Background: Notch1 is highly activated in melanoma where it plays protumorigenic functions, yet no Notch1-activating mutations have been identified in melanoma.
Results: MT1-MMP operates as a novel protease that promotes Notch1 activation in melanoma cells.
Conclusion: An MT1-MMP/Notch1 signaling pathway supports melanoma cell growth.
Significance: Notch1 emerges as a new MT1-MMP substrate that plays important biological roles in melanoma.
Keywords: Cell Proliferation, Matrix Metalloproteinase (MMP), Melanoma, Notch Receptor, Protein-Protein Interactions
Abstract
Notch1 is an evolutionarily conserved signaling molecule required for stem cell maintenance that is inappropriately reactivated in several cancers. We have previously shown that melanomas reactivate Notch1 and require its function for growth and survival. However, no Notch1-activating mutations have been observed in melanoma, suggesting the involvement of other activating mechanisms. Notch1 activation requires two cleavage steps: first by a protease and then by γ-secretase, which releases the active intracellular domain (Notch1NIC). Interestingly, although ADAM10 and -17 are generally accepted as the proteases responsible of Notch1 cleavage, here we show that MT1-MMP, a membrane-tethered matrix metalloproteinase involved in the pathogenesis of a number of tumors, is a novel protease required for the cleavage of Notch1 in melanoma cells. We find that active Notch1 and MT1-MMP expression correlate significantly in over 70% of melanoma tumors and 80% of melanoma cell lines, whereas such correlation does not exist between Notch1NIC and ADAM10 or -17. Modulation of MT1-MMP expression in melanoma cells affects Notch1 cleavage, whereas MT1-MMP expression in ADAM10/17 double knock-out fibroblasts restores the processing of Notch1, indicating that MT1-MMP is sufficient to promote Notch1 activation independently of the canonical proteases. Importantly, we find that MT1-MMP interacts with Notch1 at the cell membrane, supporting a potential direct cleavage mechanism of MT1-MMP on Notch1, and that MT1-MMP-dependent activation of Notch1 sustains melanoma cell growth. Together, the data highlight a novel mechanism of activation of Notch1 in melanoma cells and identify Notch1 as a new MT1-MMP substrate that plays important biological roles in melanoma.
Introduction
Despite a significant improvement in early detection and the increase in awareness in the general population of the danger associated with sun exposure and melanoma development, melanoma incidence has been rising for the past 30 years (1). Melanoma has now become the most common form of cancer for young adults in the 25–29 age range (2), and melanoma remains a deadly disease in its metastatic form because available therapies have failed to control, much less eradicate, the tumor.
Aggressive melanoma tumor cells have been shown to re-express embryonic stem cell pathways typical of their embryonic progenitors, the neural crest cells. However, unlike their normal counterparts, melanoma cells lack major regulatory cell cycle check points, resulting in aberrant growth and aggressive behavior that renders them more easily adaptable to microenvironmental changes and enables them to escape therapeutic insults by the rapid emergence of resistant cells (3). Given this premise, it becomes imperative to understand how these pathways operate in melanoma to develop more effective treatments.
Notch1 plays key roles in melanocyte/melanoma precursor cell homeostasis. Lack of Notch1 in the melanocytic lineage results in premature hair graying in the mouse caused by loss of melanoblasts in the embryo and premature differentiation of melanocyte stem cells in the adult mouse (4). We have previously shown that melanomas re-express and activate Notch1 and rely on its activity for tumor growth and survival (5, 6). In addition, other groups have demonstrated that Notch1 is sufficient to confer transforming properties to primary human melanocytes (7) and to convert primary melanoma cells from noninvasive to metastatic (8, 9). These data underlie a clear oncogenic role of Notch1 in melanoma. However, unlike other cancers such as T-cell lymphoblastic leukemia in which 50% of tumors show activating mutations of Notch1 (10), no Notch1-activating mutations have been observed thus far in melanoma (7), suggesting the involvement of other mechanisms. An increase in proteolytic cleavage may be one such mechanism.
Notch proteins are transmembrane receptors that are activated upon binding of membrane-associated ligands (JAGGED1 and 2 and Delta-like 1, 3, and 4) present on adjacent cells. Ligand binding allows exposure of a proteolytic cleavage site (S2), which, in numerous cell types, is recognized and cleaved by ADAM10 or -17 (a disintegrin and metalloproteinase domain 10 and 17) (tumor necrosis factor-α-converting enzyme (TACE)) (11, 12). Following this first proteolytic step, the cleavage by the γ-secretase complex happens quickly and releases the Notch intracellular domain (Notch1NIC) that is now free to enter the nucleus. There, Notch1NIC functions in a transcription complex together with the transcription factor CBF-1 (C-promoter binding factor-1) and Mastermind-Like (MAML) (13).
Although ADAM10 and -17 are generally considered the canonical proteases involved in Notch1 activation, our data point at an additional and/or alternative protease in melanoma cells capable of processing Notch1. MT1-MMP is a membrane-associated matrix metalloproteinase that functions as one of the most important, invasion-promoting, pro-tumorigenic MMP2 in cancer cell invasion (14). Melanomas express a number of MMPs (15). The MMP family comprises 25 members in humans, among which 19 are soluble and are released in the extracellular milieu and 6 are membrane-anchored. MT1-MMP is the prototypic membrane-tethered matrix metalloproteinase. Interestingly, MT1-MMP is also highly expressed in neural crest cells (16) and required for the proper specification and migration of melanophores (equivalent of melanocytes) during embryonic development of Xenopus laevis (17). Importantly, MT1-MMP is re-expressed in melanoma and often found associated with the invading tumor front (18), highlighting a role of this protease in melanoma pathogenesis.
Here we show that active Notch1 (Notch1NIC) and MT1-MMP correlate significantly in both melanoma tumors and cell lines, whereas such correlation does not exist between Notch1NIC and ADAM10 or -17. We demonstrate that the modulation of MT1-MMP expression affects Notch1 cleavage. MT1-MMP forms a complex with Notch1 at the cell membrane, implying that it could directly cleave Notch1. Importantly, MT1-MMP-dependent activation of Notch1 promotes melanoma cell growth. Collectively, these data identify Notch1 as a novel MT1-MMP substrate and support a novel mechanism of Notch1 activation in melanoma.
EXPERIMENTAL PROCEDURES
Cells and Tissue Specimens
Primary and metastatic melanoma cells were in part purchased from ATCC (American Type Culture Collection, Manassas, VA) or were gifts from Dr. Marianne Broome Powell (Stanford University) (5). The use of these cells was approved by the Case Cancer Institutional Review Board (IRB). The cell lines used in this study are, in the order they appear in the blot in Fig. 1C: immortalized human melanocytes; A375.52; WM115; WM35; UACC2842; WM266-4; UACC2521; SKMel2; D1594; K457; C32TG; SKMel24; and UACC3291. Functional assays were performed on the cell lines WM115 and WM266-4, which are syngeneic and derived from a primary and metastatic melanoma, respectively. ADAM10/17 double knock-out mouse embryonic fibroblasts were a gift from Dr. Paul Saftig (Christian-Albrechts-Universität zu Kiel, Kiel, Germany) and Dr. Carl Blobel (Weill Cornell University, New York). All cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% FCS (fetal calf serum), 1% glutamine, and 1% penicillin-streptomycin. Immortalized human melanocytes were a gift from Dr. Robert Weinberg (Harvard University) and were maintained in DMEM supplemented with 5% FCS, 1% glutamine, and 1% penicillin-streptomycin (19).
FIGURE 1.
Notch1NIC and MT1-MMP expression correlate in melanoma. A, Western blotting on nine flash-frozen, human melanoma tumors showing expression of ADAM17 (A-17), ADAM10 (A-10), MT1-MMP (MT1), and active Notch1 (NIC). β-Actin (β-Act) was used as loading control. B, correlation analysis between MT1-MMP and Notch1NIC expression of the samples in A. The right table shows R and p values for the correlation analysis of the samples in A between Notch1NIC and ADAM10 and -17. C, Western blotting on 12 human melanoma cell lines as compared with human melanocytes (M) showing expression of ADAM17 (A-17), ADAM10 (A-10), MT1-MMP (MT1), and active Notch1 (NIC). β-Actin was used as loading control. D, correlation analysis between MT1-MMP and Notch1NIC expression of the samples in C. The right table shows R and p values for the correlation analysis of the samples in C between Notch1NIC and ADAM10 and -17.
De-identified melanoma tissues were redundant diagnostic specimens obtained at surgery for clinically indicated removal at metastatic sites. The collection of such specimens followed the guidelines of the IRB-approved Human Biospecimen Resource Core in the Cleveland Clinic Department of Anatomic Pathology (IRB Protocol Number 06-050).
shRNAs and Expression Plasmids
shRNAs against human MT1-MMP (TRCN0000050855) and Notch1 (TRCN0000003359) were purchased from Sigma. The MT1-MMP lentiviral expression vectors were constructed by inserting the cDNA sequence corresponding to full-length (MT1-FL) and mutated (MT1-Mut) human MT1-MMP that were PCR-amplified from pSG-MT1-MMP (kindly provided by Dr. Motoharu Seiki, University of Tokyo, Tokyo, Japan) into the pLM-CMV-Ha-puro-PL3 lentiviral plasmid (20). The Notch1NIC construct was previously described (6). Viral particles were produced in 293-FT (fast growing variant of the 293 cell line stably expressing SV40 large T antigen) cells using the FuGENE 6 reagent (Roche Applied Science, Mannheim, Germany) as per the manufacturer's instructions. The packaging plasmids used were pMD2.G and psPAX2 that were purchased from Addgene (Cambridge, MA).
Luciferase Assays
The Hes1-reporter plasmid was a kind gift from Dr. Ryoichiro Kageyama (Kyoto University, Kyoto, Japan) (21). WM115 or WM266-4 cells (5 × 104/well, in 24-well plates) were transfected using the FuGENE 6 reagent (Roche Applied Science) as per the manufacturer's instructions. After 48 h, cells were lysed in 100 μl of lysis buffer (Promega, Madison, WI). A Renilla luciferase reporter plasmid driven by a CMV promoter was co-transfected with the HES1 reporter construct at a 1:20 ratio to assess transfection efficiency. Activities of firefly and Renilla were assessed by the Dual-Luciferase assay system (Promega), and light production was measured for 10 s in a Monolight 2010 luminometer (Molecular Devices, Sunnyvale, CA).
Notch Ligand Stimulation Assay
Notch signaling was induced in WM115 (32,000/cm2) and WM266-4 (32,000/cm2) cells plated on dishes displaying immobilized FC- or FC-JAGGED1 ligand anchored to protein-A. Plasmids expressing Fc- and FC-JAGGED1 (22, 23) were kindly provided by Dr. Aaron Proweller (Case Western Reserve University, Cleveland, OH).
Western Blot Analysis
Cells (32,000/cm2) were plated in either untreated or FC- or FC-JAGGED1-coated dishes in complete DMEM, allowed to adhere, and then collected after 24 h after seeding. WM115 and WM266-4 cells for the Western blots relative to the growth curve assays in Fig. 4 were seeded at an initial density of 16,000/cm2. The γ-secretase inhibitor dibenzazepine (10 μm) was used as control for the identification of Notch1NIC that is cleaved at Val-1744. Total protein for all assays was extracted with urea lysis buffer (9 m urea; 75 mm Tris-HCl, pH 7.5, and 100 mm 2-mercaptoethanol), and 40–50 μg/sample was separated by 8–10% SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were probed with the following antibodies: anti-Notch1-TM (C20, Santa Cruz Biotechnology, Santa Cruz, CA); anti-Notch1NIC (Val-1744) (Cell Signaling Technologies, Beverly, MA); anti MT1-MMP (clone LEM-2/15.8, Millipore, Billerica, MA); anti-ADAM10 (Abcam, Cambridge, MA); and anti-TACE (tumor necrosis factor-α-converting enzyme) (ADAM17) (eBioscience, San Diego, CA). Bands were detected using SuperSignal detection reagent (Thermo Scientific). Loading was normalized with anti-β-actin (Santa Cruz Biotechnology). Densitometric quantification of band intensity for each sample in Fig. 1, A and C, was normalized to band intensity of the respective β-actin. The resultant value was used for Pearson correlation analysis to compare the expression levels of each protease with those of Notch1NIC.
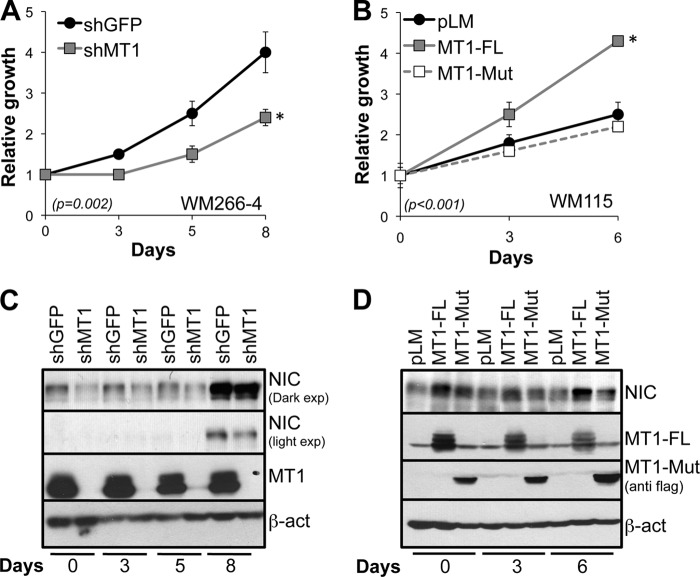
FIGURE 4.
MT1-MMP affects melanoma cell growth. A, relative growth of WM266-4 expressing a control shRNA (shGFP) or shMT1-MMP. B, relative growth of WM115 expressing a control vector (pLM) or either active (MT1-FL) or inactive (MT1-Mut) MT1-MMP. *, p < 0.001, Student's t test. C and D, expression levels of Notch1NIC (NIC) at each time point relative to the growth curves in A and B, respectively. β-act, β-actin. MT1, MT1-MMP.
Co-immunoprecipitation of Membrane Protein Lysates
Cells (WM266-4, 32,000/cm2) were harvested in ice-cold PBS containing 10 mg/ml aprotinin, 0.25 mg/ml Pefabloc, and 1 mg/ml leupeptin and subjected to three freeze-thaw cycles in dry ice/ethanol 37 °C baths followed by sonication for 3 s. Membranes were then precipitated by centrifugation (30 min, 14,000 rpm, 4 °C), and lysis of membrane pellets was performed in radioimmune precipitation buffer. Immunoprecipitation was done using either a nonspecific mouse or rabbit IgG or specific anti-MT1-MMP (clone LEM-2/15.8, Millipore, Billerica, MA) and anti Notch1 (C20, Santa Cruz biotechnology), respectively.
Real-time PCR Analysis
cDNA was synthesized from total RNA and treated with DNase I (Invitrogen), using SuperScript first-strand synthesis system for RT-PCR (Invitrogen). In all, 1 μl of cDNA was used for PCR amplification using SYBR Green PCR master mix (Bio-Rad). The following primer sets were used to amplify specific target genes: human Hes1 forward 5′-ATGGAGAAAAATTCCTCGTCCC-3′, reverse 5′-TTCAGAGCATCCAAAATCAGTGT-3′; human β-actin forward 5′-CATGTACGTTGCTATCCAGGC-3′, reverse 5′-CTCCTTAATGTCACGCACGAT-3′. PCR amplification was done in a Bio-Rad I-Cycler. β-Actin was used to normalize mRNA. Relative quantification of mRNA expression levels was determined using the relative standard curve method according to the manufacturer's instructions (Bio-Rad).
Cell Proliferation Assays
WM115 and WM266-4 cells (initial seeding density 16,000/cm2) were plated in 96-well plates in triplicate. At each time point cells were fixed with 10% buffered formalin and subjected to the crystal violet assay as described previously (24). Similarly, for the end point growth assay in Fig. 5, cells (WM266-4, WM115, V2387, and C32TG) were seeded at 32,000 cells/cm2 and analyzed 3 days later as above.
FIGURE 5.
Notch1 effects MT1-MMP-dependent melanoma cell growth. A, expression levels of MT1-MMP (MT1) and Notch1NIC in WM266-4 cells expressing shGFP or shMT1-MMP and transduced with active Notch1 (NIC). β-act, β-actin. B, relative growth at a 3-day time point of the cells in A. C, expression levels of MT1-MMP and Notch1NIC in WM115 cells expressing pLM, full-length MT1-MMP, or the catalytically inactive mutant (MT1-Mut) and transduced with a specific shRNA against Notch1 (shN1). D, relative growth at a 3-day time point of the cells in C. E, expression levels of MT1-MMP and Notch1NIC in V2387 cells expressing shGFP or shMT1-MMP and transduced with active Notch1 (NIC). F, relative growth at a 3-day time point of the cells in E. G, expression levels of MT1-MMP and Notch1NIC in C32TG cells expressing pLM, full-length MT1-MMP, or the catalytically inactive mutant (MT1-Mut) and transduced with a specific shRNA against Notch1 (shN1). H, relative growth at a 3-day time point of the cells in G. The ratio between the Notch1NIC band and the corresponding β-actin is shown below each blot.
RESULTS
The Expression of Active Notch1 Correlates with MT1-MMP in Melanoma Tumors and Cell Lines
The canonical proteases that have been shown to cleave Notch1 in a variety of cell types are the zinc-dependent proteases ADAM10 and -17 (11, 12). We therefore first tested whether a potential correlation existed between ADAM10 and -17 and active Notch1 (Notch1NIC). We used 9 flash-frozen melanoma samples and 12 human melanoma cell lines. We found that mature (active) ADAM10 and -17 were expressed by 55 and 44% of melanoma tumors and 66 and 75% of melanoma cell lines, respectively (Fig. 1, A and C). However, the expression of active ADAM10 and -17 did not correlate with the expression levels of active Notch1 (Fig. 1, B and D, right panels). This led us to speculate that a novel protease might be involved in the cleavage of Notch1 in melanoma. MT1-MMP, also a zinc-dependent membrane-bound matrix metalloproteinase, can process a variety of extracellular matrix proteins, adhesion and signaling receptors, and cytokines and growth factors; activate accessory MMPs, such as MMP2 and MMP13; and promote tumorigenesis (25–31). We therefore speculated that MT1-MMP might represent a novel protease involved in Notch1 activation in melanoma. Indeed, active MT1-MMP expression was observed in 88% of tumors and 91% of cell lines (Fig. 1, A and D), and its expression correlated significantly with that of Notch1NIC (Fig. 1, B and D), suggesting these two factors are co-deregulated in melanoma and supporting the hypothesis that MT1-MMP may be a novel protease involved in the cleavage and activation of Notch1 in melanoma cells.
Modulation of MT1-MMP Expression Affects Notch1 Processing
To determine whether MT1-MMP promotes Notch1 activation in melanoma, we inhibited MT1-MMP expression in WM266-4 cells (cell line 6 in Fig. 1D) and then assessed Notch1NIC levels, as a measure of activated Notch1. MT1-MMP knockdown reduced Notch1NIC (Fig. 2A); however, the knockdown of MMP2, a soluble MMP that requires MT1-MMP for its activation (32, 33), did not affect Notch1 cleavage. The effects on Notch1NIC appeared to be the result of cleavage defects rather than protein expression, which did not change appreciably (Fig. 2A, upper panel).
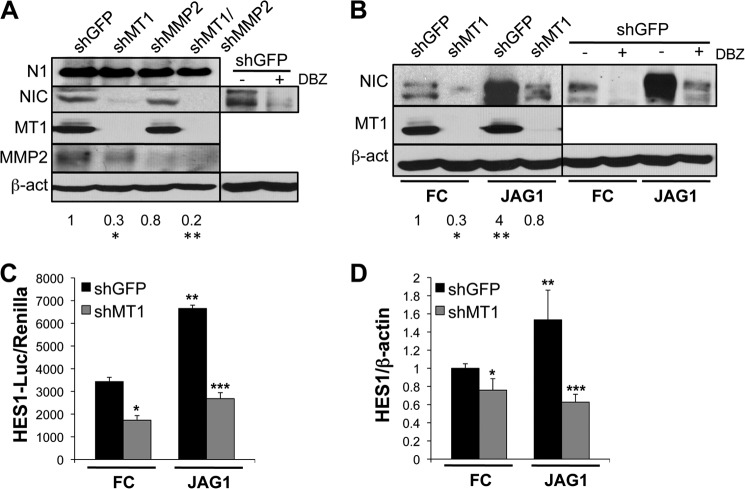
FIGURE 2.
MT1-MMP inhibition reduces Notch1 cleavage. A, expression of full-length Notch1 (N1), Notch1NIC (NIC), MT1-MMP (MT1), and MMP2 in WM266-4 cells expressing shRNAs against GFP, MT1-MMP, MMP2, or both MT1-MMP and MMP2. The ratio between the Notch1NIC band and the corresponding β-actin is shown below the blot. These values are an average of three independent experiments, and differences between treatments and control (shGFP-expressing cells) are statistically significant (*,**, p < 0.01; Student's t test). DBZ, dibenzazepine. B, Notch1NIC expression in WM266-4 cells expressing shGFP or shMT1-MMP and plated on FC- or FC-JAGGED1-coated dishes. The ratio between the Notch1NIC band and the corresponding β-actin is shown below the blot. These values are an average of three independent experiments, and differences between treatments and control (shGFP-expressing cells) are statistically significant (*, p < 0.01; **, p = 0.003; Student's t test). C, HES1-luciferase reporter of the cells in B. The luciferase signal intensity was normalized to the signal intensity of a CMV-Renilla construct. *, p < 0.001 (shGFP-FC versus shMT1-FC); **, p < 0.0001 (shGFP-FC versus shGFP-JAG1); ***, p < 0.0001 (shMT1-JAG1 versus shGFP-JAG1); Student's t test. D, quantitative RT-PCR of the cells in B. HES1 values are normalized to β-actin used as internal control. *, p = 0.04 (shGFP-FC versus shMT1-FC); **, p = 0.03 (shGFP-FC versus shGFP-JAG1); ***, p = 0.009 (shMT1-JAG1 versus shGFP-JAG1); Student's t test. Dibenzazepine (10 m) was used as control for Notch1NIC cleaved at Val-1744.
To further analyze the capacity of MT1-MMP to function as a Notch1 protease, we treated the cells with recombinant JAGGED1 (JAG1), to artificially stimulate ligand-dependent Notch1 activation. We observed an increase in Notch1NIC in JAGGED1-stimulated cells that was however reduced to the levels of controls (shGFP cells treated with the FC fragment only) in cells expressing an shMT1-MMP (Fig. 2B). The effects of MT1-MMP inhibition on Notch1 processing resulted in a decrease in luminescence signal of a Hes1-driven reporter construct and in the reduction of Hes1 mRNA levels, indicating an impairment of the Notch1 signaling cascade (Fig. 2, C and D). Again, stimulation of cells with JAGGED1 led to an increase in reporter activity and Hes1 messenger RNA that was however lost in cells expressing an shMT1-MMP.
To further confirm that MT1-MMP functions as a protease in Notch1 processing, we expressed a full-length (MT1-FL) or a mutant MT1-MMP lacking the catalytic domain (MT1-Mut) (Fig. 3B) in WM115 cells, and confirmed similar levels of expression by Western blotting using an anti-FLAG antibody (Fig. 3C). WM115 is a primary melanoma cell line syngeneic with the metastatic WM266-4 cells that express lower levels of MT1-MMP (Fig. 3A) and that therefore is suitable for comparative overexpression studies. Only full-length, catalytically active MT1-MMP increased Notch1NIC, both in nonstimulated (FC) and in ligand-stimulated cells (JAG1) (Fig. 3D). Notch1 mRNA did not change (not shown), implying that Notch1NIC increase did not depend on an increase in Notch1 expression. Active MT1-MMP, however, increased Notch1 activity, measured as the induction of a Hes1-driven reporter construct and Hes1 mRNA levels, particularly in ligand-stimulated cells (Fig. 3, E and F).
FIGURE 3.
Active MT1-MMP promotes Notch1 cleavage independently of ADAM10 or -17. A, MT1-MMP (MT1) protein expression in the syngeneic cell lines WM115 (primary melanoma) and WM2660-4 (metastatic melanoma). β-act, β-actin. B, schematic representation of the MT1-MMP constructs used in the study. SP, signal peptide; Pro, pro-peptide; CAT, catalytic domain; H, hinge domain; Pex, hemopexin domain; TM, transmembrane domain; Cyt, cytosolic tail. MT1-FL, full-length, catalytically competent MT1-MMP; MT1-Mut (ΔCAT) lacks the catalytic domain. C, expression levels of the constructs in B by a FLAG antibody. Asterisk indicates that these bands are likely autocatalytic fragments of full-length MT1-MMP. D, WM115 cells expressing MT1-FuL or MT1-Mut and seeded in control dishes (coated with FC) or stimulated with recombinant JAGGED1 (JAG1). Dibenzazepine (DBZ) (10 μm) was used as control to identify Notch1NIC (NIC) cleaved at Val-1744. E, HES1-driven reporter construct expressed in the cells in D. Values are normalized to CMV-Renilla. *, p = 0.04 (MT1-FC versus pLM-FC); **, p = 0.002 (pLM-FC versus pLM-JAG1); ***, p < 0.0001 (MT1-JAG1 versus pLM-JAG1); Student's t test. F, quantitative RT-PCR of the cells in D. HES1 values are normalized to β-actin, used as internal control. *, p = 0.002 (MT1-FC versus pLM-FC); **, p < 0.001 (pLM-FC versus pLM-JAG1); ***, p < 0.0001 (MT1-JAG1 versus pLM-JAG1); Student's t test. G, co-immunoprecipitation (IP) of Notch1 and MT1-MMP. H, wild type and ADAM10/17 double knock-out (DKO) mouse embryonic fibroblasts infected with either empty vector (pLM) or full-length, catalytically competent MT1-MMP. Western blot shows levels of expression of ADAM10 (A10), ADAM17 (A17), MT1-MMP (MT1), and Notch1NIC (NIC) in both wild type and ADAM10/17 double knock-outs. β-Actin was used as loading control.
To determine whether MT1-MMP could associate with Notch1 at the membrane, allowing potential direct cleavage of Notch1 by MT1-MMP, we performed co-immunoprecipitation studies by specifically extracting membrane proteins. We found that Notch1 and MT1-MMP were able to pull down each other (Fig. 3G) when immunoprecipitated with specific antibodies, suggesting that MT1-MMP could cleave Notch1 directly. Together, these data underscore a new interaction between Notch1 and MT1-MMP whereby the latter emerges as a novel protease involved in Notch1 activation.
MT1-MMP Affects Notch1 Cleavage Independently of ADAM10 and -17
ADAM10 and -17 are considered the canonical proteases involved in Notch1 activation in a number of cell types. Although our data point at MT1-MMP as a major protease involved in Notch1 activation in melanoma cells, we cannot a priori exclude that ADAM10 and -17 may participate in this process, as most melanoma cells express all three enzymes (Fig. 1C). To determine whether MT1-MMP could cleave Notch1 independently of ADAM10 and -17, we employed ADAM10/17 double knock-out mouse embryonic fibroblasts. Fig. 3H shows that in these cells the levels of Notch1NIC are diminished with respect to the wild type counterparts, as has been previously reported (11, 12, 34). Given that these mouse embryonic fibroblasts do not express detectable levels of MT1-MMP, we expressed the catalytically proficient, full-length MT1-MMP construct (Fig. 3B). Not only did the expression of active MT1-MMP increase the amount of Notch1 cleavage in wild type cells, but importantly, it restored Notch1 processing in the double knock-out fibroblasts, indicating that MT1-MMP is capable of cleaving Notch1 independently of ADAM10 and -17.
MT1-MMP Activation of Notch1 Modulates Melanoma Cell Growth
Previous work by us and others highlighted a key role of Notch1 in melanoma growth and survival (5–8). Therefore, we wanted to test whether MT1-MMP could affect melanoma cell growth as a consequence of activating Notch1. WM266-4 cells expressing a specific shRNA against MT1-MMP showed slower growth rates with respect to control cells (expressing shGFP; Fig. 4A). On the other hand, WM115 cells expressing active, full-length MT1-MMP showed an accelerated growth (Fig. 4B). The mutant MT1-MMP, however, did not confer any growth advantages, suggesting that the catalytic function of MT1-MMP is required for its growth-promoting activity. Interestingly, inhibition of MT1-MMP was associated with a decrease in Notch1NIC levels (Fig. 4C), whereas expression of active MT1-MMP was associated with an increase in Notch1NIC (Fig. 4D) in all time points analyzed in the growth curve. These data suggest that active Notch1 may be, at least in part, involved in the growth-dependent function of MT1-MMP. To determine whether Notch1 was indeed responsible for MT1-MMP-stimulated cell growth, WM266-4 cells were infected with a control lentiviral construct (pLM) or a construct expressing the active portion of Notch1 (Notch1NIC) that does not require catalytic activation (Fig. 5A). Cells were then incubated for 3 days, and relative growth was evaluated at the end time point (Fig. 5B). Expression of active Notch1 conferred growth advantages as described previously (6), in both control and shMT1-MMP-expressing cells, suggesting that reconstitution of Notch1 signaling can, at least in part, restore cell growth that is inhibited by MT1-MMP knockdown. Conversely, WM115 cells expressing active MT1-MMP were transduced with a specific shRNA against Notch1 (Fig. 5C), and then growth was again assessed after 3 days of incubation. Inhibition of Notch1 was sufficient to abolish the growth advantages associated with MT1-MMP expression (Fig. 5D). Similar results were obtained in two different melanoma cell lines, V2387 and C32TG, that express high and low levels of MT1-MMP, respectively (Fig. 5, E and F and G and H). These data indicate that Notch1 is an effector of MT1-MMP in modulating melanoma cell growth.
DISCUSSION
The reactivation of embryonic stem cell pathways has been proposed as a prerogative of aggressive melanomas (3, 35, 36) and is thought to be at the basis of the high plasticity of melanoma cells exemplified by their adaptability to microenvironmental changes and their capacity to counteract most therapies by the rapid emergence of resistant cells. Notch1 is an essential embryonic stem cell factor that plays key roles in the maintenance of melanocyte/melanoma precursor cell homeostasis (4). We have previously demonstrated that although in mature melanocytes the levels of Notch1 are low or undetectable, melanomas re-express Notch1, and active Notch1 is required for their growth and survival (5, 6). However, unlike T-cell lymphoblastic leukemia, melanomas do not present activating mutations in the Notch1 gene (7), suggesting that Notch1 activation occurs via other mechanisms.
In the present study we demonstrate for the first time that MT1-MMP operates as a novel protease in the activation of Notch1. Our findings indicate that MT1-MMP and active Notch1 (Notch1NIC) expression correlates significantly in both melanoma tumors and cell lines, whereas we did not find a similar correlation between Notch1NIC and ADAM10 and -17, generally recognized as the canonical proteases involved in Notch1 processing. The data suggest that MT1-MMP and Notch1 expression is concurrently deregulated in melanoma where they may play roles in the pathogenesis of the disease. Indeed, we demonstrate that MT1-MMP and Notch1 are found in a complex at the cell membrane and that MT1-MMP modulation affects Notch1 cleavage. Importantly, the activation of Notch1 downstream of MT1-MMP is partly responsible for the growth-promoting functions of the metalloproteinase, supporting an MT1-MMP/Notch1 signaling pathway regulating melanoma cell proliferation.
The Notch1 receptor is a type I transmembrane protein, which functions as a ligand-activated transcription factor (37, 38). In resting conditions, the extracellular heterodimerization (HD) and Lin/Notch repeat (LNR) domains of Notch1 located in the extracellular portion of the receptor form a molecular lock that precludes spontaneous Notch1 activation. Physiologic Notch1 signaling is triggered by interaction of the receptor with Delta-like and JAGGED ligand proteins expressed on the surface of nearby cells. This ligand-receptor interaction induces a conformational change in the HD-LNR complex and exposes the S2 site to protease cleavage. This initial cut in the extracellular portion of the receptor is required for further proteolytic processing by the presenilin γ-secretase complex at the S3 site (39) located in the transmembrane region of the receptor, which liberates the active intracellular domain. ADAM10 and -17 have been implicated in the S2 processing of Notch receptors in different organisms (39–43). Interestingly, however, work by Van Tetering et al. (34) showed that ADAM10 but not ADAM17 is essential in executing ligand-induced cleavage at the S2 site, and, importantly, suggested the presence of unknown proteases with the ability to process Notch1 signaling as well. MT1-MMP may be one such protease. In fact, our data show that expression of active MT1-MMP in ADAM10/17 double mouse embryonic fibroblasts is fully capable of restoring Notch1 processing that is inhibited by the absence of either protease.
It has been shown that Notch1 proteolytic processing by ADAM10 is strictly ligand-dependent and that ADAM17, although it cannot replace ADAM10 in activating Notch1 in response to ligand, can nonetheless activate Notch1 signaling in a ligand-independent manner (11). Our data show that MT1-MMP can increase Notch1 processing in either unstimulated or JAGGED1-stimulated cells. Although in normal culture conditions it is possible that Notch1 might be activated by cell-cell contact with neighboring cells expressing endogenous ligands, we cannot exclude the possibility that MT1-MMP may work in either a ligand-dependent or a ligand-independent manner. Further experiments are needed to fully characterize the ligand dependence of MT1-MMP.
In conclusion the data presented here highlight a novel mechanism of activation of Notch1 in melanoma cells and identify Notch1 as a new MT1-MMP substrate that plays important biological roles in melanoma.
This work was supported by funding from the Harry Lloyd Charitable Trust.
- MMP
- matrix metalloproteinase
- FL
- full length
- Mut
- mutated
- Fc
- fragment crystallizable.
REFERENCES
- 1. Howlader N., Noone A. M., Krapcho M., Neyman N., Aminou R., Waldron W., Altekruse S. F., Kosary C. L., Ruhl J., Tatalovich Z., Cho H., Mariotto A., Eisner M. P., Lewis D. R., Chen H. S., Feuer E. J., Cronin K. A., Edwards B. K. (eds). (2011) SEER Cancer Statistics Review, 1975–2008, National Cancer Institute, Bethesda, MD [Google Scholar]
- 2. Herzog C., Pappo A., Bondy M., Bleyer A., Kirkwood J. (2007) in Cancer Epidemiology in Older Adolescents and Young Adults 15–29 years of age, including SEER incidence and survival 1975–2000, SEER AYA Monograph (Bleyer A., O'Leary M., Barr R., Ries L. A. G., eds), pp. 53–63, NIH publication No. 06-5767, National Cancer Institute, Bethesda, MD [Google Scholar]
- 3. Hendrix M. J., Seftor E. A., Seftor R. E., Kasemeier-Kulesa J., Kulesa P. M., Postovit L. M. (2007) Reprogramming metastatic tumour cells with embryonic microenvironments. Nat. Rev. Cancer 7, 246–255 [DOI] [PubMed] [Google Scholar]
- 4. Osawa M., Fisher D. E. (2008) Notch and melanocytes: diverse outcomes from a single signal. J. Invest. Dermatol. 128, 2571–2574 [DOI] [PubMed] [Google Scholar]
- 5. Bedogni B., Warneke J. A., Nickoloff B. J., Giaccia A. J., Powell M. B. (2008) Notch1 is an effector of Akt and hypoxia in melanoma development. J. Clin. Invest. 118, 3660–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang K., Wong P., Zhang L., Jacobs B., Borden E. C., Aster J. C., Bedogni B. (2012) A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth. Oncogene 31, 4609–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pinnix C. C., Lee J. T., Liu Z. J., McDaid R., Balint K., Beverly L. J., Brafford P. A., Xiao M., Himes B., Zabierowski S. E., Yashiro-Ohtani Y., Nathanson K. L., Bengston A., Pollock P. M., Weeraratna A. T., Nickoloff B. J., Pear W. S., Capobianco A. J., Herlyn M. (2009) Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res. 69, 5312–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Z. J., Xiao M., Balint K., Smalley K. S., Brafford P., Qiu R., Pinnix C. C., Li X., Herlyn M. (2006) Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 66, 4182–4190 [DOI] [PubMed] [Google Scholar]
- 9. Balint K., Xiao M., Pinnix C. C., Soma A., Veres I., Juhasz I., Brown E. J., Capobianco A. J., Herlyn M., Liu Z. J. (2005) Activation of Notch1 signaling is required for β-catenin-mediated human primary melanoma progression. J. Clin. Invest. 115, 3166–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferrando A. A. (2009) The role of NOTCH1 signaling in T-ALL. Hematology Am. Soc. Hematol. Educ. Program 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bozkulak E. C., Weinmaster G. (2009) Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 29, 5679–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sulis M. L., Saftig P., Ferrando A. A. (2011) Redundancy and specificity of the metalloprotease system mediating oncogenic NOTCH1 activation in T-ALL. Leukemia 25, 1564–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miele L., Miao H., Nickoloff B. J. (2006) NOTCH signaling as a novel cancer therapeutic target. Curr. Cancer Drug Targets 6, 313–323 [DOI] [PubMed] [Google Scholar]
- 14. Sato H., Takino T., Miyamori H. (2005) Roles of membrane-type matrix metalloproteinase-1 in tumor invasion and metastasis. Cancer Sci. 96, 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hofmann U. B., Westphal J. R., Van Muijen G. N., Ruiter D. J. (2000) Matrix metalloproteinases in human melanoma. J. Invest. Dermatol. 115, 337–344 [DOI] [PubMed] [Google Scholar]
- 16. Harrison M., Abu-Elmagd M., Grocott T., Yates C., Gavrilovic J., Wheeler G. N. (2004) Matrix metalloproteinase genes in Xenopus development. Dev. Dyn. 231, 214–220 [DOI] [PubMed] [Google Scholar]
- 17. Tomlinson M. L., Guan P., Morris R. J., Fidock M. D., Rejzek M., Garcia-Morales C., Field R. A., Wheeler G. N. (2009) A chemical genomic approach identifies matrix metalloproteinases as playing an essential and specific role in Xenopus melanophore migration. Chem. Biol. 16, 93–104 [DOI] [PubMed] [Google Scholar]
- 18. Hofmann U. B., Westphal J. R., Zendman A. J., Becker J. C., Ruiter D. J., van Muijen G. N. (2000) Expression and activation of matrix metalloproteinase-2 (MMP-2) and its co-localization with membrane-type 1 matrix metalloproteinase (MT1-MMP) correlate with melanoma progression. J. Pathol. 191, 245–256 [DOI] [PubMed] [Google Scholar]
- 19. Gupta P. B., Kuperwasser C., Brunet J. P., Ramaswamy S., Kuo W. L., Gray J. W., Naber S. P., Weinberg R. A. (2005) The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat. Genet. 37, 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Razorenova O. V., Agapova L. S., Budanov A. V., Ivanov A. V., Strunina S. M., Chumakov P. M. (2005) [Retroviral reporter systems for the assessment of activity of stress-induced signal transduction pathways controlled by p53, HIF-1 and HSF-1 transcription factors]. Mol. Biol. (Mosk.) 39, 286–293 [PMC free article] [PubMed] [Google Scholar]
- 21. Nishimura M., Isaka F., Ishibashi M., Tomita K., Tsuda H., Nakanishi S., Kageyama R. (1998) Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics 49, 69–75 [DOI] [PubMed] [Google Scholar]
- 22. Proweller A., Pear W. S., Parmacek M. S. (2005) Notch signaling represses myocardin-induced smooth muscle cell differentiation. J. Biol. Chem. 280, 8994–9004 [DOI] [PubMed] [Google Scholar]
- 23. Buas M. F., Kabak S., Kadesch T. (2009) Inhibition of myogenesis by Notch: evidence for multiple pathways. J. Cell. Physiol. 218, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 25. Poincloux R., Lizárraga F., Chavrier P. (2009) Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 122, 3015–3024 [DOI] [PubMed] [Google Scholar]
- 26. Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. (2001) Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 153, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jin G., Zhang F., Chan K. M., Xavier Wong H. L., Liu B., Cheah K. S., Liu X., Mauch C., Liu D., Zhou Z. (2011) MT1-MMP cleaves Dll1 to negatively regulate Notch signalling to maintain normal B-cell development. EMBO J. 30, 2281–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koshikawa N., Mizushima H., Minegishi T., Iwamoto R., Mekada E., Seiki M. (2010) Membrane type 1-matrix metalloproteinase cleaves off the NH2-terminal portion of heparin-binding epidermal growth factor and converts it into a heparin-independent growth factor. Cancer Res. 70, 6093–6103 [DOI] [PubMed] [Google Scholar]
- 29. Sabbota A. L., Kim H. R., Zhe X., Fridman R., Bonfil R. D., Cher M. L. (2010) Shedding of RANKL by tumor-associated MT1-MMP activates Src-dependent prostate cancer cell migration. Cancer Res. 70, 5558–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Golubkov V. S., Aleshin A. E., Strongin A. Y. (2011) Potential relation of aberrant proteolysis of human protein tyrosine kinase 7 (PTK7) chuzhoi by membrane type 1 matrix metalloproteinase (MT1-MMP) to congenital defects. J. Biol. Chem. 286, 20970–20976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Golubkov V. S., Chekanov A. V., Cieplak P., Aleshin A. E., Chernov A. V., Zhu W., Radichev I. A., Zhang D., Dong P. D., Strongin A. Y. (2010) The Wnt/planar cell polarity protein-tyrosine kinase-7 (PTK7) is a highly efficient proteolytic target of membrane type-1 matrix metalloproteinase: implications in cancer and embryogenesis. J. Biol. Chem. 285, 35740–35749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. (1994) A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature 370, 61–65 [DOI] [PubMed] [Google Scholar]
- 33. Okada A., Bellocq J. P., Rouyer N., Chenard M. P., Rio M. C., Chambon P., Basset P. (1995) Membrane-type matrix metalloproteinase (MT-MMP) gene is expressed in stromal cells of human colon, breast, and head and neck carcinomas. Proc. Natl. Acad. Sci. U.S.A. 92, 2730–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Tetering G., van Diest P., Verlaan I., van der Wall E., Kopan R., Vooijs M. (2009) Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 284, 31018–31027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Postovit L. M., Costa F. F., Bischof J. M., Seftor E. A., Wen B., Seftor R. E., Feinberg A. P., Soares M. B., Hendrix M. J. (2007) The commonality of plasticity underlying multipotent tumor cells and embryonic stem cells. J. Cell. Biochem. 101, 908–917 [DOI] [PubMed] [Google Scholar]
- 36. Strizzi L., Hardy K. M., Seftor E. A., Costa F. F., Kirschmann D. A., Seftor R. E., Postovit L. M., Hendrix M. J. (2009) Development and cancer: at the crossroads of Nodal and Notch signaling. Cancer Res. 69, 7131–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gordon W. R., Vardar-Ulu D., Histen G., Sanchez-Irizarry C., Aster J. C., Blacklow S. C. (2007) Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 14, 295–300 [DOI] [PubMed] [Google Scholar]
- 38. Gordon W. R., Arnett K. L., Blacklow S. C. (2008) The molecular logic of Notch signaling–a structural and biochemical perspective. J. Cell Sci. 121, 3109–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 [DOI] [PubMed] [Google Scholar]
- 40. Pan D., Rubin G. M. (1997) Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90, 271–280 [DOI] [PubMed] [Google Scholar]
- 41. Sotillos S., Roch F., Campuzano S. (1997) The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development 124, 4769–4779 [DOI] [PubMed] [Google Scholar]
- 42. Wen C., Metzstein M. M., Greenwald I. (1997) SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development 124, 4759–4767 [DOI] [PubMed] [Google Scholar]
- 43. Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 [DOI] [PubMed] [Google Scholar]