Abstract
Realizing the potential of cell free systems will require development of ligand sensitive gene promoters that control gene expression in response to a ligand of interest. Here, we describe an approach to designing ligand sensitive transcriptional control in cell free systems that is based on the combination of a DNA aptamer that binds thrombin and the T7 bacteriophage promoter. Placement of the aptamer near the T7 promoter, and using a primarily single stranded template, results in up to a five-fold change in gene expression in a ligand concentration dependent manner. We further demonstrate that the sensitivity to thrombin concentration and the fold change in expression can be tuned by altering the position of the aptamer. The results described here pave the way for the use of DNA aptamers to achieve modular regulation of transcription in response to a wide variety of ligands in cell free systems.
Keywords: T7 promoters, cell free systems, DNA aptamer, thrombin, transcriptional regulation
Synthetic gene circuits comprised of novel genetic regulatory mechanisms have emerged as powerful tools for understanding and harnessing biological function (1). Engineered arrangements of genetic components have resulted in systems capable of predetermined functions such as bistability (2), logic control (3-5) and oscillation of gene expression (6, 7). Synthetic gene circuits also offer the opportunity to redesign biological systems for the production of biofuels and other chemicals as well as for constructing devices for sensing and responding to biomedical conditions (8, 9). In practice, the majority of synthetic gene circuits have been implemented in cell-based systems. While these demonstrations benefit from natural mechanisms to sustain a living cell, such as protein synthesis and degradation, creating predictable engineered systems can be complicated by interference from endogenous host machinery and selection pressures that act against unneeded, resource consuming systems (10, 11). Additionally, conflicts occur when sensing or generating materials that can compromise cell viability and survival. Therefore, alternative strategies to harness and understand biological complexity are a needed complement to existing cell-based approaches (12).
Cell free systems, that include both plamsid based cell free protein synthesis systems and in vitro transcription systems provide a versatile platform for understanding and applying the design elements that underlie cellular efficiency (13-15). Cell free approaches employ select cellular components, produced naturally or synthetically, to carry out defined biological processes. Issues related to plasmid compatibility, protein toxicity or maintenance of a living cell can be mostly ignored, allowing focus on defining essential system components (16) and implementing predictable dynamic behavior (17). The flexibility, simplified context, and precise specification of system components are distinct advantages of the cell free approach. A number of cell free, in vitro gene circuits have been demonstrated. For example, simplified nucleic acid templates, in which transcription was regulated by DNA hybridization, were co-opted to build bistable switches and oscillators that reasonably agree with quantitative predictions (18, 19). Additionally, expression cascades (20), negative feedback(21) and logic gates (22) have been realized using circuits involving protein intermediates in plasmid based cell free protein synthesis systems.
Well-characterized molecular tools for signal sensing and tuning gene expression are essential for the design and construction of synthetic gene circuits in cells and cell free systems (21, 22). In particular, ligand responsive gene regulation strategies are key. While a myriad of gene regulatory mechanisms are used in natural cells, ligand dependent transcriptional control strategies that function in cell free extracts remain fairly limited. This is partly due to the prevalent use of bacteriophage RNA polymerases. For example, T7 RNA polymerase is commonly used to drive transcription due to the enzyme's stability and high processivity (23, 24). While these characteristics are desirable for achieving high yield protein synthesis, synthetic biology applications can be compromised by the lack of sufficient ligand sensitive T7 promoters (20). Attempts to engineer ligand regulatable T7 promoters have used either protein based transcription factors or modified nucleic acid bases to confer ligand sensitivity (25-27). For example, control of T7 RNA polymerase can be accomplished by placing a cis acting promoter element, that binds to a repressor, downstream to the transcription start site (28). Other approaches have considered incorporation of photoresponsive elements (29) or triplex DNA formation (27, 30).
The use of nucleic acid aptamers can potentially allow regulation of gene expression in response to a wide variety of small molecules and proteins. Aptamers are single stranded DNA or RNA molecules that can be engineered to bind to specific target molecules with high affinity and specificity (31). Accordingly, RNA aptamers have found extensive application and often couple the binding event and the ensuing conformational change for regulation of transcription or translation (32-38). Nucleic acid aptamers offer several practical advantages. Aptamers can potentially be selected against any ligand of interest from a combinatorial library using an iterative affinity selection procedure (31, 39). In addition, known hybridization rules facilitate predictive and rational design of nucleic acid domains. The ease and predictability of engineering nucleic acid domains make DNA and RNA molecules particularly useful substrates for engineering flexible platforms for achieving tunable sensing and actuation (32, 40, 41).
Here we describe a new approach to using aptamers to control gene expression at the transcriptional level using viral promoters. The approach involves the insertion of a DNA aptamer sequence proximal to the T7 promoter such that binding prevents transcription (Figure 1). The required single stranded regions are easily accommodated by bacteriophage polymerases (42, 43) and employable in a cell free context. Thrombin binding DNA aptamer (TBA) was selected for demonstrating analyte specific transcriptional control. TBA is well-characterized and is known to bind to human α-thrombin with high affinity (Kd of 10-100nM) and specificity (44). Presence of additional flanking sequences and surface immobilization are not detrimental to thrombin binding, which facilitates the insertion of the aptamer sequence into the DNA template (45, 46). These features have enabled the incorporation of thrombin aptamers in several designs for biosensors and DNA circuits(46-49). We show that thrombin can be used to effectively repress expression from thrombin aptamer containing templates in a cell free context. In addition, the sensitivity of the promoter to thrombin concentration and relative expression level can be tuned.
Figure 1.
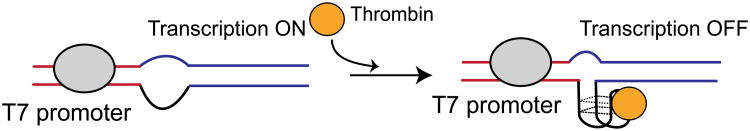
Design for aptamer mediated transcriptional regulation. A ssDNA aptamer that binds thrombin is placed downstream to the T7 promoter. The transcriptional template contains a non-complementary, ssDNA region that harbors the aptamer sequence. In the absence of thrombin, T7 RNA polymerase transcribes the template. Upon thrombin binding to the DNA aptamer, transcription from the T7-aptamer promoter is repressed.
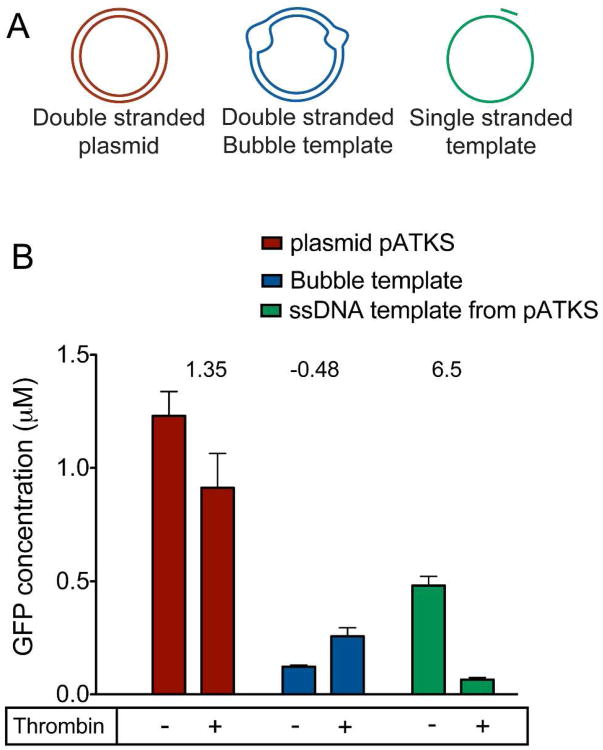
To evaluate transcriptional regulation using DNA aptamers, double stranded, largely double stranded and largely single stranded DNA templates were created. The largely double stranded template contains an unpaired “bubble” DNA region after the double stranded promoter where DNA aptamers on the template and the non-template strands are located (Supplementary Figure S1). These structures were created from hybridization of single stranded DNA templates. Several strategies for generating linear ssDNA templates such as affinity purification of biotin labeled ssDNA generated from PCR (50) and rolling circle amplification (51) were evaluated. However, these approaches resulted in only small amounts of ssDNA, which were insufficient for optimizing protein synthesis reactions (data not shown). Consequently, the ssDNA templates were generated from phagemids, which offer the advantage of producing high yield ssDNA that is long enough to code for a protein (52). Restriction digests of the resulting bubble template confirmed proper hybridization (Supplementary Figure S2). In addition, to the bubble templates, largely single stranded templates, which contain only a 19-base, double stranded T7 promoter, were generated by annealing an oligonucleotide to the ssDNA generated from the phagemid. For all templates, the aptamer sequence was placed downstream from the transcription start site along with a 4bp stem loop structure to facilitate aptamer formation and increase its stability (44). Finally, DNA encoding green fluorescent protein (GFP) was located downstream from the aptamer. Fluorescence anisotropy experiments with oligonucleotide analogues of the ssDNA and bubble DNA constructs confirmed thrombin binding to the aptamer (Supplementary Figure S1).
The ability of thrombin to repress transcription from double stranded, bubble and ssDNA templates was monitored by cell free protein synthesis reactions (Figure 2). Not surprisingly, constitutive expression from purely double stranded plasmid templates was 10 times greater than from the bubble template and about 4 times greater than from the ssDNA template. Upon incubation of the entirely double stranded template with 1.8 μM thrombin, only a modest change in gene expression is observed. This indicates that thrombin binding to the aptamer region is low, presumably due to the stable double stranded structure obscuring aptamer recognition. We also note that addition of thrombin to double stranded plasmid DNA templates without the aptamer did not result in repression from T7 promoters. This indicates that thrombin does not interfere with transcription and translation in the E. coli cell free extract. The lower baseline expression from bubble template indicates that this structure serves as poor template for gene expression. In contrast, the largely ssDNA aptamer templates are suitable structures for gene expression and a five-fold decrease in gene expression is seen when in the presence of thrombin. Further, rapid repression of expression can be observed without prior incubation with thrombin (Supplementary Figure S3).
Figure 2.
Effect of promoter design on aptamer mediated repression. Shown in A) is a schematic of templates used to test the different designs. The red structure represents the double stranded plasmid with the thrombin aptamer in a duplex form downstream from the promoter. The blue template is a “bubble template” that contains an aptamer structure both on the template (derived from pATKS) and non-template strand (generated from pANTKS) in an unpaired context downstream from the promoter. The second bubble in the diagram corresponds to the unpaired region that results from the phagemid origins of replication. The green schematic corresponds to the mostly ssDNA template derived from hybridizing an oligonucleotide to pATKS. B) Shows GFP concentrations measured after a 6 hour reaction from these templates, in the absence or presence of 1.8 μM thrombin. The numbers above the bars indicate fold change in expression upon addition of thrombin to the DNA template.
Previous reports have shown that protein binding at a position proximal to the transcription start site is essential for achieving effective repression from T7 promoters (21, 27, 28). This suggests that aptamer placement that facilitates ligand binding close to the transcriptional start site is important for specific control of gene expression. When the aptamer is present along with its complementary sequence, resulting in a fully base paired structure, addition of thrombin results in little to no change in expression. This result is presumably due to the stability of the double stranded structure that prevents significant formation of the aptamer. Placing the aptamers within unpaired, bubble regions offers a strategy to increase accessibility of the aptamer. However, as shown in Figure 2, this template was a poor substrate for transcription. T7 RNA polymerase is known to bypass gaps and discontinuities in the template strand with the aid of the non-template DNA strand (43, 53). Further, DNA topology is known to adversely affect transcription in vitro (54, 55). Several studies have shown that non-canonical DNA structures such as DNA quadruplexes present an obstacle to transcription from T7 promoters, with the effect being more pronounced when the structures are located proximal to the transcriptional start site (56-58). Collectively, structural blockages on both strands at positions near to the promoter likely lead to poor gene expression and performance of this promoter design. The final template design consisted primarily of single stranded DNA, corresponding to the template strand, and hybridization to a short, synthetic oligonucleotide to create a double stranded T7 promoter. This design proved to be the most responsive to thrombin-mediated repression of gene expression.
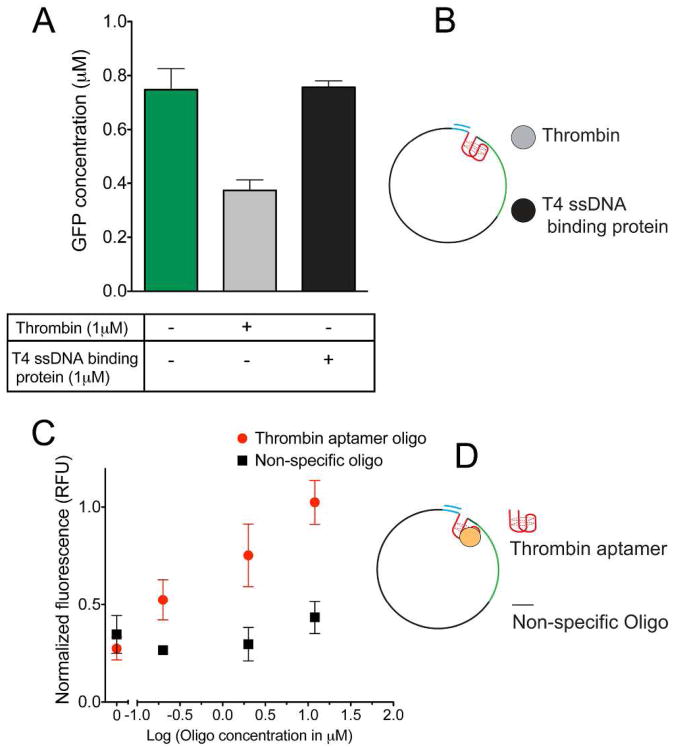
To test the specificity of thrombin dependent repression, the non-specific, single strand DNA binding T4 gene 32 protein (59) was tested for transcription repression on the single stranded T7 aptamer promoter. The addition of ssDNA binding protein did not have a significant effect on transcription demonstrating that specific protein binding to the aptamer is required for effective transcriptional repression (Figure 3A). The change in gene expression upon the addition of thrombin as a function of DNA concentration was also investigated. The highest fold change in expression from ssDNA templates (pATKS) was observed at a concentration of 20ng/μl (Supplementary Figure S5).
Figure 3.
Testing specificity of aptamer mediated transcriptional regulation. A) Depicts the response of ssDNA template to 1μM thrombin and 1μM T4 ssDNA binding protein. B) Shows the schematic of the aptamer template along with thrombin and T4ssDNA binding protein. C) Results of addition of thrombin aptamer oligo and the non-specific oligo in trans to 10 nM of the ssDNA aptamer template complexed with 2μM thrombin. Fluorescence values have been normalized to expression from ssDNA templates in the absence of thrombin and after 6 hours of expression. Error bars correspond to standard deviations of triplicate measurements. D) Schematic of the aptamer template bound to thrombin and the non-specific oligonucleotide and an oligonucleotide containing the thrombin aptamer.
The specificity of the transcriptional repression was further evaluated using exogenously added thrombin aptamer oligonucleotides to competitively inhibit gene repression when in the presence of thrombin (Figure 3B). Thrombin aptamer concentrations in excess of thrombin protein relieved thrombin mediated gene repression, whereas the addition of a non-specific DNA oligonucleotide did not affect repression. Therefore, the addition of exogenous DNA aptamer allows for “induction” of expression from these promoters in the presence of thrombin. This observation provides additional support that repression is mediated selectively by thrombin and that the DNA aptamer can bind to thrombin in a complex cell extract system. This result also highlights a mechanism for reversing thrombin mediated gene repression from aptamer templates. The presence of additional aptamer sequences can allow the turning off of gene repression or for tuning the ligand concentration required for expression. Such tools may prove useful for effectively designing gene circuit behaviour.
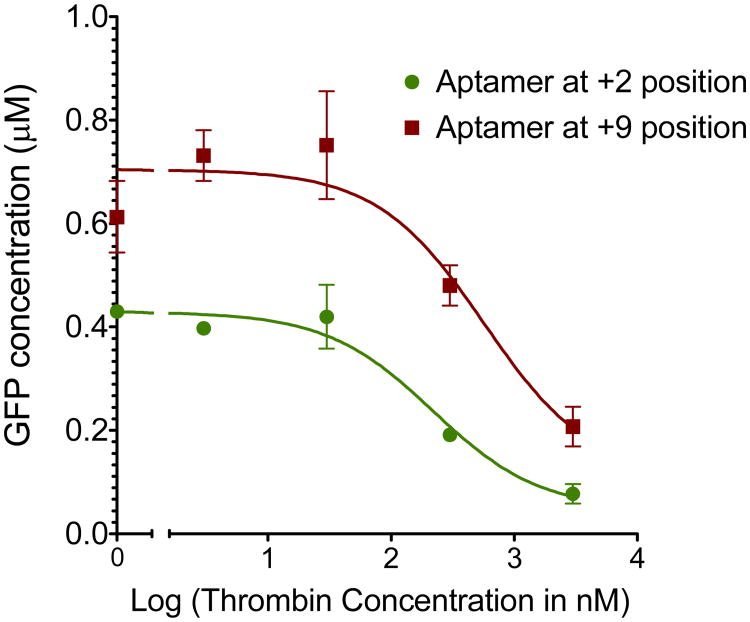
The effect of aptamer position on transcriptional efficiency was also explored by placing the thrombin aptamer at positions +2, +9 and +28, relative to the transcriptional start site (Table 1). Results from protein expression assays show that placement of the DNA aptamer further away from the transcriptional start site leads to an increase in basal gene expression levels. However, the magnitude of thrombin induced gene repression decreases. Dose response curves with the aptamer at +2 and +9 positions show a half maximal repressor concentration of 218.9 ± 0.04 nM and 567.90 ± 1.2 nM respectively (Figure 4). Dose responses of the aptamer placed at +28 position were not tested given the small fold change in the expression even to 2 μM thrombin concentration. To test if the addition of a second thrombin aptamer results in improved gene repression, a dimeric DNA aptamer template was constructed and tested. However, basal expression from the template was low (Supplementary Figure S4).
Table 1.
Effect of position of the aptamer on thrombin mediated response.
| Plasmid template | Aptamer postion | Promoter Design | Relative expression * | Fold change** |
|---|---|---|---|---|
| pATKS | +2 |
|
0.45 | 6.78 |
| pETAKS | +9 |
|
0.7 | 2.78 |
| pETA26KS | +28 |
|
1.22 | 2.01 |
Relative expression indicates expression levels normalized to expression from the ssDNA template generated from pKSGFP plasmid.
Fold change in expression indicates the change in expression upon the addition of thrombin.
Figure 4.
Effect of position of the aptamer on thrombin mediated response. Dose response curves for the +2 and +9 aptamer ssDNA constructs where the X-axis corresponds to the log of the thrombin concentration. Only nM concentrations of thrombin are needed before a logarithmic transformation is observed.
Engineered placement of the promoter offers another means for tuning gene expression in response to a ligand. Repression efficiency is expected to decrease as the aptamer is moved away from the transcription start site (28). Accordingly, placement of thrombin binding aptamer 2 bases away from the transcriptional start site results in up to a 6 to 7-fold thrombin concentration dependent change in gene expression in the ssDNA template design. While moving the aptamer 28 bases away from the transcription start site increases basal transcriptional levels, only up to a 2 fold change in gene expression upon thrombin addition is observed. T7 promoters are highly conserved in the region between -17 and +6 region. Aptamer placement at the +2 position disrupts the viral promoter sequence, while placement at the +9 and +28 positions do not. Further, the thrombin binding aptamer is known to form a DNA quadruplex structure (60, 61). Taken together with the known effects of secondary structures that can block transcription in vitro (56, 62, 63), these results indicate that disruption of the native promoter sequence and the formation of secondary DNA structures close to the transcriptional start site combine to lower basal expression levels when the aptamer is placed in the +2 position. The finding that placement of operators more proximal to the transcriptional start site achieves effective repression from T7 promoters at the expense of lower basal expression mirrors results obtained with T7lacO and T7tetO promoters (21, 28).
Cell free systems are promising platforms for implementing engineered networks from defined components (18, 19). Such systems are not constrained to native forms of biomolecules and can employ conditions that are incompatible with living cells. Moreover, the open nature of cell free systems allows users to optimize numerous parameters including the cell free extract components and their preparation, the reaction substrates and the format of the reaction such that cost effective, industrial scale protein production can be considered (64-66). Effective application of network designs in these systems will require the availability of a library of environmentally responsive promoters (67). In particular, control of gene expression at the transcriptional level will be critical for many applications. Control at the transcriptional stage offers several advantages over control at the translational or post-translational levels (10). Being the first step in gene expression, multiple downstream targets can be regulated simultaneously. In addition, signal amplification can be achieved since binding of a single transcriptional factor regulates the expression of several hundred resulting RNA and protein molecules (68). Since nucleic acid aptamers can be selected against any molecule of interest that otherwise does not have a double stranded DNA binding domain, the use of DNA aptamers for transcriptional repression in cell free systems paves the way for creation of feedback circuits with novel sensory capabilities.
Here, an effective strategy for implementing aptamer control elements has been demonstrated and relies on a simple combination of a single stranded template with an oligonucleotide to define the promoter element. Defining the placement of the aptamer sequence relative to the promoter enables engineering of gene expression levels and tuning sensitivity to ligand concentration. The approach to template synthesis offers the possibility of providing sufficient DNA substrates for larger scale cell free protein synthesis reactions. Further, tuning the strength of the T7 bacteriophage promoter in combination with changing the aptamer position can allow users to tune fold changes in response to ligand addition(69). The T7 bacteriophage promoter tolerates the single stranded template but requires placement of the aptamer near the promoter sequence. This placement can compromise promoter strength. Potentially, DNA-based aptamer control elements can be extended to other bacteriophage or bacterial promoter sequences and allow designs that lead to even greater dynamic range in gene expression control. For this study, we tested the use of thromin binding aptamer to evaluate our aptamer mediated control of gene expression. This strategy can potentially be extended to other aptamers and their associated ligands provided they function in cell extracts (70). Selection and use of DNA aptamers that work with known promoter elements will result in new approaches to regulating gene expression in response to a wide range of molecules.
Materials and Methods
Plasmid Construction
All plasmid constructions were carried out using standard techniques (71). GFP was cloned into pBluescript KS II (+) and pBluescript KS II (-) vector backbones and the aptamer sequences were then inserted downstream to T7 promoters using inverse PCR. The aptamer constructs are listed in Supplemental Table 1 and sequence information is provided in Supplementary Section S6. Plasmids will be made available upon request.
Single stranded DNA template preparation
ssDNA templates were assembled by annealing template strands generated from pBluescript KS (-) II variants with T7 promoter oligo in annealing buffer (10mM Tris-HCl pH 7.5, 50mM KCl, and 1mM MgCl2). Thrombin aptamer sequence was cloned into the phagemid vectors pBluescript KS II (+) and pBluescript KS II (-) at different locations downstream to the transcription start site. These two backbones differ from each other only with respect to the orientation of the F1 origin. ssDNA molecules were then generated using a standard procedure (71). For the preparation of double stranded templates with bubble regions, ssDNA generated from pANTKS was annealed to an oligonucleotide that is complementary to a HindIII site on the pBluescript KS II (+) DNA backbone prior to digestion with HindIII restriction endonuclease. The resulting template was purified and annealed to the single stranded DNA template derived from pBluescript KS II (-) by slow cooling from 95° C to room temperature in a thermocycler in annealing buffer. The resulting construct contains a mismatched bubble region corresponding to the F1 origin region. The efficiency of annealing and dsDNA generation was verified by digesting the DNA template using restriction endonucleases. For preparation of the largely single stranded templates, the oligonucleotide 5′TAATACGACTCACTATAGG 3′ was hybridized to the single stranded DNA template generated from plasmids pATKS, pETAKS and pETA26KS in the presence of annealing buffer.
Cell free protein synthesis experiments (CFPS)
The Promega S30 T7 High-Yield Expression System kit (Promega TM306) was used for CFPS experiments. The S30 premix and the cell extract were mixed in proportions recommended by the manufacturer and template was used at 20 ng/μl concentration per reaction. DNA concentrations used were based on previous optimizations (21). Reactions were set up in Corning CLS3820 plates following manufacturer's instructions except that the final reaction volume was scaled to 15 μL. Samples were incubated at 30°C with shaking and fluorescence measurements (485/20 nm excitation, 528/20 nm emission) were made every 7 minutes in a Biotek Synergy 2 plate reader. Error bars associated with the fluorescence measurements represent standard deviation of three replicates. Values indicated in the graphs represent GFP concentrations obtained after 6 hours of cell free protein synthesis reactions.
Thrombin dependent gene repression was tested by incubating the DNA templates with thrombin (diluted from stock solution into 10mM Tris-HCl pH 7.5 and 50mM KCl) along with 0.01%Tween-20 for one hour at room temperature, followed by the addition of the cell extract. Thrombin protein stocks were supplied in 50% glycerol solution. Thrombin concentrations used in this study ensured that the final glycerol concentration in cell free protein synthesis reactions were less than 0.5% (72). Human α-Thrombin was purchased from Haematologic Technologies, Essex Junction, VT. The thrombin aptamer sequence used to relieve repression was 5′ GGTTGGTGTGGTTGG 3′ and the non-specific oligonucleotide sequence was 5′ GGCGTAACTGATCGATCGT 3′. To test the effect of different thrombin aptamer and non-specific oligonucleotide concentrations on thrombin mediated repression, different oligonucleotide amounts were heat denatured and slowly cooled to room temperature in the presence of 10mM Tris-HCl pH 7.5, 5mM KCl and 1mM MgCl2 before they were added to cell free protein synthesis reaction.
Supplementary Material
Acknowledgments
Authors acknowledge Dr. Robert Standaert and Dr Ana Kitazono for helpful discussions. SI and MJD were supported by National Institutes of Health Grant EB000657. MJD also acknowledges support by the in-house research program of the Center for Nanophase Materials Sciences, which is sponsored at Oak Ridge National Laboratory by the Division of Scientific User Facilities, U.S. Department of Energy. This work was performed at the Oak Ridge National Laboratory, managed by UT-Battelle, LLC, for the U.S. DOE under Contract No. DE-AC05- 00OR22725.
Footnotes
Supporting Information Available: This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Haseltine EL, Arnold FH. Synthetic gene circuits: design with directed evolution. Annu Rev Biophys Biomol Struct. 2007;36:1–19. doi: 10.1146/annurev.biophys.36.040306.132600. [DOI] [PubMed] [Google Scholar]
- 2.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 3.Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 4.Wang B, Kitney RI, Joly N, Buck M. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nat Commun. 2011;2:508. doi: 10.1038/ncomms1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhan J, Ding B, Ma R, Ma X, Su X, Zhao Y, Liu Z, Wu J, Liu H. Develop reusable and combinable designs for transcriptional logic gates. Mol Syst Biol. 2010;6:388. doi: 10.1038/msb.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 7.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabinovitch-Deere CA, Oliver JW, Rodriguez GM, Atsumi S. Synthetic Biology and Metabolic Engineering Approaches To Produce Biofuels. Chem Rev. 2013 doi: 10.1021/cr300361t. [DOI] [PubMed] [Google Scholar]
- 9.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: new engineering rules for an emerging discipline. Mol Syst Biol. 2006;2(2006):0028. doi: 10.1038/msb4100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 12.Noireaux V, Maeda YT, Libchaber A. Development of an artificial cell, from self-organization to computation and self-reproduction. Proc Natl Acad Sci U S A. 2011;108:3473–3480. doi: 10.1073/pnas.1017075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol Syst Biol. 2008;4:220. doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eric Hodgman C, Jewett MC. Cell-free synthetic biology: Thinking outside the cell. Metab Eng. 2011 doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson ML. Cell-free synthetic biology: a bottom-up approach to discovery by design. Mol Syst Biol. 2006;2:69. doi: 10.1038/msb4100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doktycz MJ, Simpson ML. Nano-enabled synthetic biology. Mol Syst Biol. 2007;3:125. doi: 10.1038/msb4100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol Syst Biol. 2011;7:466. doi: 10.1038/msb.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, White KS, Winfree E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol Syst Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Winfree E. Synthetic in vitro transcriptional oscillators. Mol Syst Biol. 2011;7:465. doi: 10.1038/msb.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noireaux V, Bar-Ziv R, Libchaber A. Principles of cell-free genetic circuit assembly. Proc Natl Acad Sci U S A. 2003;100:12672–12677. doi: 10.1073/pnas.2135496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karig DK, Iyer S, Simpson ML, Doktycz MJ. Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Res. 2012;40:3763–3774. doi: 10.1093/nar/gkr1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin J, Noireaux V. An E. coli Cell-Free Expression Toolbox: Application to Synthetic Gene Circuits and Artificial Cells. Acs Synthetic Biology. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- 23.Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studier FW, Moffatt BA. Use of Bacteriophage-T7 Rna-Polymerase to Direct Selective High-Level Expression of Cloned Genes. Journal of Molecular Biology. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 25.Sakaguchi-Mikami A, Taniguchi A, Sode K, Yamazaki T. Construction of a novel glucose-sensing molecule based on a substrate-binding protein for intracellular sensing. Biotechnol Bioeng. 2010 doi: 10.1002/bit.23006. [DOI] [PubMed] [Google Scholar]
- 26.Bello-Roufai M, Roulon T, Escude C. Ligand-mediated transcription elongation control using triplex-based padlock oligonucleotides. Chem Biol. 2004;11:509–516. doi: 10.1016/j.chembiol.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Skoog JU, Maher LJ., 3rd Repression of bacteriophage promoters by DNA and RNA oligonucleotides. Nucleic Acids Res. 1993;21:2131–2138. doi: 10.1093/nar/21.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez PJ, Guillerez J, Sousa R, Dreyfus M. On the mechanism of inhibition of phage T7 RNA polymerase by lac repressor. J Mol Biol. 1998;276:861–875. doi: 10.1006/jmbi.1997.1576. [DOI] [PubMed] [Google Scholar]
- 29.Liu M, Asanuma H, Komiyama M. Azobenzene-tethered T7 promoter for efficient photoregulation of transcription. J Am Chem Soc. 2006;128:1009–1015. doi: 10.1021/ja055983k. [DOI] [PubMed] [Google Scholar]
- 30.Maher LJ., 3rd Inhibition of T7 RNA polymerase initiation by triple-helical DNA complexes: a model for artificial gene repression. Biochemistry. 1992;31:7587–7594. doi: 10.1021/bi00148a021. [DOI] [PubMed] [Google Scholar]
- 31.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 32.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 33.Qi L, Lucks JB, Liu CC, Mutalik VK, Arkin AP. Engineering naturally occurring trans-acting non-coding RNAs to sense molecular signals. Nucleic Acids Res. 2012;40:5775–5786. doi: 10.1093/nar/gks168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An CI, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunka DH, Stockley PG. Aptamers come of age - at last. Nat Rev Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 37.Goldfless SJ, Belmont BJ, de Paz AM, Liu JF, Niles JC. Direct and specific chemical control of eukaryotic translation with a synthetic RNA-protein interaction. Nucleic Acids Res. 2012;40:e64. doi: 10.1093/nar/gks028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodrigo G, Landrain TE, Jaramillo A. De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells. Proc Natl Acad Sci U S A. 2012;109:15271–15276. doi: 10.1073/pnas.1203831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 40.Liang JC, Smolke CD. Rational design and tuning of ribozyme-based devices. Methods Mol Biol. 2012;848:439–454. doi: 10.1007/978-1-61779-545-9_27. [DOI] [PubMed] [Google Scholar]
- 41.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rong M, Durbin RK, McAllister WT. Template strand switching by T7 RNA polymerase. J Biol Chem. 1998;273:10253–10260. doi: 10.1074/jbc.273.17.10253. [DOI] [PubMed] [Google Scholar]
- 44.Macaya RF, Waldron JA, Beutel BA, Gao H, Joesten ME, Yang M, Patel R, Bertelsen AH, Cook AF. Structural and functional characterization of potent antithrombotic oligonucleotides possessing both quadruplex and duplex motifs. Biochemistry. 1995;34:4478–4492. doi: 10.1021/bi00013a041. [DOI] [PubMed] [Google Scholar]
- 45.Baldrich E, Restrepo A, O'Sullivan CK. Aptasensor development: elucidation of critical parameters for optimal aptamer performance. Anal Chem. 2004;76:7053–7063. doi: 10.1021/ac049258o. [DOI] [PubMed] [Google Scholar]
- 46.Yang L, Ellington AD. Real-time PCR detection of protein analytes with conformation-switching aptamers. Anal Biochem. 2008;380:164–173. doi: 10.1016/j.ab.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamaguchi N, Ellington A, Stanton M. Aptamer beacons for the direct detection of proteins. Anal Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 48.Han D, Zhu Z, Wu C, Peng L, Zhou L, Gulbakan B, Zhu G, Williams KR, Tan W. A logical molecular circuit for programmable and autonomous regulation of protein activity using DNA aptamer-protein interactions. J Am Chem Soc. 2012;134:20797–20804. doi: 10.1021/ja310428s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dittmer WU, Reuter A, Simmel FC. A DNA-based machine that can cyclically bind and release thrombin. Angew Chem Int Ed Engl. 2004;43:3550–3553. doi: 10.1002/anie.200353537. [DOI] [PubMed] [Google Scholar]
- 50.Mitchell LG, Merril CR. Affinity generation of single-stranded DNA for dideoxy sequencing following the polymerase chain reaction. Anal Biochem. 1989;178:239–242. doi: 10.1016/0003-2697(89)90631-3. [DOI] [PubMed] [Google Scholar]
- 51.Lin CX, Wang X, Liu Y, Seeman NC, Yan H. Rolling circle enzymatic replication of a complex multi-crossover DNA nanostructure. Journal of the American Chemical Society. 2007;129:14475–14481. doi: 10.1021/ja0760980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrington C, Perrino FW. Initiation of RNA-primed DNA synthesis in vitro by DNA polymerase alpha-primase. Nucleic Acids Res. 1995;23:1003–1009. doi: 10.1093/nar/23.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou W, Reines D, Doetsch PW. T7 RNA polymerase bypass of large gaps on the template strand reveals a critical role of the nontemplate strand in elongation. Cell. 1995;82:577–585. doi: 10.1016/0092-8674(95)90030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ditlevson JV, Tornaletti S, Belotserkovskii BP, Teijeiro V, Wang G, Vasquez KM, Hanawalt PC. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 2008;36:3163–3170. doi: 10.1093/nar/gkn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Broxson C, Beckett J, Tornaletti S. Transcription arrest by a G quadruplex forming-trinucleotide repeat sequence from the human c-myb gene. Biochemistry. 2011;50:4162–4172. doi: 10.1021/bi2002136. [DOI] [PubMed] [Google Scholar]
- 57.Wolffe AP, Drew HR. Initiation of transcription on nucleosomal templates. Proc Natl Acad Sci U S A. 1989;86:9817–9821. doi: 10.1073/pnas.86.24.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Portugal J, Rodriguez-Campos A. T7 RNA polymerase cannot transcribe through a highly knotted DNA template. Nucleic Acids Res. 1996;24:4890–4894. doi: 10.1093/nar/24.24.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alberts BM. Function of gene 32-protein, a new protein essential for the genetic recombination and replication of T4 bacteriophage DNA. Fed Proc. 1970;29:1154–1163. [PubMed] [Google Scholar]
- 60.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci U S A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J Mol Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 62.Neaves KJ, Huppert JL, Henderson RM, Edwardson JM. Direct visualization of G-quadruplexes in DNA using atomic force microscopy. Nucleic Acids Res. 2009;37:6269–6275. doi: 10.1093/nar/gkp679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Droge P, Pohl FM. The influence of an alternate template conformation on elongating phage T7 RNA polymerase. Nucleic Acids Res. 1991;19:5301–5306. doi: 10.1093/nar/19.19.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: Applications come of age. Biotechnology advances. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zawada JF, Yin G, Steiner AR, Yang J, Naresh A, Roy SM, Gold DS, Heinsohn HG, Murray CJ. Microscale to manufacturing scale-up of cell-free cytokine production--a new approach for shortening protein production development timelines. Biotechnol Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnology and bioengineering. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 67.Jewett MC, Forster AC. Update on designing and building minimal cells. Curr Opin Biotechnol. 2010;21:697–703. doi: 10.1016/j.copbio.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buskirk AR, Liu DR. Creating small-molecule-dependent switches to modulate biological functions. Chem Biol. 2005;12:151–161. doi: 10.1016/j.chembiol.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 69.Temme K, Hill R, Segall-Shapiro TH, Moser F, Voigt CA. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Javaherian S, Musheev MU, Kanoatov M, Berezovski MV, Krylov SN. Selection of aptamers for a protein target in cell lysate and their application to protein purification. Nucleic Acids Res. 2009;37:e62. doi: 10.1093/nar/gkp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1982. [Google Scholar]
- 72.Siuti P, Retterer ST, Choi CK, Fowlkes JD, Doktycz MJ. Cell Free Translation in Engineered Picoliter Volume Containers. Annu ORNL Biomed Sci Eng Cent Conf. 2009;2009:1–4. doi: 10.1109/BSEC.2009.5090477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.