Abstract
DAXX is a multifunctional protein that regulates a wide range of cellular signaling pathways for both cell survival and apoptosis. Regulation of DAXX gene expression remains largely obscure. We recently reported that berberine (BBR), a natural product derived from a plant used in Chinese herbal medicine, downregulates DAXX expression at the transcriptional level. Here, we further investigate the mechanisms underlying the transcriptional suppression of DAXX by BBR. By analyzing and mapping the putative DAXX gene promoter, we identified the core promoter region (from −161 to −1), which contains consensus sequences for the transcriptional factors Sp1 and Ets1. We confirmed that Sp1 and Ets1 bound to the core promoter region of DAXX and stimulated DAXX transcriptional activity. In contrast, BBR bound to the DAXX core promoter region and suppressed its transcriptional activity. Following studies demonstrated a possible mechanism that BBR inhibited the DAXX promoter activity through blocking or disrupting the association of Sp1 or Ets1 and their consensus sequences in the promoter. Downregulation of DAXX by BBR resulted in inhibition of MDM2 and subsequently, activation of p53, leading to cancer cell death. Our results reveal a novel possible mechanism: by competitively binding to the Sp1 and Ets1 consensus sequences, BBR inhibits the transcription of DAXX, thus inducing cancer cell apoptosis through a p53-dependent pathway.
Keywords: DAXX, Sp1, Ets1, BBR, neuroblastoma
The death-domain-associated protein (DAXX) was originally identified as a protein that specifically binds to the death domain of the transmembrane death receptor FAS (also called CD95) normally located in the cytoplasm and it potentiates FAS-induced apoptosis [1]. A subsequent study showed that although DAXX specifically enhances FAS function, the human homolog of DAXX (hDAXX) does not bind FAS and instead is found in the nucleus, where it localizes to promyelocytic leukemia protein (PML) oncogenic domains (PODs) [2]. Later studies demonstrated that DAXX is a cytoplasm-nuclear shuttle protein and possesses multiple cellular functions in the regulation of either pro-apoptosis or anti-apoptosis, depending on the cell type and the signaling pathway it regulates [3,4]. For example, in primary cells DAXX appears to be a pro-apoptotic protein, by regulating stress kinases such as JUK and p38 [1,5], while in many cancer cells DAXX serves as an anti-apoptotic factor by inhibiting the p53 function through stabilizing of MDM2 [6,7]. It is now clear that DAXX exhibits each of its potential biological activities mainly by relying on the cellular localization of the protein and the availability of various interacting partners. When it localizes in the nucleus, DAXX is a transcriptional co-regulator that can associate with many transcription factors and it participates in regulation, either activating or repressive, of many target genes [8-10].
The distinct biological activities of DAXX may also be associated with its expression levels. DAXX is ubiquitously expressed throughout the body, with particularly high expression levels in the thymus and testes [1]. Several studies have demonstrated that the expression level of DAXX is post-translationally modified. Peptidyl-prolyl isomerase Pin1 inhibits DAXX-involved apoptosis by inducing phosphorylation on ser178 of DAXX, a location mediating DAXX ubiquitination and degradation [11]. The ubiquitination of DAXX can also be regulated by p14ARF and MDM2 [12]. In addition, the viral E1B-55K protein binds to DAXX and induces its degradation through a proteasome-dependent pathway [13]. Despite studies showing that regulation of the DAXX protein is via post-translational modification, any regulation of DAXX expression at the transcriptional and translational levels is currently poorly understood.
Berberine (BBR), a benzyl-tetra isoquinoline alkaloid that is extracted from plants of the Berberidaceae family, has been extensively used for many centuries in traditional Chinese and Native American medicine [14]. Recent evidence suggests that, besides its traditional use as an antimicrobial, BBR possesses several new potential therapeutic functions, including anti-tumor activity [15]. The anti-tumor activity of BBR has been demonstrated in a wide variety of neoplasms, but the mechanisms of action are not completely clear yet for each individual type of cancer. One previous study shows that BBR inhibits human neuroblastoma cell growth through the induction of p53-dependent apoptosis [16]. We recently studied the molecular mechanism of action by which BBR induces p53-dependent apoptosis, in acute lymphoblastic leukemia (ALL). There, we found that BBR transmits a non-stress kind of signaling that is able to degrade MDM2 by inhibiting the transcription of the DAXX gene, which results in the activation of p53 and brings on leukemic cell death [7]. In that study, we did not characterize how BBR inhibits DAXX transcription.
It is known that BBR is a DNA intercalator that plays numerous pharmacological functions by associating with DNA [17-19]. A recent study demonstrates that following administration of BBR, it can rapidly distribute into the nucleus of living cells, where it alters the spatial conformation of DNA and can suppress nearby gene transcription by inhibiting the association between the TATA binding protein (TBP) and the TATA box in the promoter of those genes [20]. In our present study, we evaluated the mechanism of action of BBR in the suppression of DAXX transcription. We found that BBR becomes bound to the core promoter region of DAXX and that action inhibits transcriptional activity possibly by blocking the association of Sp1 and Ets1 with their consensus sequences in the DAXX promoter region. Any inhibition of DAXX expression resulted in the degradation of MDM2, followed by the activation of p53 and then apoptosis of neuroblastoma cells, in a manner similar to its apoptotic effect in ALL cells.
MATERIALS AND METHODS
Cell lines and reagents
The human neuroblastoma cell lines SK-N-SH and NB-1691 were obtained from Dr. H. Findley (Emory University). These were grown in standard culture medium (RPMI 1640 containing 10% FBS, 2 mmol/L of L-glutamine, 50 units/ml of penicillin and 50 ug/ml of streptomycin) in incubators set at 37°C with a humidified atmosphere containing 5% CO2. The BBR, purchased from Sigma-Aldrich, was dissolved in 50% ethanol to create a 10 mM stock solution that was stored in small aliquots at −20°C until use. For this study, the working concentration of BBR was from 10-100μM.
Construction of plasmids
To date, there have been no studies of the DAXX gene promoter reported. We searched the 5′-flank of the untranslated DNA sequence of the DAXX gene by navigating the Human Genome at the dedicated web site (http://www.ncbi.nlm.nih.gov/Genomes/index.html). A 695 bp fragment in this region, spanning from −695 to −1, was generated by PCR using the following primers: 5′-CGTTGTGCTCATTTGTACGC-3′ and 5′-TTCCTTCCCACTCCCACCGC-3′. Next, we cloned the fragment into the promoterless luciferase vector pGL3 basic (Promega) at the Hind III and Nco I sites, to produce a full-length DAXX-p 695 construct. In addition, various 5′-3′ or 3′-5′ deletion constructs and site-directed mutants of the DAXX promoter fragments were made. For the construction of the putative DAXX promoter deletion constructs DAXX-p515, 275, 161, 65, 534 and 630, we first determined their corresponding PCR primer pairs from within the sequence of the 695 bp fragment, and then used them to make the different constructs. Mutant DAXX-p 161 constructs were made using mutated nested primers at the residues of SP1 and Ets1 binding sites. Mutated promoter fragments were made by site-directed mutagenesis using an in vitro site-directed mutagenesis system (Promega). Constructs, including different deleted or mutated fragments, were then ligated to the pGL3 basic vector. DNA sequencing was performed to confirm that the sequence of the PCR products were correct as compared with the DAXX promoter that is published in the Human Genome data base.
The Sp1 expression plasmid was kindly provided by Dr. R. Tjian (University of California). The Ets-1 expression plasmid was generated by inserting a cDNA fragment that had been synthesized by RT-PCR into the pcDNA3.1+ vector (Invitrogen) at the Nhe I and Hind III sites. The RT-PCR was performed using total RNA that was extracted from SK-N-SH cells and a primer pair (5′-GCGCGCTAGCAACTTGCTACCATCCCGT-3′ and 5′-GCGCAAGCTTTGCCATCACTCGTCGGCA-3′) that were designed based on the Ets-1 gene. The DAXX expression plasmid was generated similarly in the pcDNA3.1+ vector, using a primer pair (5′-TGAAATCCCCACCACTTCCTCCCTC-3′ and 5′-GAGAGGCAGTGTTTTCAGCATTTGT-3′). We also constructed a pSUPER-Sp1 siRNA plasmid and a pSUPER-Ets-1 siRNA plasmid by inserting a 19-nt Sp1 (TGGTGGTGCCTTTTCACAG) sequence and a 19-nt Ets-1 (GATATGGAATGTGCAGATG) sequence, respectively, into the expression plasmid pSUPER-neo vector, which was purchased from OligoEngine (Seattle, WA). As a control, we inserted a 19-nt scrambled sequence (GAGGCTATTATACTGTGAT) into pSUPER-neo.
Transfection and reporter assay
For the gene transfection and reporter assays, SK-N-SH and NB-1691 cells that were in an exponential growth stage were transiently transfected with the DAXX promoter-luciferase constructs, as were described above, or co-transfected with these constructs and the Sp1 or Ets1 expression plasmids, plus the siRNA plasmids to suppress expression, by way of electroporation at 320V, 975 microfarads, using a Gene Pulser II system (Bio-Rad). Briefly, 2×106 cells were mixed with the corresponding plasmids plus the pRL (Renilla luciferase)-CMV vector (to provide an internal control), and electrophoresed. Transfected cells were re-suspended in 3 ml of RPMI containing 5% FBS. Then 24h after a given transfection, the cells were treated with BBR for another 4h. Next, cell extracts were prepared with a 1×lysis buffer. After centrifugation, 20μl aliquots of the supernatant were mixed with 50μl of luciferase assay reagent II (Promega), in order to measure the FL activity. Next, the RL activity was determined by adding Stop & Glo® reagent to the same sample. These luciferase activities were analyzed on a Microplate Luminometer (Turner Designs) and normalized to values for Renilla luciferase.
Fluorescence titration assay
The binding properties that BBR has with the DAXX promoter were examined by a fluorescent titration assay. The 161-nt DAXX core promoter DNA fragment, synthesized as described above, was labeled with fluorescein using the 5′ Endtag Nucleic Acid Labeling System (Vector Laboratories, Burlingame, CA, USA). Briefly, 0.5 nmols of DNA and 1 unit of alkaline phosphatase were incubated for 30 minutes at 37 °C in 10μl of 1x universal reaction buffer. After dephosphorylation, a sulfur-modified phosphate group from ATP-ˠ-S was added to the 5′ end of the DNA, using T4 polynucleotide kinase. Through a thiol-reactive labeling system, fluorescein was then added to the 5′ end of the DNA, by incubating it with fluorescein maleimide at 65 °C for 30 min. For the BBR titration assay, the fluorescein-labeled DNA was prepared in 10 mM of Hepes buffer (pH 7.2) to achieve a final concentration of 100 nM. All the BBR solutions were prepared in the same buffer containing the fluorescein-labeled DNA, so that the DNA concentration was kept constant during titration. The steady-state fluorescence of the DNA-BBR mixture was acquired on a PTI Quanta-Master spectrometer (Photon Technology International, Birmingham, NJ) using a 3 ml cuvette. The slit widths for excitation and emission were adjusted to minimize photobleaching of the sample while achieving sufficient fluorescent signal intensity. The fluorescence measurements as a function of BBR concentration were fitted with the hyperbolic function F = Ff + (Fb − Ff)[ligandf]/(Kd +[ligandf], where F is the observed fluorescence, Ff is the fluorescence of unbound DNA, Fb is the fluorescence from the DNA-berberine complex, [ligandf] is the concentration of BBR and Kd is the dissociation constant.
Chromatin immunoprecipitation (ChIP) assay
The CHIP assay was performed to analyze the DNA binding properties of SP1 and Ets-1 proteins to the DAXX promoter and explore any changes in them following BBR treatment. Briefly, SK-N-SH cells that were treated with or without BBR were incubated with a solution of 1% formaldehyde at room temperature for 10 min with mild shaking to cross-link the ETS-1 or SP1 proteins with the DAXX promoter region. Then, 1×106 of those incubated cells were washed twice with cold phosphate-buffered saline and resuspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) with 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml aprotinin, and 1 μg/ml pepstatin A. After a brief sonication, the cellular lysates obtained were cleared by centrifugation and were diluted 10-fold with dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl at pH 8.1, and 167 mM NaCl) containing the same protease inhibitors as mentioned above. Anti-SP1 and anti-ETS-1 or control antibodies were then added at 4°C overnight, with rotation. The resulting immunoprecipitated complexes were collected by a protein A/G plus-agarose column. Precipitants were sequentially washed once with a low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, and 150 mM NaCl),a high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.1, and 500 mM NaCl), and finally a LiCl wash buffer (0.25M LiCl, 1% Nonidet P-40, 1% deoxycholate, 1mM EDTA, 10 mM Tris-HCl at pH 8.1), followed by two washes with 1xTE. After the final wash, 250 μl of elution buffer (1% SDS, 0.1 M NaHCO3) was added, and the precipitants were incubated at room temperature for 15 min, with rotation. We added 5M NaCl to reverse the formaldehyde cross-linking when heated at 65°C for 4h. After a precipitation with ethanol, the resulting pellets were re-suspended and treated with proteinase K. The DNA was recovered by a phenol/chloroform extraction and ethanol precipitation. Pellets were re-suspended in TE buffer and subjected to q-PCR, performed with a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA), using a primer pair for amplification of the DAXX core promoter (−161 to −1).
Electrophoretic mobility shift assay (EMSA)
The binding of Sp1 and Ets1 to the DAXX promoter and the effects of BBR on their binding were also tested by EMSA. A nuclear protein extraction from SK-N-SH cells was prepared using a kit (NEPERTM from Pierce). Nuclear protein was incubated for 15 min in a binding buffer (10mM Tris-HCl, 50mM NaCl, 0.5mM EDTA, 10% glycerol, 1mM dithiothreitol, and 7.5 mM MgCl) plus 0.1 μg of poly(dI-dC) carrier, and a 32P-labeled DAXX core promoter (wt or a mutation within the Sp1 or the Ets1 binding sites). Anti-Sp1, anti-Ets1 or normal IgG antibodies that were additional controls were used in experiments to supershift the specific complexes of interest, by pretreating the extract for 1 h at 4°C with these antibodies, in the presence or absence of BBR. The samples were electrophoresed on a 5% polyacrylamide gel, then dried and an overlying X-ray film was developed with an intensifying screen at −70°C.
Reverse transcription-polymerase chain reaction (RT-PCR)
RT-PCR was performed to analyze the effects of BBR on DAXX mRNA expression. Total RNA was prepared from SK-N-SH or NB-1691 cells, with or without BBR treatment, using the Ultraspec RNA purification Reagent (Biotecx). RT-PCR was conducted using an Access RT-PCR Kit (Promega) according to the manufacturer’s protocols. The oligonucleotide primers that were used to analyze the DAXX transcripts were: Forward: 5′-TGAAATCCCCACCACTTCCTCCCTC-3′ and Reverse: 5′-GAGAGGCAGTGTTTTCAGCATTTGT-3′. GAPDH was used as an internal control. The RT-PCR products were visualized with ethidium bromide staining under UV light, after electrophoresis on a 1.5% agarose gel.
Western blot assay
Cells were lysed for 30 min at 4° C in a lysis buffer that was composed of 150 mM NaCl, 50 mM Tris at pH 8.0, 5 mM EDTA, 1% (v/v) Nonidet p-40, 1 mM PMSF, 20 μg/ml aprotinin and 25 μg/ml leupeptin. Equal amounts of protein extracts (10μg) were resolved by SDS-PAGE. Following transfer of the gel to a nitrocellulose filter, it was blocked for 1 h at room temperature with a buffer containing 20 mM Tris-HCl at pH 7.5, 500 mM NaCl, and 5% non-fat milk; incubated with specific antibodies for 3 h at room temperature; washed; and then incubated with a HRP-labeled secondary antibody for 1 h. Finally, the blots were developed using a chemiluminescent detection system (ECL, Amersham Life Science, Buckinghamshire, England).
Flow cytometry
Flow cytometry was performed to analyze the presence of apoptosis through quantitative detection of apoptotic cells using annexin-V staining. Cells with or without treatment were washed once with PBS and then stained with FITC-annexin-V, according to the manufacturer’s instructions. The stained cells were analyzed by FACScan (Becton Dickinson), using WinList software (Verity Software House, Inc).
RESULTS
Identification of the DAXX core promoter region
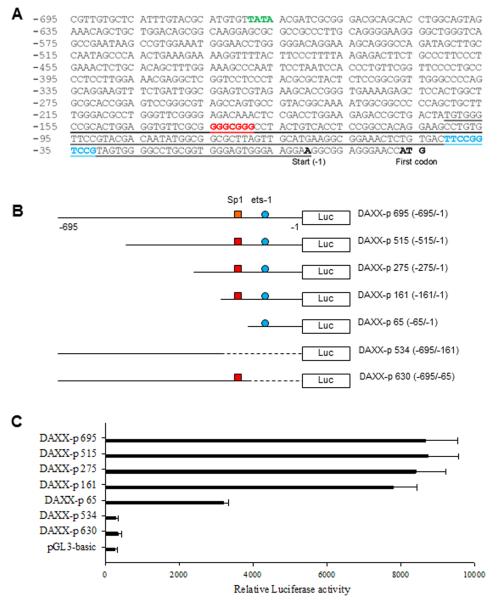
By navigating the Human Genome, we found the nucleotide sequence of the proximal 5′-flank UTR of the DAXX gene. This DNA sequence contains a TATA box (Fig. 1A), implying it has promoter activity. To determine the promoter activity of the sequence and identify the core promoter region, we constructed the full-length and a series of deleted or mutated fragments of this sequence into the pGL3 basic vector (Fig. 1B) and performed transfections and a luciferase activity assay in the neuroblastoma cell line SK-N-SH. Our results demonstrated that the sequence in the proximal 5′-flank UTR of the DAXX gene had promoter activity, as it promoted luciferase expression (Fig. 1C). As is also shown in Fig. 1C, our construct designated DAXX-p 161 expressed a maximum luciferase activity similar to that of the full-length promoter DAXX-p 695, while the 3′-5′ deleted construct DAXX-p 534 (−695/−161) showed no promoter activity, as compared with control (transfection of pGL3-basic vector), suggesting the core promoter region was located from −1 to −161. This core promoter region of the DAXX gene contains consensus sequences for the transcriptional factors Sp1 (Fig. 1A, red) and Ets1 (Fig. 1A, blue).
Fig. 1.
Characterization of the DAXX core promoter region. A, nucleotide sequence of 5′-flank UTR of the DAXX gene contains a TATA box and the Sp1 (red) and Ets1 (blue) consensus sequences in the core promoter region (underlined). The cDNA start site and the first codon (bold and black) are indicated. B, schematic representation of DAXX promoter-luciferase reporter plasmids: DAXX-p 695 containing the full length promoter with a putative Sp1 response element and an Ets-1 binding site; DAXX-p 515, DAXX-p 275, DAXX-p 161, DAXX-p 65, DAXX-p 534 and DAXX-p 630 containing a series of 5′-3′ or 3′-5′ deleted promoters. C, transient transfection and luciferase assay of DAXX promoter activity in SK-N-SH. Cells were transfected with 5 μg of each DAXX promoter construct, as shown in (A), and the PGL3 basic vector served as a control. Data represent the mean ± SD of three independent experiments normalized to RL activity (co-transfection with pRL-CMV as an internal control).
Regulation of the DAXX promoter activity by Sp1, Ets1 and BBR
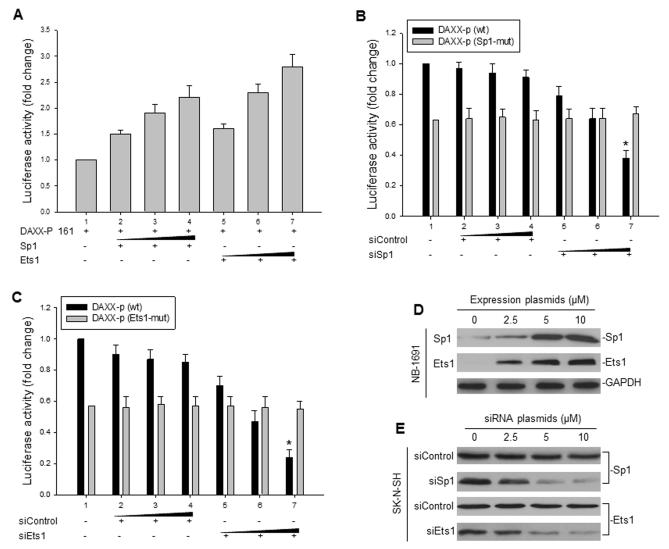
To investigate whether the DAXX promoter is regulated by Sp1 or Ets1, the DAXX promoter p161 (DAXX-p 161) luciferase construct was co-transfected with either the expression plasmid containing Sp1 or Ets1. For this, we used the neuroblastoma cell line NB-1691, which expresses either no or very low levels of Sp1 and Ets1. Fig. 2A shows that enforced expression of Sp1 or Ets1 increased the DAXX promoter activity in NB-1691 cells. In addition, we performed co-transfections and reporter assays in the neuroblastoma cell line SK-N-SH, which normally expresses high levels of Sp1 and Ets1, executing knockdown of Sp1 and Ets1 expression: Silencing of Sp1 expression by specific Sp1 siRNA decreased the DAXX promoter activity in those cells, but we found that it did not affect the activity of the DAXX core promoter when the Sp1 response element was mutated, although it expressed a relative low level of promoter activity as compared with the wt promoter (Fig. 2B). Similar results were seen in the knockdown of Ets1: Treatment with Ets1 siRNA reduced the wt DAXX core promoter activity, but not the Ets1 response element-mutated promoter activity (Fig. 2C). Similarly, the Ets1 response element-mutated promoter also expressed a lower level of activity than wt promoter. Western blot was used to detect the basal levels of endogenous Sp1 and Ets1 expression in NB-1691 and SK-N-SH cells, enforced expression of transfected Sp1 and Ets1 in NB-1691 cells and downregulation of Sp1 and Ets1 by siRNA in SK-N-SH cells (Fig. 2D and E).
Fig. 2.
Regulation of the DAXX promoter’s activity by Sp1 and Ets1. A, co-transfection and luciferase reporter assay demonstrating the effects of enforced Sp1 and Ets-1 expression on DAXX promoter activity in NB-1691 cells. Cells were co-transfected with 5 μg of the DAXX-p 161 construct with either 10 μg of neo vector as a control (lane 1) or increasing amounts (2.5, 5, and 10 μg) of Sp1 plasmid (lanes 2-4), or increasing amounts (2.5, 5, and 10 μg) of the Ets1 plasmid (lanes 5-7). Transfection and luciferase activity assays were performed as described in Fig 1. B, the effect of Sp1 knockdown on DAXX promoter activity in SK-N-SH cells. Cells were co-transfected with 5 μg wild-type DAXX-p 161 [DAXX-p (wt)], or with a variation of DAXX-p 161 that had a Sp1 binding site mutation [DAXX-p (Sp1-mut)], as well as either 10 μg of pSUPER vector as a control (lane 1), with increasing amounts (2.5, 5, and 10 μg) of siControl (lanes 2-4) or increasing amounts (2.5, 5, and 10 μg) of siSp1 (lanes 5-7) plasmids, *p<0.01. C, a similar reporter assay as in (B) for testing the effect of Ets-1 knockdown on the DAXX promoter activity in SK-N-SH. DAXX-p (Ets1-mut) was the DAXX promoter with the Ets1 binding site mutation, * p<0.01. D and E, Western blot showing the expression of transfected Sp1 and Ets1 in NB-1691 (D) and the expression of endogenous Sp1 and Ets1 in SK-N-SH cells treated with siRNA against Sp1 (siSp1) and siRNA for Ets1 (siEts1). The siControl was a control siRNA (E).
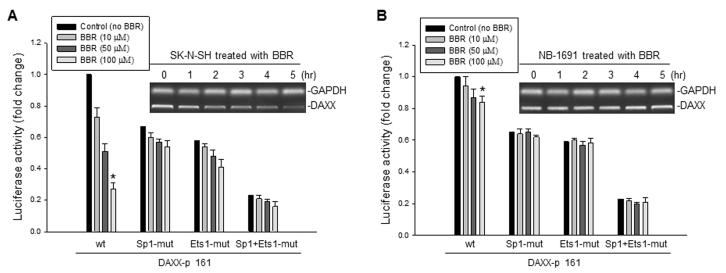
To determine whether the DAXX promoter activity can be regulated by BBR, the DAXX promoter luciferase constructs that included wt and mutations of the Sp1 or Ets1 consensus sequences, or both, were transfected into SK-N-SH or NB-1691 cells, respectively. Subsequent BBR treatment remarkably inhibited the wt DAXX promoter activity in SK-N-SH cells in a dose-dependent manner. Inhibition of the DAXX promoter activity by BBR became reduced when the Sp1 and Ets1 response elements were mutated, although they expressed lower levels of basal promoter activities due to the key response elements mutated (Fig. 3A). The expression of endogenous DAXX mRNA was also downregulated by BBR in SK-N-SH cells (Fig. 3A, insert). In contrast, both the transfected DAXX promoter activity and endogenous DAXX mRNA were not inhibited by BBR in the NB-1691 cells (Fig 3B). Because SK-N-SH cells express Sp1 and Ets1 and the NB-1691cells do not express these proteins, our results suggested that BBR-regulated inhibition of DAXX transcription will be closely associated with the presence of Sp1 and Ets1.
Fig. 3.
The effect of BBR on the DAXX promoter’s activity and on DAXX mRNA expression. A, SKN-SH cells were transfected with 5μg of various DAXX promoter luciferase plasmids (wt or mutations at the binding sites of Sp1, Ets1 or both these mutations). At 24 hr post-transfection, the cells were treated with different concentrations of BBR for another 4 hr, and then cell extracts were prepared to test for relative luciferase activity. The expression of endogenous DAXX mRNA in SKN-SH cells that were treated with 100 μM of BBR for different times, as indicated, was detected by RT-PCR (insert), * p<0.01. B, a similar assay to (A) was performed using the NB-1691 cells, * p>0.5.
BBR downregulates DAXX transcription via blocking of the binding of Sp1 and Ets1 to their promoters
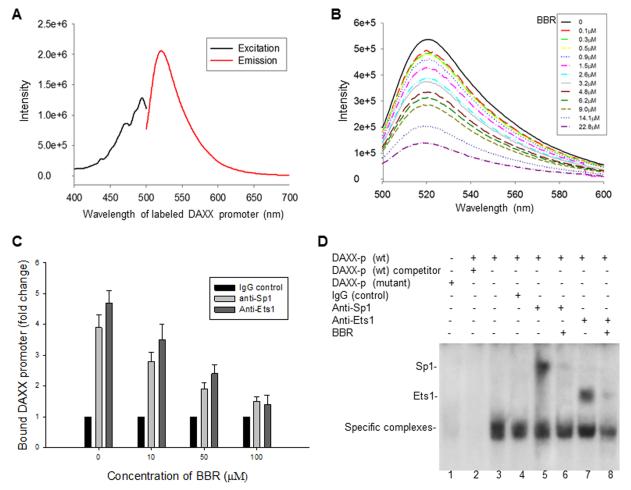
Because we found that BBR inhibits the DAXX promoter activity, we tested to see if BBR can physically interact with the DAXX promoter. We performed an in vitro fluorescence titration assay for possible binding between BBR and the DAXX promoter. The fluorescence-labeled DAXX core promoter showed an excitation of 490 nm and emission of 515 nm (Fig. 4A); and the fluorescent titration assay proved that BBR does bind to the DAXX promoter (Fig. 4B) with a Kd of 0.41 μM.
Fig. 4.
Effects of BBR on the binding activities of Sp1 and Ets1 with the DAXX promoter. A, excitation and emission spectrum of the fluorescence-labeled DAXX promoter. B, fluorescent titration assay for the binding of BBR to the DAXX promoter. C, CHIP assay to detect the effects of BBR on the binding of Sp1 and Ets1 with the DAXX promoter. Lysates from SK-N-SH cells that were treated with different concentrations of BBR were precipitated with anti-Sp1 or anti-Ets1 antibodies. Precipitation of the same lysates with a normal mouse IgG served as a control. The DNA isolated from the precipitated complexes was subjected to qPCR. D. the effects of BBR on the binding activities of Sp1 and Ets1 with the DAXX promoter, as detected by EMSA. Nuclear extracts from SK-N-SH cells, with or without BBR treatment, were incubated to form binding reactions with 32P-labeled wt or mutant probes (both Sp1 and Ets1 binding sites mutated). Samples were run on a non-denaturing 5% polyacrylamide gel and imaged by autoradiography. Lane 1: labeled mutant probe, lane 2: labeled wt probe and a 25-fold molar excess of non-labeled wt probe, lane 3: labeled wt probe only. In the reactions run in lanes 4 to 8, cellular extracts were preincubated with 2 μg of antibodies, as indicated, for 1 h at 4 °C before adding the probes. The specific protein-DNA complexes and supershifts with antibodies are indicated.
To further determine whether Sp1 and Ets1 bind to the proposed consensus sequences found within the DAXX core promoter, as well as the possible effects BBR might have on their binding abilities, we performed a CHIP analysis. After cross-linking and an immunoprecipitation with anti-Sp1 or anti-Ets1 antibodies, we performed qPCR using the primers to the fragment containing the core DAXX promoter region. Assaying these immunocomplexes showed that Sp1 and Ets1were indeed bound to the DAXX promoter and that their binding capacity was significantly reduced by BBR treatment in a dose-dependent manner (Fig. 4C).
We also performed EMSA to confirm the observed effects of BBR on the binding of both Sp1 and Ets1 to the DAXX promoter. The EMSA was carried out using nuclear extracts from SK-N-SH cells with or without BBR treatment, that were incubated in binding reactions with 32P-labeled wt or mutant probe (where both Sp1 and Ets1 binding sites were mutated). One large DNA-protein complex was specifically observed with wt probe use (Fig. 4D, lanes 3 to 8), but not with the mutant probe (Fig. 4D, lane 1). The binding of cellular extracts to the wt DAXX promoter was effectively competed for with cold wt oligonucleotide (Fig. 4D, lane 2). Results from a supershift assay with anti-Sp1 and anti-Ets1 antibodies further proved that Sp1 and Ets-1 did specifically bind to the DAXX core promoter (Fig. 4D, lanes 5 and 7, respectively). We found that BBR treatment remarkably diminished or abrogated the binding of Sp1 and Ets1 to the DAXX promoter (Fig. 4D, lanes 6 and 8).
Downregulation of DAXX by BBR results in inhibition of MDM2, followed by activation of p53 and subsequent apoptosis
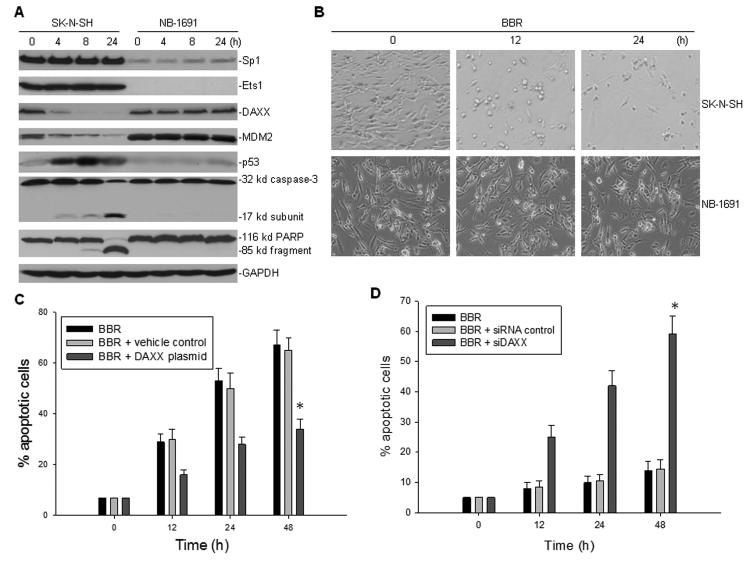
To test whether BBR inhibits DAXX transcription in SK-N-SH cells likely through downregulation of Sp1 and/or Ets1 expression, we performed Western blot assays. We detected no effect of BBR on the expression of Sp1 nor Ets1 in both SK-N-SH and NB-1691 cells (Fig. 5A). Consistent with the inhibition of DAXX transcription, expression of the DAXX protein was downregulated by BBR in SKN-SH cells, but not NB-1691 cells.
Fig. 5.
Effect of the BBR-regulated inhibition of DAXX on neuroblastoma cell growth and apoptosis. A, western blot showing the expression of proteins, as indicated, in SK-N-SH and NB-1691 neuroblastoma cells that were treated with 100 μM BBR for different times. B, representative light microscopy photographs showing cell growth of SK-N-SH and NB-1691 treated with 100 μM BBR for different times, as indicated. C, enforced expression of DAXX inhibited SK-N-SH cells to BBR. Cells transiently transfected with DAXX expression plasmid or control plasmid (vehicle) were treated with 100 μM BBR for the indicated time and apoptosis was detected by annexin V staining and flow cytometry, *p<0.01. D, knockdown of DAXX by siRNA sensitized NB-1691 cells to BBR. Cells were treated with BBR (100 μM) plus siRNA (100nM) for the indicated time and apoptosis was detected as in (C), *p<0.01.
We previously demonstrated in detail that in ALL, BBR-regulated inhibition of DAXX results in instability and degradation of the MDM2 oncoprotein, which leads to activation of p53 and apoptosis of those BBR-treated cancer cells [7]. Here, we performed similar experiments in SK-N-SH and NB-1691 neuroblastoma cells that were treated with BBR. Western blot results showed that the expression of MDM2 was indeed downregulated following BBR-induced inhibition of DAXX, which resulted in the accumulation and activation of p53 in SK-N-SH cells but not in the NB-1691 cells (Fig. 5A). Correspondingly, this observed activation of p53 led to activation of caspase-3 and cleavage of the death substrate PARP in SK-N-SH cells, but not in NB-1691 cells. BBR was able to induce obvious inhibition of cell growth (Fig. 5B) and apoptosis in the SK-N-SH cells (Fig. 5C), but not in the NB-1691 cells (Fig. 5D). Since BBR induces apoptosis of SK-N-SH cells, which is possibly due to downregulation of DAXX, we tested to see whether rescue of DAXX can inhibit SK-N-SH to BBR. We transfected DAXX expression plasmid into SK-N-SH and found that apoptosis induced by BBR was significantly reduced in SK-N-SH transfected with DAXX but not in same cells transfected with control plasmid (Fig. 5C). In contrast, knock-down of DAXX by siRNA sensitized NB-1691 to BBR. As seen in Fig. 5D, a flow-cytometry apoptosis assay revealed there was a notably increased percentage of apoptosis of NB-1691 cells treated with combined BBR and siDAXX, as compared to those receiving BBR plus a siRNA control.
DISCUSSION
In this study, we report the identification of the promoter region of the DAXX gene and analysis of the regulation of its transcriptional activity in neuroblastoma cells, as well as disruption of that regulation by RNA interference and by BBR, a natural agent with anticancer properties.
By first searching the Human Genome and then performing deletion mapping and reporter assays, we have identified the DAXX core promoter region that is located between −1 and −161 of the proximal 5′-flank UTR of the DAXX gene. We found that the core promoter of DAXX contains consensus sequences for the transcriptional factors Sp1 and Ets1, and demonstrated that both Sp1 and Ets1 indeed bound to the DAXX core promoter to stimulate its activity. Interestingly, we also found that BBR inhibits the DAXX promoter activity by blocking or disrupting the binding of both Sp1 and Ets1 to the DAXX promoter, which resulted in reduced DAXX protein expression that increased MDM2 degradation, leading to the activation of p53 and cancer cell death by apoptosis.
For the first time, we have described the promoter of the DAXX gene and the portion of its regulation that is associated with control by Sp1 and Ets1, as well as the ability of BBR to disrupt that regulation in neuroblastoma cells. For this study, we chose to use the SK-N-SH and NB-1691 neuroblastoma cell lines because in preliminary testing they had different responses to BBR treatment, although both cell lines have a similar expression of DAXX. These two cell lines express MDM2 and have a wild-type p53, which provide a good model to study the DAXX/MDM2/p53 signaling pathway in neuroblastoma. Induction of DAXX promoter activity by addition of Sp1 and Ets1 was detected in the NB-1691 cells that express no or very low levels of Sp1 and Ets1, experimentally accomplished by enforced expression of these transcriptional factors. Because the SK-N-SH cell line already expresses high levels of Sp1 and Ets1, when we enforced further expression of these proteins in gene transfection and reporter assays, we did not further increase the DAXX promoter activity (data not shown). However, knockdown of this Sp1 and Ets1 expression in SK-N-SH cells by siRNA remarkably inhibited the DAXX promoter activity, suggesting that Sp1 and Ets1 are important transactivators of DAXX. This notion was further confirmed by our CHIP and EMSA assays, as well as by mutagenesis analyses that proved the binding of Sp1 and Ets1 to the DAXX core promoter region and showed that mutations of the binding sites abrogated Sp1 and Ets1 binding and transactivation activity.
Previously, Sp1 was reported to regulate the expression of numerous genes implicated in the control of a diverse array of cellular processes, such as in cell growth [21, 22], differentiation [23] and apoptosis [24]. Ets1 is known as a proto-oncogene that was initially shown to be essential for normal development during the earlier stages of embryogenesis, while in later stages it becomes important in organ formation and tissue remodeling [25]. The oncogenic properties of Ets1 were identified in diverse cancer-related functions such as development of drug resistance and in metastasis [26-28]. Ets1 is a member of the Ets family of transcription factors that share a common 85-amino-acid DNA-binding domain [29] and are thus able to bind to unique DNA sequences, either alone or by association with other proteins [30].
Cooperative interactions between Ets1 and Sp1 have been shown to play a critical role in regulating the transcription of many genes, including CTP: phosphocholine cytidylyltransferase α [31], α11 integrin chain [32], platelet-derived growth factor D-chain [33], hepatocyte PA1 [34], β-1,4-galactosyltransferase V [35], Runx2 [36] and Guanylyl cyclase/atrial natriuretic peptide receptor [37]. Our data were consistent with the previous studies indicating that Ets1 and Sp1 are transcriptional activators that can stimulate transcription, most likely through cooperative interactions, in this case to enhance expression of the DAXX gene. From our studies, one question that remained to be answered is why the NB-1691 cell line expresses similarly basal level of DAXX as the SK-N-SH cells, while both these lines expressed such distinct levels of Sp1 and Ets1. This suggests that additional mechanisms may exist for the regulation of DAXX expression in different cancer cells such as these.
We recently reported that BBR downregulates DAXX expression at the transcriptional level in ALL [7]. Here, we further investigate the mechanisms underlying the transcriptional suppression of DAXX by BBR in the neuroblastoma cell line SK-N-SH. Although BBR and siRNAs designed specifically against Sp1 and Ets1 all similarly inhibited the DAXX promoter activity in these neuroblastoma cells, we found that BBR did not inhibit the expression of endogenous Sp1 and Ets1. Instead, it blocked or disrupted the binding of Sp1 and Ets1 to the DAXX promoter. A recent study characterized the transcriptional repression function of BBR well: BBR inhibits the association between TBP and the TATA box in the promoter of genes, suppressing transcription of any genes containing the TATA box in a non-specific way [20]. The DAXX gene promoter contains a TATA box at −669 to −666 upstream of the core promoter region. In this study, we did not test to see whether BBR suppresses DAXX transcription also, through inhibiting that TATA box function; however, our deletion mapping showed that BBR did not suppress the activity of the DNA fragment DAXX-p 534 (−695/−161), which contains a TATA box and has a deletion of the core promoter region. This experimental observation suggested that even though BBR has a role related to TATA boxes, the presence of the core promoter is apparently critical for BBR’s ability to act to inhibit DAXX transcription. Our studies, focused on the core promoter region, demonstrated a more specific functional role for BBR in the suppression of DAXX transcription via inhibition of the functionality of Sp1 and Ets1 response elements.
BBR has previously been reported to induce cancer cell apoptosis through the p53 pathway [16,38,39]. Also, we previously demonstrated that BBR induces the activation of p53 in ALL cells through a mechanism differing from that triggered by genotoxic reagents such as radiation and the chemotherapeutic drug doxorubicin. Genotoxic reagents induce an immediate activation of p53 through the ATM pathway [40], while BBR induces p53 activation via inhibition of the p53 inhibitor MDM2. BBR primarily inhibits the expression of DAXX, which results in the dissociation of MDM2 from the MDM2-DAXX-HAUSP complexes. The dissociated MDM2 protein becomes unstable due to its self-ubiquitination and degradation, leading to subsequent activation of the tumor suppressor p53, which induces cancer cell death [6,7]. In this study, we also observed that there was inhibition of MDM2 followed by activation of p53 and cell apoptosis, after there was BBR-induced downregulation of DAXX in these neuroblastoma cells. We further found that the BBR-induced neuroblastoma cell apoptosis was specifically related to the expression levels of Sp1 and Ets1 by the cells: BBR induced potent apoptosis in the SK-N-SH cell line that expresses high levels of both Sp1 and Ets1, while it induced no apoptosis in the neuroblastoma line NB-1691, which expresses no or very low levels of Sp1 and Ets1, even though this cell line has otherwise a similar MDM2 expression and a similar wt p53 status. These differential observations of cytotoxicity suggested that the BBR-regulated inhibition of DAXX transcription that occurred by its interacting with and blocking of Sp1 and Ets1 was critical for the drug’s induction of apoptosis in neuroblastoma. Due to the mechanism of action for the regulation of DAXX discovered and described herein, we expect that the drug BBR will be effective against those neuroblastomas having wt p53 and expression of Sp1 and Ets1.
Acknowledgements
This study was supported by National Institutes of Health (NIH) grants (R01 CA123490) and (R01 CA143107) and National Natural Science Foundation of China (No.81072168) and (No.30900545).
Footnotes
Conflict of interest The authors confirm that there are no conflicts of interest.
REFERENCES
- 1.Yang X, Khosravi-Far R, Chang HY, et al. DAXX, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–76. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torii S, Egan DA, Evans RA, et al. Human DAXX regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs) EMBO J. 1999;18:6037–49. doi: 10.1093/emboj/18.21.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen A, Wang PY, Yang YC, et al. SUMO regulates the cytoplasmonuclear transport of its target protein DAXX. J Cell Biochem. 2006;98:895–911. doi: 10.1002/jcb.20703. [DOI] [PubMed] [Google Scholar]
- 4.Salomoni P, Khelifi AF. DAXX: death or survival protein? Trends Cell Biol. 2006;16(2):97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Raoul C, Estévez AG, Nishimune H, et al. Motoneuron death triggered by a specific pathway downstream of Fas. Potentiation by ALS-linked SOD1 mutations. Neuron. 2002;35:1067–83. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 6.Tang J, Qu LK, Zhang J, et al. Critical role for Daxx in regulating MDM-2. Nat Cell Biol. 2006;8:855–62. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Gu L, Li J, et al. Degradation of MDM2 by the interaction between berberine and DAXX leads to potent apoptosis in MDM2-overexpressing cancer cells. Cancer Res. 2010;70:9895–904. doi: 10.1158/0008-5472.CAN-10-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang YS, Shih HM. Daxx positively modulates beta-catenin/TCF4-mediated transcriptional potential. Biochem Biophys Res Commun. 2009;386:762–8. doi: 10.1016/j.bbrc.2009.06.126. [DOI] [PubMed] [Google Scholar]
- 9.Croxton R, Puto LA, de Belle I, et al. Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-kappaB. Cancer Res. 2006;66:9026–35. doi: 10.1158/0008-5472.CAN-06-1047. [DOI] [PubMed] [Google Scholar]
- 10.Wethkamp N, Klempnauer KH. Daxx is a transcriptional repressor of CCAAT/enhancer-binding protein beta. J Biol Chem. 2009;284:28783–94. doi: 10.1074/jbc.M109.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryo A, Hirai A, Nishi M, et al. A suppressive role of the prolyl isomerase Pin1 in cellular apoptosis mediated by the death-associated protein Daxx. J Biol Chem. 2007;282:36671–81. doi: 10.1074/jbc.M704145200. [DOI] [PubMed] [Google Scholar]
- 12.Ivanchuk SM, Mondal S, Rutka JT. p14ARF interacts with DAXX: effects on HDM2 and p53. Cell Cycle. 2008;7:1836–50. doi: 10.4161/cc.7.12.6025. [DOI] [PubMed] [Google Scholar]
- 13.Schreiner S, Wimmer P, Sirma H, et al. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J Virol. 2010;84:7029–38. doi: 10.1128/JVI.00074-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diogo CV, Machado NG, Barbosa IA, et al. Berberine as a promising safe anti-cancer agent - is there a role for mitochondria? Curr Drug Targets. 2011;12:850–9. doi: 10.2174/138945011795528930. [DOI] [PubMed] [Google Scholar]
- 15.Stermitz FR, Lorenz P, Tawara JN, et al. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc Natl Acad Sci U S A. 2000;97:1433–7. doi: 10.1073/pnas.030540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi MS, Yuk DY, Oh JH, et al. Berberine inhibits human neuroblastoma cell growth through induction of p53-dependent apoptosis. Anticancer Res. 2008;28:3777–84. [PubMed] [Google Scholar]
- 17.Krey AK, Hahn FE. Berberine: complex with DNA. Science. 1969;166:755–7. doi: 10.1126/science.166.3906.755. [DOI] [PubMed] [Google Scholar]
- 18.Kuo CL, Chou CC, Yung BY. Berberine complexes with DNA in the berberine-induced apoptosis in human leukemic HL-60 cells. Cancer Lett. 1995;93:193–200. doi: 10.1016/0304-3835(95)03809-b. [DOI] [PubMed] [Google Scholar]
- 19.Bhadra K, Maiti M, Kumar GS. Berberine-DNA complexation: new insights into the cooperative binding and energetic aspects. Biochim Biophys Acta. 2008;1780:1054–61. doi: 10.1016/j.bbagen.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Kheir MM, Chai Y, et al. Comprehensive study in the inhibitory effect of berberine on gene transcription, including TATA box. PLoS One. 2011;6:e23495. doi: 10.1371/journal.pone.0023495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santiago FS, Ishii H, Shafi S, et al. Yin Yang-1 inhibits intimal thickening by repressing p21WAF1/ Cip1 transcription and p21WAF1/Cip1-Cdk4-Cyclin D1 assembly. Circ Res. 2007;101:146–55. doi: 10.1161/CIRCRESAHA.106.145235. [DOI] [PubMed] [Google Scholar]
- 23.Opitz OG, Rustgi AK. Interaction between Sp1 and cell cycle regulatory proteins is important in transactivation of a differentiation-related gene. Cancer Res. 2000;60:2825–30. [PubMed] [Google Scholar]
- 24.Tasseva G, Cole L, Vance JE. N-Myc and SP regulate phosphatidylserine synthase-1 expression in brain and glial cells. J Biol Chem. 2011;286:1061–73. doi: 10.1074/jbc.M110.158709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maroulakou IG, Papas TS, Green JE. Differential expression of ets-1 and ets-2 protooncogenes during murine embryogenesis. Oncogene. 1994;9:1551–65. [PubMed] [Google Scholar]
- 26.Kars MD, Işeri OD, Gündüz U. Drug resistant breast cancer cells overexpress ETS1 gene. Biomed Pharmacother. 2010;64:458–62. doi: 10.1016/j.biopha.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Khanna A, Mahalingam K, Chakrabarti D, et al. Ets-1 expression and gemcitabine chemoresistance in pancreatic cancer cells. Cell Mol Biol Lett. 2011;16:101–13. doi: 10.2478/s11658-010-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alipov G, Nakayama T, Ito M, et al. Overexpression of Ets-1 proto-oncogene in latent and clinical prostatic carcinomas. Histopathology. 2005;46:202–8. doi: 10.1111/j.1365-2559.2005.02059.x. [DOI] [PubMed] [Google Scholar]
- 29.Leng RX, Pan HF, Chen GM, et al. The dual nature of ETS1: focus to the pathogenesis of systemic lupus erythematosus. Autoimmun Rev. 2011;10:439–43. doi: 10.1016/j.autrev.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Li R, Pei H, Watson DK, et al. EAP1/DAXX interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene. 2000;19:745–53. doi: 10.1038/sj.onc.1203385. [DOI] [PubMed] [Google Scholar]
- 31.Sugimoto H, Okamura K, Sugimoto S, et al. Sp1 is a co-activator with Ets-1, and Net is an important repressor of the transcription of CTP:phosphocholine cytidylyltransferase alpha. J Biol Chem. 2005;280:40857–66. doi: 10.1074/jbc.M503578200. [DOI] [PubMed] [Google Scholar]
- 32.Lu N, Heuchel R, Barczyk M, et al. Tandem Sp1/Sp3 sites together with an Ets-1 site cooperate to mediate alpha11 integrin chain expression in mesenchymal cells. Matrix Biol. 2006;25:118–29. doi: 10.1016/j.matbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Liu MY, Eyries M, Zhang C, et al. Inducible platelet-derived growth factor D-chain expression by angiotensin II and hydrogen peroxide involves transcriptional regulation by Ets-1 and Sp1. Blood. 2006;107:2322–9. doi: 10.1182/blood-2005-06-2377. [DOI] [PubMed] [Google Scholar]
- 34.Nakatsuka H, Sokabe T, Yamamoto K, et al. Shear stress induces hepatocyte PAI-1 gene expression through cooperative Sp1/Ets-1 activation of transcription. Am J Physiol Gastrointest Liver Physiol. 2006;291:G26–34. doi: 10.1152/ajpgi.00467.2005. [DOI] [PubMed] [Google Scholar]
- 35.Sato T, Furukawa K. Sequential action of Ets-1 and Sp1 in the activation of the human beta-1,4-galactosyltransferase V gene involved in abnormal glycosylation characteristic of cancer cells. J Biol Chem. 2007;282:27702–12. doi: 10.1074/jbc.M611862200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Hassan MQ, Xie RL, et al. Co-stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats) J Biol Chem. 2009;284:3125–35. doi: 10.1074/jbc.M807466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar P, Garg R, Bolden G, et al. Interactive roles of Ets-1, Sp1, and acetylated histones in the retinoic acid-dependent activation of guanylyl cyclase/atrial natriuretic peptide receptor-A gene transcription. J Biol Chem. 2010;285:37521–30. doi: 10.1074/jbc.M110.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi MS, Oh JH, Kim SM, et al. Berberine inhibits p53-dependent cell growth through induction of apoptosis of prostate cancer cells. Int J Oncol. 2009;34:1221–30. [PubMed] [Google Scholar]
- 39.Kim S, Han J, Kim NY, et al. Effect of berberine on p53 expression by TPA in breast cancer cells. Oncol Rep. 2012;27:210–5. doi: 10.3892/or.2011.1480. [DOI] [PubMed] [Google Scholar]
- 40.Morgan SE, Kastan MB. p53 and ATM: cell cycle, cell death, and cancer. Adv Cancer Res. 1997;71:1–25. doi: 10.1016/s0065-230x(08)60095-0. [DOI] [PubMed] [Google Scholar]