Background: HJURP is a molecular chaperone essential for the deposition of the centromere marker CENP-A.
Results: Mis18β binds with and specifies the centromere localization of HJURP.
Conclusion: Mis18β governs centromere assembly via the Mis18β-HJURP-CENP-A axis.
Significance: Our finding reveals a novel mechanism underlying CENP-A incorporation into the centromere.
Keywords: Cell Division, Centromeres, Chromatin Structure, Histone Chaperone, Phosphorylation
Abstract
The centromere is essential for precise and equal segregation of the parental genome into two daughter cells during mitosis. CENP-A is a unique histone H3 variant conserved in eukaryotic centromeres. The assembly of CENP-A to the centromere is mediated by Holliday junction recognition protein (HJURP) in early G1 phase. However, it remains elusive how HJURP governs CENP-A incorporation into the centromere. Here we show that human HJURP directly binds to Mis18β, a component of the Mis18 complex conserved in the eukaryotic kingdom. A minimal region of HJURP for Mis18β binding was mapped to residues 437–460. Depletion of Mis18β by RNA interference dramatically impaired HJURP recruitment to the centromere, indicating the importance of Mis18β in HJURP loading. Interestingly, phosphorylation of HJURP by CDK1 weakens its interaction with Mis18β, consistent with the notion that assembly of CENP-A to the centromere is achieved after mitosis. Taken together, these data define a novel molecular mechanism underlying the temporal regulation of CENP-A incorporation into the centromere by accurate Mis18β-HJURP interaction.
Introduction
Chromosome movements in mitosis are orchestrated by dynamic interactions between spindle microtubules and the kinetochore, a multiprotein complex assembled onto centromeric DNA of the chromosome (1, 2). Centromere provides the platform for the assembly of a proteinaceous complex called the kinetochore and mediates the interaction between chromosomes and spindle microtubules. In eukaryotic organisms, the centromere is epigenetically specified by nucleosomes containing a unique histone H3 variant, CENP-A, with the exception of budding yeast (1–4). CENP-A orthologues present in all known eukaryotes. It is the most important epigenetic marker of the centromere (5–9) and is required for the correct localization of most centromere- and kinetochore-related proteins (5, 10–17). Very recently, Cleveland and colleagues (17) unambiguously demonstrated that CENP-A is the epigenetic marker to maintain and propagate the centromere identity and function through gene targeting in human cells and fission yeast. Biochemically, the centromere-targeting domain of CENP-A is sufficient to direct its centromere incorporation, whereas the assembly of a functional kinetochore requires the N-terminal and C-terminal tails of CENP-A (17).
Unlike typical histone proteins, the deposition of CENP-A to the centromere occurs in early G1 phase (18). Several proteins have been reported to be essential to the deposition and maintenance of CENP-A, including CENP-C, CENP-N, RbAp46/48, Mis18 complex (Mis18α, Mis18β, and Mis18BP1), RSF (remodeling and spacing factor) complex, and HJURP3 (3, 19–28). Among these proteins, HJURP is of great importance for CENP-A deposition. In yeast, the HJURP homologue Scm3 is required for the deposition of centromeric Cse4 nucleosome (29–33). Down-regulation of HJURP impairs the deposition of newly synthesized CENP-A in mammalian cells (26–27). As a molecular chaperone of CENP-A, HJURP forms a prenucleosomal complex with the CENP-A/H4 heterodimer via its highly conserved Scm3 domain (25, 34–36). However, although the Scm3 domain is sufficient to facilitate the formation of CENP-A nucleosomes in vitro, the Scm3 domain itself is not sufficient to localize on the centromere (22). The Mis18 complex, composed of Mis18α, Mis18β, and Mis18BP1, is required for the deposition of CENP-A (19–20). A recent study showed that the centromere localization of HJURP is dependent on the Mis18 complex (22). However, the mechanism by which Mis18 directs HJURP recruitment to centromere is still unclear.
In this study, we report that Mis18β is a novel binding partner of HJURP. A minimal region of HJURP (residues 437–460) is required for the direct interaction with Mis18β. A central fragment of HJURP (residues 401–490) is sufficient for the centromere recruitment of HJURP. In contrast, a mutant of HJURP lacking residues 437–460 failed to localize on the centromere. Depletion of Mis18β by siRNA perturbed the recruitment of HJURP to the centromere. Furthermore, we demonstrated that phosphorylation of HJURP by CDK1 reduced its binding affinity to Mis18β. Our findings provide novel insights into how centromere deposition of CENP-A occurs in a cell cycle-dependent manner.
MATERIALS AND METHODS
Plasmid Construction
The cDNA of RbAp46, Mis18α, and CENP-N were amplified from the human testis cDNA library (Clontech) using a standard PCR protocol. The cDNA of Mis18β, Mis18BP1 (Knl-2), and RbAp48 were purchased from GeneCopoeia. The cDNA of HJURP was purchased from Proteintech Group. The cDNA of CENP-C was a gift from Dr. Andrea Musacchio (European Institute of Oncology, Milan, Italy). Constructs of AD and BD fusion proteins were generated by subcloning the indicated cDNAs into pGADT7 and pGBKT7 (Clontech), respectively. Constructs of 3×FLAG, GFP, GST, and His6-tagged proteins were generated by subcloning the indicated cDNAs into p3XFLAG-myc-CMV-24 (Sigma), pEGFP-C1 (Clontech), pGEX-5X-3 (GE Healthcare), and pET-28a (Novagen), respectively.
To generate mCherry-Mis18β, cDNA of GFP in GFP-Mis18β was replaced by cDNA of mCherry. All constructs of HJURP truncations were generated by ligating restriction enzyme-digested PCR products and corresponding vectors. Point mutations were introduced using the QuikChange site-directed mutagenesis kit (Agilent). All plasmids were verified by sequencing.
Yeast Two-hybrid Assay
Yeast two-hybrid assays were performed as described previously (37, 38). Briefly, the plasmids of BD-tagged protein and AD-tagged protein were co-transformed into yeast strain AH109. The co-transformed yeast were grown on SD medium with X-α-Gal but lacking Leu, Trp, His, and Ade.
Recombinant Protein Production and Pull-down Assays
Recombinant protein preparation and pull-down assays were performed as described (39). Briefly, the GST fusion protein in bacteria in the soluble fraction was purified by using glutathione-agarose chromatography and chromatography, and histidine-tagged protein was purified using nickel-nitrilotriacetic acid-agarose beads. In the purification of GST-tagged proteins and pull-down assays, we used PBS containing 0.2% Triton X-100.
λ-Phosphatase Treatment
Mitotic cells were lysed in 1× NEBuffer for PMP (50 mm HEPES, 100 mm NaCl, 2 mm dithiothreitol, 0.01% Brij 35, pH 7.5) supplemented with 1 mm MnCl2, complete protease inhibitor (Roche Applied Science), and 0.2% Triton-X-100 on ice for 15 min. Then the cell lysate was treated with λ-phosphatase (New England Biolabs) for 30 min at 30 °C.
Immunoprecipitation
For FLAG immunoprecipitation, 293T cells transiently transfected to express FLAG fusion proteins were lysed in IP buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, protease inhibitors (Roche Applied Science), 0.2% Triton X-100) on ice for 10 min. The cell lysates were centrifuged at 12,000 × g for 15 min. To prepare nuclear extract, cells were homogenized in 0.1× IP buffer. Homogenized extracts were centrifuged at 350 × g for 5 min. The pellet was suspended in IP buffer, sonicated, and centrifuged at 12,000 × g for 15 min to produce a chromatin-free extract. Cell lysates or nuclear extracts were incubated with anti-FLAG M2 affinity beads (Sigma) at 4 °C for 3 h. After washing with IP buffer three times, proteins associated with beads were eluted by boiling in SDS sample buffer. Protein samples were separated by SDS-PAGE and analyzed by Western blotting. For GFP tag immunoprecipitation, GFP-Trap_A (Chromotek) was used to incubate with cell lysate of GFP-HJURP stable cell line in 1× NEBuffer for PMP (New England Biolabs).
In Vitro Kinase Assay
Recombinant GST-HJURP-C1 and GST-HJUP-C1-S3A were incubated with 2 units of CDK1-cyclin B (New England Biolabs) in kinase buffer (50 mm Tris-HCl, 10 mm MgCl2, 1 mm EGTA, 2 mm dithiothreitol, 0.01% Brij 35, pH 7.5) containing 100 μm ATP at 30 °C for 30 min. Protein samples were separated by SDS-PAGE. The phosphorylated proteins were detected by staining the gel by the Diamond Phosphoprotein Gel Stain Kit (Invitrogen). Then the level of protein was visualized by Coomassie Blue staining of the same gel.
Cell Culture and Cell Cycle Synchronization
HeLa and 293T cells, from the American Type Culture Collection (Manassas, VA) were maintained as subconfluent monolayers in Dulbecco's modified Eagle's medium (Invitrogen) with 10% FBS (Hyclone, Logan, UT) and 100 units/ml penicillin plus 100 μg/ml streptomycin (Invitrogen) at 37 °C with 8% CO2. To obtain the cell lines stably expressing GFP-HJURP and GFP-HJURP-S3D, respectively, HeLa cells were transfected with GFP-HJURP or GFP-HJURP-S3D plasmid using Lipofectamine2000 (Invitrogen). Both cell lines were selected in complete medium containing 800 μg/ml G418 and then maintained in complete medium containing 200 μg/ml G418. For synchronization in prometaphase, HeLa cells were treated for 16 h with 50 ng/ml nocodazole. For synchronization in early G1 phase, HeLa cells were treated for 16 h with 2 mm thymidine, washed three times with PBS, and released in complete medium for 8 h and then assayed.
Transfection and siRNA Treatment
Plasmids were transfected into HeLa and 293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The following siRNAs were purchased from Qiagen: si-HJURP (SI00121674); si-Mis18α (5′-CAGAAGCUAUCCAAACGUGTT-3′); and si-Mis18β (5′-AGGCAGUACUUACAACCUUTT-3′). A scramble RNA duplex (5′-UUCUCCGAACGUGUCACGU-3′) was used as a control. All siRNAs were transfected into HeLa cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Antibodies
The following antibodies were used: anti-FLAG-M2 (Sigma-Aldrich, F1804; 1:1,000); anti-α-tubulin (DM1A, Sigma-Aldrich T9026; 1:5,000); human anti-centromere antibody (ACA; a gift from Dr. Don Cleveland, University of California at San Diego, La Jolla, CA; 1:500); anti-Mis18α (Abcam; 1:1000); anti-Mis18β (Proteintech; 12142-1-AP; 1:500); anti-HJURP (Proteintech; 15283–1-AP; 1:500); anti-GFP mAb (Sigma; G6539; 1:1,000); anti-GST (Cell Signaling; 2625; 1:1,000); anti-CENP-A (Cell Signaling; 2186; 1:100); and MPM-2 (Millipore; 05-368; 1:500).
Immunofluorescence Microscopy and Live Cell Imaging
HeLa cells were grown on coverslips and fixed with 3.7% (v/v) formaldehyde (Sigma) in PBST (PBS containing 0.2% Triton X-100) for 10 min. After fixation, cells were blocked with BSA (1% in PBS) for 1 h. Then cells were incubated with primary antibody for 1 h. After three washes in PBST, cells were incubated with secondary antibody for 1 h, followed by DNA staining with DAPI for just 1 min. After three washes in PBS, coverslips were mounted in Vecta-Shield medium. Secondary antibodies used here were rhodamine-conjugated goat anti-mouse IgG (Jackson ImmunoResearch; 1:400), rhodamine-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch; 1:400), and Alexa647-conjugated goat anti-human (IgG + IgM) (Molecular Probes; 1:400).
Deconvolution images were collected using a Deltavision wide field deconvolution microscope system built on an Olympus IX-71 inverted microscope base. For imaging, a ×100, 1.35 numerical aperture lens was used, and optical sections were taken at intervals of 0.3 μm. Images for display were generated by projecting single optical sections as described previously (38).
For live cell imaging, cells were cultured in MatTek glass-bottom microwell dishes (MatTek) in CO2-independent medium (Invitrogen) containing 10% (v/v) FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. A z-series of three images at 0.5-μm intervals was acquired every 5 min using the same equipment. Image stacks were deconvolved and projected with SoftWorx (Applied Precision) and mounted onto figures with Photoshop and Illustrator (Adobe).
Fluorescence Intensity Quantification
Quantification of the level of centromere-localizing signal was conducted as described by Liu et al. (37). In brief, the average pixel intensities from at least 100 centromeres from 10 cells were measured, and background pixel intensities were subtracted. The pixel intensities at each centromere were then normalized against ACA pixel values to account for any variations in staining or image acquisition. The values of specific siRNA-treated cells were then plotted as a percentage of the values obtained from cells transfected with a control scramble siRNA duplex. The values of GFP-HJURP full-length and deletion mutant were expressed as arbitrary units.
Data Analyses
To determine significant differences between means, unpaired t tests assuming unequal variance were performed; differences were considered significant when p was <0.05.
RESULTS
Identifying Mis18β as a Binding Partner of HJURP
To identify proteins that recruit HJURP to centromeres through direct interaction, we tested the interaction between HJURP and factors that are essential for CENP-A deposition and maintenance via a yeast two-hybrid assay because most recombinant centromeric proteins are either less soluble or unstable (37, 38). As shown in Fig. 1A, our assay showed that HJURP interacts with Mis18β but not Mis18α, Mis18BP1, RbAp46, RbAp48, CENP-C, and CENP-N. To confirm this interaction, a GST pull-down assay using purified GST-HJURP and His-Mis18β was performed (Fig. 1B). Indeed, HJURP directly binds to Mis18β in vitro, as judged by Western blotting of Mis18β in the GST-HJURP-bound fraction (Fig. 1B, bottom, lane 5). No Mis18β was retained on GST-Sepharose beads, suggesting that the interaction between Mis18β and HJURP is specific. To test if Mis18β and HJURP form a cognate complex, we carried out immunoprecipitation using cell lysates from 293T cells transiently transfected to express FLAG-HJURP protein. As shown in Fig. 1C, FLAG immunoprecipitation successfully pulled down FLAG-HJURP based on Western blotting using an anti-FLAG antibody (top, lane 4). Western blotting analysis with an anti-Mis18β antibody confirmed the existence of Mis18β protein in the FLAG-HJURP immunoprecipitates (Fig. 1C, bottom, lane 4), confirming the existence of HJURP-Mis18β complex in cells.
FIGURE 1.
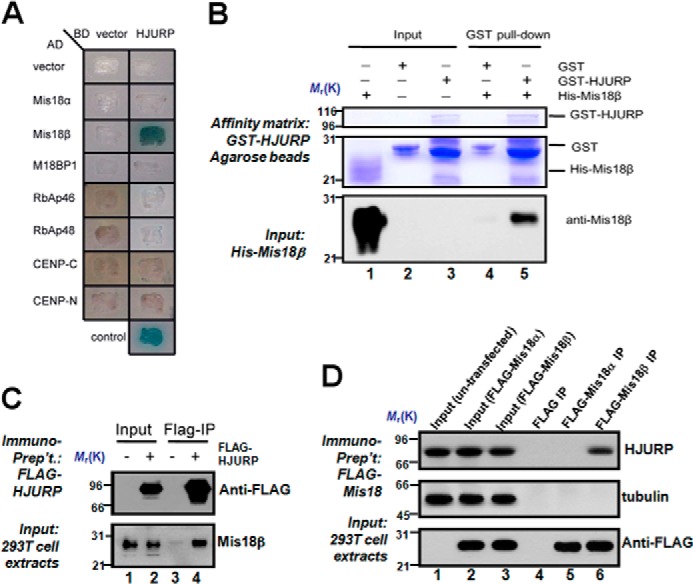
HJURP interacts with Mis18β in vitro and in vivo. A, BD-HJURP, together with different AD constructs, were co-transformed into yeast cells and cultured on a selective plate containing X-α-Gal but lacking uracil, tryptophan, leucine, and histidine. Control is the cells cotransformed with BD-p53 and AD-T as a positive control. B, GST-HJURP directly binds to His-Mis18β in vitro. Purified His-Mis18β was incubated with purified GST-HJURP immobilized on glutathione-agarose. After exclusive wash, proteins associated with glutathione-agarose were separated by SDS-PAGE and analyzed by Coomassie Blue staining or Western blotting with anti-Mis18β antibody. C, nuclear extracts from 293T cells expressing FLAG-HJURP were subjected to anti-FLAG immunoprecipitation followed by Western blotting using antibodies against Mis18β and FLAG, respectively. Lysate of untransfected 293T cells was used as a negative control. D, nuclear extracts from 293T cells expressing FLAG-Mis18α or FLAG-Mis18β were subjected to anti-FLAG immunoprecipitation followed by Western blotting using antibodies against HJURP, tubulin, and FLAG, respectively. Lysate of untransfected 293T cells was used as a negative control (lane 1).
There are two Mis18 isoforms (α and β) in mammalian cells (19). To probe whether HJURP forms a cognate complex with Mis18 protein in an isoform-selective manner, we carried out immunoprecipitation using lysates from 293T cells transiently transfected to express FLAG-Mis18α and FLAG-Mis18β, respectively. As shown in Fig. 1D, both FLAG-Mis18α and FLAG-Mis18β proteins expressed at a comparable level in 293T cells judged by Western blotting analysis (bottom, lanes 2 and 3). Anti-FLAG immunoprecipitation brought down equivalent amount of FLAG-Mis18α and FLAG-Mis18β proteins (bottom, lanes 5 and 6). As predicted, HJURP protein is only present in the Mis18β immunoprecipitates (top, lane 6), which is consistent with the outcome from an early yeast two-hybrid assay. The absence of tubulin protein in any of the FLAG immunoprecipitates confirmed the specificity of the immunoprecipitation. Thus, we conclude that HJURP directly binds to Mis18β and forms a cognate complex with Mis18β in cells.
Mapping the Binding Interface between HJURP and Mis18β
To delineate the structure-functional relationship of the HJURP-Mis18β interaction, we sought to ask which region of HJURP is required for interaction with Mis18β. To this end, a series of HJURP truncations as illustrated in Fig. 2A were generated, and then the yeast two-hybrid assay was carried out using Mis18β as bait. HJURP-C (residues 401–748) mediates the interaction with Mis18β (Fig. 2B). In contrast, HJURP-N (residues 1–400) interacts with Mis18β weakly in our assay. Next, a GST pull-down assay was carried using GST-Mis18β as affinity matrix to absorb lysates of cells expressing different GFP-HJURP truncations. Indeed, Mis18β pulls down HJURP-C strongly (Fig. 2C, lane 15). On the contrary, it pulls down HJURP(1–200) less efficiently (Fig. 2C, lane 12). To pinpoint the minimal region of HJURP required for binding to Mis18β, interactions of Mis18β with a series of smaller HJURP fragments were tested via a yeast two-hybrid assay. The results showed that the minimal region of HJURP required for binding to Mis18β is the fragment encompassing residues 401–490 of HJURP (Fig. 2D). In addition, both C2a(421–580) and C2b(437–580) fragments interact with Mis18β. However, the C2c(461–580) fragment failed to interact with Mis18β (Fig. 2D). This result suggests that the region of amino acids 437–460 is essential for HJURP-Mis18β interaction. Consistent with our prediction, deletion of residues 437–460 from HJURP disrupted HJURP-Mis18β interaction (Fig. 2E). However, deleting residues 437–446 or 447–460 from HJURP did not disrupt HJURP-Mis18β interaction. Thus, we conclude that the region containing residues 437–460 of HJURP is the minimal region required for its interaction with Mis18β.
FIGURE 2.
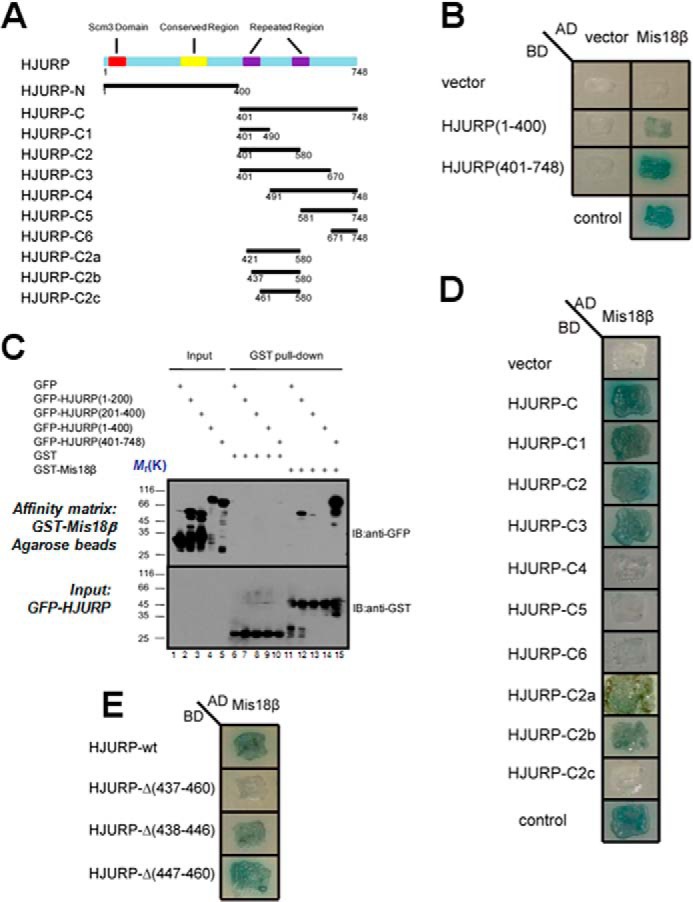
Identification of structural determinant of HJURP essential for binding to Mis18β. A, schematic representations of the domain organization of HJURP and constructs of truncations. B, AD-Mis18β, together with two BD constructs as indicated, were co-transformed into yeast cells and cultured on a selective plate containing X-α-Gal but lacking uracil, tryptophan, leucine, and histidine. Control is the cells co-transformed with BD-p53 and AD-T as a positive control. C, purified GST-Mis18β was immobilized on glutathione-agarose and incubated with extracts of 293T cells expressing GFP-tagged HJURP fragments. After extensive wash, proteins associated with glutathione-agarose were eluted and separated by SDS-PAGE followed by transfer to nitrocellulose membrane for Western blotting analyses (IB) with anti-GFP and anti-GST antibody, respectively. D and E, AD-Mis18β, together with different BD constructs of different HJURP truncations, were co-transformed into yeast cells and cultured on selective plate containing X-α-Gal but lacking uracil, tryptophan, leucine, and histidine. Control is the cells co-transformed with BD-p53 and AD-T as a positive control. Note that deletion of 24 amino acids (aa 437–460) from HJURP abolished the interaction between HJURP and Mis18β.
HJURP Is Recruited to Centromere via a Direct Binding to Mis18β
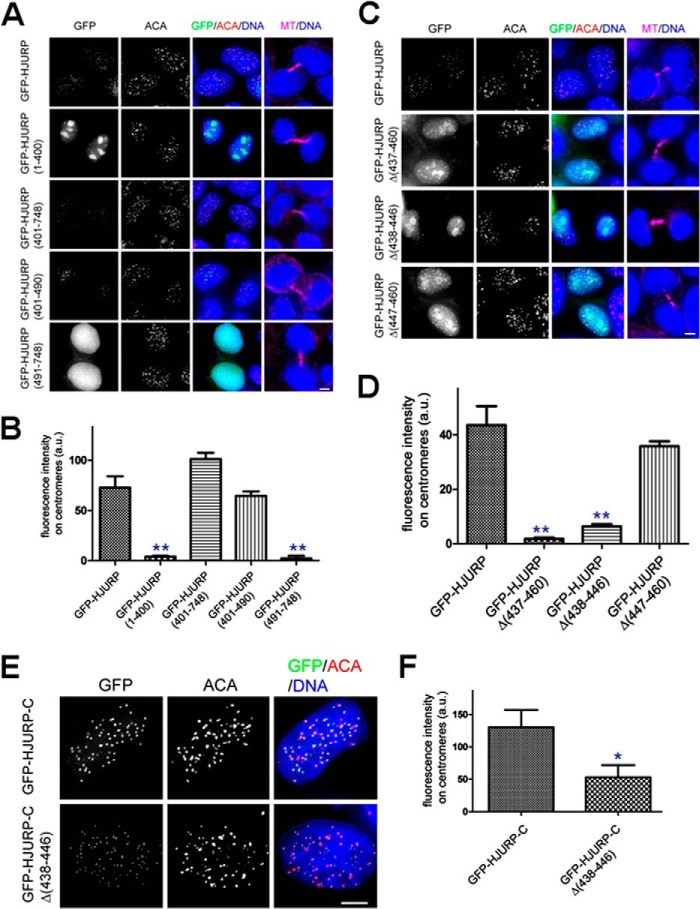
To map the region of HJURP responsible for its centromere localization, GFP-tagged HJURP truncations were transiently transfected to express in HeLa cells, and their localization was examined by immunofluorescence microscopy. GFP-tagged full-length HJURP localizes on centromeres in early G1 phase (Fig. 3A). Consistent with a recent report (40), the C-terminal HJURP(401–748) localizes to centromeres (Fig. 3A), whereas the N-terminal HJURP(1–400) failed to localize to centromeres (Fig. 3A). Interestingly, HJURP-C1(401–490), the fragment essential for binding Mis18β, is capable of localizing to the centromere (Fig. 3A; see Fig. 3B for quantitative analysis). However, the HJURP-C4 fragment (aa 491–748) failed to localize to centromeres. Collectively, the deletion analyses demonstrate that the deletion mutant HJURP-C1 (aa 401–490) is essential and sufficient for localizing HJURP to the centromere. Because our biochemical characterization demonstrated that the HJURP-C1 fragment of HJURP physically interacts with Mis18β, we reason that HJURP is recruited to the centromere through a direct interaction with Mis18β. If this is true, mutation of HJURP unable to bind Mis18β would not be able localize to the centromeres. Consistent with our speculation, an immunofluorescence assay demonstrated that HJURP deletion mutant (HJURPΔ437–460) failed to localize onto centromeres (Fig. 3, C and D), which is consistent with the outcome from our yeast two-hybrid assay (Fig. 2E).
FIGURE 3.
Characterization of the structural determinants underlying localization of Mis18β to the centromere. A, representative immunofluorescence images of early G1 cells expressing wild type GFP-HJURP and different HJURP deletion mutants. B, bar graph showing quantification of GFP-HJURP deletion mutant fluorescence intensities at centromeres for immunofluorescence assay described in A. Error bars, S.E. More than 70 centromeres were quantified per condition. **, p < 0.01 compared with those of full-length GFP-HJURP-expressing cells from three independent experiments. C, representative immunofluorescence images of early G1 cells expressing wild type GFP-HJURP and different HJURP truncation mutants. Briefly, 36 h after transfection, cells were fixed and co-stained for ACA (red), microtubule (magenta), and DNA (blue). Cells with midbody were selected to show they were in early G1 phase. Scale bar, 5 μm. D, bar graph showing quantification of GFP-HJURP mutant fluorescence intensities at centromeres for immunofluorescence assay described in C. Error bars, S.E. More than 70 centromeres were quantified per condition. **, p < 0.01 compared with those of full-length GFP-HJURP-expressing cells from three independent experiments. E, representative immunofluorescence images of early G1 cells expressing different GFP-HJURP constructs. 36 h after transfection, cells were fixed and co-stained for ACA (red) and DNA (blue). Scale bar, 5 μm. F, bar graph showing quantification of GFP-HJURP mutant fluorescence intensities at centromeres for the immunofluorescence assay described in E. Error bars, S.E. More than 70 centromeres were quantified per condition. *, p < 0.05 compared with those of GFP-HJURP-C-expressing cells from three independent experiments. a.u., arbitrary units.
Interestingly, our yeast two-hybrid assay showed that a shorter deletion, such as HJURPΔ447–460 or HJURPΔ438–446, remains active in binding to Mis18β (Fig. 2E). Because GFP-HJURPΔ447–460, but not GFP-HJURPΔ438–446, is localized on centromeres, (Fig. 3, C and D), we conclude that the region containing amino acids 438–446 is critical for HJURP localization via its interaction with Mis18β. Interestingly, deletion of residues 438–446 from HJURP-C did not completely abolish its ability to localize to the centromere, although its fluorescence intensity was significantly impaired (Fig. 3, E and F; p < 0.01).
Given the possibility that HJURP can form dimer (40), we tested whether the low level centromere localization of GFP-HJURPΔ437–460 is due to heterodimerization between endogenous HJURP and exogenous GFP-HJURPΔ437–460. To this end, we carried out a co-immunoprecipitation assay using lysates from 293T cells transiently transfected to express FLAG-HJURP and deletion mutants of GFP-HJURP. As shown in Fig. 4A, the removal of amino acids 437–460 from GFP-HJURP did not alter its interaction with FLAG-HJURP (Fig. 4A, lane 7). Thus, we conclude that HJURP dimerization facilitates its localization to centromere.
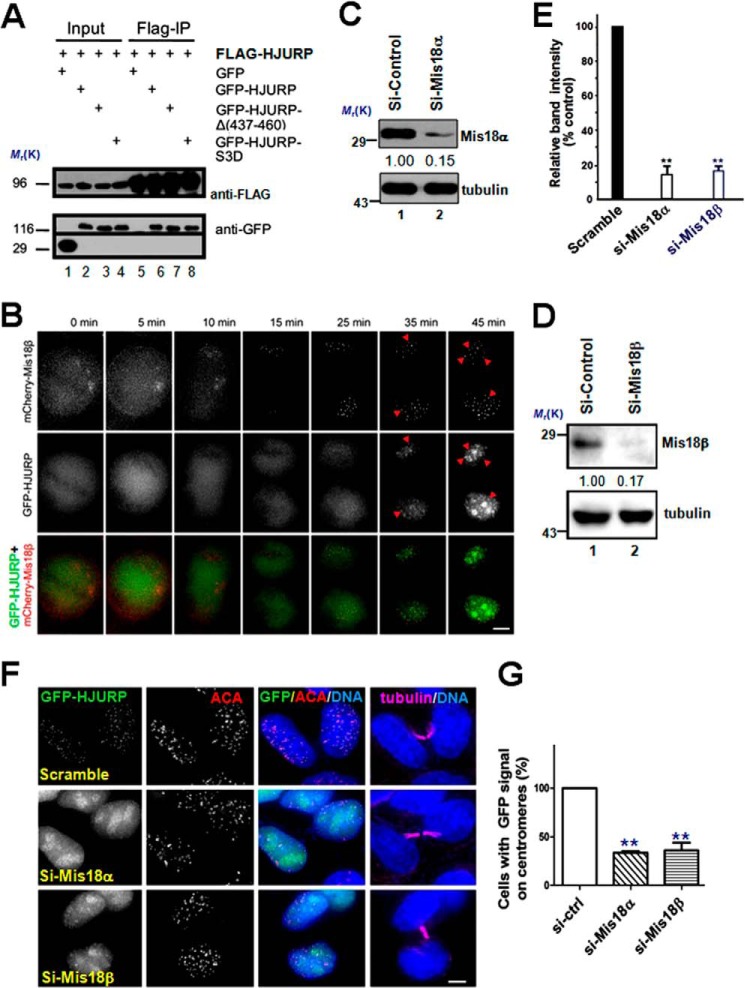
FIGURE 4.
HJURP is recruited to centromeres through binding to Mis18β. A, extracts of 293T cells co-expressing FLAG-HJURP and different GFP-HJURP constructs as indicated were subjected to anti-FLAG immunoprecipitation and then followed by immunoblotting using anti-FLAG and anti-GFP antibody, respectively. B, representative image stills of live cell imaging of HeLa cell co-expressing GFP-HJURP and mCherry-Mis18β. The arrowheads pinpoint the co-localization sites. Scale bar, 5 μm. C, immunoblots of lysates from cells transfected with siRNA against Mis18α or a scramble sequence siRNA (a negative control; lane 1) probed with the indicated antibodies. 48 h after siRNA transfection, cells were collected and assayed by Western blotting of Mis18α and tubulin, respectively. D, immunoblots of lysates from cells transfected with siRNA against Mis18β or a scramble sequence siRNA (a negative control; lane 1) probed with the indicated antibodies. 48 h after siRNA transfection, cells were collected and assayed by Western blotting of Mis18β and tubulin, respectively. E, quantification of Mis18α and Mis18β protein suppression by siRNA. Values represent the means ± S.E. (error bars) from three independent experiments (**, p < 0.01 compared with that of scramble-transfected cells). F, Representative images of HeLa cells stably expressing GFP-HJURP cells treated with different siRNAs, as indicated. 48 h after transfection, cells were fixed and co-stained for ACA (red), microtubule (magenta), and DNA (blue). Cells with midbody were selected to show they were in early G1 phase. Scale bar, 5 μm. G, bar graph showing the quantification of percentage of cells with GFP-HJURP-positive centromeres for siRNA treatments described in F. The data represent mean ± S.E. of three independent experiments. More than 53 early G1 phase cells (midbody-positive) were analyzed per condition. **, p < 0.01 compared with that of scramble transfected cells.
Both Mis18 complex assembly and HJURP localization to the centromere occur in late mitosis and early G1 phase of the following cycle (20, 26–27). If Mis18β recruits HJURP to centromeres, Mis18 complex should concentrate to centromeres before HJURP. Consistent with an earlier report (27), live cell imaging showed that mCherry-Mis18β started to appear at the centromeres during anaphase B. However, GFP-HJURP exhibits a diffused localization in cytoplasm (Fig. 4B), demonstrating the temporal order of centromere localization. Thus, the centromere loading sequence of Mis18 complex and HJURP is consistent with our hypothesis that HJURP is recruited to centromeres by Mis18β.
To test the requirement of Mis18β for HJURP centromere targeting, we sought to suppress Mis18α and Mis18β protein levels using siRNA. As shown in Fig. 4, C and D, both siRNAs against Mis18α and Mis18β effectively suppress the protein expression of Mis18α and Mis18β, respectively. Quantitative analyses show that our siRNA protocol gives typically ∼87 ± 5% and 83 ± 4% reduction in the amount of Mis18α and Mis18β protein, respectively, although the levels of tubulin showed no fluctuation in siRNA-treated cells (Fig. 4E; p < 0.01). When we treated GFP-HJURP stable cell line with siRNA against Mis18α or Mis18β, the centromere localization of HJURP largely disappeared (Fig. 4, F and G). Because Mis18α, Mis18β, and Mis18BP1 are mutually dependent for their centromeric localization (20), it is not surprising that depletion of Mis18α or Mis18BP1 exhibits the same perturbation effect on HJURP localization that was also seen in the Mis18β-depleted cells. Thus, we conclude that HJURP is recruited to centromeres by the Mis18 complex through its direct interaction with Mis18β.
Regulation of HJURP to Centromeres by CDK1 Phosphorylation
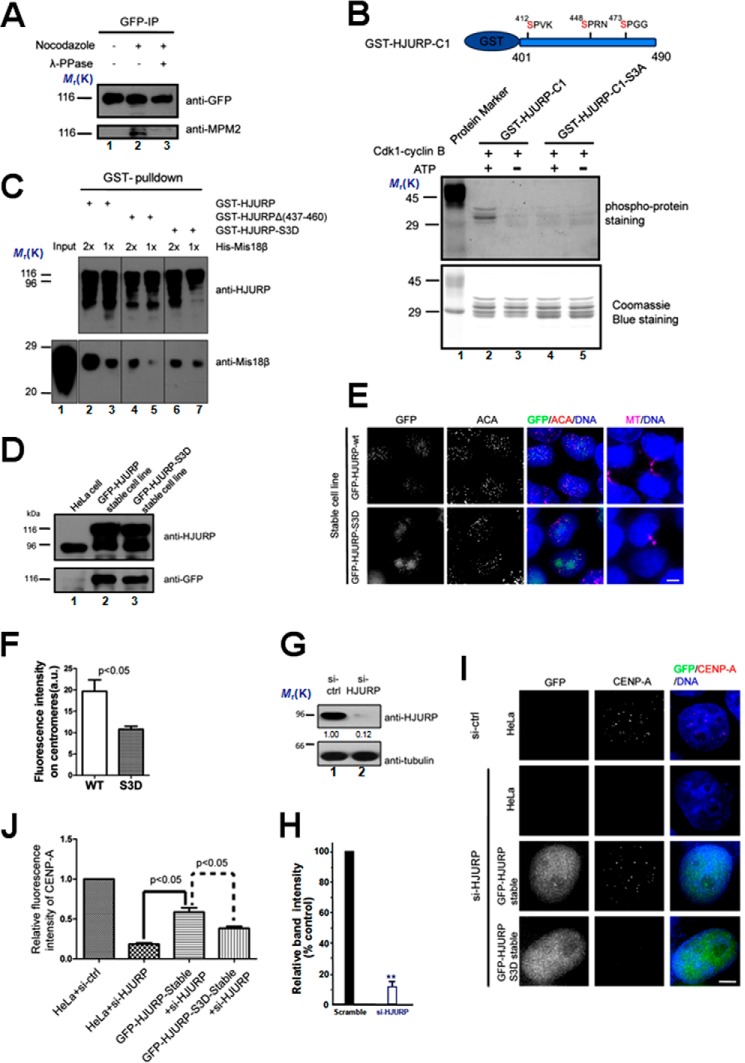
It has been reported that the temporal restriction of CENP-A incorporation is mediated by CDK1 activity. One possible molecular pathway of this restriction is phosphorylation of Mis18BP1 inhibiting its centromere recruitment via CENP-C (41–42). Because HJURP plays a key role in CENP-A incorporation, its recruitment to centromeres may also be temporally controlled by CDK1 activity. Using a mitotic phosphorylation-specific antibody, MPM2, we showed that HJURP is highly phosphorylated in mitosis (Fig. 5A). A phosphoproteome screen has identified at least eight phosphorylation sites of HJURP during mitosis (43). Three phosphorylation sites that conform to CDK1 consensus sites (Ser-412, Ser-448, and Ser-473) localized in the HJURP-C1 (aa 401–490) fragment. Because HJURP-C1 (aa 401–490) mediates interaction with Mis18β, we hypothesize that CDK1 may control the temporal interaction between HJURP and Mis18β through phosphorylation of these sites. To address this possibility, an in vitro kinase assay using purified GST-HJURP(401–490) as substrate was performed (Fig. 5B). Our data showed that CDK1 can phosphorylate GST-HJURP(401–490) in vitro. When Ser-412, Ser-448, and Ser-473 were mutated to alanines, phosphorylation of HJURP-3A was undetectable (Fig. 5B, top panel, lane 4). To examine if the phosphorylation of HJURP affects the binding between HJURP and Mis18β, different GST-tagged HJURP constructs were used as affinity matrix to pull down His-Mis18β. Indeed, compared with wild type HJURP, HJURP-3D, in which Ser-412, Ser-448, and Ser-473 were mutated to aspartic acid to mimic the phosphorylation state, pulled down a much lesser amount of Mis18β (Fig. 5C).
FIGURE 5.
HJURP recruitment to centromeres is regulated by CDK1. A, immunoprecipitation was performed using GFP antibody-coupled Sepharose beads using lysates from HeLa cells stably expressing GFP-HJURP treated as indicated. Nocodazole was added to block cells in mitosis. Extract of mitotic cells were treated with λ-phosphatase (λ-PPase) before immunoprecipitation to dephosphorylate proteins. The immunopurified products were probed with anti-GFP antibody and MPM2 antibody, respectively. B, an in vitro kinase assay was performed using CDK1 as kinase and purified GST-tagged HJURP fragment or the fragment harboring point mutations as illustrated above. The top panel shows the gel staining with a Pre-Q diamond phosphor-protein staining kit. The bottom panel shows Coomassie Blue-stained gel. C, different purified GST fusion HJURP mutants were used as affinity matrix to pull down purified His-Mis18β. The pull-downs were examined by anti-HJURP and anti- Mis18β antibody, respectively. D, immunoblots of lysates from normal HeLa cells, HeLa cells stably expressing GFP-HJURP-WT, and HeLa cells stably expressing GFP-HJURP-3D probed with the indicated antibodies. E, representative images of cells stably expressing GFP-HJURP or GFP-HJURP-S3D. Cells were fixed and co-stained for ACA (red), microtubule (magenta), and DNA (blue). Cells with midbody were selected to show that they were in early G1 phase. Scale bar, 5 μm. F, bar graph showing quantification of GFP fluorescence intensity at centromeres for the immunofluorescence assay described in D. The data represent mean ± S.E. (error bars) of three independent experiments. More than 70 centromeres (from 10 cells) were quantified for each group. Student's t test was used to calculate the p value for comparison of HJURP-3D and HJURP-WT. G, immunoblots of lysates from cells transfected with siRNA against HJURP or a scramble sequence siRNA (a negative control) probed with the indicated antibodies. 48 h after siRNA transfection, cells were collected and assayed by Western blotting. H, quantification of HJURP protein suppression by siRNA. Values represent the means ± S.D. (error bar) from three independent experiments. I, representative images of different cells as indicated treated with a control siRNA (si-ctrl) or siRNA against HJURP twice. 96 h after transfection, cells were fixed and stained for CENP-A (red) and DNA (blue). Scale bar, 5 μm. J, bar graph showing quantification of CENP-A fluorescence intensity at centromeres for siRNA treatments described in I. The data represent mean ± S.E. of three independent experiments. More than 200 centromeres (from 20 cells) were quantified per condition. Student's t test was used to calculate p value for comparison between the indicated groups.
Next, we examine the localization of phosphomimetic HJURP-3D in HeLa cells. For this purpose, we generated HeLa cell lines stably expressing GFP-HJURP and GFP-HJURP-S3D, respectively (Fig. 5D). Although GFP-HJURP-S3D can still localize at centromeres in early G1 phase, its signal intensity significantly diminished, compared with cells expressing GFP-HJURP-WT (Fig. 5, E and F). It is likely that there are additional phosphorylation sites on HJURP or Mis18β that affect their interactions. The other possibility is that HJURP-3D may be recruited to centromeres by dimerization with endogenous HJURP. Next, we asked if the phosphorylation of HJURP affects the centromere incorporation of CENP-A. Our siRNA against HJURP effectively inhibits HJURP protein expression at 48 h after transfection, which yielded a typical 87 ± 5% suppression of HJURP protein (Fig. 5, G and H; p < 0.01).
As predicted, suppression of HJURP resulted in a significant reduction of CENP-A level at the centromere (Fig. 5, I and J). Ectopically expression of wild type HJURP rescued the assembly of CENP-A efficiently (Fig. 5J). However, expression of HJURP-3D was unable to recover the efficient incorporation of CENP-A (Fig. 5, I and J). Thus, we conclude that the temporal control of HJURP phosphorylation in mitosis by CDK1 is essential for achieving a correct assembly of CENP-A protein to centromeres for faithful mitosis.
DISCUSSION
Faithful genome segregation depends on the functions of the eukaryotic centromere, which is characterized by a unique histone 3 variant, CENP-A. Given the crucial role of CENP-A in centromere specification, discovering the mechanism of CENP-A nucleosome assembly is of great importance for our understanding of epigenetic inheritance of centromere. Earlier studies show that the Mis18 complex is required for newly CENP-A assembly in human cells, and the loading time of the Mis18 complex is slightly earlier than CENP-A loading (18, 20, 27). However, the precise molecular mechanism by which the Mis18 complex directs CENP-A nucleosome assembly has remained elusive. Recent studies show that CENP-C can bind to Mis18BP1, a component of the Mis18 complex, to facilitate CENP-A nucleosome assembly (41, 44). Among all of the proteins that are required for CENP-A loading, only CENP-C and CENP-N can directly bind to CENP-A nucleosome (23, 24).
Here, we delineate the molecular mechanism by which Mis18 complex recruits prenucleosomal CENP-A complex. We believe that our findings fit the known knowledge and fill a missing gap of the CENP-A assembly pathway. Based on our own findings and the previous publications in the field, we propose a roadmap of CENP-A assembly (Fig. 6). This model demonstrates a special relationship between old CENP-A and new CENP-A. The old CENP-A nucleosome recruits CENP-C through direct interaction between the CENP-A C-terminal tail and CENP-C. CENP-C recruits the Mis18 complex at late anaphase when CDK1 activity is sharply reduced. As a consequence, the Mis18 complex recruits prenucleosomal CENP-A complex through directly binding between Mis18β and HJURP. Scm3, the homolog of HJURP in fission yeast, can physically interact with Mis18 in vitro (33–34). This indicates that the mechanism of HJURP-mediated CENP-A protein assembly is conserved from fission yeast to humans.
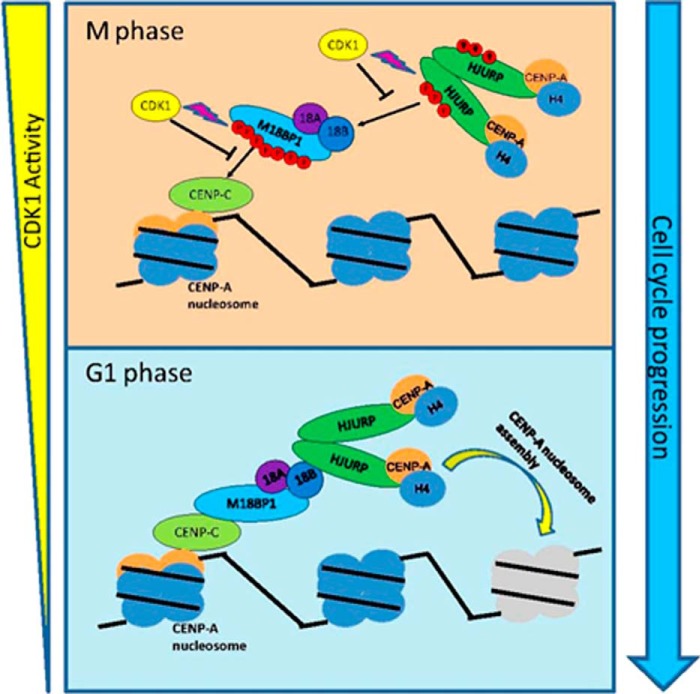
FIGURE 6.
A schematic model illustrating the pathway of HJURP recruitment to centromeres. During G2 and M phase, high CDK activity phosphorylates Mis18BP1 and HJURP. The phosphorylation of Mis18BP1 prevents its binding with CENP-C, and phosphorylated HJURP prevents HJURP-Mis18β interaction. Therefore, CENP-A incorporation is inhibited. When cells exited mitosis, CDK activity decreased sharply. Mis18BP1 and HJURP were dephosphorylated by phosphatase. Therefore, CENP-A nucleosome recruits CENP-C, and CENP-C recruits Mis18 complex at late anaphase by directly binding with Mis18BP1. In turn, Mis18 complex recruits prenucleosomal CENP-A complex through direct binding between Mis18β and HJURP.
It is reported that the cell cycle control of CENP-A nucleosome assembly to the centromere is mediated by CDK activity, and inhibition of CENP-A nucleosome assembly by CDK activity is probably exerted through phosphorylation of CENP-A assembly factors, including Mis18BP1 (42). However, aberrant centromere targeting of the non-phosphorylatable mutant Mis18BP1S24A did not result in premature CENP-A nucleosome assembly. This indicates that other factors directing CENP-A assembly are also under the CDK phospho-regulation (42). Consistent with this notion, our data indicate that phosphorylation of HJURP by CDK1 alleviates the interaction affinity between HJURP and Mis18β and inhibits premature CENP-A assembly. Given the fact that GFP-HJURP-S3D remains localizing to the centromeres to a certain extent, we reason that other CDK1 phosphorylation sites on HJURP or other CENP-A loading factors may also play an important role in regulating CENP-A assembly. It would be of great interest down the road to identify and characterize those regulatory mechanisms.
The kinetochore is a complex structure that functions as a molecular machine to power chromosome movement and as a signaling device to govern chromosome segregation and cell cycle control. We have recently developed an expanded palette of optical sensors to visualize and quantify dynamic phosphorylation (45), methylation (46), and acetylation (39) in live cell mitosis. The dynamics of centromere methylation is tightly coupled to mitotic phosphorylation to ensure chromosome plasticity during cell division (46). It has been reported that Mis18 deficiency leads to perturbed histone methylation and liberation of DNMT3A/B from the centromere (47). Given the cross-talk and interdependence between lysine 9 methylation and phosphorylation of serine 10 seen in histone 3 during the cell division cycle (48), it would be of great interest to probe whether and how methylation and phosphorylation gradients govern HJURP-mediated CENP-A loading to the centromere during mitosis and whether the mitotic phosphorylation also affects the assembly dynamics and plasticity of other histone fold-containing proteins, such as CENP-S/X complex, in centromere plasticity (we refer to centromere plasticity as reversible assembly and disassembly of functional centromere) control (49).
In summary, we demonstrate that Mis18β governs the centromere loading of CENP-A by recruiting CENP-A chaperone HJURP. We have also demonstrated that the phosphorylation of HJURP by CDK1 inhibits the centromere deposition of CENP-A, providing mechanistic insight into a better understanding of spatiotemporal dynamics underlying CENP-A deposition during early G1 phase. Our findings provide novel insights for better understanding the molecular pathway underlying HJURP-directed CENP-A loading to the centromere.
Acknowledgments
We thank Dr. Don Hill for proofreading and all members of our laboratories for helpful discussions during the course of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants DK56292, CA164133, 8G12MD007602, and G12RR03034. This work was also supported by Chinese 973 Projects 2010CB912103, 2012CB917204, and 2013CB911203; Chinese Academy of Science Grants KSCX1-YW-R-65 and KSCX2-YW-R-195; MOST Grant 2009DFA31010; MOE Grants 20113402130010 and IRT13038; Chinese Natural Science Foundation Grants 91313303, 31320103904, 90508002, 91129714, 81270466, 31271518, 31371363, and 90913016; China Fellowship 2012M510210; and Fundamental Research Funds for Central Universities Grants WK2340000032 and WK2060190018.
- HJURP
- Holliday junction recognition protein
- IP
- immunoprecipitation
- AD
- activation domain
- BD
- binding domain
- ACA
- anti-centromere antibody.
REFERENCES
- 1. Black B. E., Jansen L. E., Foltz D. R., Cleveland D. W. (2010) Centromere identity, function, and epigenetic propagation across cell divisions. Cold Spring Harb. Symp. Quant. Biol. 75, 403–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cleveland D. W., Mao Y., Sullivan K. F. (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 [DOI] [PubMed] [Google Scholar]
- 3. Allshire R. C., Karpen G. H. (2008) Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9, 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morris C. A., Moazed D. (2007) Centromere assembly and propagation. Cell 128, 647–650 [DOI] [PubMed] [Google Scholar]
- 5. Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. (2001) Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palmer D. K., O'Day K., Wener M. H., Andrews B. S., Margolis R. L. (1987) A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104, 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meluh P. B., Yang P., Glowczewski L., Koshland D., Smith M. M. (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94, 607–613 [DOI] [PubMed] [Google Scholar]
- 8. Henikoff S., Ahmad K., Platero J. S., van Steensel B. (2000) Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Black B. E., Brock M. A., Bédard S., Woods V. L., Jr., Cleveland D. W. (2007) An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 104, 5008–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006) The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469 [DOI] [PubMed] [Google Scholar]
- 11. Guse A., Carroll C. W., Moree B., Fuller C. J., Straight A. F. (2011) In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477, 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu S. T., Rattner J. B., Jablonski S. A., Yen T. J. (2006) Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175, 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Régnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., Brown W. (2005) CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25, 3967–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goshima G., Kiyomitsu T., Yoda K., Yanagida M. (2003) Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160, 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Howman E. V., Fowler K. J., Newson A. J., Redward S., MacDonald A. C., Kalitsis P., Choo K. H. (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. U.S.A. 97, 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amor D. J., Bentley K., Ryan J., Perry J., Wong L., Slater H., Choo K. H. (2004) Human centromere repositioning “in progress”. Proc. Natl. Acad. Sci. U.S.A. 101, 6542–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fachinetti D., Folco H. D., Nechemia-Arbely Y., Valente L. P., Nguyen K., Wong A. J., Zhu Q., Holland A. J., Desai A., Jansen L. E., Cleveland D. W. (2013) A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jansen L. E., Black B. E., Foltz D. R., Cleveland D. W. (2007) Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. (2004) Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729 [DOI] [PubMed] [Google Scholar]
- 20. Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. (2007) Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β and M18BP1. Dev. Cell 12, 17–30 [DOI] [PubMed] [Google Scholar]
- 21. Maddox P. S., Hyndman F., Monen J., Oegema K., Desai A. (2007) Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176, 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barnhart M. C., Kuich P. H., Stellfox M. E., Ward J. A., Bassett E. A., Black B. E., Foltz D. R. (2011) HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194, 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carroll C. W., Milks K. J., Straight A. F. (2010) Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189, 1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carroll C. W., Silva M. C., Godek K. M., Jansen L. E., Straight A. F. (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11, 896–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shuaib M., Ouararhni K., Dimitrov S., Hamiche A. (2010) HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl. Acad. Sci. U.S.A. 107, 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dunleavy E. M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. (2009) HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 [DOI] [PubMed] [Google Scholar]
- 27. Foltz D. R., Jansen L. E., Bailey A. O., Yates J. R., 3rd, Bassett E. A., Wood S., Black B. E., Cleveland D. W. (2009) Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137, 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perpelescu M., Nozaki N., Obuse C., Yang H., Yoda K. (2009) Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 185, 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Camahort R., Li B., Florens L., Swanson S. K., Washburn M. P., Gerton J. L. (2007) Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26, 853–865 [DOI] [PubMed] [Google Scholar]
- 30. Mizuguchi G., Xiao H., Wisniewski J., Smith M. M., Wu C. (2007) Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129, 1153–1164 [DOI] [PubMed] [Google Scholar]
- 31. Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., Baker R. E. (2007) Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. U.S.A. 104, 10571–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams J. S., Hayashi T., Yanagida M., Russell P. (2009) Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 33, 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pidoux A. L., Choi E. S., Abbott J. K., Liu X., Kagansky A., Castillo A. G., Hamilton G. L., Richardson W., Rappsilber J., He X., Allshire R. C. (2009) Fission yeast Scm3. A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33, 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanchez-Pulido L., Pidoux A. L., Ponting C. P., Allshire R. C. (2009) Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137, 1173–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu H., Liu Y., Wang M., Fang J., Huang H., Yang N., Li Y., Wang J., Yao X., Shi Y., Li G., Xu R. M. (2011) Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bassett E. A., DeNizio J., Barnhart-Dailey M. C., Panchenko T., Sekulic N., Rogers D. J., Foltz D. R., Black B. E. (2012) HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell 22, 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu D., Ding X., Du J., Cai X., Huang Y., Ward T., Shaw A., Yang Y., Hu R., Jin C., Yao X. (2007) Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J. Biol. Chem. 282, 21415–21424 [DOI] [PubMed] [Google Scholar]
- 38. Zhu M., Wang F., Yan F., Yao P. Y., Du J., Gao X., Wang X., Wu Q., Ward T., Li J., Kioko S., Hu R., Xie W., Ding X., Yao X. (2008) Septin 7 interacts with centromere-associated protein E and is required for its kinetochore localization. J. Biol. Chem. 283, 18916–18925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xia P., Wang Z., Liu X., Wu B., Wang J., Ward T., Zhang L., Ding X., Gibbons G., Shi Y., Yao X. (2012) EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16564–16569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zasadzińska E., Barnhart-Dailey M. C., Kuich P. H., Foltz D. R. (2013) Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 32, 2113–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moree B., Meyer C. B., Fuller C. J., Straight A. F. (2011) CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 194, 855–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Silva M. C., Bodor D. L., Stellfox M. E., Martins N. M., Hochegger H., Foltz D. R., Jansen L. E. (2012) Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 22, 52–63 [DOI] [PubMed] [Google Scholar]
- 43. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dambacher S., Deng W., Hahn M., Sadic D., Fröhlich J., Nuber A., Hoischen C., Diekmann S., Leonhardt H., Schotta G. (2012) CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3, 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chu Y., Yao P. Y., Wang W., Wang D., Wang Z., Zhang L., Huang Y., Ke Y., Ding X., Yao X. (2011) Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J Mol. Cell. Biol. 3, 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chu L., Zhu T., Liu X., Yu R., Bacanamwo M., Dou Z., Chu Y., Zou H., Gibbons G. H., Wang D., Ding X., Yao X. (2012) SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J Mol. Cell. Biol. 4, 331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim I. S., Lee M., Park K. C., Jeon Y., Park J. H., Hwang E. J., Jeon T. I., Ko S., Lee H., Baek S. H., Kim K. I. (2012) Roles of Mis18α in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol. Cell. 46, 260–273 [DOI] [PubMed] [Google Scholar]
- 48. Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z. W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D., Jenuwein T. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 [DOI] [PubMed] [Google Scholar]
- 49. Tao Y., Jin C., Li X., Qi S., Chu L., Niu L., Yao X., Teng M. (2012) The structure of the FANCM-MHF complex reveals physical features for functional assembly. Nat. Commun. 3, 782–791 [DOI] [PMC free article] [PubMed] [Google Scholar]