Abstract
Nonribosomal peptide synthetases (NRPSs) are versatile engines of bioactive natural product biosynthesis that function according to the multiple carrier thiotemplate mechanism. C-terminal thioesterase (TE) domains of these giant modular proteins typically catalyze product release by hydrolysis or macrocylization. We now report an unprecedented, dual-function TE involved in nocardicin A biosynthesis, the paradigm monocyclic β-lactam antibiotic. Contrary to expectation, a stereodefined series of potential peptide substrates for the nocardicin TE domain failed to undergo hydrolysis. The stringent discrimination against peptide intermediates was dramatically overcome by prior monocyclic β-lactam formation at an L-seryl site. Kinetic data are interpreted such that the TE domain acts as a gatekeeper to hold the assembling peptide on an upstream domain until β-lactam formation takes place and then rapidly catalyzes epimerization, not previously observed as a TE catalytic function, and thioesterase cleavage to discharge a fully fledged pentapeptide β-lactam harboring nocardicin G, the universal precursor of the nocardicins.
Introduction
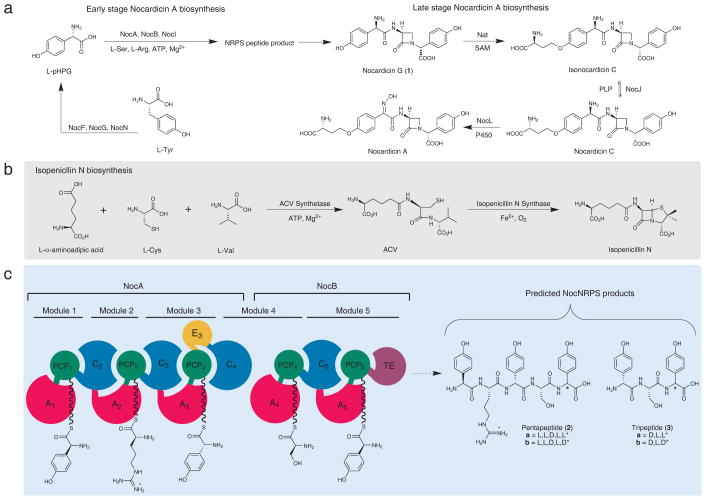
Nocardicin A, isolated from fermentation of Nocardia uniformis ssp. tsuyamanensis, is a monocyclic β-lactam antibiotic that possesses both modest activity against Gram-negative bacteria and β-lactamase resistance.1 In addition to nocardicin A, several minor nocardicin metabolites have been isolated that differ in the presence or absence of an ether-linked homoseryl side-chain and amine oxidation state. Of particular note, nocardicin G (1), the simplest member of the nocardicin family and apparently derived from a tripeptide core having the D,L,D-stereochemistry, has been demonstrated to be incorporated intact into nocardicin A (Fig 1a).2 Earlier studies demonstrated that the β-lactam ring of nocardicin A originates from L-serine (L-Ser), that no change in oxidation state takes place at the seryl β-carbon,3 and that this center undergoes clean stereochemical inversion during C–N bond formation.4 Having also observed partial overall retention of the seryl α-hydrogen, intramolecular nucleophilic displacement (SNi) of an activated seryl hydroxyl in a hypothetical peptide precursor has been proposed to account for β-lactam ring formation. In contrast, the bicyclic β-lactam isopenicillin N is assembled as a δ-(L-α-aminoadipoyl)-L-cysteinyl-D-valine (ACV) tripeptide by the NRPS ACV synthetase and then oxidatively cyclized separately to form the fused β-lactam and thiazolidine rings (Fig. 1b).5–8 It was thought by analogy, therefore, that nocardicin G could be constructed initially as a linear tripeptide (3a/b, Fig. 1c) by the nocardicin NRPS(s) and then modified through seryl activation and SNi displacement (inversion) to form the β-lactam core.4,9 The gene cluster responsible for the production of nocardicin A was reported in 2004.10 Although the majority of the enzymes encoded by the biosynthetic cluster has been functionally identified,11–15 those involved in the early stages of nocardicin peptide assembly have only recently yielded to experiment. Prominent among them is a pair of NRPSs, NocA and NocB, which together comprise five modules (Fig. 1c). Despite the tripeptide architecture of the nocardicins, however, all five modules of NocA and NocB are essential to antibiotic synthesis.16
Figure 1. Isopenicillin N and Nocardicin A biosyntheses.
(a) Isopenicillin N biosynthesis from NRPS ACV synthetase and isopenicillin N synthase. (b) Overall biosynthesis of nocardicin A from early stage peptide assembly initiated by pHPG biosynthesis to late stage nocardicin G modification. (c) Domain and module architecture of NRPSs NocA and NocB and predicted peptide products.
Each module of an NRPS contains a peptidyl carrier protein (PCP) domain, which is post-translationally modified by coenzyme A-derived phosphopantetheine (P-pant) attachment mediated by a phosphopantetheinyl transferase.17,18 Chain initiation is carried out by amino acid selection and activation by an ATP-dependent adenylation (A) domain followed by transfer of the resulting aminoacyl adenylate to the P-pant thiolate on a partner PCP to form an aminoacyl thioester (aa-S-PCP).19 These building blocks are ordered according to the sequence of the NRPS modules and oligomerized commonly through amide linkages by condensation (C) domains.20 Apart from these core domains, other editing domains can be present in NRPSs, for example, those catalyzing L- to D- epimerization (E domains) and N-methylation (M domains).21 The termination module found in these megasynthetases typically contains a thioesterase (TE) as the most downstream domain, which catalyzes the release of the mature peptide product from its cognate PCP domain.22,23 The pivotal discovery that NocI, a member of the MbtH superfamily, essential accessory proteins for some A domains24, is required for NocA1, NocA2 and NocA4 activity enabled analysis of domain specificity and thus determination of the overall predicted peptide product of NocA/B to be L-pHPG-L-Arg-D-pHPG-L-Ser-L-pHPG (2a, Fig. 1c) where L-pHPG is L-(p-hydroxyphenyl)glycine and L-Arg is L-arginine.25 Although all of the α-amino acids activated by the respective A domains are L-amino acids, the internal pHPG of the pentapeptide was predicted to be epimerized to its D-diastereomer based on the presence of an E domain in module 3 (Fig. 1c).
While these recent studies have provided vital clues toward unraveling the early peptide construction steps to nocardicin A, they also define central unresolved questions. First, while a pentapeptide is predicted from NocA/B, the naturally occurring nocardicins A-G contain only three amino acid residues. With the recent identification of L-pHPG and L-Arg as specifically activated by modules 1 and 2 of NocA, respectively, it would seem that these two amino acid residues of the presumed pentapeptide precursor are removed in the course of antibiotic biosynthesis while the C-terminal tripeptide is further elaborated by way of β-lactam formation and C-terminal epimerization to form nocardicin G (1). Second, despite the presence of an E domain in module 3, no E domain is embedded in module 5 leaving unanswered the question of how the C-terminal D-pHPG stereochemistry arises. Finally, the timing of these events is unclear. β-Lactam formation and epimerization at the carboxyl terminus must logically occur after the last pHPG unit is added in module 5, but cleavage of a pentapeptide to a tripeptide intermediate can take place before or after one or both of these reactions.
To address the roles that NocA and NocB play in the biosynthesis of the nocardicins, we focused on the function of the TE domain. Here we present the in vitro reconstitution of the excised TE domain and the detailed biochemical characterization of a series of potential peptide substrates and modified peptides bearing seryl residues that are O-phosphorylated, O-acetylated or cyclized to an integrated β-lactam ring. We demonstrate that the NocB thioesterase (NocTE) shows acute specificity for β-lactam-containing peptide substrates, and is responsible for catalyzing epimerization of the C-terminal pHPG residue to the D-configuration characteristic of the nocardicins. A β-lactam-containing pentapeptide is the preferred product released from NocTE, whose two N-terminal L-amino acid residues are readily proteolyzed to yield nocardicin G (1).
Results
In vitro reconstitution of TE activity
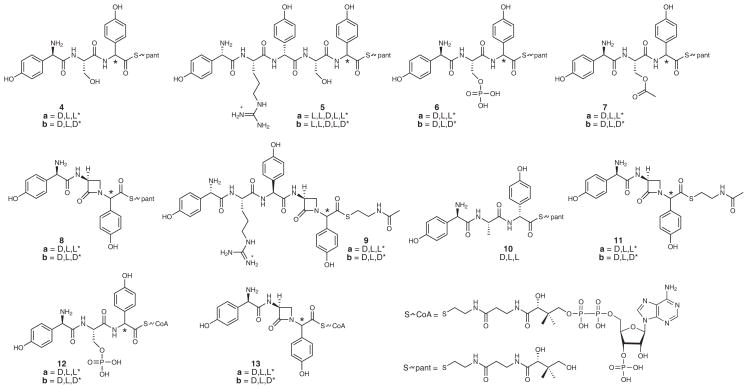
To identify the final product released from the nocardicin NRPSs, the purified, recombinant NocTE was reconstituted in vitro and supplemented with a variety of pantetheine (pant) and N-acetylcysteamine (SNAC) thioester peptides varying in length, serine modification and C-terminal pHPG stereochemistry (Fig. 2). Their syntheses have been fully described elsewhere,26 apart from 10, 12 and 13, which are detailed in Supplementary Note, Fig. 2. This experimental approach was chosen because the peptidyl-S-PCP found in the natural biosynthetic template has been successfully replaced by SNAC and pantetheine thioesters in other related systems.27,28
Figure 2. Substrates synthesized to probe the specificity of NocTE.
Substrates vary in peptide length, C-terminal pHPG stereochemistry and O-seryl modification.
First, thioester-containing linear tripeptides 4a and 4b and pentapeptides 5a and 5b were incubated with purified NocTE. These four peptidyl thioester substrates were the most likely candidates for thioesterase processing, presuming the nocardicin megasynthetases assemble a peptide product in the expected canonical fashion. Surprisingly, after 3 h incubation of 20 μM NocTE with either the pantetheinyl tripeptide 4a or 4b or pantetheinyl pentapeptide 5a or 5b, less than 5% of each substrate was hydrolyzed to its free acid (Supplementary Results, Supplementary Fig. 1a–c). This analysis was quantified by comparison to a negative control in which the active site serine of the NocTE was replaced with alanine (TE*S1779A). Of particular note, the C-terminal D-pHPG diastereomer of the pentapeptide 5b and tripeptide 4b thioesters were found to be the poorest substrates, providing little to no observable hydrolysis above background. This result suggested that, although C-terminal L-pHPG thioesters preferably acylate NocTE, the pentapeptides (2a/b) and tripeptides (3a/b), regardless of C-terminal stereochemistry, are not the products of NocA/B as previously proposed.10,16,29
Next, pantetheinyl tripeptides containing modified seryl residues, O-phosphoryl 6a/b and O-acetyl 7a/b, were examined as possible substrates for the TE. These substrates were envisioned as plausible hydroxyl-activated peptides for SNi β-lactam ring formation, and –OX displacement within the TE active site.10 It was found, however, that incubation of the excised NocTE with either pantetheinyl 6a/b or 7a/b tripeptides failed to produce nocardicin G (1) and <5% thioester hydrolysis was observed when compared to the TE*S1779A negative control (Supplementary Fig. 2).
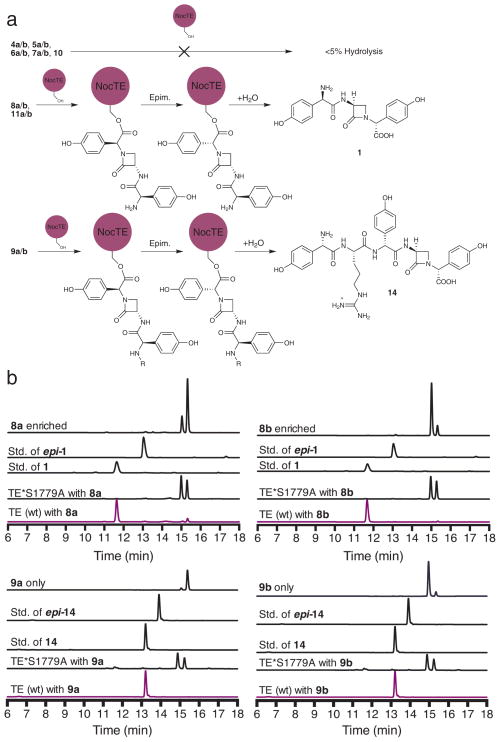
Following the disheartening failure of all predicted or proposed linear and activated peptidyl thioester substrates for the nocardicin TE domain, pantetheinyl β-lactams epi-nocardicin G (8a) and nocardicin G (8b) were examined next as potential substrates for the NocTE, which presumes prior β-lactam formation. TE catalysis, indeed, was observed (Fig. 3a). After 3 h, >99% of both β-lactam substrates 8a and 8b had been converted to the same epimer, nocardicin G (1), containing cleanly the D,L,D-configuration and no observable epi-nocardicin (epi-1, D,L,L; Fig. 3b). TE turnover of β-lactam-containing substrates revealed that azetidinone formation precedes TE release, implying, therefore, that it occurs on a peptide precursor covalently tethered to an upstream PCP domain.
Figure 3. In vitro substrate profiling of NocTE.
(a) Schematic of NocTE reactivity towards various substrates. Top: Linear and activated penta- and tri- peptides are poor substrates for NocTE. Lower: Pre-formation of β-lactam at the seryl position results in NocTE epimerization and hydrolysis activity. (b) HPLC analysis of β-lactam-containing thioester substrates with excised NocTE. All of the β-lactam-containing substrates were processed by NocTE and only one product was formed, containing a C-terminal D-pHPG. Top traces show NocTE incubated with 8a and 8b, forming 1 and bottom traces show NocTE incubated with 9a and 9b, forming 14.
To shed light on the length of the peptide product released by the nocardicin NRPSs, L-pHPG-L-Arg-epi-nocardicin G-SNAC (9a) and L-pHPG-L-Arg-nocardicin G-SNAC (9b) substrates were synthesized, varying from the nocardicin analogs 11a and 11b by the addition of the N-terminal dipeptide L-pHPG-L-Arg in keeping with the incorporation of all 5 amino acids activated by NocA and NocB. After 3 h incubation with NocTE, these substrates strikingly were also efficiently converted to L-pHPG-L-Arg-nocardicin G (14) containing a C-terminal D-pHPG and no observable L-pHPG-L-Arg-epi-nocardicin G (epi-14, L,L,D,L,L; Fig. 3b).
To more deeply probe the necessity for prior β-lactam ring formation, D-pHPG-L-Ala-L-pHPG-pant (10) was incubated with NocTE to determine its behavior compared to 8a or 8b. Substitution of alanine for serine was envisioned as a neutral potential substrate similar in size and polarity to the β-lactam, but conformationally distinct. After 3 h incubation of 10 with NocTE, <10% hydrolysis was observed (Supplementary Fig. 1d). Again, as in the case of the serine-containing peptide substrates 4a, 4b, 5a, and 5b, the alanine tripeptide 10 containing a C-terminal L-pHPG was even more poorly hydrolyzed than the D-pHPG diastereomer relative to the TE*S1779A control. In sum, these findings with potential peptide substrates underscore the binding selectivity of the NocTE domain.
NocTE domain does not form the β-lactam ring
With insight now that stringent discrimination by the NocTE against peptide intermediates can be decisively overcome by prior monocyclic β-lactam formation, we set out to more rigorously test the proposed role that NocTE may play in mediating β-lactam ring formation by investigating peptidyl-S-PCP5 delivery of a suitably activated substrate. Although SNAC and pantetheine thioesters can typically serve as artificial substrate mimics for the PCP-phosphopantetheine moiety, studies of peptidyl-SNAC substrates for fengycin and mycosubtilin peptide cyclases failed to elicit activity from their respective TEs.30 In these cases activity was only established upon loading peptidyl-4′-phosphopantetheine onto the upstream PCP by an Sfp-catalyzed transfer of a synthetic acyl-CoA onto the corresponding apo-PCP. To replicate this approach, the CoA thioesters (12a/b, Fig. 2), corresponding to 6a/b, were prepared (Supplementary Note). The apo-PCP5-TE didomain was genetically excised and covalently modified with an O-phosphoryl tripeptidyl CoA 12a/b through an Sfp-mediated phosphopantetheine transfer reaction to produce the holo-O-phosphoryl tripeptidyl-S-PCP5-TE didomain (Supplementary Fig. 3a). As was seen with the pantetheinyl analogs 6a/b, incubation of this holo-construct failed to elicit thioesterase activity or produce nocardicin G (Supplementary Fig. 3b).
As a positive control, epi-nocardicin G/nocardicin G CoA (13a/b, Fig 2) was also synthesized (Supplementary Note) and loaded onto the apo-PCP5-TE construct with Sfp (Supplementary Fig 4a). As had occurred with the incubation of NocTE with 8a and 8b, the holo-epi-nocardicin G/nocardicin G-S-PCP5-TE construct rapidly produced nocardicin G (1) with no observable epi-nocardicin-G (epi-1) formed (Supplementary Fig. 4b). This result, together with the failure of O-phosphoryl and O-acetyl pantetheinyl tripeptides 6a/b and 7a/b and holo-PCP5-TE covalently modified with 12a/b, reasonably eliminates the cyclodehydration potential of the TE domain with an O-phosphoryl or O-acetyl substrate primed for SNi displacement.
1H-NMR Studies of NocTE epimerization activity
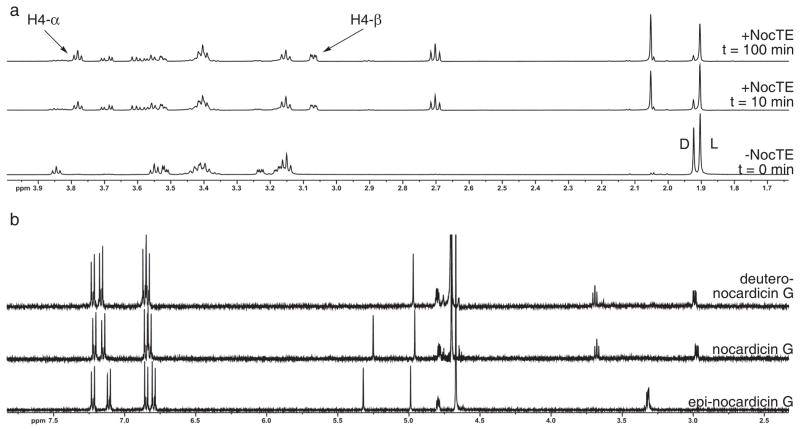
Coordination of the two catalytic activities of the NocTE was further characterized by reconstitution of the reaction of epi-nocardicin G-SNAC (11a) to nocardicin G (1) in D2O, and examination by NMR spectroscopy. To conduct this analysis, NocTE was dialyzed into 50 mM ammonium bicarbonate buffer pH 7.5, and lyophilized to dryness overnight. The dry NocTE was resuspended in deuterated assay buffer, pD 7.2. The reaction was initiated by the addition of 1 μM NocTE to freshly prepared 10 mM 1:1 epi-nocardicin G-SNAC (11a):noc G-SNAC (11b) in deuterated assay buffer and the 1H-spectrum was monitored as a function of time (Fig. 4a).
Figure 4. 1H-NMR spectrometric analysis of NocTE with epi-nocardicin G/nocardicin G-SNAC.
(a) 1H-NMR experiment demonstrating NocTE turnover of epi-nocardicin G/nocardicin G-SNAC. The bottom spectrum labeled “-NocTE” shows the substrates 11a/b before NocTE addition. “D” and “L” correspond to the N-acetyl peaks of SNAC of the nocardicin G-SNAC (11b) and epi-nocardicin G-SNAC (11a) thioesters, respectively. Arrows highlight the two C-4 diastereotopic hydrogen resonances (H-4α/β) of 1, whose appearance was monitored throughout the experiment. (b) 1H-NMR comparison of synthetic epi-nocardicin G and nocardicin G with isolated deutero-nocardicin G from NocTE conversion of substrate 11a/b in D2O.
Upon incubation with NocTE, the appearance of the diagnostic, diastereotopic C-4 hydrogens of the β-lactam ring corresponding to the D,L,D-diastereomer was monitored (Fig 4a). These resonances appear at 3.80 ppm (t, J = 5.7 Hz, 1H) and 3.11 ppm (dd, J = 5.7, 2.3 Hz, 1H). Not observed were the resonances for the corresponding C-4 hydrogens in epi-nocardicin G, which appear as a multiplet centered at 3.32 ppm. The spectral dispersion at 500 MHz provided a clear separation of the two potential hydrolyzed products. The progression of the reaction was also evaluated by the disappearance of the singlet corresponding to the acetyl group of epi-nocardicin G-SNAC (11a) and nocardicin G-SNAC (11b) at 1.92 and 1.90 ppm, respectively, along with the disappearance of the C-4 diastereotopic hydrogens of the epi-nocardicin G-SNAC.
In accord with the in vitro experiments monitored by HPLC analysis (Fig. 3b), we observed the exclusive formation of nocardicin G and no detectable formation of the diastereomeric epi-nocardicin G (Fig. 4a). If NocTE were only catalyzing hydrolysis of each diastereomer, the formation of both nocardicin G and epi-nocardicin G at the thermodynamic equilibrium of 1.1: 1 11b: 11a would be expected.26 Interestingly, the depletion of nocardicin G-SNAC (12b) occurred more rapidly and could be visualized most strikingly from comparison of the integration of the singlets corresponding to the acetyl group of the substrates in the no-enzyme control spectrum to the first collected spectrum at t = 10 min (Fig. 4a). The singlet at 1.92 ppm, corresponding to the N-acetyl moiety of nocardicin G-SNAC (11b), disappears more rapidly and equilibrates to a steady-state of approximately 1:4.3 (1.92:1.90 ppm) from a starting ratio of ~1: 1 throughout the the experiment.
To further verify this result, the NMR experiment was quenched and the products were analyzed by HPLC to isolate the β-lactam produced from the remaining starting material and HSNAC by-product (Supplementary Fig. 5a). 1H-NMR spectral comparison to synthetic standards of nocardicin G and epi-nocardicin G revealed that the α-hydrogen resonance corresponding to the C-terminal pHPG (5.25 ppm) was absent in the isolated nocardicin G while the N-terminal pHPG α-hydrogen resonance (4.96 ppm) remained fully intact (Fig. 4b). MS analysis confirmed the identity of the product as monodeutero-nocardicin G (Supplementary Fig. 5c).
Epimerase function of NocTE
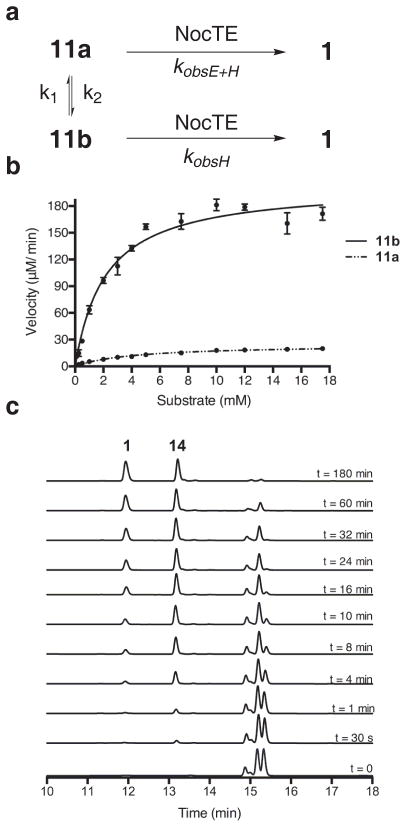
More precise characterization of NocTE as a dual function epimerase/ hydrolase was achieved by determining the steady-state rate of epi-nocardicin G-SNAC (11a) turnover to 1 in the presence of NocTE. This transformation requires enzyme acylation, C-terminal pHPG epimerization and hydrolysis. The rate of uncatalyzed epimerization of nocardicin G-SNAC (11b) to epi-nocardicin G-SNAC (11a) in buffered aqueous solution at pH 7.5 has been previously determined to be 3.3(1) x 10−2 min−1 (k1 + k2) with a half life of ~21 min.26 Dynamic equilibrium is achieved at a ratio of 1.1: 1 11b to 11a, indicating that the C-terminal D-diastereomer is the slightly more thermodynamically favored thioester species. Thus, the rate of uncatalyzed epimerization of 11b to 11a, k1, was calculated to be 1.57(5) x 10−2 min−1 while the reverse reaction, k2, was determined to be 1.73(6) x 10−2 min−1.
Steady-state kinetic parameters were identified for the NocTE catalyzed epimerization and hydrolysis of epi-nocardicin G-SNAC (11a) and nocardicin G-SNAC (11b) by measuring free thiol production using Ellman’s reagent.31,32 The substrates nocardicin G-SNAC (11b) and epi-nocardicin G-SNAC (11a) were >99% diasteromerically pure as determined by 1H-NMR and 13C-NMR spectroscopy.26 Owing to limited solubility at high concentrations of the SNAC thioesters in aqueous buffer, DMSO was present at 5% (v/v), which had no measureable effect on NocTE activity (data not shown). As a further control, HPLC analysis verified that uncatalyzed epimerization of 11a and 11b dissolved in DMSO did not occur on the time scale of the experiment (data not shown) and, thus, allowed for the accurate measurement of NocTE-catalyzed epimerization. Finally, the NocTE catalyzed reactions were initiated with the addition of substrate rather than enzyme to strictly limit non-enzymatic background epimerization once the substrate was introduced to pH 7.5 buffer. The measured reaction rates were corrected for the rate of background hydrolysis in the absence of TE and the data were fit to the non-linear Michaelis-Menten equation to determine kobsE+H, kobsH and Km (Fig. 5a). KobsE+H is defined as the observed rate of NocTE conversion of stereopure 11a to 1 and kobsH is defined as the observed rate of NocTE conversion of stereopure 11b to 1 (Fig. 5a).
Figure 5. Kinetic analysis and determination of NocTE as a dual epimerase/hydrolase.
(a) Depiction of NocTE catalyzed conversion of epi-nocardicin G-SNAC (11a) and nocardicin G-SNAC (11b) to nocardicin G (1). (b) Plots of initial velocities vs. 11a and 11b concentration. Experiments were conducted in triplicate; data represents mean values ± standard deviation. (c) HPLC analysis of time points in competition experiments between 11b and 9b in the presence of NocTE, showing the appearance of 1 and 14.
The rate of NocTE catalyzed conversion of epi-nocardicin G-SNAC (11a) to nocardicin G (1) was determined to be kobsE+H = 24.5(7) min−1 with a Km = 4.2(3) mM (Fig. 5b). The kobsE+H rate, in comparison to the uncatalyzed rate of 11a to 11b substrate epimerization, k2, is over 1,400 x faster. This rate enhancement relative to spontaneous chemical epimerization, established the NocTE domain as a dual epimerase/hydrolase, a previously uncharacterized function of NRPS TE domains.
To distinguish whether epi-nocardicin G-SNAC is being epimerized as the PCP-bound thioester, and then transferred to the active site Ser1779 of NocTE, or as an oxyester after acyl transfer, the rate of epimerization of nocardicin G-SNAC to epi-nocardicin G-SNAC was measured in assays with TE*S1779A or TE*S1779C, NocTE mutants lacking their catalytic serine. Comparison of k1 and k2 to epimerization in the presence of TE*S1779A or TE*S1779C indicated no observable rate change (Supplementary Fig. 6). These results further suggest that NocTE catalyzed epimerization occurs on a peptidyl-O-TE bound intermediate and not as its cognate PCP bound thioester.
The rate of NocTE catalyzed conversion of nocardicin G-SNAC (11b) by the NocTE to nocardicin G was determined to be kobsH = 220(5) min−1 with a Km = 2.6(2) mM by direct measurement of the rate of turnover of NocTE incubated with >99% diasteromerically pure nocardicin G-SNAC (11b) with excised NocTE. Thus, hydrolysis is ~10x faster than epimerization, which, therefore, is partially rate-determining in the overall reaction sequence. An alternative estimate of TE specificity was obtained by direct calculation of the specificity parameter kobs/Km.. The kobsE+H/Km for NocTE catalyzed turnover of epi-nocG-SNAC 11a was calculated to be 5.8(4) mM−1 min−1 while the kobsH/Km for nocardicin G-SNAC 11b is calculated to be 84(8) mM−1min−1, a 14-fold preference for the C-terminal D-pHPG β-lactam-containing thioester stereochemistry.
Finally, with these observations in hand, a competition experiment was conducted to more clearly identify the peptide length preferentially processed by the NocTE domain. A 1:1 mixture of nocardicin G-SNAC (11b) and L-pHPG-L-Arg-nocardicin G-SNAC (9b) was incubated with NocTE and the reaction was monitored by HPLC over time. HPLC analysis clearly showed production of L-pHPG-L-Arg-nocardicin G (14) after only 30 s with little to no observable nocardicin G (1) formation (Fig. 5c). After 4 min, virtually no 9b remained compared to 11b. Similar results were obtained in a separate competition experiment between 9a and 11a (Supplementary Fig. 7). These data show at least a 20-fold preference of the NocTE domain for the pentapeptide β-lactam-containing intermediate, L-pHPG-L-Arg-nocardicin G (14), in harmony with the previously unexpected requirement that all five modules in NocA/B be functional to observe antibiotic synthesis.16
Discussion
The function of a TE domain to epimerize peptide products is a previously unknown NRPS strategy compared to that of canonical E domains. These domains have distinct primary functions and active site architectures, but may rely on related mechanisms to catalyze the epimerization of α-centers. In typical E domains, epimerization occurs on the PCP bound peptide thioester and rapidly produces an equilibrium of ~ 60% D-/40% L-a.a.-S-PCP while the downstream C domain selects and incorporates the D-isomer into the growing peptide chain.33 In the case of E domains embedded in elongation modules, epimerization occurs at the peptidyl-S-PCP stage after condensation but before peptide transfer.34 In both scenarios, it is the presence of the carrier protein thioester linkage that enables epimerization of the α-center through stabilization of the carbanion generated. The pKa of an α-hydrogen adjacent to a simple thioester is ~21 compared to a value of ~25 for the corresponding oxyester, as would be encountered in a TE domain.35 Additional anion stabilization is provided, however, by the presence of an aryl substituent at the α-carbon. This unique structural feature of a pHPG residue lowers the pKa by an additional 6–7 units36 and brings sufficiently rapid epimerization into the range of a physiological base for its oxyester in the NocTE active site. This effect is further enhanced by a ~10-fold amplification of the exchange rate afforded by the adjacent preformed β-lactam.26
Like NocTE, other NRPS domains have been characterized to perform epimerization reactions in addition to their canonical functions. In comparative examples to nocardicin A, such as in the assembly of arthrofactin, syringomycin, syringopeptin and ramoplanin, it has been established that C domains in their corresponding NRPSs were responsible for catalyzing both epimerization and amide bond formation.37 Alignment of these dual C/E domains revealed a unique N-terminal sequence of about 50 amino acids ending in an elongated His motif exhibiting the consensus sequence HHI/LXXXXGD and is distinct from the His motif associated with amide bond formation.37 Upon primary sequence comparison of NocC5 with these characterized dual functioning C/E domains, the conserved N-terminal sequence conferring epimerization activity was not found. Epimerization occurs in these dual C/E domains through non-covalent interaction with the aminoacyl-S-PCP or peptidyl-S-PCP, by again exploiting the α-carbanion stabilizing properties of the adjacent thioester. In examples of pyocein and yersiniabactin biosynthesis, encoded A domains within their respective NRPSs were found to have 310-360 residue inserts that confer epimerization activity.38 These inserts are also used to facilitate proton transfer on the acyl-S-PCP intermediate after chain elongation through non-covalent interactions with the “E” insert within the A domain.
In TE domains where a catalytic triad is present the peptidyl intermediate is first covalently transferred to the active site serine, followed by intermolecular or intramolecular nucleophilic attack on the oxyester to form the corresponding linear peptide or macrocyclic product.39 Primary sequence analysis of NocTE does not reveal any suspect E domain-like catalytic motif or new signature sequence (Supplementary Fig. 8). It is postulated that the epimerization activity of NocTE is conferred by the catalytic triad itself and enabled by the anomalously low pKa of the pHPG α-hydrogen. TE mediated hydrolysis and macrocyclization reactions are thought to occur by a mechanism in which the histidinium ion formed in the initial attack of the active site serine is deprotonated by the thiolate of the departing carrier protein–pantetheine arm as the tetrahedral transition state collapses to the enzyme–acyl intermediate. The neutral histidine is then free to deprotonate water or a nucleophilic group internal to the substrate and complete the catalytic cycle.40–42 The evidence gathered shows that epimerization takes place on the peptide β-lactam O-acyl TE species, not on the peptidyl-S-PCP as has been established in all other characterized E, C/E and A/E domains. Given the covalent attachment of substrate to the active site serine, the triad histidine is the proximal base. Analogous mechanisms have been advanced for Dieckmann reactions catalyzed by TE domains in fungal, non-reducing polyketide synthases,43 as well as those mediating pyrone formation44 and coupled Dieckmann/retro-Claisen reaction.45 In contrast to canonical E domains, which rely on a downstream C domain to specifically incorporate the correct stereoisomer from an equilibrating D/L-pool, NocTE performs this resolution autonomously. Unlike most type I thioesterases, which exhibit a degree of relaxed selectivity with structurally related substrates, the NocTE is evolutionarily optimized for the steric constraints imposed by an integrated β-lactam ring. Related peptide substrate analogs were only very poorly hydrolyzed.
Experiments, even with peptides bearing activated O-seryl moieties for SNi displacement, while negative, render unlikely a role for the TE domain in monocyclic β-lactam synthesis. It can be deduced, however, from the data reported here that β-lactam formation precedes NocTE catalyzed epimerization and hydrolysis and must occur elsewhere on the NRPS. One possibility is that azetidinone formation takes place on a the peptidyl-S-PCP bound intermediate, presumably in module 5 since it is the C-terminal pHPG nitrogen that is required for β-lactam ring formation, and is catalyzed in cis by the NRPS (Fig. 6a). A second, perhaps more compelling, potential mechanism has been proposed4 in which cyclodehydration could occur, and would invoke a presently unknown auxiliary enzyme-assisted reaction acting in trans with the NocNRPS (Fig. 6b). Such a process is not without precedent.21 A particularly relevant example is the biosynthesis of coronamic acid in which two enzymes transform an L-allo-isoleucine-S-PCP to a cyclopropane-containing-S-PCP bound intermediate.46 By extension, cyclodehydration of a peptidyl-S-PCP5 intermediate in nocardicin biosynthesis could be catalyzed by an auxiliary enzyme through activation of the seryl hydroxyl and subsequent β-lactam formation through deprotonation of the C-terminal amide, an efficient transformation precedented in synthetic models.47 The β-lactam intermediate can then undergo transacylation to NocTE, where epimerization occurs, and the propeptide-β-lactam intermediate released. The evidence provided by the competition experiment suggests that it is the pentapeptide β-lactam (14) that is the product of NocTE and presumably removal of the N-terminal L-pHPG-L-Arg residues occurs by rapid cellular proteolysis to give nocardicin G (1). Thus, simple editing of the programmed NRPS pentapeptide product to a modified tripeptide is proposed to be the last step(s) in formation of the central β-lactam-containing biosynthetic intermediate, nocardicin G (1). In contrast to the dazzlingly efficient oxidative reaction to penicillin using an entirely conventional NRPS tripeptide, ACV (Fig. 1b), the non-oxidative strategy to the structurally simpler monocyclic β-lactam in the nocardicins has arisen from the appearance of new NRPS synthetic capabilities.
Figure 6. Possible pathways to monocyclic β-lactam formation.
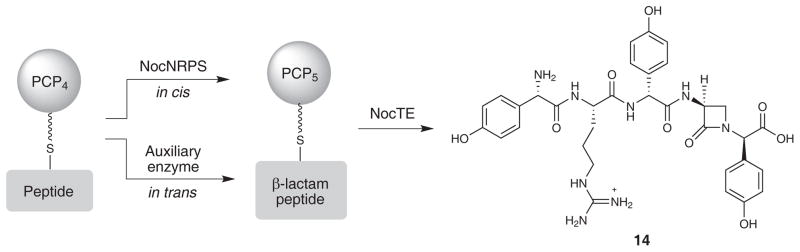
Top, pathway represents in cis β-lactam formation, invoking an external enzyme responsible for cyclodehydration. Bottom, pathway represents in trans β-lactam formation catalyzed by a domain within the NocNRPS. Upon β-lactam formation NocTE catalyzes C-terminal epimerization and hydrolysis of 14.
Online Methods
General Methods
DNA sequencing of PCR-amplified fragments was conducted at the Johns Hopkins Core Sequencing Facility (Baltimore, MD). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). DNA polymerase (Herculase II) was purchased from Agilent Technologies (Santa Clara, CA). Subcloning vector pCRBlunt-TOPO was purchased from Invitrogen (Grand Island, NY). The pET28b expression vector and Rosetta 2 cells were purchased from Novagen (Darmstadt, Germany). TALON® metal affinity resin was purchased from Clontech (Mountain View, CA). Amicon Ultra filtration devices were purchased from Millipore (Billerica, MA).
HPLC analyses of enzymatic reactions were performed on an Agilent model 1200 equipped with a multi-wavelength UV-Vis detector and a reverse phase Phenomenex Luna 5u phenyl/hexyl analytical column (250 x 4.60 mm ID). Analytical Method A (water + ACN + 0.1% TFA): 0–5 min isocratic 93% water + 7% ACN +0.1% TFA, 5–22 min gradient 7% to 50% ACN + 0.1% TFA, 22–25 min gradient 50% to 7% ACN + 0.1% TFA, 25–35 min isocratic 93% water + 7% ACN + 0.1% TFA. Flow rate = 1.0 mL/min. Analytical Method B (water + ACN + 0.1% TFA): 0–20 min gradient 7% to 50% ACN + 0.1% TFA, 20–25 min gradient 50% to 7% ACN + 0.1% TFA, 25–35 min isocratic 93% water + 7% ACN + 0.1% TFA. Flow rate = 1.0 mL/min.
Molecular Cloning of Protein Constructs
PCP5-TE-pET28
This construct was obtained from PCR amplification of the pMG0531 cosmid containing the entire nocardicin NRPS gene cluster.10 This template was PCR amplified with Herculase II Fusion DNA polymerase by using the following oligonucleotide primers (underlined: restriction site):
forward: 5′-GCGTAACATATGGGCGAGCTGGTGGTGCTCGACGCG-3′ and
reverse: 5′-GTAAGCGGCCGCTCACCGCTCTCCTCCCAGCGCGCGGCG-3′.
The PCR products were subcloned into pCRBlunt-TOPO and sequence verified. The pCRBlunt-PCP4-TE construct was digested with NdeI and NotI and ligated with T4 DNA ligase into a similarly digested pET28b vector to create the N-terminal 6x His-tagged expression construct.
PCP5-TE*S1779A-pET28b
The gene for the full-length PCP-TE didomain containing a serine to alanine mutation in the catalytic triad of the TE domain was constructed by splicing by overlap extension (SOE) mutagenesis,48 using Herculase II DNA polymerase. In the first round of PCR amplification, PCP5-TE-pET28b was used as template in a reaction with the following oligonucleotide primers (underlined: restriction site, bold: mutation):
forward: 5′-GCGTAACATATGGGCGAGCTGGTGGTGCTCGACGCG-3′
reverse: 5′-CGCCGCCGAAGGCCCAGCCGCCG-3′
forward: 5′-CGGCGGCTGGGCCTTCGGCGGCG-3′
reverse: 5′-GTAAGCGGCCGCTCACCGCTCTCCTCCCAGCGCGCGGCG-3′.
The products of the first round of PCR were combined, ligated and amplified in a second round of PCR amplification with the following oligonucleotide primers:
forward: 5′-GCGTAACATATGGGCGAGCTGGTGGTGCTCGACGCG-3′
reverse: 5′-GTAAGCGGCCGCTCACCGCTCTCCTCCCAGCGCGCGGCG-3′.
The PCR product was subcloned into pCRBlunt-TOPO and sequenced. The pCRBlunt-PCP5-TE*S1779A mutant construct was digested with NdeI and NotI and ligated with T4 DNA ligase into a similarly digested pET28b vector to create an N-terminal 6x His-tagged expression construct.
NocTE-pET28b
Cloning of the intact gene into an expression vector was achieved by PCR amplification of the desired TE monodomain fragment from the PCP5-TE-pET28b construct. This template was PCR amplified with Herculase II using the following oligonucleotide primers (underlined: restriction site): forward: 5′-GGGATACATATGGTCGAGGGCTCCGGGTC-3′ and reverse: 5′-GGATAAAGCTTTCACCGCTCTCCTCCCAG-3′. The PCR products were subcloned into a pCRBlunt-TOPO vector and verified to be the desired insert through sequencing. The pCRBlunt-PCP-NocTE construct was digested with NdeI and HindIII and ligated with T4 DNA ligase into a similarly digested pET28b vector to create an N-terminal 6x-His tagged expression construct.
TES*1779A-pET28b
Cloning of the intact gene into the expression vector was achieved by PCR amplification of the desired TE mono domain fragment from the PCP5-TE*S1779A-pET28b construct with Herculase II by using the following oligonucleotide primers (underlined: restriction site):
forward: 5′-GGGATACATATGGTCGAGGGCTCCGGGTC-3′
reverse: 5′-GGATAAAGCTTTCACCGCTCTCCTCCCAG-3′.
The PCR products were ligated into pCRBlunt-TOPO and sequence verified. The pCRBlunt-TE construct was digested with NdeI and HindIII and ligated with T4 DNA ligase into a similarly digested pET28b vector to create an N-terminal 6x His-tagged construct.
TE*S1779C-pET28b
This construct was created analogously to the PCP5-TE*S1779A-pET28b construct by SOE mutagenesis using the wild-type TE-pET28b plasmid as the template and the following two sets of primers for the first round of PCR amplification (underlined: restriction site, bold: mutation):
forward: 5′-GGGATACATATGGTCGAGGGCTCCGGGTC-3′
reverse: 5′-CGCCGCCGAAGCACCAGCCGCCG-3′
forward: 5′-CGGCGGCTGGTGCTTCGGCGGCG-3′
reverse: 5′-GGATAAAGCTTTCACCGCTCTCCTCCCAG-3′
The products of these two reactions were combined, ligated and PCR amplified with the following oligonucleotide primers:
forward: 5′-GGGATACATATGGTCGAGGGCTCCGGGTC-3′
reverse: 5′-GGATAAAGCTTTCACCGCTCTCCTCCCAG-3′.
The full-length PCR product was subcloned into pCRBlunt-TOPO and sequenced verified. The pCRBlunt-TE*S1779C mutant construct was digested with NdeI and HindIII and ligated with T4 DNA ligase into a similarly digested pET28b vector to create and N-terminal 6x His-tagged construct.
Protein Expression and Purification
Expression and purification of NocTE, PCP5-TE, PCP5-TE*S1779A, TE*S1779A, and TE*S1779C
Each desired pET28b protein construct was transformed into Rosetta 2 cells, plated onto Luria-Bertaini (LB)-agar containing 50 μg/mL kanamycin and 50 μg/mL chloramphenicol and incubated at 37 °C overnight. One colony per starter culture was selected and grown at 37 °C at 200 rpm overnight in 10 mL LB medium supplemented with 50 μg/mL kanamycin and 50 μg/mL chloramphenicol. One liter of 2XYT medium containing 50 μg/mL kanamycin and 50 μg/mL chloroamphenicol was inoculated with 10 mL of starter culture and cells were grown at 37 °C, at 180 rpm to an OD600 of 0.6–0.8. The temperature of the culture was cold-shocked at 4 °C for 1 hour and then placed on a shaker equilibrated to 18 °C. 1 mM of isopropyl α-D-thiogalactopyranoside (IPTG) was added and the culture was grown for 18 h at 180 rpm.
The cells were harvested by centrifugation (5,000 x g, 15 min, 4°C) and stored at −80 °C. Cells were thawed in lysis buffer (50 mM phosphate, 300 mM NaCl, pH 8.0) and disrupted by sonication (60% amplitude, pulse 9 sec on/off, 3 min) on ice. Cell debris was removed by centrifugation (25,000 x g, 30 min, 4 °C) and the clarified cell lysate was incubated with 2 mL of 50% suspension/L of cell culture of cobalt TALON® metal affinity resin for 1–2 h at 4 °C in a batch-binding format. The resin was pelleted (750 x g, 4 °C, 5 min) and the supernatant was removed. The resin was re-suspended in lysis buffer and loaded onto a gravity column. The desired protein was then eluted with a stepwise gradient of imidazole (20–200 mM) in lysis buffer. Fractions containing the purified protein, as determined by SDS-PAGE with Coomassie staining, were pooled and dialyzed against 2 x 3 L of assay buffer, pH 7.5. Protein concentrations were quantified by Bradford assay. Images of SDS-PAGE gels of all protein constructs described can be found in Supplementary Figure 9.
Synthesis of peptidyl-pantetheine, peptidyl-SNAC, and peptidyl-S-CoA substrates
Syntheses of peptidyl-pantetheine, peptidyl-SNAC, were conducted as described previously.26 General methods and syntheses of 10, and peptidyl-S-CoAs 13a/b and 14a/b are described in Supplementary Note. Standards used for comparison in these studies (including 1, epi-1, 2a/b, 3a/b, 14 and epi-14) were obtained using standard deblocking methods from the corresponding protected precursor followed by HPLC purification. Characterization data of standards are reported in Supplementary Note.
In vitro NocTE assay with peptidyl-S-pantetheine substrates
To test the in vitro activities of NocTE or other mutant proteins, 1 mM of desired thioester substrate was added to a 300 μL solution containing 20 μM of freshly purified protein in 50 mM KiPO4 buffer, pH 7.5. Reactions were incubated at room temperature for 3 h and acidified to pH 2 with an aqueous solution of TFA. The enzyme was removed with an Amicon Ultra 0.5 ML 10 kMWCO filtration device and the flow-through was directly analyzed. Enzymatic reactions were analyzed using HPLC Analytical Method A.
1H-NMR time course in D2O
NocTE was isolated, purified and dialyzed into 2 x 2 L of 20 mM NH3CO3 solution over 12 h at 4 °C. The protein was concentrated to 7 mL, flash frozen and lyophilized to dryness. Lyophilized protein was resuspended in 600 μL of 50 mM PO4, 25 mM NaCl, pD = 7.2 in deuterium oxide. Insoluble material was removed by centrifugation and the protein was concentrated to 15 mg/mL. β-Lactam-containing substrates 11a/b (~1:1 11a:11b) were dissolved in 50 mM phosphate (tribasic), 25 mM NaCl, pD = 7.2 in deuterium oxide to a final concentration of 10 mM. The reaction was initiated by the addition of 1 μM of NocTE. Substrate turnover was monitored for 3 h by 1H-NMR spectroscopy on a Varian 500 MHz spectrometer equipped with an inverse probe. Spectra were acquired at approximately 1 per min, 1 scan/spectrum.
Determination of NocTE kinetic parameters
Hydrolysis of SNAC-thioesters 11a and 11b was followed spectrophotometrically by observation of the formation of 3-thio-5-nitrobenzoate (DNTB) at 412 nm (ε = 13,600 M−1 cm−1).49 Each assay contained 50 mM KiPO4, pH = 7.5, a variable amount of substrate, 1 μM NocTE, and 3 mM DNTB dissolved in assay buffer in a total volume of 60 μL. Stock solutions of 11a and 11b were dissolved in DMSO such that the desired final concentration in the assay was achieved with the addition of 5 μL of DMSO substrate solution. The reactions were initiated by the addition of substrate and the rate of free thiol formation was observed continuously at an interval time of 30 s. Rates of hydrolysis were calculated from the initial linear portion of the curves. In all cases, rates were corrected for the background rate of chemical hydrolysis in the absence of enzyme. The concentration range of the substrates analyzed was 0.1 to 20 mM. Experiments were conducted in triplicate; data presented as mean values ± standard deviation. Michaelis-Menten curves were obtained by fitting the data to a nonlinear regression using Prism 6.0, GraphPad software (La Jolla, CA).
Determination of rate of epimerization of epi-nocardicin G-SNAC 11a to nocardicin G-SNAC 11b in the presence of TE*S1779A or TE*S1779C
To measure the rate of chemical epimerization of epi-nocardicin G-SNAC (11a) to nocardicin G-SNAC (11b), a 500 μL solution containing 1.0 mM 11a, and either 10 μM or TE*S1779A or TE*S1779C in 50 mM KiPO4 pH 7.5 was prepared. 25 μL of the reaction solution was aliquoted into 15 μL of a 0.2 % TFA aqueous solution on ice at t = 1, 2, 5, 10, 15, 20, 30, 45, 65, 80, 120 and 180 min. Quenched reactions were analyzed by HPLC using Analytical Method B, monitoring absorption at 272 nm. The rate of chemical equilibrium (k1 + k2) was calculated by fitting the data to a non-linear one-phase decay curve using Prism 6.0, GraphPad software (La Jolla, CA). Data reflect a single experiment.
Loading of CoA substrates 12a/b and 13a/b onto apo-PCP5-TE
In a total volume of 300 μL containing 10 mM MgCl2 and 50 mM HEPES pH 7.5, 100 μM of the apo-PCP5-TE construct and 150 μM of either 12a/b or 13a/b were combined. Reactions were initiated by the addition of 2 μM of Sfp and the reaction mixture was allowed to incubate at room temperature for 45 min. The reactions were quenched with TFA to pH 2. Precipitated proteins were removed with an Amicon Ultra 0.5 ML 10 kMWCO filtration device. The filtrate was directly analyzed for the formation of product using HPLC Analytical Method A, monitoring the absorption at 272 nm.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grant AI014937. We warmly thank K.A. Moshos, C.J. Hastings, and J.W. Li for their helpful discussion, chemical advice and encouragement and R.F Li for guidance with molecular biology. We also thank K. Belecki and I.P. Mortimer for HRMS data and Prof. C.T. Walsh (Harvard University) for providing the pET29-Sfp expression plasmid. A. Majumdar is graciously acknowledged for his assistance with the 1H-NMR arrayed D2O-exchange experiment.
Footnotes
Author contributions:
C.A.T. and N.M.G developed the hypothesis and designed the study. N.M.G. performed all syntheses and experiments reported. Both authors analyzed and discussed the results. N.M.G and C.A.T prepared the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Aoki H, Sakai H, Kohsaka M, Konomi T, Hosoda J. Nocardicin A, a New Monocyclic β-Lactam Antibiotic. I Discovery, Isolation and Characterization. J Antibiot. 1976;29:492–500. doi: 10.7164/antibiotics.29.492. [DOI] [PubMed] [Google Scholar]
- 2.Townsend CA, Wilson BA. The Role of Nocardicin G in Nocardicin A Biosynthesis. J Am Chem Soc. 1988;110:3320–3321. [Google Scholar]
- 3.Townsend CA, Brown AM. Nocardicin A: Biosynthetic Experiments with Amino Acid Precursors. J Am Chem Soc. 1983;105:913–918. [Google Scholar]
- 4.Townsend CA, Brown AM, Nguyen LT. Nocardicin A: Stereochemical and Biomimetic Studies of Monocyclic β-Lactam Formation. J Am Chem Soc. 1983;105:919–927. [Google Scholar]
- 5.Fawcett PA, et al. Synthesis of δ-(α-Aminoadipyl)cysteinylvaline and its Role in Penicillin Biosynthesis. Biochem J. 1976;157:651–660. doi: 10.1042/bj1570651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samson SM, et al. Isolation, sequence determination and expression in Escherichia coli of the isopenicillin N synthetase gene from Cephalosporium acremonium. Nature. 1985;318:191–194. doi: 10.1038/318191a0. [DOI] [PubMed] [Google Scholar]
- 7.Banko G, Demain AL, Wolfe S. δ-(L-α-Aminoadipyl)-L-cysteinyl-D-valine synthetase (ACV synthetase): A Multifunctional Enzyme with Broad Substrate Specificity for the Synthesis of Penicillin and Cephalosporin Precursors. J Am Chem Soc. 1987;109:2858–2860. [Google Scholar]
- 8.Roach PL, et al. Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation. Nature. 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- 9.Townsend CA, Brown AM. Nocardicin A biosynthesis: stereochemical course of monocyclic β-lactam formation. J Am Chem Soc. 1982;104:1748–1750. [Google Scholar]
- 10.Gunsior M, et al. The Biosynthetic Gene Cluster for a Monocyclic β-Lactam Antibiotic, Nocardicin A. Chem Biol. 2004;11:927–938. doi: 10.1016/j.chembiol.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Reeve AM, Breazeale SD, Townsend CA. Purification, Characterization, and Cloning of an S-Adenosylmethionine-dependent 3-Amino-3-carboxypropyltransferase in Nocardicin Biosynthesis. J Biol Chem. 1998;273:30695–30703. doi: 10.1074/jbc.273.46.30695. [DOI] [PubMed] [Google Scholar]
- 12.Kelly WL, Townsend CA. Role of the Cytochrome P450 NocL in Nocardicin A Biosynthesis. J Am Chem Soc. 2002;124:8186–8187. doi: 10.1021/ja025926g. [DOI] [PubMed] [Google Scholar]
- 13.Kelly WL. Mutational Analysis and Characterization of Nocardicin C-9′ Epimerase. J Biol Chem. 2004;279:38220–38227. doi: 10.1074/jbc.M405450200. [DOI] [PubMed] [Google Scholar]
- 14.Kelly WL, Townsend CA. Mutational Analysis of nocK and nocL in the Nocardicin A Producer Nocardia uniformis. J Bacteriol. 2005;187:739–746. doi: 10.1128/JB.187.2.739-746.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidsen JM, Townsend CA. Identification and Characterization of NocR as a Positive Transcriptional Regulator of the β-Lactam Nocardicin A in Nocardia uniformis. J Bacteriol. 2009;191:1066–1077. doi: 10.1128/JB.01833-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidsen JM, Townsend CA. In Vivo Characterization of Nonribosomal Peptide Synthetases NocA and NocB in the Biosynthesis of Nocardicin A. Chem Biol. 2012;19:297–306. doi: 10.1016/j.chembiol.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambalot RH, et al. A new enzyme superfamily-the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 18.Stachelhaus T, Hüser A, Marahiel MA. Biochemical characterization of peptidyl carrier protein (PCP), the thiolation domain of multifunctional peptide synthetases. Chem Biol. 1996;3:913–921. doi: 10.1016/s1074-5521(96)90180-5. [DOI] [PubMed] [Google Scholar]
- 19.Meier JL, Burkart MD. The chemical biology of modular biosynthetic enzymes. Chem Soc Rev. 2009;38:2012–2045. doi: 10.1039/b805115c. [DOI] [PubMed] [Google Scholar]
- 20.Marahiel MA, Stachelhaus T, Mootz HD. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 21.Walsh CT, et al. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr Opin Struct Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 22.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 23.Du L, Lou L. PKS and NRPS release mechanisms. Nat Prod Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 24.Baltz RH. Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J Ind Microbiol Biotechnol. 2011;38:1747–1760. doi: 10.1007/s10295-011-1022-8. [DOI] [PubMed] [Google Scholar]
- 25.Davidsen JM, Bartley DM, Townsend CA. Non-ribosomal Propeptide Precursor in Nocardicin A Biosynthesis Predicted from Adenylation Domain Specificity Dependent on the MbtH Family Protein NocI. J Am Chem Soc. 2013;135:1749–1759. doi: 10.1021/ja307710d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudelli NM, Townsend CA. Stereocontrolled Syntheses of Peptide Thioesters Containing Modified Seryl Residues as Probes of Antibiotic Biosynthesis. J Org Chem. 2013;78:6412–6426. doi: 10.1021/jo4007893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trauger JW, Kohli RM, Mootz HD, Marahiel MA, Walsh CT. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature. 2000;407:215–218. doi: 10.1038/35025116. [DOI] [PubMed] [Google Scholar]
- 28.Ehmann DE, Trauger JW, Stachelhaus T, Walsh CT. Aminoacyl-SNACs as small-molecule substrates for the condensation domains of nonribosomal peptide synthetases. Chem Biol. 2000;7:765–772. doi: 10.1016/s1074-5521(00)00022-3. [DOI] [PubMed] [Google Scholar]
- 29.Hamed RB, et al. The enzymes of β-lactam biosynthesis. Nat Prod Rep. 2013;30:21–107. doi: 10.1039/c2np20065a. [DOI] [PubMed] [Google Scholar]
- 30.Sieber SA, Walsh CT, Marahiel MA. Loading Peptidyl-Coenzyme A onto Peptidyl Carrier Proteins: A Novel Approach in Characterizing Macrocyclization by Thioesterase Domains. J Am Chem Soc. 2003;125:10862–10866. doi: 10.1021/ja0361852. [DOI] [PubMed] [Google Scholar]
- 31.Gokhale RS, Hunziker D, Cane DE, Khosla C. Mechanism and specificity of the terminal thioesterase domain from the erythromycin polyketide synthase. Chem Biol. 1999;6:117–125. doi: 10.1016/S1074-5521(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 32.Lu H, Tsai SC, Khosla C, Cane DE. Expression, Site-Directed Mutagenesis, and Steady State Kinetic Analysis of the Terminal Thioesterase Domain of the Methymycin/Picromycin Polyketide Synthase. Biochemistry. 2002;41:12590–12597. doi: 10.1021/bi026006d. [DOI] [PubMed] [Google Scholar]
- 33.Stachelhaus T, Walsh CT. Mutational Analysis of the Epimerization Domain in the Initiation Module PheATE of Gramicidin S Synthetase. Biochemistry. 2000;39:5775–5787. doi: 10.1021/bi9929002. [DOI] [PubMed] [Google Scholar]
- 34.Linne U, Marahiel MA. Control of Directionality in Nonribosomal Peptide Synthesis: Role of the Condensation Domain in Preventing Misinitiation and Timing of Epimerization. Biochemistry. 2000;39:10439–10447. doi: 10.1021/bi000768w. [DOI] [PubMed] [Google Scholar]
- 35.Amyes TL, Richard JP. Generation and Stability of a Simple Thiol Ester Enolate in Aqueous Solution. J Am Chem Soc. 1992;114:10297–10302. [Google Scholar]
- 36.Bordwell FG. Equilibrium Acidities in Dimethyl Sulfoxide Solution. Acc Chem Res. 1988;21:456–463. [Google Scholar]
- 37.Balibar CJ, Vaillancourt FH, Walsh CT. Generation of D Amino Acid Residues in Assembly of Arthrofactin by Dual Condensation/Epimerization Domains. Chem Biol. 2005;12:1189–1200. doi: 10.1016/j.chembiol.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Patel HM, Tao J, Walsh CT. Epimerization of an L-Cysteinyl to a D-Cysteinyl Residue during Thiazoline Ring Formation in Siderophore Chain Elongation by Pyochelin Synthetase from Pseudomonas aeruginosa. Biochemistry. 2003;42:10514–10527. doi: 10.1021/bi034840c. [DOI] [PubMed] [Google Scholar]
- 39.Kopp F, Marahiel MA. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat Prod Rep. 2007;24:735–749. doi: 10.1039/b613652b. [DOI] [PubMed] [Google Scholar]
- 40.Giraldes JW, et al. Structural and mechanistic insights into polyketide macrolactonization from polyketide-based affinity labels. Nat Chem Biol. 2006;2:531–536. doi: 10.1038/nchembio822. [DOI] [PubMed] [Google Scholar]
- 41.Smith S. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. FASEB J. 1994;8:1248–1259. [PubMed] [Google Scholar]
- 42.Tsai SC, et al. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc Natl Acad Sci USA. 2001;98:14808–14813. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korman TP, et al. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc Natl Acad Sci USA. 2010;107:6246–6251. doi: 10.1073/pnas.0913531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newman AG, Vagstad AL, Belecki K, Scheerer JR, Townsend CA. Analysis of the cercosporin polyketide synthase CTB1 reveals a new fungal thioesterase function. Chem Commun (Camb) 2012;48:11772–11774. doi: 10.1039/c2cc36010a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vagstad AL, Hill EA, Labonte JW, Townsend CA. Characterization of a fungal thioesterase having Claisen cyclase and deacetylase activities in melanin biosynthesis. Chem Biol. 2012;19:1525–1534. doi: 10.1016/j.chembiol.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 47.Salituro GM, Townsend CA. Total Syntheses of (−)-Nocardicins A-G: a Biogenetic Approach. J Am Chem Soc. 1990;112:760–770. [Google Scholar]
- 48.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 49.Collier HB. Letter: A Note on the Molar Absorptivity of Reduced Ellman’s Reagent, 3-Carboxylato-4-Nitrothiophenolate. Anal Biochem. 1973;56:310–311. doi: 10.1016/0003-2697(73)90196-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.