Abstract
Stressors after injury from a multitude of factors can lead to cell death. We have identified four fibronectin (FN) peptides, two from the first FN type III repeat (FNIII1), one from the 13th FN type III repeat (FNIII13), and one from FN variable region (IIICS), that when tethered to a surface acted as platelet-derived growth factor-BB (PDGF-BB) enhancers to promote cell survival. One of the FNIII1 peptides and its smallest (14mer) bioactive form (P12) were also active in solution. Specifically, P12 bound PDGF-BB (KD = 200nM), enhanced adult human dermal fibroblast (AHDF) survival under serum starvation, oxidative or endoplasmic reticulum (ER) stressors, and limited burn injury progression in a rat hot comb model. Furthermore, P12 inhibited ER stress-induced c-Jun N-terminal kinase (JNK) activation. Although many growth factors have been found to bind FN directly or indirectly, this is the first report to identify peptide sequences of growth factor-binding sites in FN. The finding of these novel peptides further delineated how the extracellular matrix protein FN can support cell survival. Since the peptide P12 is active in either soluble form or tethered to a substrate, it will have multifactorial uses as a bioactive in tissue engineering.
Keywords: fibronectin, growth factor, PDGF-BB, cell stress, cell survival
Introduction
Tissue injury after stroke, heart attack, burns or other trauma is a dynamic process that extends in size and severity over several days, leading to excess mortality and morbidity (Brait et al., 2010; Piper and Garcia-Dorado, 2012; Shupp et al., 2010). After injury the central necrotic tissue is surrounded by ischemic, less severely affected tissue that potentially remains salvageable. However, since the ischemic tissue is exposed to various stresses such as nutrition depletion, reactive oxygen species (ROS), cytokines, and hypoxia, tissue cells in this region often die, by either apoptosis or necroptosis (Galluzzi and Kroemer, 2008; Hirth et al., 2013; Lanier et al., 2011), unless effective post-injury therapy is given (Yuan, 2009). Hence, preventing cell death in potentially salvageable tissue of an acutely damaged organ is a major goal for therapies of these injuries.
Fibronectin (FN) (Figure1a), an extracellular protein found in blood and connective tissue, plays a crucial role in cell growth, survival, migration, angiogenesis and wound healing (Yamada and Clark, 1996). In the absence of tissue cell-derived FN, plasma FN can support neuronal survival and reduce brain injury following transient focal cerebral ischemia (Sakai et al., 2001). Although FN probably supports cell survival in many ways, one mechanism is the interaction of RGD peptide in the FN 10th type-III repeat with integrin receptors. For example, the FN integrin receptor α5β1 supports Chinese hamster ovary cell survival and up-regulates Bcl-2 expression (Zhang et al., 1995). In addition, FN-ligated α5β1 and αvβ3 integrins promote brain capillary endothelial cell survival and proliferation via MAP kinase signaling (Wang and Milner, 2006). FN-dependent survival signaling transits through PI3 kinase and NF-kappa B pathways in human bronchial epithelial cells (Han and Roman, 2006).
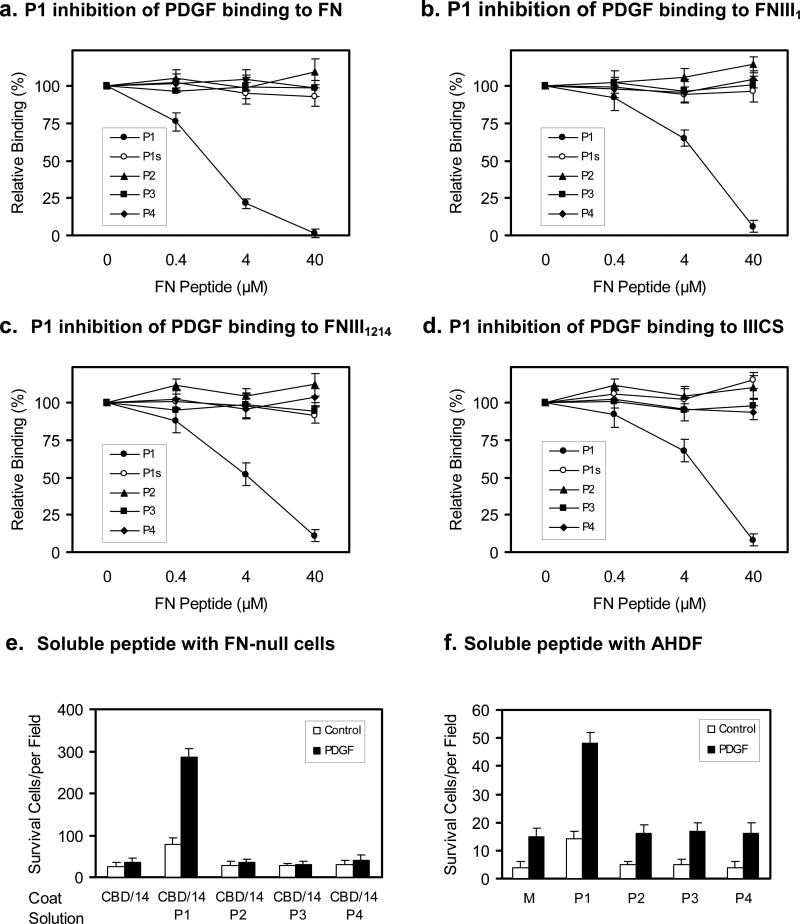
Figure 1. Peptide from FNIII1 binds PDGF-BB.
a. Schematic of human FN. FN type I repeats are shown as thin rectangles, FN type II repeats as ovals, and FN type III repeats as thick rectangles. PDGF-BB and cell-binding sites are indicated (arrows). b. Sequence of FNIII1 (FN609-718). Sequences of P1 (FN634-658) and P2 (FN680-704) are underlined. c. Equilibrium-binding of PDGF-BB with anastellin (FN630-704). d. Kinetic-binding of PDGF-BB with FN630-704. Increasing concentrations (6.25-200nM) of FN630-704 were injected across the biosensor chip coupled with PDGF-BB (first arrow) followed 120s later by a continuing flow of buffer (second arrow). Chip without PDGF-BB was used as a reference. KD was calculated by averaging the koff divided by kon. Sensorgrams are representative of three different experiments.
Platelet-derived growth factors (PDGFs) are major mitogens and survival factors for most mesenchymal cell types including fibroblasts (Romashkova and Makarov, 1999). PDGF occurs as five isoforms, classic PDGF-AA, AB, and BB isoforms, and the more recently described PDGF-CC and -DD isoforms (Fredriksson et al., 2004). The most potent PDGF isoforms, -AB and –BB, are secreted either by platelets (PDGF-AB), macrophages (PDGF-BB), or epidermal cells (PDGF-BB) in response to injury. PDGFB knockout in mice is embryonic lethal while knockouts of other isoforms are not (Betsholtz, 2004).
Survival signals from FN and growth factors converge at the level of the focal contact (Plopper et al., 1995) and are co-stimulatory (Miyamoto et al., 1996) for progression through the cell cycle (Assoian and Schwartz, 2001). For example, FN can bind vascular endothelial growth factor (VEGF) and enhance its bioactivity (Wijelath et al., 2006). It was more recently demonstrated that FNIII12-14 bound a large number of growth factors (Martino and Hubbell, 2010) including PDGF-BB, which we confirmed (Lin et al., 2011). Furthermore, we demonstrated that FN contains three growth factor-binding (FN-GFB) domains, i.e. FNIII1, FNIII13, and IIICS, respectively, which worked co-operatively with PDGF-BB to sustain optimal cell metabolism, and prevent autophagy and apoptosis (Lin et al., 2011). Here we identify four peptide sequences within these domains that bind PDGF-BB. In doing so, we discovered a 14mer peptide that bound PDGF-BB and promoted cell survival, whether bound to a surface or presented to cells in fluid-phase. Attributes of these FN-GFB peptides have important implications in tissue engineering.
Results
First FN type III repeat has two PDGF binding sites
To define peptide sequences that bind PDGF-BB and support cell survival in FN-GFB domains (Lin et al., 2011), we first focused on the first type III repeat of FN (FNIII1)(Figure 1b) because the bioactive peptide, anastellin (FN630-704), resides within this domain and has been reported to promote FN fibrillogenesis (Ohashi and Erickson, 2005) and anti-tumor activity (Yi and Ruoslahti, 2001). To study whether anastellin interacted with PDGF-BB (125I-labeled), anastellin was expressed with a Cys-tag at the C-terminus for sulfhydryl coupling to SulfoLink beads. From equilibrium binding assays, anastellin-PDGF-BB interaction gave a KD of 90nM (Figure 1c). This finding was confirmed by kinetic binding assays using plasmon surface resonance (Figure 1d, KD=22.9nM).
To determine which peptide sequence(s) within anastellin bind(s) PDGF-BB, FN630-667 and FN668-704, representing the first and second halves of anastellin, were synthesized with cysteine at their C-termini. ELISA equilibrium binding assays demonstrated that PDGF-BB bound FN630-667 with a KD of 210nM and FN668-704 with a KD = 80nM. Plasmon surface resonance confirmed binding of PDGF-BB to FN630-667 and FN668-704 (Figure S1). These results demonstrated that FNIII1 contains two PDGF-BB binding sequences. Although it was known that FNIII1 domain bound PDGF-BB (Lin et al., 2011), the finding of two growth factor-binding sequences in this domain was a surprise.
Four peptides from three FN-GFB domains bound PDGF-BB
To determine whether similar peptide sequences existed in the two halves of anastellin, the two halves were analyzed by PRATT software (Swiss Institute of Bioinformatics), a program that interactively generates conserved patterns from a series of unaligned proteins. Two sequences - FN634-658 (QPSHISKYILRWRPKNSVGRWKEAT) from the first half of anastellin and FN680-704 (QLISHQQYGHQEVTRFDFTTTSTST) from the second half of anastellin - were found, and designated P1 and P2 (Figure S2a), respectively. To study whether these two 25 amino acid peptides interacted with PDGF-BB, peptides were synthesized with a cysteine at the C-terminus for coupling to SulfoLink beads. ELISA equilibrium-binding assay using 125I-PDGF-BB showed P1 and P2 binding with KDs of 250nM and 110nM, respectively (Figure S2b and c). Interestingly, the P1 binding data fit a sigmoid binding curve (Figure S2f) better than the classic hyperbola curve shown in Figure S2b. The sigmoid curve had a Hills-coefficient of 1.38 indicating modest co-operative binding between two sites. Since PDGF-BB is an anti-parallel homodimer, these data suggested that P1 binds both ends in a co-operative manner.
Next we searched for other growth factor-binding sequences in FN as well as all other proteins in the human proteome database using ScanProsite (Swiss Institute of Bioinformatics), a program that scans a sequence against PROSITE, or a pattern against the UniProt Knowledgebase (Swiss-Prot and TrEMBL). Peptide sequences with similar patterns were found within FNIII13 and IIICS, two domains that we previously reported had PDGF-BB binding activity (Lin et al., 2011). These two FN peptides were designated P3 (FN1853-1877) and P4 (FN2043-2076), respectively (Figure S2a). Equilibrium-binding assays with cys-tagged P3 and P4 demonstrated that 125I-PDGF-BB bound P3 and P4 with a KD of 87nM and 56nM, respectively (Figure S2d and e). Thus, four PDGF-BB-binding peptides with similar sequence patterns were found within the three FN-GFB domains. Furthermore, a fifth similar sequence with growth factor-binding activity was found in the provisional matrix protein, vitronectin, but none other in the entire human proteome database.
FN-GFB peptides coupled to CBD/14 supports survival and decreases autophagy and apoptosis of FN-null cells in the presence of PDGF-BB
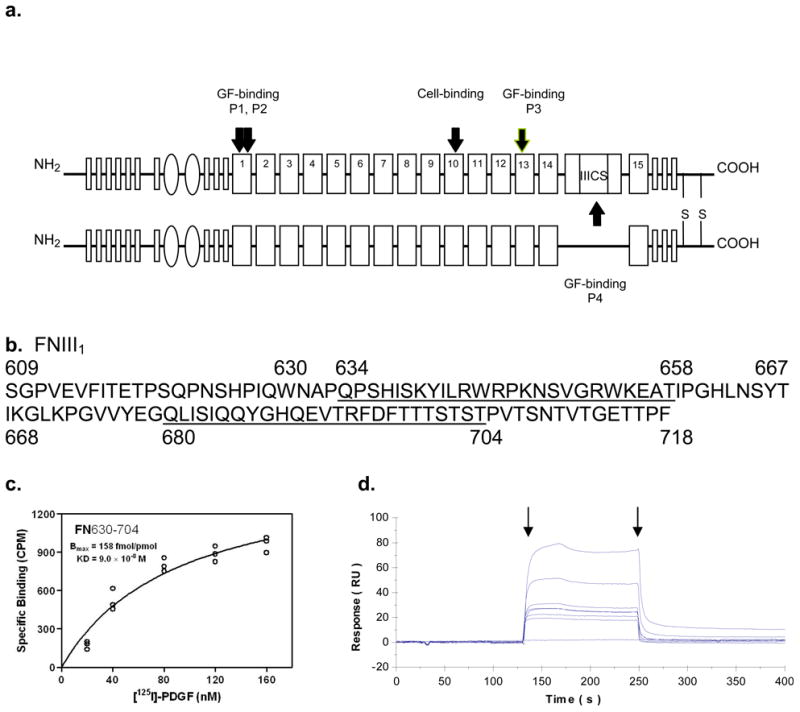
Previously, we showed that FN cell-binding domain (FNIII8-11) did not support FN-null fibroblast survival in the presence of 1nM PDGF-BB without the presence of a FN-GFB domain, i.e. FNIII1, FNIII13, or IIICS (Lin et al., 2011). FNIII8-11 (Cell-Binding Domain) fused to the FN 14th type III repeat (CBD/14) has been reported to prevent cell spreading, thereby inducing anoikis and apoptosis (Dai et al., 2005). However, CBD/14 in the presence either of FNIII1, FNIII13, or IIICS promoted optimal FN-null cell survival and response to PDGF-BB, results similar to those shown previously (Lin et al., 2011). This finding led us to investigate whether FN-GFB peptides when coupled to CBD/14 supported FN-null cell survival. To address this question, cell survival assays were carried out in serum-free medium on wells precoated with FN or with glutathione S-transferase (GST)-CBD/14 fusion product ± tethered FN-GFB peptides. As shown previously GST markedly improves protein adsorption to tissue culture plate surfaces (Lin et al., 2011). Results showed that FN-null cells could not survive on CBD/14 even in the presence of PDGF-BB (Figure 2a). When cultured in serum-free DMEM plus 1nM PDGF-BB, cells survived on surfaces pre-coated with CBD/14 coupled to any one of the four FN-GFB peptides (Figure 2a), but did not survive on CBD/14 coupled to scrambled peptides (Figure 2b). Cell attachment was similar under all conditions (Figure S3a). Not surprisingly, FN-null cell metabolism was robust on FN, mediocre on CBD/14 even in the presence of PDGF-BB, but almost equivalent to FN when FN-GFB peptides were coupled to CBD/14 and cultured with PDGF-BB (Figure S3b).
Figure 2. FN-GFB peptide coupled to CBD/14 supported FN-null fibroblast responsiveness to PDGF-BB.
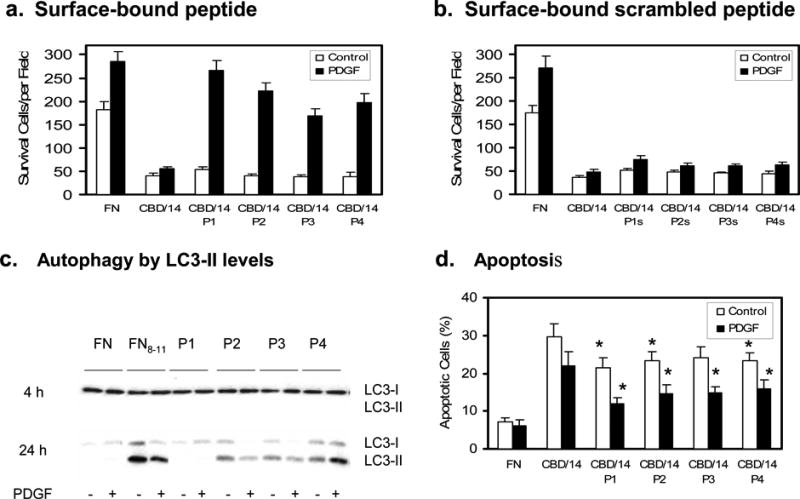
Cells were plated in 96-well (a,b,d) or 6-well (c) plates precoated with 0.125 μM FN or glutathione S-transferase (GST)-tagged CBD/14±tethered peptide and cultured in serum-free DMEM±PDGF-BB at 37°C for 3d. a, b. Viable spread cells in five 10xfields were counted in three wells at 3d (mean ± SD, n = 15). c. LC3-II was detected by a size shift on Western blot using a polyclonal antibody specific for LC3 (Zhang et al 2007). d. Apoptosis was determined by TUNEL at 3d. Percent apoptosis was calculated from (positive-cells/total-cells)×100. Fifty cells were counted in 3 replicate plates. Data points indicate mean ± SD. Each panel represents at least 3 experiments. *p<0.05 compared with CBD/14.
FN-null cells cultured on FN- CBD/14 increased autophagy at 24 hours compared with those growing on FN even in the presence of PDGF-BB as judged by quantification of total cellular Light Chain 3 (LC3), an inducible autophagosome component (Figure S3c) and Western blot analysis for LC3-II, a cleaved LC3 product that is bound to autophagosomes (Figure 2c) (Klionsky et al., 2012). When CBD/14 was coupled to FN-GFB peptides, the accumulations of LC3 or LC3-II in FN-null cells were significantly decreased (Figure S3b and Figure 2c, respectively). Thus, PDGF-BB could decrease autophagy in FN-null cells when plated on CBD/14 coupled to FN-GFB peptides, but not in cells on CBD/14 alone. This led us to investigate whether FN-GFB peptides also promoted the ability of PDGF-BB to limit apoptosis in FN-null cells cultured on CBD/14. FN-null cells showed little apoptosis up to 24 hours in all conditions (data not shown). FN-null cells cultured on FN showed low levels of apoptosis at 3 days, even in the absence of PDGF-BB (~ 7%), while PDGF-BB failed to prevent apoptosis in FN-null cells plated on CBD/14 (30% apoptosis at 3 days without PDGF-BB; 23% apoptosis with PDGF-BB) (Figure 2d). When FN-null cells were plated on CBD/14 coupled with a FN-GFB peptide, apoptosis was significantly decreased in the presence of PDGF-BB (Figure 2d).
Only P1 inhibited PDGF-BB binding to FN and FN-GFB domains
Next, we studied whether soluble FN-GFB peptides inhibited PDGF-BB binding to FN or FN-GFB domains (FNIII1, FNIII13, or IIICS). Binding assays of 125I-PDGF-BB to FN or FN-GFB domains were carried out in the presence of FN-GFB peptides at various concentrations. At 4μM, P1 inhibited over 75% binding of PDGF-BB to FN and at 40μM, the binding was nearly completely inhibited (Figure 3a). Scrambled P1 (P1s), with the same amino acid composition and molar concentrations as P1, failed to inhibit PDGF-BB binding to FN (Figure 3a). Authentic P1 also inhibited PDGF-BB binding to FNIII1, which contains P1 sequences (Figure 3b), as well as PDGF-BB binding to the other FN-GFB domains. Specifically, P1 inhibited PDGF-BB binding to FNIII12-14 and IIICs in a concentration dependent manner (Figure 3c and 3d, respectively). At 40μM, P1 inhibited nearly all the binding of PDGF to these domains. In contrast, scrambled P1 showed no inhibition of PDGF-BB binding to FN or any FN-GFB domain (Figure 3b-3d). Surprisingly, P2, P3, and P4 also failed to inhibit PDGF-BB binding to FN (Figure 3a) or any FN-GFB domain, even their parent domain (Figure 3b-d).
Figure 3. Soluble P1 inhibited PDGF-BB binding to FN and FN-GFB-domains, and supported FN-null cell and AHDF survival.
a-d. 125I-radiolabeled PDGF-BB was incubated with FN or GST-tagged FN-GFB-domain-coated wells. After washing, radioactivity of bound PDGF-BB was determined by a γ-counter. e. Fluid-phase P1 supported FN-null cell survival on CBD/14. FN-null fibroblasts were plated at 4,000 cells/well(96-well-plates) pre-coated with 0.125μM GST-tagged CBD/14 in serum-free DMEM with FN-GFB peptide in solution±1nM PDGF-BB at 37°C for 3d. Viable cells in five-10xfields were counted in three wells (mean±SD, n=15). f. Fluid-phase P1 supported AHDF survival under serum-starvation conditions. AHDF were plated at 1,000 cells/well(96-well plates) in serum-free DMEM and cultured overnight, then peptide(10μM)±PDGF-BB(1nM) was added, cells cultured for 6d, and viable cells counted. Data shown are mean±SE (n=4).
The ability of P1 to inhibit PDGF-BB binding to FN and to the three FN-GFB domains was not surprising since FN-GFB peptides within these domains shared a similar sequence pattern. Furthermore, P1 when added in solution significantly enhanced the survival of FN-null cells on CBD/14 in the presence of PDGF-BB (Figure 3e). Similar results were found for the survival of adult human dermal fibroblasts (AHDF) growing in serum-deprived medium for 6 days (Figure 3f). Consistent with the lack of effect on FN-null cell survival, P2, P3, and P4 showed little effect on AHDF survival even in the presence of PDGF-BB (Figure 3f). Together the inhibition experiments and the experiments with individual FN-GFB peptides in solution demonstrated that P1 had special growth factor-binding properties, like co-operative binding shown in Figure S2f, which potentiated its ability to promote cell survival.
P12 is the shortest FN-derived peptide to enhance FN-null cell survival
To determine the minimal amino acid sequence that enhanced cell survival, we investigated whether P11, a 20 residue peptide from the first third of anastellin that terminates after RWRPK, a vasoactive peptide described by the Hocking group (Hocking et al., 2008), had PDGF-BB binding and FN-null cell survival activity (Table S1). P11 as well as P5, which overlaps with P1 and P11, bound PDGF-BB and promoted PDGF-BB survival signals (Table S1). Next, a series of peptides were obtained by progressively trimming P5, starting from either N- or C-termini (Table S2) and were tested for bioactivity. Only P12, a peptide with one residue deletion from the N-terminus of P5, supported FN-null cell survival in the presence of PDGF-BB (Figure S4a) and inhibited PDGF-BB binding to FN (Figure S4b). Real-time binding studies showed that P12 interacted with PDGF-BB with a KD of 200nM and tissue culture investigations demonstrated that P12 could collaborate with PDGF-BB to enhance AHDF survival in serum-free medium (Figure S5). These results demonstrated that P12, a 14-mer peptide, was the smallest peptide with P1 activity.
P12 enhanced AHDF survival under stress conditions
The unique ability of soluble P12 to enhance FN-null cell survival led us to investigate whether P12 could also enhance tissue cell survival under stress conditions. AHDF were challenged with reactive oxygen species (ROS) stress or endoplasmic reticulum (ER) stress. AHDF in the presence of serum were challenged with hypoxanthine/xanthine oxidase (HO/XO) or hydrogen peroxide (H2O2). The HO/XO system generates constant low levels of ROS, while H2O2 is a bolus ROS insult. AHDF were sensitive to both HO/XO (Figure S6a) and H2O2(Figure S6b). In HO/XO system, cell death was inhibited when P12 was introduced 20 hours earlier or at the same time with oxidative stress. With H2O2, P12 only showed protective effect on AHDF survival when AHDF were pre-incubated with P12 prior to H2O2 challenge. P12 in the presence of PDGF-BB or serum also promoted cell survival when ER stress was induced by tunicamycin (Tun) as judged by the XTT assay (Figure 4).
Figure 4. P12 enhanced AHDF survival under tunicamycin (Tun)-induced endoplasmic reticulum (ER) stress and suppressed Tun-induced JNK activation.
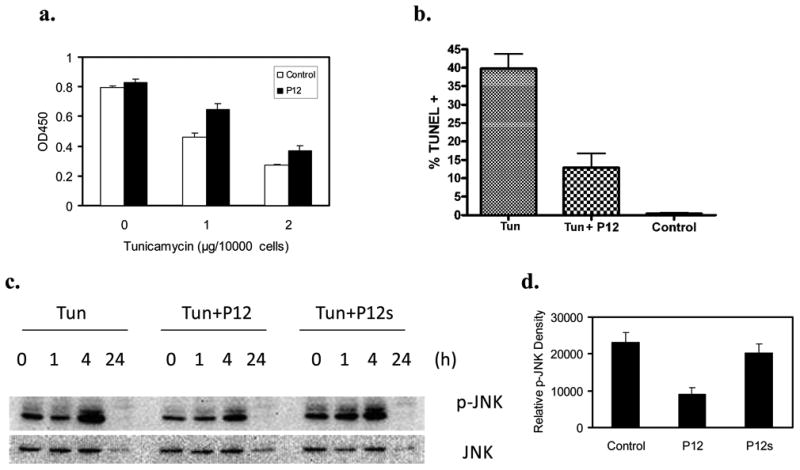
a. AHDF at 1000 cells/well were cultured in serum-free DMEM + 1nM PDGF-BB in 96-well plates overnight and challenged with Tun at 1 or 2μg/104cells for 4h. Cell viability was determined by XTT assay. Histograms show mean±SD, n=4. b. AHDF were incubated in serum-free DMEM + 1nM PDGF-BB with 1μg Tun/104cells±10μM P12 for 20h. Apoptosis was determined by TUNEL assay. c-d. AHDF were cultured in complete DMEM and exposed to 5μg/ml Tun (1μg/104cells)±10μM P12 or 10μM scrambled P12 (P12s) for the indicated times. c. p-JNK levels were determined by immunoblotting. d. Quantitative analysis of p-JNK band at 4h from 3 immunoblots, mean±SD.
Since ER stress is coupled to Jun N-terminal kinase (JNK) activation (Urano et al., 2000), which can lead to cell death by apoptosis (Lin et al., 2013; Urano et al., 2000), P12 effects on (JNKs) activation were investigated. After AHDFs were exposed to Tun in the presence of PDGF-BB ± P12 or scrambled P12, phosphorylated JNK (p-JNK) was determined by immunoblotting (Figure 4a, b). In the presence of 1nM PDGF-BB, but absence of P12, Tun significantly increased p-JNK by 4h. However, in the presence of both PDGF-BB and P12, p-JNK induction was markedly inhibited, while scrambled P12 (P12s) showed a minimal effect.
P12 limited burn-injury progression in a rat hot comb model
The effect of P12 on burn-injury progression was assessed in a rat hot comb burn model as described in the Methods section and in our previous publication (Singer et al., 2011). Each hot comb contained four brass prongs that left three 5mm non-burned interspaces. Burn injury progression across the interspaces was assessed both visually and histologically. When P12 was infused 1h after burns, injury progression into the interspaces between burns was inhibited in a dose response fashion (Figure 5).
Figure 5. P12 intravenous (IV) infusion 1 and 24 hrs after burn inhibited injury progression.
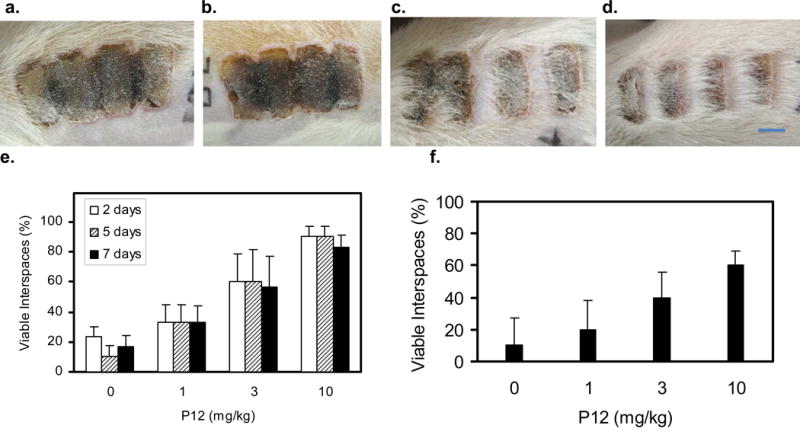
a., b. Burn at 7d after injury without treatment. c.,d. Burn at 7d after injury that had been treated with 10mg/kg P12. e. Necrotic interspaces 2, 5 and 7d after injury in rats treated or not treated with P12. f. Necrotic interspaces 7d after injury determined by histologic analysis. 0=infusion with lactated Ringer's buffer. Variance of interspace necrosis among rats within a specific treatment arm was no more than variance among interspaces on a given rat; therefore, interspace data for all 5 rats per treatment arm were pooled. n = 30 interspaces (6 interspaces/rat) in 5 rats per treatment arm. Bar = 10mm.
Discussion
From our results P12, a novel peptide from the 1st type III repeat of FN, appeared to act as a co-factor of PDGF-BB, enhancing its ability to promote survival of cells under stress. In addition, we found a similar peptide in vitronectin (Lin and Clark, unpublished data). Although it was previously established by us (Lin et al., 2011) and others (Martino and Hubbell, 2010; Wijelath et al., 2006) that FN can directly bind growth factors, and thereby enhance their activity, this is the first report that localized growth factor-binding and -enhancing activity to specific peptides within FN domains. We propose naming this new class of bioactive peptides that act as co-factors of growth factors, particularly promoting their cell survival activity, epiviosamines from the Greek word epivios, the adjective of the verb epiviono, which means “to survive in the face of adversity.” The prefix epivios has been combined with “amine,” used to indicate a substance made from peptides. Historically, vita, the Latin word for life, was combined with amine to make the word vitamin for the same reason. However, when it was discovered that vitamins were not peptides the “e,” from amine, was dropped.
Interestingly, the FN peptide P12, which had the most robust PDGF-BB binding activity and which was active in solution, was found within the amino-terminus of anastellin, a 76mer peptide that promotes FN fibrillogenesis (Ohashi and Erickson, 2005), inhibits endothelial cell growth, and has anti-angiogenesis activity (Yi and Ruoslahti, 2001). Preliminary investigations of P12 bioactivity on human dermal microvascular endothelial cells (HDMEC) demonstrated that P12 did not inhibit HDMEC metabolism when the cells were cultured with 10ng/ml VEGF in endothelial cell growth medium, while anastellin did inhibit (McTigue, Tonnesen and Clark, unpublished observations). In contrast, fibroblast survival was not inhibited by anastellin in solution and was enhanced when anastellin was adsorbed on the culture dish. Furthermore, the carboxyterminus of P12 contains RWRPK, the carboxy-terminus of vasoactive peptides previously described by the Hocking group (Hocking et al., 2008). Although P12 has vasodilation activity on the microvascular bed of a hamster cheek pouch (manuscript in preparation, Frame, Lin and Clark), RWRPK does not collaborate with PDGF-BB to promote survival of FN-null cells. In fact, the Hocking group demonstrated the RWRPK by itself inhibits cell growth (Hocking et al., 2008). Thus anastellin, P12, and RWRPK are nested peptides within the FN first type III repeat that have distinctly different biological activities.
The Hubbel group previously demonstrated that FNIII12-14 bound a large number of growth factors (Martino and Hubbell, 2010) including PDGF-BB as we confirmed (Lin et al., 2011). Furthermore, we have confirmed that FNIII12-14 as well as FNIII1 and IIICS bind multiple, but not all, growth factors. Often only a few members of a growth factor family bind FN. From the differential binding of FN-GFB domains to growth factors, we have delineated peptide sequences within these growth factors to which P1, P2, P3 and P4 bind (Lin and Clark, manuscript in preparation). In fact, preliminary computer modeling done by Wallqist and Rollins at Ft. Detrick demonstrated a probable P12 docking site at the interface of PDGF-BB and the PDGF-BB receptor (PDGFR-β) that contains the putative PDGF-BB, binding-partner peptide for P12. Since there are two sites for P12 binding to the PDGF-BB antiparallel dimer, the co-operative binding pattern observed for P1 interaction with PDGF-BB is not surprising.
More recently, the Hubbell group demonstrated that multiple recombinant domains of FN including the central cell-binding domain (FNIII9-10) and the promiscuous FNIII12-14 growth factor-binding domain greatly enhanced the regenerative effects of growth factors in a diabetic mouse model and a critical-size rat bone defect (Martino et al., 2011). In addition, the Hocking group recently demonstrated that FN recombinant fusion products consisting of the latter half of FNIII1 fused to FNIII8-10 enhanced wound healing in a diabetic mouse model (Roy et al., 2013). Taken together, these two groups have used FN domains that contain 3 out of the 4 peptides we have determined are the growth factor-binding sites in FN. The ability of P12 to limit burn injury progression adds further evidence that FN growth factor-binding sites have utility in tissue survival and healing.
JNKs play a critical role in cell apoptosis initiated by both extrinsic and intrinsic pathways (Verma and Datta, 2012). To date, three JNKs, namely JNK1, JNK2, JNK3 encoded by three distinct genes have been identified (Johnson and Nakamura, 2007). In response to specific stimuli such as heat shock, reperfusion injury, ER and oxidative stress, JNK proteins are activated by phosphorylation at its Thr- or Tyr-residues of a TXY motif. JNKs in turn activate apoptotic signaling either through the up-regulation of pro-apoptotic genes via the transactivation of specific transcription factors including C-Jun, or by directly modulating the activities of mitochondrial pro- and anti-apoptotic proteins through phosphorylation. Here we demonstrate that P12 promoted the ability PDGF-BB to suppress JNK activation. Therefore, the down-regulation of the JNK pro-apoptotic pathway may be responsible in part for the decreased apoptosis observed of cells under stress that were treated with P12. The ability of P12 to limit burn injury progression in a rat hot comb model is consistent with this hypothesis.
With elucidation of more epiviosamines like P12 we will increase our understanding of how extracellular matrix proteins can enhance growth factor signaling in such a way to sustain cell survival in the setting of tissue stress (Macri et al., 2007). In addition, the findings described here will increase the number of small peptide bioactives that can be added to tissue-engineered products to induce tissue survival and regeneration (Clark, 2013).
Materials and Methods
Materials
Recombinant PDGF-BB was obtained from Biosource (Camarillo, CA), and 125I-PDGF-BB from Amersham Biosciences (Piscataway, NJ). Human plasma FN was purchased from Chemicon (Temecula, CA) and found to be 99% pure and intact by SDS-PAGE. SulfoLink coupling beads was acquired from Pierce (Rockford, IL). Fatty acid-free bovine serum albumin (BSA) was purchased from ICN Biomedicals Inc (Aurora, OH). Ni-NTA agarose beads were acquired from Qiagen (Valencia, CA). IPTG and L-Broth were purchased from Fisher Scientific (Fair Lawn, NJ) and fetal bovine serum from Hyclone (Logan, Utah). Culture plates were from BD Bioscience (Billerica, MA). The Cell proliferation kit II (XTT) was from Roche Diagnostics (Indianapolis, IN). LC3 polyclonal antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA) and Novus Biologicals (Littleton, CO). BIAcore 2000, CM5 sensor chips, N-hydroxysuccinimide, N-ethyl-N'-(3-diethylaminopropyl)carbodiimide hydrochloride, and 1 M ethanolamine (pH8.5) were purchased from BIAcore Inc. (Piscataway, NJ). Peptides were synthesized by Bio-synthesis Inc. (Lewisville, TX) and PEGDVS was acquired from NEKTAR Transforming Therapeutics (Huntsville, AL).
Expression and purification of FN functional domains
Methods for cloning, expression and purification of recombinant human FN domains and fusion products of FN domains with glutathione sulfate transferase (GST) were previously published (Wang et al., 2005). GST tag on the N-terminus of the recombinant proteins facilitated FN domain adsorption onto tissue culture plastic for cell function studies. We also previously described cloning, expression and purification of cys-tagged FN domains (Ghosh et al., 2006). Cys-tagged FN domains allowed linkage to SulfoLink agarose beads with minimal impact on protein conformation.
Equilibrium binding of PDGF-BB with FN or peptides
Cys-tagged FN peptides were conjugated to agarose beads via –SH groups, using the manufacturer's protocol (SulfoLink Coupling Gel). To block non-specific binding, agarose beads, conjugated with or without FN or FN peptides, were incubated with 2% BSA at room temperature for 2h. For equilibrium binding, 20μl of conjugated agarose beads were incubated with varying concentrations of 125I-labeled PDGF-BB in binding buffer (DMEM+1%BSA) at room temperature for 2h with rotation. After washing, the radioactivity bound to the agarose beads was quantified using a γ-counter. The binding constant of PDGF-BB with FN or FN domains was determined using Prism 4 nonlinear regression software (GraphPad, San Diego, CA).
Kinetic binding assay of PDGF-BB with peptides
Using the BIAcore 2000 plasmon resonance system, kinetic binding constants were determined by passing varying concentrations of analytes dissolved in 20mM Hepes, 150mM NaCl, 3.4mM EDTA and 0.05% Tween-20, pH7.4 over chip surfaces coupled with either PDGF-BB or purported FN-GFB partner, which were immobilized on the Sensor Chips as previously described (Lin et al., 2011). All kinetic experiments were carried out at 20C at a flow rate of 30μl/min. For mass transport experiments, each analyte was injected at a fixed concentration and run at flow rates ranging from 5 to 75μl/min. All analytes were injected over a PDGF-coupled surface as well as over a control surface for 120s, followed by 300s of dissociation in running buffer. Regeneration of the sensor chip for subsequent injections was accomplished by one pulse of 0.05% SDS (30 s). Sensorgrams were prepared and globally fitted using nonlinear least-squares analysis and numerical integration of differential rate equations following the BIAcore manual and using kinetic models available in BIAevaluation software.
Cell culture
Mouse FN-null fibroblasts generated as described previously (Saoncella et al., 1999) and AHDF (passages 5 - 13, Clonetics, San Diego, CA) were maintained in DMEM supplemented with 100U/ml penicillin, 100μg/ml streptomycin, and 10% fetal bovine serum at 37°C and 5% CO2/95% air in a humidified atmosphere. For cell function assays, fibroblasts were grown to approximately 80% confluence, harvested, and transferred to tissue culture plates in assay appropriate numbers.
Mouse FN-null cell survival assays on FN domains
Cell survival experiments were performed in 96-well plates that had been coated with 0.125μM of FN, CBD/14, or CBD/14 coupled with various peptides as previously described (Lin et al., 2011). After blocking with 2% BSA for 2h to exclude further protein-binding, plates were washed with PBS. For each well, 4,000 FN-null cells were plated and incubated at 37°C for 4h. Then, 25μl of 5% BSA in DMEM was added to each well ± PDGF-BB or FN peptides and wells were incubated for times indicated. Cell survival was assayed by counting cells and metabolism by XTT following the manufacturer's protocol.
TUNEL assay
Apoptotic cells were identified by nick end-labeling using the In Situ Cell Death Detection Kit and TMR red as previously described (Lin et al., 2011).
Analysis of the autophagosome protein LC3
ELISA for total LC3 and Western blots of LC3I and LC3II were performed as previously described (Lin et al., 2011).
Rat hot comb burn model
Animal experiments were approved by the IACUC at Stony Brook University. Sprague-Dawley rats with a weight around 350 grams were used. After removing hair, 2 burns were created on the back of each animal using a brass comb with four rectangular prongs preheated in boiling water and applied for 30 seconds resulting in four rectangular 10 × 20 mm full thickness burns separated by three 5 × 20 mm unburned interspaces (zone of ischemia). Forty comb burns with 120 unburned interspaces were created and distributed among control and three doses of P12 (1, 3, 10 mg/kg administered in 1 ml of Lactated Ringers solution). An observer blinded to the treatment determined necrotic interspaces at 2, 5, and 7 days. Full-thickness biopsies from interspaces 7 days after injury were processed, stained with hematoxylin and eosin, and evaluated for evidence of necrosis by a board certified dermatopathologist (SAM) blinded to the protocol.
Supplementary Material
Figure 1. FNIII1 has two PDGF-BB binding sites, P1 and P2, and P1 shows a binding curve suggestive of co-operatively. Real-time interactions of PDGF-BB with FNIII1-derived peptides were determined by plasmon surface resonance (Biacore2000). FNIII1-derived peptide FN630-667 (a) or FN668-704 (b) was injected across a biosensor chip to which PDGF-BB had been conjugated. Chip without PDGF-BB was used as a reference. Sensorgrams are representative of three different experiments.
Supplement Figure 2. FN contains four PDGF-BB binding sequences. a. Amino acid sequence alignments of four FN growth factor-binding peptides P1-P4. b-e. Equilibrium binding of PDGF-BB with FN-derived peptides P1 (b), P2 (c), P3 (d), and P4 (e). Cys-tagged peptides were conjugated to agarose beads via-SH group according to manufacturer’s protocol and incubated with 125I-radiolabeled PDGF-BB. After washing, radioactivity of bound PDGF-BB was determined by a γ-counter. f. Binding of I125-labeled PDGF-BB to P1 in an ELISA-like assay gave a sigmoid curve rather than a hyperbola suggesting co-operative binding. The Hills-coefficient was 1.38 indicative of modest co-operative binding between two sites.
Supplement, Figure 3. FN-GFB peptide coupled to CBD/14 supported FN-null fibroblast responsiveness to PDGF-BB but did not affect cell attachment. Mouse FN-null cells were plated in 96- well cell culture plates precoated with 0.125 μM FN or glutathione S-transferase (GST)-tagged CBD/14 coupled with or without GFBP in serum-free DMEM ± 1nM PDGF-BB at 37°C. a. Cell attachment at 4h. Data points indicate mean±SD. Results represent three different experiments. b. Metabolic response at 3d as judged by XTT assay over 4 hours. c. Total cellular LC3, a component of autophagosomes, was detected at 24h by rabbit polyclonal antibodies using ELISA (Jager 2004). Data points indicate mean ± SD. Results in panels b. and c. represent at least three different experiments. OD, Optical density. * p<0.05 and ** p<0.01 compared with FN8-11 or FN8-11/PDGF, respectively.
Supplement, Figure 4. P12 is the shortest peptide derived from FN to support cell survival and to inhibit PDGF-BB binding to FN. a. Mouse FN-null fibroblasts were plated at 4,000 cells/well in 96-well plates that had been precoated with 0.125μM glutathione transferase (GST)- tagged CBD/14 and then cultured at 37°C in DMEM±FN-GFB peptide in solution with or without PDGF-BB for 3d. Viable spread cells in five 10x fields were visually counted in each of three replicate wells (mean ± SD, n = 15). b. FN-coated wells in a 96-well plate were incubated with 125I-radiolabeled PDGF-BB ± 40μM FN-derived peptide. After washing, radioactivity of bound PDGF-BB was determined by a γ-counter. Data shown are mean ± SD (n=6).
Supplement Figure 5. P12 interaction with PDGF-BB enhances its activity. a. Real time interaction of P12 with PDGF-BB was determined by plasmon surface resonance (Biacore2000). P12 was coupled to chip as a ligand and varying concentrations of PDGF-BB (analyte) were injected across biosensor chip (6.25-200 nM, 30 μl minute-1). Chip without P12 was used as a reference. Sensorgrams are representative of three different experiments. Arrows indicate the beginning and the end of the analyte injection. b. Adult human dermal fibroblasts (AHDF) growth curves with 1nM PDGF-BB demonstrated a more robust response in the presence of increasing amounts of P12. AHDF were cultured at 500 cells/well of a 96-well plate in serumfree DMEM containing 1%BSA with 1nM PDGF-BB ± P12 at the concentrations shown for up to 7d. Cell metabolism as a reflection of cell number was determined by XTT assay. c. P12 dose response demonstrated 5μM P12 maximally enhanced 1nM PDGF-BB survival activity on AHDF. These data were extrapolated from Supplement Figure 5b by calculating percent viable cells at 5d with various P12 concentrations added to 1nM PDGF-BB in serum-free DMEM and 1%BSA compared to cell cultures with 1nM PDGF and DMEM and 1%BSA without P12.
Supplement Figure 6. P12 supported AHDF survival under ROS stress. P12 protected AHDF from oxidative stress-mediated cell death. AHDF were grown in complete DMEM at 2000 cells per well in 96-well plate. Cells were challenged with hypoxanthine/xanthine oxidase (HX/XO) (a) or hydrogen peroxide (H2O2) (b) overnight ± 10 μM P12. Cell viability was determined by visual counts in five fields. Histograms showed mean ± SD, n=4.
Supplement Table 1 Bioactivities and fibronectin/growth factor binding inhibition
Supplement Table 2 Bioactivities* and fibronectin/growth factor binding inhibition**
Acknowledgments
This work was support by NIH/NA10143 Merit Award (RAFC), NIH RC2 AR059384 (RAFC), and the Armed Forces Institute of Regenerative Medicine W81XWH-08-2-0034 (RAFC). We thank Deane F. Mosher for providing the FN-null cells.
Footnotes
Conflict of Interest: Richard Clark and Fubao Lin co-discovered P12. Richard Clark is President and Founder of NeoMatrix Formulations, a biotechnology company engaged in preclinical studies for P12 treatment of burns. Marcia Tonnesen is Secretary of NMF.
References
- Assoian RK, Schwartz MA. Coordinate signaling by integrins and receptor tyrosine kinases in the regulation of G1 phase cell-cycle progression. Curr Opin Genet Dev. 2001;11:48–53. doi: 10.1016/s0959-437x(00)00155-6. [DOI] [PubMed] [Google Scholar]
- Betsholtz C. Insight into the physiological functions of PDGF through genetic studies in mice. Cytokine Growth Factor Rev. 2004;15:215–228. doi: 10.1016/j.cytogfr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Brait VH, Jackman KA, Walduck AK, Selemidis S, Diep H, Mast AE, Guida E, Broughton BR, Drummond GR, Sobey CG. Mechanisms contributing to cerebral infarct size after stroke: gender, reperfusion, T lymphocytes, and Nox2-derived superoxide. J Cereb Blood Flow Metab. 2010;30:1306–1317. doi: 10.1038/jcbfm.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RAF. Wound repair: basic biology to tissue engineering. In: Lanza RP, Langer R, Chick WL, editors. Principles of Tissue Engineering. Elsevier; Amsterdam, Boston: 2013. [Google Scholar]
- Dai R, Iwama A, Wang S, Kapila YL. Disease-associated fibronectin matrix fragments trigger anoikis of human primary ligament cells: p53 and c-myc are suppressed. Apoptosis. 2005;10:503–512. doi: 10.1007/s10495-005-1880-5. [DOI] [PubMed] [Google Scholar]
- Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004;15:197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Kroemer G. Necroptosis: a specialized pathway of programmed necrosis. Cell. 2008;135:1161–1163. doi: 10.1016/j.cell.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Ghosh K, Ren XD, Shu XZ, Prestwich GD, Clark RA. Fibronectin functional domains coupled to hyaluronan stimulate adult human dermal fibroblast responses critical for wound healing. Tissue Eng. 2006;12:601–613. doi: 10.1089/ten.2006.12.601. [DOI] [PubMed] [Google Scholar]
- Han SW, Roman J. Fibronectin induces cell proliferation and inhibits apoptosis in human bronchial epithelial cells: pro-oncogenic effects mediated by PI3-kinase and NF-kappa B. Oncogene. 2006;25:4341–4349. doi: 10.1038/sj.onc.1209460. [DOI] [PubMed] [Google Scholar]
- Hirth DA, McClain SA, Singer AJ, Clark RAF. Endothelial necrosis at 1h post-burn predicts progression of tissue injury. Wound Repair Regen. 2013 doi: 10.1111/wrr.12053. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking DC, Titus PA, Sumagin R, Sarelius IH. Extracellular matrix fibronectin mechanically couples skeletal muscle contraction with local vasodilation. Circ Res. 2008;102:372–379. doi: 10.1161/CIRCRESAHA.107.158501. [DOI] [PubMed] [Google Scholar]
- Johnson GL, Nakamura K. The c-jun kinase/stress-activated pathway: regulation, function and role in human disease. Biochim Biophys Acta. 2007;1773:1341–1348. doi: 10.1016/j.bbamcr.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NY, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ET, Banhegyi G, Bartholomew CR, Bassham DC, Bast RC, Jr, Batoko H, Bay BH, Beau I, Bechet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier ST, McClain SA, Lin F, Singer AJ, Clark RA. Spatiotemporal progression of cell death in the zone of ischemia surrounding burns. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2011;19:622–632. doi: 10.1111/j.1524-475X.2011.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Ren XD, Pan Z, Macri L, Zong WX, Tonnesen MG, Rafailovich M, Bar-Sagi D, Clark RA. Fibronectin growth factor-binding domains are required for fibroblast survival. The Journal of investigative dermatology. 2011;131:84–98. doi: 10.1038/jid.2010.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Zhu J, Tonnesen MG, Clark RAF. Novel fibronectin peptides bind PDGF-BB and enhance cell survival under stress. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.420. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macri L, Silverstein D, Clark RAF. Growth factor binding to the pericellular matrix and its importance in tissue engineering. Adv Drug Deliv Rev. 2007;59:1366–1381. doi: 10.1016/j.addr.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Martino MM, Hubbell JA. The 12th-14th type III repeats of fibronectin function as a highly promiscuous growth factor-binding domain. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:4711–4721. doi: 10.1096/fj.09-151282. [DOI] [PubMed] [Google Scholar]
- Martino MM, Tortelli F, Mochizuki M, Traub S, Ben-David D, Kuhn GA, Muller R, Livne E, Eming SA, Hubbell JA. Engineering the growth factor microenvironment with fibronectin domains to promote wound and bone tissue healing. Sci Transl Med. 2011;3:100ra189. doi: 10.1126/scitranslmed.3002614. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Teramoto H, Gutkind JS, Yamada KM. Integrins can collaborate with growth factors for phosphorylation of receptor tyrosine kinases and MAP kinase activation: roles of integrin aggregation and occupancy of receptors. J Cell Biol. 1996;135:1633–1642. doi: 10.1083/jcb.135.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T, Erickson HP. Domain unfolding plays a role in superfibronectin formation. J Biol Chem. 2005;280:39143–39151. doi: 10.1074/jbc.M509082200. [DOI] [PubMed] [Google Scholar]
- Piper HM, Garcia-Dorado D. Reducing the impact of myocardial ischaemia/reperfusion injury. Cardiovasc Res. 2012;94:165–167. doi: 10.1093/cvr/cvs133. [DOI] [PubMed] [Google Scholar]
- Plopper GE, McNamee HP, Dike LE, Bojanowski K, Ingber DE. Convergence of integrin and growth factor receptor signaling pathways within the focal adhesion complex. Mol Biol Cell. 1995;6:1349–1365. doi: 10.1091/mbc.6.10.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romashkova JA, Makarov SS. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- Roy DC, Mooney NA, Raeman CH, Dalecki D, Hocking DC. Fibronectin Matrix Mimetics Promote Full-Thickness Wound Repair in Diabetic Mice. Tissue engineering Part A. 2013 doi: 10.1089/ten.tea.2013.0024. online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Johnson KJ, Murozono M, Sakai K, Magnuson MA, Wieloch T, Cronberg T, Isshiki A, Erickson HP, Fassler R. Plasma fibronectin supports neuronal survival and reduces brain injury following transient focal cerebral ischemia but is not essential for skin-wound healing and hemostasis. Nat Med. 2001;7:324–330. doi: 10.1038/85471. [DOI] [PubMed] [Google Scholar]
- Saoncella S, Echtermeyer F, Denhez F, Nowlen JK, Mosher DF, Robinson SD, Hynes RO, Goetinck PF. Syndecan-4 signals cooperatively with integrins in a Rho-dependent manner in the assembly of focal adhesions and actin stress fibers. Proc Natl Acad Sci U S A. 1999;96:2805–2810. doi: 10.1073/pnas.96.6.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shupp JW, Nasabzadeh TJ, Rosenthal DS, Jordan MH, Fidler P, Jeng JC. A review of the local pathophysiologic bases of burn wound progression. J Burn Care Res. 2010;31:849–873. doi: 10.1097/BCR.0b013e3181f93571. [DOI] [PubMed] [Google Scholar]
- Singer AJ, Taira BR, Lin F, Lim T, Anderson R, McClain SA, Clark RA. Curcumin reduces injury progression in a rat comb burn model. Journal of burn care & research : official publication of the American Burn Association. 2011;32:135–142. doi: 10.1097/BCR.0b013e318203337b. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Verma G, Datta M. The critical role of JNK in the ER-mitochondrial crosstalk during apoptotic cell death. J Cell Physiol. 2012;227:1791–1795. doi: 10.1002/jcp.22903. [DOI] [PubMed] [Google Scholar]
- Wang J, Milner R. Fibronectin promotes brain capillary endothelial cell survival and proliferation through alpha5beta1 and alphavbeta3 integrins via MAP kinase signalling. J Neurochem. 2006;96:148–159. doi: 10.1111/j.1471-4159.2005.03521.x. [DOI] [PubMed] [Google Scholar]
- Wang R, Clark RA, Mosher DF, Ren XD. Fibronectin's central cell-binding domain supports focal adhesion formation and Rho signal transduction. J Biol Chem. 2005;280:28803–28810. doi: 10.1074/jbc.M501421200. [DOI] [PubMed] [Google Scholar]
- Wijelath ES, Rahman S, Namekata M, Murray J, Nishimura T, Mostafavi-Pour Z, Patel Y, Suda Y, Humphries MJ, Sobel M. Heparin-II domain of fibronectin is a vascular endothelial growth factor-binding domain: enhancement of VEGF biological activity by a singular growth factor/matrix protein synergism. Circ Res. 2006;99:853–860. doi: 10.1161/01.RES.0000246849.17887.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KM, Clark RAF. Provisional matrix. In: Clark RAF, editor. Molecular and Cellular Biology of Wound Repair. Plenum; New York: 1996. pp. 51–93. [Google Scholar]
- Yi M, Ruoslahti E. A fibronectin fragment inhibits tumor growth, angiogenesis, and metastasis. Proc Natl Acad Sci U S A. 2001;98:620–624. doi: 10.1073/pnas.98.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J. Neuroprotective strategies targeting apoptotic and necrotic cell death for stroke. Apoptosis. 2009;14:469–477. doi: 10.1007/s10495-008-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Vuori K, Reed JC, Ruoslahti E. The alpha 5 beta 1 integrin supports survival of cells on fibronectin and up-regulates Bcl-2 expression. Proc Natl Acad Sci U S A. 1995;92:6161–6165. doi: 10.1073/pnas.92.13.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.