nat Genet. 2011;43(10):1001–1004. doi: 10.1038/ng.938. [DOI] [PubMed] [Google Scholar]
- Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162(3):435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Wang JJ, Rogers S, Klein R, Kuller LH, Larsen EK, Wong TY. Early age-related macular degeneration, cognitive function, and dementia: the Cardiovascular Health Study. Arch Ophthalmol. 2009;127(5):667–673. doi: 10.1001/archophthalmol.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4(7):534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Casserly I, Topol E. Convergence of atherosclerosis and Alzheimer’s disease: inflammation, cholesterol, and misfolded proteins. Lancet. 2004;363(9415):1139–1146. doi: 10.1016/S0140-6736(04)15900-X. [DOI] [PubMed] [Google Scholar]
- Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, Tan PL, Conley YP, Kanda A, Kopplin L, Li Y, Augustaitis KJ, Karoukis AJ, Scott WK, Agarwal A, Kovach JL, Schwartz SG, Postel EA, Brooks M, Baratz KH, Brown WL, Brucker AJ, Orlin A, Brown G, Ho A, Regillo C, Donoso L, Tian L, Kaderli B, Hadley D, Hagstrom SA, Peachey NS, Klein R, Klein BE, Gotoh N, Yamashiro K, Ferris Iii F, Fagerness JA, Reynolds R, Farrer LA, Kim IK, Miller JW, Corton M, Carracedo A, Sanchez-Salorio M, Pugh EW, Doheny KF, Brion M, Deangelis MM, Weeks DE, Zack DJ, Chew EY, Heckenlively JR, Yoshimura N, Iyengar SK, Francis PJ, Katsanis N, Seddon JM, Haines JL, Gorin MB, Abecasis GR, Swaroop A. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107(16):7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon N, O’Colmain B, Klaver CC, Klein R, Munoz B, Friedman DS, Kempen J, Taylor HR, Mitchell P. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004;122(4):477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7(2):180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Dentchev T, Milam AH, Lee VM, Trojanowski JQ, Dunaief JL. Amyloid-beta is found in drusen from some age-related macular degeneration retinas, but not in drusen from normal retinas. Mol Vis. 2003;9:184–190. [PubMed] [Google Scholar]
- Ding JD, Lin J, Mace BE, Herrmann R, Sullivan P, Bowes Rickman C. Targeting age-related macular degeneration with Alzheimer’s disease based immunotherapies: anti-amyloid-beta antibody attenuates pathologies in an age-related macular degeneration mouse model. Vision Res. 2008;48(3):339–345. doi: 10.1016/j.visres.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Kazim AS, Brown AJ, Yang H. An essential role of Hrs/Vps27 in endosomal cholesterol trafficking. Cell Rep. 2012;1(1):29–35. doi: 10.1016/j.celrep.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Ebrahimi KB, Handa JT. Lipids, lipoproteins, and age-related macular degeneration. J Lipids 2011. 2011 doi: 10.1155/2011/802059. 802059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308(5720):421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. European journal of human genetics : EJHG. 2009;17(1):100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. Jama. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- Fillit H, Ding WH, Buee L, Kalman J, Altstiel L, Lawlor B, Wolf-Klein G. Elevated circulating tumor necrosis factor levels in Alzheimer’s disease. Neurosci Lett. 1991;129(2):318–320. doi: 10.1016/0304-3940(91)90490-k. [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, Igo RP, Jr, Buitendijk GH, Sim X, Weeks DE, Guymer RH, Merriam JE, Francis PJ, Hannum G, Agarwal A, Armbrecht AM, Audo I, Aung T, Barile GR, Benchaboune M, Bird AC, Bishop PN, Branham KE, Brooks M, Brucker AJ, Cade WH, Cain MS, Campochiaro PA, Chan CC, Cheng CY, Chew EY, Chin KA, Chowers I, Clayton DG, Cojocaru R, Conley YP, Cornes BK, Daly MJ, Dhillon B, Edwards AO, Evangelou E, Fagerness J, Ferreyra HA, Friedman JS, Geirsdottir A, George RJ, Gieger C, Gupta N, Hagstrom SA, Harding SP, Haritoglou C, Heckenlively JR, Holz FG, Hughes G, Ioannidis JP, Ishibashi T, Joseph P, Jun G, Kamatani Y, Katsanis N, C NK, Khan JC, Kim IK, Kiyohara Y, Klein BE, Klein R, Kovach JL, Kozak I, Lee CJ, Lee KE, Lichtner P, Lotery AJ, Meitinger T, Mitchell P, Mohand-Said S, Moore AT, Morgan DJ, Morrison MA, Myers CE, Naj AC, Nakamura Y, Okada Y, Orlin A, Ortube MC, Othman MI, Pappas C, Park KH, Pauer GJ, Peachey NS, Poch O, Priya RR, Reynolds R, Richardson AJ, Ripp R, Rudolph G, Ryu E, Sahel JA, Schaumberg DA, Scholl HP, Schwartz SG, Scott WK, Shahid H, Sigurdsson H, Silvestri G, Sivakumaran TA, Smith RT, Sobrin L, Souied EH, Stambolian DE, Stefansson H, Sturgill-Short GM, Takahashi A, Tosakulwong N, Truitt BJ, Tsironi EE, Uitterlinden AG, van Duijn CM, Vijaya L, Vingerling JR, Vithana EN, Webster AR, Wichmann HE, Winkler TW, Wong TY, Wright AF, Zelenika D, Zhang M, Zhao L, Zhang K, Klein ML, Hageman GS, Lathrop GM, Stefansson K, Allikmets R, Baird PN, Gorin MB, Wang JJ, Klaver CC, Seddon JM, Pericak-Vance MA, Iyengar SK, Yates JR, Swaroop A, Weber BH, Kubo M, Deangelis MM, Leveillard T, Thorsteinsdottir U, Haines JL, Farrer LA, Heid IM, Abecasis GR. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta LB, Vitali M, Zanola A, Venturelli E, Fenoglio C, Galimberti D, Scarpini E, Finazzi D. Polymorphisms in the LOC387715/ARMS2 putative gene and the risk for Alzheimer’s disease. Dementia and geriatric cognitive disorders. 2008;26(2):169–174. doi: 10.1159/000151050. [DOI] [PubMed] [Google Scholar]
- Giganti M, Beer PM, Lemanski N, Hartman C, Schartman J, Falk N. Adverse events after intravitreal infliximab (Remicade) Retina. 2010;30(1):71–80. doi: 10.1097/IAE.0b013e3181bcef3b. [DOI] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT, Hageman GS, Dean M, Allikmets R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38(4):458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308(5720):419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Hamilton G, Proitsi P, Williams J, O’Donovan M, Owen M, Powell J, Lovestone S. Complement factor H Y402H polymorphism is not associated with late-onset Alzheimer’s disease. Neuromolecular medicine. 2007;9(4):331–334. doi: 10.1007/s12017-007-8013-y. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The Gene Ontology (GO) database and informatics resource. Nucleic acids research. 2004;32(Database issue):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Umehara T, Saito K, Harada T, Watanabe S, Yabuki T, Kigawa T, Takahashi M, Kuwasako K, Tsuda K, Matsuda T, Aoki M, Seki E, Kobayashi N, Guntert P, Yokoyama S, Muto Y. Structural insight into the zinc finger CW domain as a histone modification reader. Structure. 2010;18(9):1127–1139. doi: 10.1016/j.str.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Hill JM, Bhattacharjee PS, Neumann DM. Apolipoprotein E alleles can contribute to the pathogenesis of numerous clinical conditions including HSV-1 corneal disease. Exp Eye Res. 2007;84(5):801–811. doi: 10.1016/j.exer.2006.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Johnson AD, O’Donnell CJ. PRIMe: a method for characterization and evaluation of pleiotropic regions from multiple genome-wide association studies. Bioinformatics. 2011;27(9):1201–1206. doi: 10.1093/bioinformatics/btr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L, Schachat AP, He Q, Leske MC. Hypertension, cardiovascular disease, and age-related macular degeneration. Age-Related Macular Degeneration Risk Factors Study Group. Arch Ophthalmol. 2000;118(3):351–358. doi: 10.1001/archopht.118.3.351. [DOI] [PubMed] [Google Scholar]
- Itzhaki RF, Lin WR. Herpes simplex virus type I in brain and the type 4 allele of the apolipoprotein E gene are a combined risk factor for Alzheimer’s disease. Biochem Soc Trans. 1998;26(2):273–277. doi: 10.1042/bst0260273. [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77(3):389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Holmans PA, Hamshere ML, Harold D, Moskvina V, Ivanov D, Pocklington A, Abraham R, Hollingworth P, Sims R, Gerrish A, Pahwa JS, Jones N, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Peters O, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Ruther E, Carrasquillo MM, Pankratz VS, Younkin SG, Hardy J, O’Donovan MC, Owen MJ, Williams J. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PloS one. 2010;5(11):e13950. doi: 10.1371/journal.pone.0013950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic acids research. 2006;34(Database issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, Voight BF, Bonnycastle LL, Jackson AU, Crawford G, Surti A, Guiducci C, Burtt NP, Parish S, Clarke R, Zelenika D, Kubalanza KA, Morken MA, Scott LJ, Stringham HM, Galan P, Swift AJ, Kuusisto J, Bergman RN, Sundvall J, Laakso M, Ferrucci L, Scheet P, Sanna S, Uda M, Yang Q, Lunetta KL, Dupuis J, de Bakker PI, O’Donnell CJ, Chambers JC, Kooner JS, Hercberg S, Meneton P, Lakatta EG, Scuteri A, Schlessinger D, Tuomilehto J, Collins FS, Groop L, Altshuler D, Collins R, Lathrop GM, Melander O, Salomaa V, Peltonen L, Orho-Melander M, Ordovas JM, Boehnke M, Abecasis GR, Mohlke KL, Cupples LA. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly SP, Lawlor BA, Kenny RA. Blood pressure and dementia - a comprehensive review. Ther Adv Neurol Disord. 2009;2(4):241–260. doi: 10.1177/1756285609103483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishan AU, Modjtahedi BS, Martins EN, Modjtahedi SP, Morse LS. Lipids and age-related macular degeneration. Surv Ophthalmol. 2011;56(3):195–213. doi: 10.1016/j.survophthal.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Kivipelto M, Helkala EL, Laakso MP, Hanninen T, Hallikainen M, Alhainen K, Soininen H, Tuomilehto J, Nissinen A. Midlife vascular risk factors and Alzheimer’s disease in later life: longitudinal, population based study. BMJ. 2001;322(7300):1447–1451. doi: 10.1136/bmj.322.7300.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaver CC, Ott A, Hofman A, Assink JJ, Breteler MM, de Jong PT. Is age-related maculopathy associated with Alzheimer’s Disease? The Rotterdam Study. Am J Epidemiol. 1999;150(9):963–968. doi: 10.1093/oxfordjournals.aje.a010105. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Linton KL, DeMets DL. The Beaver Dam Eye Study: the relation of age-related maculopathy to smoking. Am J Epidemiol. 1993;137(2):190–200. doi: 10.1093/oxfordjournals.aje.a116659. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Tomany SC, Cruickshanks KJ. The association of cardiovascular disease with the long-term incidence of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology. 2003;110(6):1273–1280. doi: 10.1016/S0161-6420(03)00599-2. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005 Mar 10;308(5720):385–389. doi: 10.1126/science.1109557. Epub 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M, Masaki R, Yamamoto A, Kitamura N. Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J Biol Chem. 1997;272(33):20538–20544. doi: 10.1074/jbc.272.33.20538. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, European Alzheimer’s Disease Initiative I, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, Destefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannfelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltunen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013 doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fur I, Laumet G, Richard F, Fievet N, Berr C, Rouaud O, Delcourt C, Amouyel P, Lambert JC. Association study of the CFH Y402H polymorphism with Alzheimer’s disease. Neurobiology of aging. 2010;31(1):165–166. doi: 10.1016/j.neurobiolaging.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A, Abecasis GR. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38(9):1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtlen P, Lam TT, Nork TM, Streit T, Urech DM. Relative contribution of VEGF and TNF-alpha in the cynomolgus laser-induced CNV model: comparing the efficacy of bevacizumab, adalimumab, and ESBA105. Invest Ophthalmol Vis Sci. 2010;51(9):4738–4745. doi: 10.1167/iovs.09-4890. [DOI] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87(1):139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue MW, Schu M, Vardarajan BN, Buros J, Green RC, Go RC, Griffith P, Obisesan TO, Shatz R, Borenstein A, Cupples LA, Lunetta KL, Fallin MD, Baldwin CT, Farrer LA. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol. 2011;68(12):1569–1579. doi: 10.1001/archneurol.2011.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhatadze NJ. Tumor necrosis factor locus: genetic organisation and biological implications. Hum Immunol. 1998;59(9):571–579. doi: 10.1016/s0198-8859(98)00056-1. [DOI] [PubMed] [Google Scholar]
- Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, Seddon JM. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38(9):1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39(10):1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- Marino MW, Dunn A, Grail D, Inglese M, Noguchi Y, Richards E, Jungbluth A, Wada H, Moore M, Williamson B, Basu S, Old LJ. Characterization of tumor necrosis factor-deficient mice. Proc Natl Acad Sci U S A. 1997;94(15):8093–8098. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay GJ, Patterson CC, Chakravarthy U, Dasari S, Klaver CC, Vingerling JR, Ho L, de Jong PT, Fletcher AE, Young IS, Seland JH, Rahu M, Soubrane G, Tomazzoli L, Topouzis F, Vioque J, Hingorani AD, Sofat R, Dean M, Sawitzke J, Seddon JM, Peter I, Webster AR, Moore AT, Yates JR, Cipriani V, Fritsche LG, Weber BH, Keilhauer CN, Lotery AJ, Ennis S, Klein ML, Francis PJ, Stambolian D, Orlin A, Gorin MB, Weeks DE, Kuo CL, Swaroop A, Othman M, Kanda A, Chen W, Abecasis GR, Wright AF, Hayward C, Baird PN, Guymer RH, Attia J, Thakkinstian A, Silvestri G. Evidence of association of APOE with age-related macular degeneration: a pooled analysis of 15 studies. Hum Mutat. 2011;32(12):1407–1416. doi: 10.1002/humu.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, Kawarabayashi T, Shoji M, Tomita N, Arai H, Asada T, Harigaya Y, Ikeda M, Amari M, Hanyu H, Higuchi S, Ikeuchi T, Nishizawa M, Suga M, Kawase Y, Akatsu H, Kosaka K, Yamamoto T, Imagawa M, Hamaguchi T, Yamada M, Moriaha T, Takeda M, Takao T, Nakata K, Fujisawa Y, Sasaki K, Watanabe K, Nakashima K, Urakami K, Ooya T, Takahashi M, Yuzuriha T, Serikawa K, Yoshimoto S, Nakagawa R, Kim JW, Ki CS, Won HH, Na DL, Seo SW, Mook-Jung I, St George-Hyslop P, Mayeux R, Haines JL, Pericak-Vance MA, Yoshida M, Nishida N, Tokunaga K, Yamamoto K, Tsuji S, Kanazawa I, Ihara Y, Schellenberg GD, Farrer LA, Kuwano R. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PloS one. 2013;8(4):e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. Faseb J. 2000;14(7):835–846. [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43(5):436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang CP, Baum L, Chan WM, Lau TC, Poon PM, Lam DS. The apolipoprotein E epsilon4 allele is unlikely to be a major risk factor of age-related macular degeneration in Chinese. Ophthalmologica. 2000;214(4):289–291. doi: 10.1159/000027506. [DOI] [PubMed] [Google Scholar]
- Proitsi P, Lupton MK, Dudbridge F, Tsolaki M, Hamilton G, Daniilidou M, Pritchard M, Lord K, Martin BM, Johnson J, Craig D, Todd S, McGuinness B, Hollingworth P, Harold D, Kloszewska I, Soininen H, Mecocci P, Velas B, Gill M, Lawlor B, Rubinsztein DC, Brayne C, Passmore PA, Williams J, Lovestone S, Powell JF. Alzheimer’s disease and age-related macular degeneration have different genetic models for complement gene variation. Neurobiology of aging. 2012;331843(8):e9–e17. doi: 10.1016/j.neurobiolaging.2011.12.036. [DOI] [PubMed] [Google Scholar]
- Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang LS, Valladares O, Lin CF, Larson EB, Graff-Radford NR, Evans D, De Jager PL, Crane PK, Buxbaum JD, Murrell JR, Raj T, Ertekin-Taner N, Logue M, Baldwin CT, Green RC, Barnes LL, Cantwell LB, Fallin MD, Go RC, Griffith P, Obisesan TO, Manly JJ, Lunetta KL, Kamboh MI, Lopez OL, Bennett DA, Hendrie H, Hall KS, Goate AM, Byrd GS, Kukull WA, Foroud TM, Haines JL, Farrer LA, Pericak-Vance MA, Schellenberg GD, Mayeux R. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4,and the risk of late-onset Alzheimer disease in African Americans. Jama. 2013;309(14):1483–1492. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Tang MX, Luchsinger J, Mayeux R. Relation of plasma lipids to Alzheimer disease and vascular dementia. Arch Neurol. 2004;61(5):705–714. doi: 10.1001/archneur.61.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Weber BH. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14(21):3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, Arase N, Shiratori I, Tanaka S, Kawaguchi Y, Spear PG, Lanier LL, Arase H. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132(6):935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DW, Klein ML, Humpert A, Majewski J, Schain M, Weleber RG, Ott J, Acott TS. Lack of an association of apolipoprotein E gene polymorphisms with familial age-related macular degeneration. Arch Ophthalmol. 2003;121(5):679–683. doi: 10.1001/archopht.121.5.679. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr, Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM, Consortium C, Consortium G, Consortium E. Genome-wide analysis of genetic loci associated with Alzheimer disease. Jama. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla SY, Singh YK, Shukla D. Role of nectin-1, HVEM, and PILR-alpha in HSV-2 entry into human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2009;50(6):2878–2887. doi: 10.1167/iovs.08-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nature reviews Genetics. 2013;14(7):483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Abe-Dohmae S, Iwamoto N, Yokoyama S. Roles of ATP-binding cassette transporter A7 in cholesterol homeostasis and host defense system. J Atheroscler Thromb. 2011;18(4):274–281. doi: 10.5551/jat.6726. [DOI] [PubMed] [Google Scholar]
- The Eye Disease Case-Control Study Group. Risk factors for neovascular age-related macular degeneration. The Eye Disease Case-Control Study Group. Arch Ophthalmol. 1992;110(12):1701–1708. doi: 10.1001/archopht.1992.01080240041025. [DOI] [PubMed] [Google Scholar]
- Tobinick EL, Gross H. Rapid cognitive improvement in Alzheimer’s disease following perispinal etanercept administration. J Neuroinflammation 5, 2. 2008a doi: 10.1186/1742-2094-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobinick EL, Gross H. Rapid improvement in verbal fluency and aphasia following perispinal etanercept in Alzheimer’s disease. BMC Neurol. 2008b;8:27. doi: 10.1186/1471-2377-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie D, Ferguson RA, Fishman K, Frankola KA, Van Praag H, Holloway HW, Luo W, Li Y, Caracciolo L, Russo I, Barlati S, Ray B, Lahiri DK, Bosetti F, Greig NH, Rosi S. Tumor necrosis factor-alpha synthesis inhibitor 3,6’-dithiothalidomide attenuates markers of inflammation, Alzheimer pathology and behavioral deficits in animal models of neuroinflammation and Alzheimer’s disease. J Neuroinflammation. 2012;9:106. doi: 10.1186/1742-2094-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen R, Ikram MK, Vingerling JR, Witteman JC, Hofman A, de Jong PT. Blood pressure, atherosclerosis, and the incidence of age-related maculopathy: the Rotterdam Study. Invest Ophthalmol Vis Sci. 2003;44(9):3771–3777. doi: 10.1167/iovs.03-0121. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Yao PJ. Clathrin-mediated endocytosis and Alzheimer’s disease: an update. Ageing Res Rev. 2009;8(3):147–149. doi: 10.1016/j.arr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht AM, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357(6):553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- Yu Y, Bhangale TR, Fagerness J, Ripke S, Thorleifsson G, Tan PL, Souied EH, Richardson AJ, Merriam JE, Buitendijk GH, Reynolds R, Raychaudhuri S, Chin KA, Sobrin L, Evangelou E, Lee PH, Lee AY, Leveziel N, Zack DJ, Campochiaro B, Campochiaro P, Smith RT, Barile GR, Guymer RH, Hogg R, Chakravarthy U, Robman LD, Gustafsson O, Sigurdsson H, Ortmann W, Behrens TW, Stefansson K, Uitterlinden AG, van Duijn CM, Vingerling JR, Klaver CC, Allikmets R, Brantley MA, Jr, Baird PN, Katsanis N, Thorsteinsdottir U, Ioannidis JP, Daly MJ, Graham RR, Seddon JM. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011;20(18):3699–3709. doi: 10.1093/hmg/ddr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg M, Landgren S, Andersson ME, Palmer MS, Gustafson DR, Skoog I, Minthon L, Thelle DS, Wallin A, Bogdanovic N, Andreasen N, Blennow K, Zetterberg H. Association of complement factor H Y402H gene polymorphism with Alzheimer’s disease. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2008;147B(6):720–726. doi: 10.1002/ajmg.b.30668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.