Abstract
The pathogenesis of the cutaneous T-cell lymphoma (CTCL), mycosis fungoides (MF) is unclear. MicroRNA (miRNA) are small non-coding RNA that target mRNA leading to reduced mRNA translation. Recently, specific miRNA were shown to be altered in CTCL. We identified significantly reduced expression of miR-223 in early stage MF skin, and the levels of miR-223 diminished further in advanced stage disease. CTCL peripheral blood mononuclear cells and cell lines also had reduced miR-223 as compared to controls. Elevated expression of miR-223 in these cell lines reduced cell growth and clonogenic potential, whereas inhibition of miR-223 increased cell numbers. Investigations into putative miR-223 targets with oncogenic function, including E2F1 and MEF2C, and the predicted miR-223 target, TOX, revealed all three are targeted by miR-223 in CTCL. E2F1, MEF2C, and TOX proteins were decreased with miR-223 overexpression, while miR-223 inhibition led to increased protein levels in CTCL. In addition, we showed the 3′-UTR of TOX mRNA was a genuine target of miR-223. Therefore, reduced levels of miR-223 in MF/CTCL lead to increased expression of E2F1, MEF2C, and TOX, which likely contribute to the development and/or progression of CTCL. Thus, miR-223 and its targets may be useful for the development of new therapeutics for MF/CTCL.
Introduction
MicroRNA (miRNA) are small, 18–22bp, non-coding RNA that negatively regulate protein translation (Bartel, 2004; Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). They elicit their effects on protein translation through the binding of the 3′ untranslated region (3′-UTR) of mRNA, primarily causing inhibition of translation or mRNA cleavage (Bartel, 2004; Llave et al., 2002; Reinhart et al., 2000). Since the discovery that a reduction of miR-15/16 may be a causal factor in the development of chronic lymphocytic leukemia (Calin et al., 2002), there has been tremendous interest in the role miRNA have in the development and progression of malignancies, as well as the use of miRNA in diagnostic signatures and cancer biomarkers.
Mycosis fungoides (MF) is the most common form of cutaneous T-cell lymphoma (CTCL) and is increasing in incidence (Weinstock and Horm, 1988). It classically presents as patches and plaques, and progresses to tumors, with eventual blood and visceral involvement. We have few therapeutic options in advanced MF, and the 5-year survival dramatically reduces from 94% in Stage IA to 48% in Stage IIB, and 18% in Stage IV (Agar et al., 2010). Additionally, little is known about the pathogenesis of MF. Although there have been efforts made to identify potential causative infectious agents, genetic mutations, and chronic antigenic stimulants, there is no clear understanding of the etiology. Recently, studies have identified altered miRNA expression in MF and CTCL. Ballabio et al, reported decreased levels of miR-342 and miR-223 in Sézary syndrome (SS), a leukemic form of CTCL (Ballabio et al., 2010). Another group identified a ‘diagnostic signature’ of miRNA (miR-155, -203 and -205) in CTCL to help differentiate it from benign dermatoses (Ralfkiaer et al., 2011). It is clear that miRNA are altered in MF/CTCL, but it is unknown what role these alterations have in the oncogenesis or progression of the disease. In a malignancy with increasing incidence, poor 5-year survival, and limited therapeutic options, it is imperative that we make efforts to further understand MF/CTCL.
miR-223 is an intergenic miRNA under the control of its own dedicated promoter (Pulikkan et al., 2010). Interestingly, it has been shown to be elevated in immature or blastic T-cell malignancies (Mavrakis et al., 2011), but was found to have reduced expression in CTCL, a mature T-cell malignancy (Ballabio et al., 2010; Narducci et al., 2011; Ralfkiaer et al., 2011). However, other groups have not detected a significant alteration in miR-223 expression between CTCL and controls (Qin et al., 2012; van Kester et al., 2011). Based upon these data, the precise alteration and role that miR-223 has in CTCL has not been firmly established.
miR-223 has been demonstrated to target potential oncogenic transcription factors, E2F1 (Pulikkan et al., 2010) and the myocyte enhancing factor 2C (MEF2C) (Johnnidis et al., 2008) in myeloid cells. A proposed miR-223 target, the thymocyte selection-associated high mobility group box (TOX) that is necessary for the development of CD4+ T-lymphocytes, was recently found to be overexpressed in early stage MF (Aliahmad et al., 2012; Lewis et al., 2005; Wang, 2008; Wang and El Naqa, 2008; Zhang et al., 2012). Here we show that miR-223 is expressed at decreased levels in MF and CTCL, and that miR-223 targets TOX, E2F1, and MEF2C in CTCL. Moreover, increasing miR-223 led to reduced levels of TOX and oncogenic proteins, E2F1 and MEF2C, with an associated decrease in CTCL growth and clonogenic potential, indicating miR-223 may be an inhibitor of CTCL development and/or progression.
Results
Decreased miR-223 levels in MF/CTCL
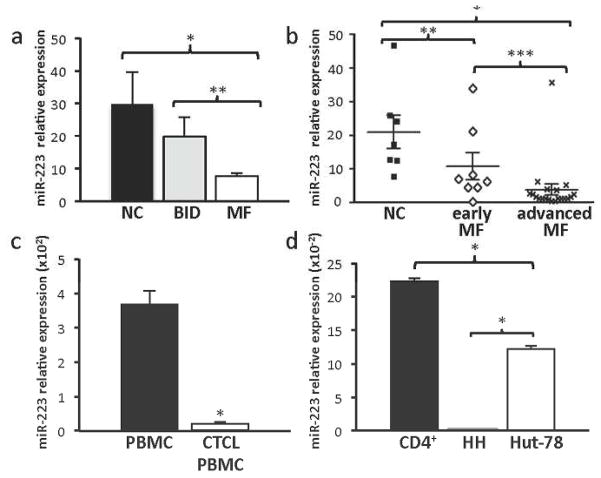
There is evidence that miR-223 levels are reduced in CTCL, but its expression in MF has yet to be determined. To evaluate miR-223 levels in MF, we analyzed 28 tissue samples of MF (n=8 early stage I-IIA, n=20 advanced stage IIB-IV), 6 tissue samples of benign inflammatory dermatoses (BID), and 7 tissue samples of normal skin controls (NC). BID samples were evaluated as they contain activated T cells and therefore, serve as a comparison control to malignant activated T cells in MF. Quantitative real-time polymerase chain reaction (qRT-PCR) revealed a significant decrease of miR-223 in MF as compared to normal controls and BID (t-test, *p<0.001, **p=0.004, Figure 1a). There was no significant difference in miR-223 expression between the subgroups of BID (t-test, p=0.16). There was even greater reduction of miR-223 in advanced MF as compared to early stage MF (t-test, *p<0.001, **p=0.022, ***p=0.036, Figure 1b). Peripheral blood mononuclear cells (PBMC) from patients with leukemic MF and SS (n=6) also demonstrated reduced miR-223 levels versus a pooled collection of PBMCs from Red Cross donors (t-test, *p<0.001, Figure 1c). These data indicate miR-223 is expressed at a reduced level in both the diseased skin and blood of MF patients compared to normal controls, and that miR-223 diminishes as the clinical stage advances.
Figure 1. Reduced expression of miR-223 in MF.
miR-223 expression levels relative to control small RNA RNU24 levels by qRT-PCR. a) Skin biopsies from subjects with MF (n=8, early stage IA-IIA; n=20, advanced stage IIB-IV), normal controls (NC, n=7) (*p<0.001), and BID (n=6) (**p=0.04). b) Skin biopsies from normal controls and patients with early or late stage MF, (*p<0.001, **p=0.022, ***p=0.036. c) Pooled control Red Cross PBMC and CTCL PBMC (n=6, *p<0.001). d) CTCL cell lines (HH and Hut-78) and CD4+ normal controls (n=2, *p<0.001).
To determine whether the decreased miR-223 levels observed in patients with MF were similar in CTCL cell lines, we measured miR-223 levels in two CTCL cell lines, HH and Hut-78. We determined miR-223 levels in these two CTCL cell lines were decreased compared to levels in CD4+ T-cells from normal controls (n=2, Figure 1d). Specifically, Hut-78 had a greater than 45% reduction in miR-223 expression compared to control (t-test, *p<0.001), and the miR-223 levels were almost undetectable in the HH cells (t-test, *p<0.001). Therefore, miR-223 is reduced in CTCL lines as well as patient samples, making the CTCL lines a viable tool to test the effects of miR-223.
miR-223 inhibits CTCL cell growth and clonogenic potential
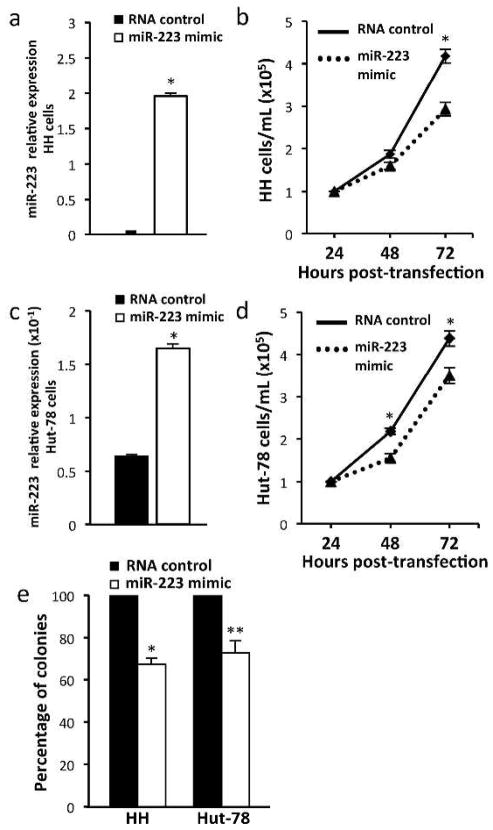
Although miRNA are altered within MF/CTCL, it is unknown what role these changes have in the malignancy. To assess the consequences of restoring miR-223 levels on CTCL growth, we transfected miR-223 mimic into both HH and Hut-78 cells. The increase in miR-223 levels from the mimic was determined by qRT-PCR (Figure 2a). Using the Trypan Blue Dye exclusion assay, we measured a significant decrease (30%) in viable cell numbers 72 hours post-transfection in the HH cells transfected with miR-223 mimic as compared to an RNA control (n=5, t-test, *p<0.001, Figure 2b). Similarly, with increased levels of miR-223 from the mimic (Figure 2c), the Hut-78 cells showed a 29% and 20% decrease in viable cell numbers at 48 and 72 hours, respectively, following miR-223 transfection (n=3, t-test, *p<0.001, Figure 2d). No difference in the numbers or percentage of dead cells was detected when comparing miR-223 mimic and RNA control samples at any of the times evaluated.
Figure 2. Increased miR-223 inhibits CTCL cell growth and decreases clonogenic potential.
a/c) miR-223 levels after transfection with the miR-223 mimic in HH and Hut-78 cells. b) Number of viable HH (n=5) and d) Hut-78 (n=3) cells transfected with miR-223 mimic or control RNA were determined at intervals by Trypan Blue Dye Exclusion assay (*p<0.001). e) Methylcellulose clonogenic assays. Colony numbers in HH and Hut-78 cells transfected with miR-223 mimic are relative to cells transfected with RNA control (n=3; *p<0.001, **p=0.003).
To assess the effect of increased levels of miR-223 on CTCL clonogenic capabilities, we transfected HH and Hut-78 cells with either miR-223 mimic or RNA control and performed methylcellulose assays. HH cells transfected with miR-223 mimic had a 32.5% reduction in colony formation and Hut-78 cells had a 27.1% reduction in colony number as compared to RNA control transfected cells (n=4, t-test, *p<0.001, **p=0.003, Figure 2e). We did not detect a difference in colony size between the mimic and RNA control transfected cells.
To further assess the impact of altered miR-223 levels on CTCL growth, an inhibitor of miR-223 was utilized to block its function. Transfection of HH and Hut-78 cells with the miR-223 inhibitor led to increased cell growth, as measured by Trypan Blue Dye exclusion and MTS assays (Figure 3a and data not shown). Trypan Blue Dye exclusion assay showed a 24% increase in viable cell numbers in the HH cells transfected with miR-223 inhibitor versus the cells transfected with negative control (n=3, t-test, *p<0.001, Figure 3a). The Hut-78 cells also showed a statistically significant increase in viable cell numbers (18%) in miR-223 inhibitor transfected cells by MTS assay (n=3, t-test, **p=0.013, Figure 3b). Together our results indicate miR-223 inhibits CTCL cell growth and clonogenic potential, but does not appear to impact apoptosis.
Figure 3. Inhibition of miR-223 enhances CTCL growth.
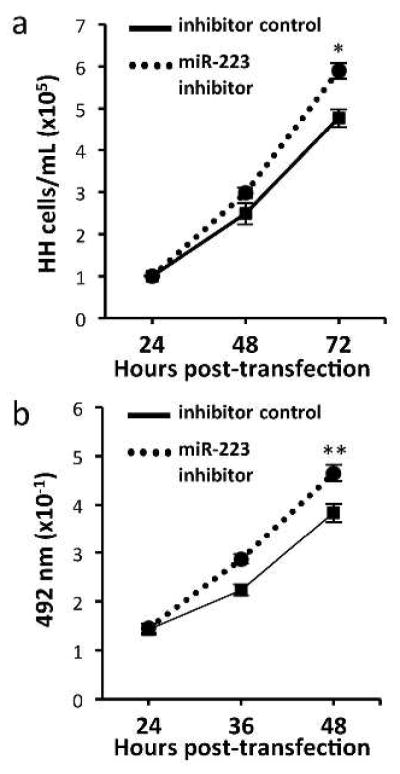
a) Number of viable HH cells transfected with miR-223 inhibitor or control (n=3) were determined at 24 hour intervals by Trypan Blue Dye Exclusion assay (*p<0.001). b) Hut-78 cells transfected with miR-223 inhibitor or control (n=3) were evaluated by MTS assay every 12 hours (**p=0.013).
TOX is a direct target of miR-223
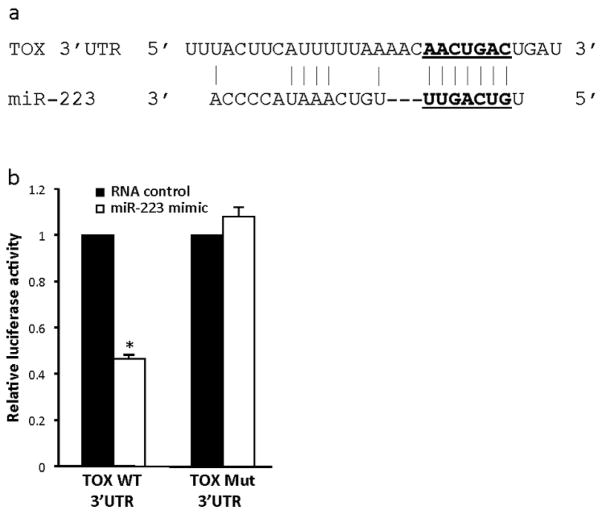
TOX, a protein normally expressed in thymocytes with unclear function, was recently found to have aberrant increased expression in MF, suggesting a possible role in MF pathogenesis (Zhang et al., 2012). By utilizing online miRNA target databases (TargetScan.org and miRDB.org), we identified TOX as a potential miR-223 target. The predicted highly conserved miR-223 binding site (seed sequence) is within the 3′-UTR of TOX (Figure 4a). To test whether TOX is a direct target of miR-223, we generated a luciferase expression plasmid with the 3′-UTR of TOX containing the putative miR-223 binding site. We also generated a similar plasmid with a mutated seed sequence, in which miR-223 could not bind. NIH-3T3 cells were co-transfected with the wild-type or mutated TOX 3′-UTR luciferase expression plasmid and either miR-223 mimic or control RNA. The cells with the miR-223 mimic and the TOX wild-type 3′-UTR had a significant reduction (53.6%) in luciferase activity, compared to the RNA control transfected cells (n=3, t-test, *p<0.001, Figure 4b). Moreover, miR-223 mimic did not induce a significant change in luciferase activity in cells with the mutated 3′-UTR of TOX (Figure 4b). These results indicate that miR-223 binds to the 3′-UTR of TOX and demonstrates TOX is a direct target of miR-223.
Figure 4. TOX is a direct target of miR-223.
a) miR-223 binding site within the 3′-UTR of TOX and miR-223 sequence (seed sequence underlined). b) Luciferase activity was measured in NIH-3T3 cells transfected with miR-223 mimic or RNA control and a pMIR-REPORT plasmid with the wild-type 3′-UTR of TOX or a 3′-UTR of TOX with a mutated miR-223 seed sequence. Luciferase activity is relative to β-galactosidase activity, which controlled for transfection efficiency for each (n=3; *p<0.001).
miR-223 targets E2F1, MEF2C, and TOX in CTCL
We have identified TOX as a direct miR-223 target, and miR-223 has been previously shown to target the oncogenic transcription factors E2F1 and MEF2C in myeloid cells (Johnnidis et al., 2008; Pulikkan et al., 2010). To determine whether these miR-223 targets are actually targeted by miR-223 in CTCL and thus, could contribute to the pathogenesis of this disease, we first used qRT-PCR to assess levels of E2F1, MEF2C, and TOX mRNA in CTCL lines. We detected significantly higher levels of E2F1, MEF2C, and TOX in HH cells compared to CD4+ controls (n=3; t-test, *p<0.001, **p=0.007, ***p=0.006, Figure 5a). In addition, increased levels of E2F1 and TOX mRNA were observed in Hut-78 cells (n=3, t-test, *p<0.001, Figure 5a). Immunohistochemistry also revealed increased TOX protein expression in both early and advanced MF patient samples as compared to normal and inflammatory controls (Figure 5b). There is notable staining of TOX in lymphocytes within both Pautrier’s microabscesses and MF tumors.
Figure 5. Endogenous expression of miR-223 mRNA targets is increased in CTCL.
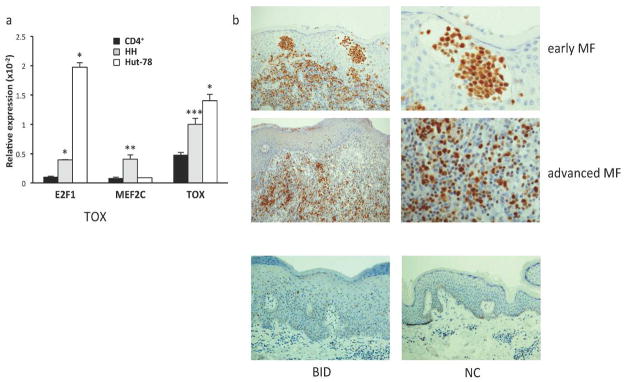
a) qRT-PCR for miR-223 targets E2F1, MEF2C, and TOX mRNA levels relative to β-actin in HH, Hut-78, and CD4+ cell controls (n=2; *p<0.001, **p=0.007, ***p=0.006). b) Immunohistochemical staining of TOX in early stage MF (Stage Ia-IIa; n=10), advanced MF (stage IIb-IV, n=5), BID (n=8) and NC (n=3) (representative photomicrographs of each shown).
To more directly test the effects of miR-223 on its targets in CTCL, we transfected HH cells with miR-223 mimic or control RNA and evaluated mRNA and protein levels by qRT-PCR and Western blot, respectively, for E2F1, MEF2C, and TOX. miR-223 mimic transfected cells had a statistically significant reduction in E2F1 (24.5%), MEF2C (31.1%), and TOX (16.4%) mRNA versus RNA control transfected cells (n=6, t-test, *p=0.005, **p=0.003, ***p=0.019, Figure 6a). We also assessed protein levels of these targets. Western blots showed decreased protein expression of E2F1, MEF2C, and TOX in HH cells transfected with miR-223 mimic compared to the RNA control transfected cells (Figure 6b). Conversely, when miR-223 inhibitor was transfected into Hut-78 cells, we observed an increase in protein expression of all three miR-223 targets (Figure 6c). These results suggest miR-223 targets E2F1, MEF2C, and TOX in CTCL cells, and this likely contributes to MF pathogenesis.
Figure 6. Modulation of miR-223 alters E2F1, MEF2C, and TOX expression.
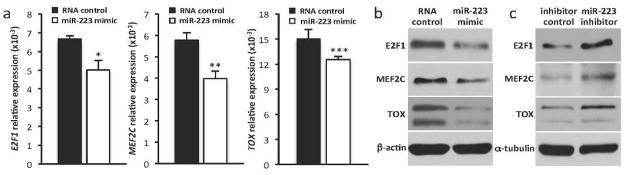
a) qRT-PCR of the miR-223 targets E2F1, MEF2C, and TOX relative to β-actin control in HH cells 48 hours after transfection with miR-223 mimic or RNA control (n=4, *p=0.005, **p=0.003, ***p=0.019). Western blots of E2F1, MEF2C, TOX and β-actin or α-tubulin control in b) HH cells after transfection with miR-223 mimic or RNA control and c) Hut-78 cells after transfection with miR-223 inhibitor or inhibitor control.
Discussion
miRNA have been shown to be altered within a wide variety of malignancies, including CTCL and MF (Ballabio et al., 2010; Ralfkiaer et al., 2011). It remains unclear if these miRNA are innocent bystanders, occurring secondarily from other primary oncogenic mutations, or if they function as initiators and/or drivers of the disease. Our current study focused on miR-223 expression in early and advanced stage MF and CTCL, and identifying how it may affect oncogenesis or disease progression. Most of the research on miR-223 has centered on granulopoesis/myelopoesis, but less is known about miR-223 within T-cell malignancies (Fazi et al., 2007; Fazi et al., 2005; Fukao et al., 2007). We demonstrated miR-223 was significantly reduced in both early and late stage MF skin samples compared to normal and inflammatory skin controls, and that the levels decreased further as the clinical stage advanced. Also there were lower levels of miR-223 in CTCL PBMC and cell lines compared with controls. The fact that miR-223 was consistently reduced across a variety of CTCL, and that it diminished as disease clinically progressed suggests miR-223 has an important role in the development and/or maintenance of MF/CTCL. Indeed, CTCL cell growth was slowed by increasing miR-223 levels, while inhibiting miR-223 increased growth. Clonogenic assays with miR-223 and CTCL cells revealed similar findings. Our data indicate miR-223 is retarding CTCL growth by targeting pro-proliferative genes, such as E2F1 and MEF2C, resulting in decreased proliferation. However, we cannot exclude the possibility that elevated miR-223 levels may halt proliferation entirely in some CTCL cells, potentially by inducing senescence. Therefore, our results indicate alteration of a single miRNA affects cell growth and clonogenic potential in CTCL and provides evidence that miR-223 is likely to have an integral part in disease development and/or progression.
To address how miR-223 is capable of affecting CTCL cell growth, we investigated potential and putative mRNA targets. The oncogenic transcription factor E2F1 has been shown to be a target of miR-223 in acute myeloid leukemia cells (Pulikkan et al., 2010). E2F1 is a member of the E2F family of transcription factors and is essential for cell cycle progression (Johnson and Degregori, 2006). E2F1 functions as an oncogene and is amplified and/or overexpressed in many malignancies (Chen et al., 2009). We demonstrated that E2F1 levels could be modulated by miR-223 in CTCL cells. MEF2C was also shown to be targeted by miR-223 in myeloid cells (Johnnidis et al., 2008). MEF2C is a transcription factor elevated in a subset of the T-cell malignancy T-cell acute lymphoblastic leukemia. It has been shown to work synergistically with the RAS and MYC oncogenes to induce in vitro transformation of immortalized fibroblasts (Homminga et al., 2011). We showed that miR-223 was able to regulate MEF2C protein expression. Our data suggest reduced levels of miR-223 in MF/CTCL allow for increased expression of the oncogenic transcription factors E2F1 and MEF2C, and may help to explain the dysregulated cell growth in CTCL.
TOX is a small DNA-binding protein that is tightly regulated in the thymus during positive-selection, and is necessary for CD4+ T-lymphocyte development. Once T cells have matured, TOX is normally no longer expressed (Aliahmad et al., 2012). However, TOX has recently been shown to be overexpressed in mature CD4+ lymphocytes in MF (Zhang et al., 2012), and we have also found that TOX is increased in MF as compared to normal and inflammatory controls. We have demonstrated TOX is a direct target of miR-223, and modulating miR-223 in CTCL cells led to significant changes in TOX protein expression. While the function of TOX has yet to be established, it has been hypothesized that TOX may influence E protein transcription factors and/or Id2, both of which are involved in lymphocyte proliferation and differentiation (Aliahmad et al., 2012). Loss of E proteins in T cell development mimics TOX overexpression in thymocytes (Jones and Zhuang, 2007). Elevated levels of Id2 have been found in CTCL (Cotta et al., 2008), and it has been suggested that Id2 hastens cell cycle progression through interactions with the Rb family (Lasorella et al., 1996). Therefore, miR-223 regulation of TOX should have significant ramifications for MF/CTCL growth.
Overall, MF/CTCL have been shown to have aberrant miRNA expression, including reduced miR-223 levels. Our study indicates that “correcting” this aberrancy leads to slowed CTCL cell proliferation and reduced clonogenic potential, which is mitigated through the effects of miR-223 target mRNA/protein. Our data provide evidence that miR-223 contributes to the oncogenesis and/or progression of MF/CTCL, and that miR-223 or its targets should be evaluated for the development of new therapeutics.
Materials & Methods
Tissue, blood, and cell lines
Frozen, banked PBMC and 5 to 6 mm bisected skin biopsies from MF/CTCL patients were identified, retrospectively. The diagnosis of MF/CTCL was confirmed through clinical and histologic evaluation as well as flow cytometric analysis. Skin biopsies from patients with benign inflammatory dermatoses (eg. Psoriasis n=3, spongiotic dermatitis n=3) were identified retrospectively through clinical and histologic reports. Control normal skin was obtained through discarded normal skin from surgery and 6 mm biopsies were performed, bisected and frozen. Dr. Utpal Dave kindly provided control PBMC RNA from pooled Red Cross donors. The HH (CRL-2105) and Hut-78 (TIB-161) CTCL cell lines and NIH-3T3 cells were cultured as described by the American Type Culture Collection (Manassas, VA). CD4+ cells from normal donors were obtained from Sanguine Biosciences (Santa Monica, CA). This study was approved by the Vanderbilt Institutional Review Board.
RNA isolation and quantitative real-time PCR (qRT-PCR)
RNA was isolated from skin biopsies, PBMC, and cell lines using Trizol (Invitrogen, Grand Island, NY) according to the manufacturer’s protocol, with one exception. To enhance the isolation of small RNA, the isopropanol precipitation step was performed overnight at −20°C. Sequences for β-actin, E2F1, MEF2C, and TOX-specific qRT-PCR primer pairs were obtained from the Primer Bank (Harvard Medical School) and synthesized by Eurofins MWG Operon (Huntsville, AL). cDNA was generated and qRT-PCR was performed with SybrGreen (SABiosciences, Valencia, CA) in triplicate, as previously reported (Wang et al., 2008) The data are expressed in 2−ΔCt using β-actin as an internal reference. TaqMan qRT-PCR for miRNA used TaqMan MicroRNA Assay (Applied BioSystems, Grand Island, NY) in triplicate and was compared with the expression of RNU24, an internal small RNA control.
Transfection
HH cells (2x106/sample) were prepared utilizing the Nucleofector Kit V (Lonza, Basel, Switzerland) and transfected with the X-005 program in the Nucleofector II instrument (Lonza). Hut-78 cells (#/sample) were prepared with the Nucleofector Kit R (Lonza) and transfected with the V-001 program. The miRIDIAN miR-223 mimic RNA oligomer and control RNA (100 nM), and the miRIDIAN miR-223 hairpin inhibitor and inhibitor control (500 nM) were utilized for transfection (Dharmacon, ThermoScientificBio, Waltham, MA).
Cell viability and growth
Cell numbers and cell viability following transfection of miR-223 mimic and inhibitor were determined by Trypan Blue Dye exclusion assay and with the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (MTS, Promega, Madison, WI).
Clonogenic Assays
CTCL cells (5 x 103/mL) were plated in duplicate in methylcellulose medium containing Iscove’s MDM with L-glutamine and 25 mM HEPES (Gibco, Grand Island, NY), 20% FBS, and 2.6% methylcellulose (per protocol MethoCult H4100, Stemcell Technologies, Vancouver, Canada). Twenty-four hours prior to plating, CTCL cells were transfected with 100 nM miR-223 mimic or RNA control as described above. After 7 days (HH) or 10 days (Hut-78), colonies consisting of more than 30 cells were quantified, and colony numbers were compared between the cells transfected with miR-223 mimic versus RNA control.
Luciferase Vector Generation and Luciferase Assay
A 60mer of the 3′-UTR of TOX containing the miR-223 seed sequence and a 60mer with a mutated (base substitutions) seed sequence designed to prevent miR-223 from binding was cloned into pMIR-REPORT (Invitrogen). NIH-3T3 cells were transfected with 100 ng of pMIR-REPORT wild-type TOX 3′-UTR or mutant TOX 3′-UTR and 150 nM of miR-223 mimic or control RNA using Lipofectamine 2000 (Invitrogen). The cells were also co-transfected with 100 ng of pMIR-REPORT vector expressing a β-galactosidase control for normalizing transfection efficiency. Luciferase reporter gene activity was assayed 24 hours after transfection using the luciferase assay kit (Promega). β-galactosidase activity was assayed by combining 50 μL cellular lysate with 50 μL β-galactosidase assay buffer (0.2 M sodium phosphate buffer (pH 7.3), 2 mM MgCl2, 0.1 M β-mercaptoethanol, ortho-nitrophenyl-β-D-galactopyranoside) for 1 hour, followed by absorbance evaluation at 405 nm.
Immunohistochemistry
Slides were placed on the Leica Bond Max IHC stainer (Leica Microsystems, Inc., Buffalo Grove, IL). Slides were deparaffinized and heat induced antigen retrieval was performed using the Epitope Retrieval 2 solution for 20 minutes. Slides were incubated with anti-TOX (Sigma-Aldrich, Inc., St. Louis, MO. Catalog # HPA018322) at a 1:500 dilution for 1 hour.
Western Blotting
Cell pellets were lysed with RIPA buffer (50 mM Tris, 150 mM sodium chloride, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 1% sodium deoxycholate) at 48 or 72 hours after transfection (see above) with miR-223 mimic or RNA control or miR-223 inhibitor or inhibitor control, respectively, and total cellular proteins western blotted, as previously described (Alt et al., 2003). Antibodies specific for E2F1 (KH-95, Santa Cruz Biotechnology, Santa Cruz, CA), MEF2C (4B10, Abcam, Cambridge, MA), TOX (HPA018322, Sigma-Aldrich), β-actin (Sigma-Aldrich), and α-tubulin (T6074, Sigma-Aldrich) were utilized.
Acknowledgments
We would like to acknowledge members of the Eischen laboratory who provided thoughtful advice for this project, including Dr. Mick Edmonds, Brian Grieb, and Alexia Melo. This work was funded in part by the American Cancer Society Institutional Research Grant (IRG-58-009-510), as well as the Vanderbilt University Medical Center Department of Medicine/Dermatology. Dr. McGirt is a recipient of the Dermatology Foundation Physician-Scientist Career Development Award and the NIH supported Vanderbilt Clinical Oncology Research Career Development Program (K12CA090625). Clare Adams is supported by F31CA165728, and Dr. Eischen is supported by R01CA148950. The project was also supported by CTSA award No. UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Abbreviations
- MF
mycosis fungoides
- CTCL
cutaneous T-cell lymphoma
- MEF2C
myocyte-enhancing factor 2C
- TOX
thymocyte selection-associated high mobility group box
- miRNA
microRNA
- SS
Sézary syndrome
- qRT-PCR
quantitative real-time polymerase chain reaction
- PBMC
peripheral blood mononuclear cells
- UTR
untranslated region
Footnotes
Conflicts of Interest
The authors state no conflicts of interest.
References
- Agar NS, Wedgeworth E, Crichton S, Mitchell TJ, Cox M, Ferreira S, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:4730–9. doi: 10.1200/JCO.2009.27.7665. [DOI] [PubMed] [Google Scholar]
- Aliahmad P, Seksenyan A, Kaye J. The many roles of TOX in the immune system. Current opinion in immunology. 2012;24:173–7. doi: 10.1016/j.coi.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt JR, Greiner TC, Cleveland JL, Eischen CM. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. The EMBO journal. 2003;22:1442–50. doi: 10.1093/emboj/cdg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio E, Mitchell T, van Kester MS, Taylor S, Dunlop HM, Chi J, et al. MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood. 2010;116:1105–13. doi: 10.1182/blood-2009-12-256719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature reviews Cancer. 2009;9:785–97. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta CV, Leventaki V, Atsaves V, Vidaki A, Schlette E, Jones D, et al. The helix-loop-helix protein Id2 is expressed differentially and induced by myc in T-cell lymphomas. Cancer. 2008;112:552–61. doi: 10.1002/cncr.23196. [DOI] [PubMed] [Google Scholar]
- Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer cell. 2007;12:457–66. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–31. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, et al. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–31. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- Homminga I, Pieters R, Langerak AW, de Rooi JJ, Stubbs A, Verstegen M, et al. Integrated transcript and genome analyses reveal NKX2–1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer cell. 2011;19:484–97. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Degregori J. Putting the Oncogenic and Tumor Suppressive Activities of E2F into Context. Current molecular medicine. 2006;6:731–8. doi: 10.2174/1566524010606070731. [DOI] [PubMed] [Google Scholar]
- Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–70. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Iavarone A, Israel MA. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Molecular and cellular biology. 1996;16:2570–8. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–6. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43:673–8. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sezary syndrome. Cell death & disease. 2011;2:e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Muller-Tidow C, Bohlander SK, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–78. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Buermans HP, van Kester MS, van der Fits L, Out-Luiting JJ, Osanto S, et al. Deep-sequencing analysis reveals that the miR-199a2/214 cluster within DNM3os represents the vast majority of aberrantly expressed microRNAs in Sezary syndrome. The Journal of investigative dermatology. 2012;132:1520–2. doi: 10.1038/jid.2011.481. [DOI] [PubMed] [Google Scholar]
- Ralfkiaer U, Hagedorn PH, Bangsgaard N, Lovendorf MB, Ahler CB, Svensson L, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL) Blood. 2011;118:5891–900. doi: 10.1182/blood-2011-06-358382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- van Kester MS, Ballabio E, Benner MF, Chen XH, Saunders NJ, van der Fits L, et al. miRNA expression profiling of mycosis fungoides. Molecular oncology. 2011;5:273–80. doi: 10.1016/j.molonc.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2008;27:1590–8. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
- Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–7. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–32. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- Weinstock MA, Horm JW. Mycosis fungoides in the United States. Increasing incidence and descriptive epidemiology. JAMA : the journal of the American Medical Association. 1988;260:42–6. [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Yu R, Huang Y, Su M, Xiao C, et al. Molecular markers of early-stage mycosis fungoides. The Journal of investigative dermatology. 2012;132:1698–706. doi: 10.1038/jid.2012.13. [DOI] [PubMed] [Google Scholar]