Background: Excitation-contraction coupling in striated muscle requires intracellular Ca2+ release through ryanodine receptor/Ca2+-release channels (RyRs).
Results: S-Palmitoylation is a previously unidentified post-translational modification of skeletal muscle RyR1. Diminishing S-palmitoylation significantly diminishes RyR1 activity including stimulus-dependent Ca2+ release.
Conclusion: S-Palmitoylation provides a previously unidentified mechanism to regulate Ca2+ flux in skeletal muscle.
Significance: S-Palmitoylation is likely to regulate Ca2+ flux in many cell types.
Keywords: Calcium Intracellular Release, Post-translational Modification, Protein Palmitoylation, Ryanodine Receptor, Skeletal Muscle
Abstract
The ryanodine receptor/Ca2+-release channels (RyRs) of skeletal and cardiac muscle are essential for Ca2+ release from the sarcoplasmic reticulum that mediates excitation-contraction coupling. It has been shown that RyR activity is regulated by dynamic post-translational modifications of Cys residues, in particular S-nitrosylation and S-oxidation. Here we show that the predominant form of RyR in skeletal muscle, RyR1, is subject to Cys-directed modification by S-palmitoylation. S-Palmitoylation targets 18 Cys within the N-terminal, cytoplasmic region of RyR1, which are clustered in multiple functional domains including those implicated in the activity-governing protein-protein interactions of RyR1 with the L-type Ca2+ channel CaV1.1, calmodulin, and the FK506-binding protein FKBP12, as well as in “hot spot” regions containing sites of mutations implicated in malignant hyperthermia and central core disease. Eight of these Cys have been identified previously as subject to physiological S-nitrosylation or S-oxidation. Diminishing S-palmitoylation directly suppresses RyR1 activity as well as stimulus-coupled Ca2+ release through RyR1. These findings demonstrate functional regulation of RyR1 by a previously unreported post-translational modification and indicate the potential for extensive Cys-based signaling cross-talk. In addition, we identify the sarco/endoplasmic reticular Ca2+-ATPase 1A and the α1S subunit of the L-type Ca2+ channel CaV1.1 as S-palmitoylated proteins, indicating that S-palmitoylation may regulate all principal governors of Ca2+ flux in skeletal muscle that mediates excitation-contraction coupling.
Introduction
Post-translational protein modifications, which provide the principal cellular mechanism for regulating protein function, target multiple amino acids. Cysteine (Cys) residues are distinctive targets in that they are acted on through multiple chemistries that mediate separate, reversible, and dynamic modifications including S-nitrosylation, S-oxidation, and S-palmitoylation. These modifications have been shown to operate on thousands of proteins and thereby to regulate a large array of signal transduction pathways. The ryanodine receptor/Ca2+-release channels (RyR1–3),2 localized to the sarcoplasmic reticulum (SR) of skeletal and cardiac striated muscle as well as to the dendritic compartment of neurons, provide a laboratory for elucidating the regulation of and interplay between Cys-based post-translational modifications. RyRs are unusually large proteins (approximately 5000 amino acids) that contain approximately 100 Cys residues. RyR1, the predominant form in skeletal muscle, is S-nitrosylated under physiological conditions at a single Cys residue (Cys-3635) (1, 2). Physiological S-nitrosylation, which reflects production of nitric oxide by a nitric-oxide synthase localized to the sarcolemma, is coupled to muscle pO2 through O2-dependent S-oxidation: at relatively high pO2 (ambient O2), S-oxidation of a small set of thiols on average (∼6), which does not include Cys-3635 (3), allosterically abrogates S-nitrosylation of Cys-3635 (1) and enhances RyR1 activity (1, 4). This average set of thiols represents substoichiometric S-oxidation of a larger set of Cys residues that constitute an allosteric network, the constituents of which have been identified recently (5). pO2-coupled S-oxidation is mediated by O2-dependent production of H2O2 by an SR-resident NADPH oxidase, Nox4 (4), and represents at least in part the formation of intra-RyR1 subunit disulfide linkages (5).
We report here that RyR1 is modified by an additional form of Cys-directed post-translational modification, S-palmitoylation, the thioester-linked addition to a Cys thiol of a 16-carbon saturated fatty acid. Mass spectrometric analysis identified 18 S-palmitoylated RyR1 Cys residues, which are distributed widely within RyR1 in multiple functional domains. Removing palmitate from RyR1 significantly diminishes RyR1 activity both in situ (intact SR vesicles) and in vitro (purified RyR1), and moreover, in cultured myofibers, inhibiting palmitoylation significantly diminishes electrically evoked Ca2+ release through RyR1. Thus, RyR1 is subject to multisite, activity-governing S-palmitoylation.
EXPERIMENTAL PROCEDURES
Preparation of SR Vesicles and Purification of RyR1
SR vesicles were prepared from rabbit hind-limb muscle essentially as described (4, 6). In brief, tissue was homogenized in buffer containing 20 mm HEPES, pH 7.4, 2 mm EDTA, 0.2 mm EGTA, 0.3 m sucrose, and protease inhibitors (100 nm aprotinin, 20 μm leupeptin, 1 μm pepstatin, 0.2 mm PMSF, 1 mm benzamidine). Homogenates were centrifuged at 9200 × g for 20 min, and the resultant supernatant was centrifuged at 100,000 × g for 1 h. The resultant membrane-enriched microsomal pellet was resuspended and fractionated on a continuous 20–45% sucrose gradient by centrifugation at 100,000 × g for 14 h. Fractions containing SR vesicles were eluted, and SR vesicles were collected by centrifugation at 120,000 × g, resuspended, aliquoted, and stored under liquid nitrogen. Unless otherwise noted, the heavy SR fraction was used for subsequent analysis. RyR1 was purified from CHAPS-solubilized SR vesicles by sucrose density gradient centrifugation as described (4, 7). Protein concentrations were determined with a bicinchoninic acid-based assay.
Acyl-RAC (Resin-assisted Capture of S-Acylated Proteins)
The acyl-RAC method was applied essentially as described (8, 9) with minor modifications. SR vesicles were solubilized in thiol-blocking buffer (100 mm HEPES, 1.0 mm EDTA, 2.5% SDS, 50 mm N-ethylmaleimide, pH 7.5) at a final protein concentration of 1 mg/ml and incubated at 50 °C for 1 h with frequent vortexing. Following acetone precipitation, the pellet was rinsed four times with 70% acetone, repelleted, and rinsed four more times with 70% acetone before resuspension in 150 μl of binding buffer (100 mm HEPES, 1.0 mm EDTA, 1% SDS, pH 7.5). When employed, an equal volume of freshly prepared 1 m NH2OH (pH adjusted to 7.0 with NaOH) was added followed by ∼40 μl of prewashed thiopropyl-Sepharose (Amersham Biosciences). Binding was carried out on a rotator at room temperature for 4 h. Resins were pelleted at 1000 × g following each of 3–5 washes with binding buffer and either eluted in 40 μl of Laemmli loading buffer (2.1% SDS, 66 mm Tris, 26% glycerol w/v, 50 mm DTT) at 42 °C for 10 min for Western blotting or silver staining, or processed for mass spectrometric analysis. Western blotting for RyR1, SERCA 1A, and the α1S subunit of CaV1.1 employed, respectively, mouse monoclonal antibodies 34C, VE121Gp, and 1A (Pierce).
Liquid Chromatography and Tandem Mass Spectrometry (LC-MS/MS)
Following acyl-RAC, thiopropyl-Sepharose was suspended in 1 ml of 50 mm NH4HCO3, 1 mm EDTA, 1 mm CaCl2 containing 0.5 μg of trypsin (Promega) and rotated at 37 °C for 12 h. Following five washes with 1 ml of wash buffer containing 500 mm NaCl, 1% Nonidet P-40 and five washes with 1 ml of 50 mm NH4HCO3, the resin was resuspended in 50 μl of 5 mm TCEP in 50 mm NH4HCO3, pH 8.0, and heated at 60 °C for 30 min with frequent vortexing. The resin was then pelleted by centrifugation at 2000 × g for 2 min, the eluate was removed, and eluted peptides were labeled with 15 mm iodoacetamide in 50 mm NH4HCO3 for 45 min at room temperature in the dark. Peptides were dried under reduced pressure and resuspended in 0.5% trifluoroacetic acid, 5% acetonitrile. Residual iodoacetamide and TCEP were removed with a C18 spin column (Pierce) according to the manufacturer's instructions, made 0.1% with respect to formic acid, and analyzed by LC-MS/MS. Peptides were separated via capillary liquid chromatography with a Waters nanoAquity system (Waters Corp., Milford, MA). The mobile phase A (aqueous) contained 0.1% formic acid in 5% acetonitrile, and mobile phase B (organic) contained 0.1% formic acid in 85% acetonitrile. Separation was achieved using a C18 column (BEH300, 75 μm × 20 cm; Waters Corp.) and a 180-min gradient of 6–45% mobile phase B at a flow rate of 300 nl/min. Mass spectrometric analysis was performed using a hybrid linear ion trap Orbitrap Velos mass spectrometer (LTQ-Orbitrap Velos; Thermo, Waltham, MA). A survey scan was carried out at 60,000 resolution, followed by 10 data-dependent collision-induced dissociation fragmentations. Peptide identification was achieved by searching against the rabbit RyR1 protein sequence (access no. P11716.1) or the nonredundant rabbit database from the National Centre for Biotechnology Information (NCBInr, 2011). Protein identification using Sequest (10) or ProLuCID (11), and DTASelect (12, 13) was carried out with the Integrated Proteomics Pipeline (IP2; Integrated Proteomics Applications, San Diego) or MassMatrix (14). Mass accuracy was limited to 10 ppm for precursor ions and 0.6 Da for product ions, with tryptic enzyme specificity and up to two missed cleavages. Variable modifications included cysteine alkylation by iodoacetamide (57 Da) or N-ethylmaleimide (125 Da), and methionine oxidation (16 Da). Identified spectra were checked manually to ensure quality.
Assay of RyR1 Activity by [3H]Ryanodine Binding
RyR1 activity was assayed by [3H]ryanodine binding essentially as described (4, 15). SR vesicles derived from rabbit hind-limb muscle were incubated for 2 h at room temperature in 100 mm HEPES, pH 7, 1 mm EDTA with or without 0.5 m NH2OH (pH 7.0 by titration with NaOH), and collected by centrifugation at 100,000 × g for 1 h at 4 °C. The pellets were washed three times with 100 mm phosphate buffer, pH 7.4, and resuspended in [3H]ryanodine binding buffer comprising 20 mm imidazole, 125 mm KCl, pH 7.0, 1 mm CaCl2, 0.3 mm Pefabloc (Roche Applied Science), and 30 μm leupeptin and containing 5 nm [3H]ryanodine (PerkinElmer Life Sciences). Nonspecific binding was determined by co-incubation with a 1000-fold excess of unlabeled ryanodine. After incubation overnight at room temperature, samples were diluted with 20 volumes of H2O at 4 °C and distributed evenly on Whatman GF/B filters soaked with 2% (w/w) polyethyleneimine. Filters were washed three times under vacuum with 5 ml of buffer/wash (1 mm Pipes, 0.1 m KCl, pH 7.0), and the radioactivity remaining on the filters was measured by liquid scintillation counting. We also employed [3H]ryanodine binding to assay the activity of RyR1 purified from CHAPS-solubilized SR vesicles by sucrose density gradient centrifugation, as above. Fractions containing RyR1 were pooled, and 35 μg of protein was added to 1 ml of [3H]ryanodine binding buffer and incubated overnight at room temperature. Binding was terminated by the addition of a 10-fold excess of cold water, and the resultant solution was spotted on filters and processed as above.
Assay of Intracellular Ca2+ Release in Primary Myocytes
Single muscle fibers were isolated from mouse (C57BL/6) flexor digitorum brevis muscle as described (4, 16). In particular, muscle fiber bundles were separated by blunt dissection and incubated in Tyrode's solution containing 2 mg/ml collagenase (type II; Worthington) for 2 h at 37 °C with gentle agitation and then transferred to DMEM supplemented with BSA (0.2% w/v), 50 units/ml penicillin, and 50 mg/ml streptomycin. Individual myofibers were generated by trituration, collected by centrifugation at 1000 × g for 1 min, and resuspended in DMEM. Myofibers were plated and incubated overnight in DMEM (37 °C; 5% CO2/95% O2). Prior to assay, myofibers were incubated for 1, 2, or 4 h (37 °C; 5% CO2/95% O2) with or without 2-bromopalmitate (2-BP, 1 μm) and subsequently washed three times with Tyrode's solution. Myofibers were then loaded with Fluo 3-AM (7 μm) for 45 min, collected by centrifugation at 1000 × g for 1 min, and resuspended in Tyrode's solution. Myofibers were then placed in a sealed chamber designed for field stimulation (Warner Instruments) that was superfused with a continuous flow of Tyrode's solution externally sparged with room air and visualized using an inverted Axiovert microscope (Zeiss). Ca2+ transients were evoked by electrical stimulation at 1 Hz (50-ms square-wave pulse, 40–50 V). Evoked Ca2+ transients were recorded with a CCD camera system (PentaMAX; Princeton Instruments), and the amplitudes of transients were calculated as the difference between peak and baseline levels normalized with respect to baseline fluorescence (F/Fo).
Assay of RyR1 Free Thiol Content
The thiol-specific fluorescent agent monobromobimane (MBB; Molecular Probes) (17) was used to determine the free thiol content of RyR1 derived from control versus NH2OH-treated SR vesicles. SR vesicles were prepared as above, and 3 mg total vesicle protein was incubated for 4 h in 100 mm HEPES, pH 7, 1 mm EDTA, 50 μm MBB, with or without 500 mm NH2OH. Vesicles were then collected by centrifugation at 100,000 × g and solubilized in CHAPS prior to isolation of RyR1 by sucrose density gradient centrifugation, as above. This fraction was then subjected to SDS-PAGE, the resultant gel was silver-stained, and the band containing RyR1 (verified by Western blotting) was excised. The resultant gel fragment was destained, and RyR1 was digested in-gel with 0.5 μg of trypsin in 100 μl of 50 mm NH4HCO3, 1 mm Ca2+. MBB fluorescence of the eluted peptides was measured on a fluorescence spectrometer.
Assay of Palmitate Turnover on Ryr1 (18–20)
The ω-alkynyl-palmitate analog 17-octadecynoic acid (18–20) (17-ODYA; Cayman Chemical) was dissolved in dimethyl sulfoxide, and a working stock solution of 100 μm 17-ODYA was prepared in serum-free DMEM supplemented with 5% fatty acid-free BSA and sonicated for 15 min followed by rotation for 15 min at room temperature. For each experiment (n = 4), gastrocnemius and flexor digitorum brevis muscles were removed bilaterally from one or two C57BL/6 mice, and myofibers were prepared and incubated overnight as described above. Myofibers were then replated in serum-free DMEM containing 5% fatty acid-free BSA and incubated at 37 °C under 5% CO2 for 1 h, and then replated in DMEM/fatty acid-free BSA containing 100 μm 17-ODYA with or without 1 μm 2-BP and incubated for 2 h at 37 °C/5% CO2. Myofibers were then washed twice with ice-cold phosphate-buffered saline and lysed in Nonidet P-40 Cell Lysis Buffer (Invitrogen) containing 1% SDS, 1 mm PMSF (Sigma-Aldrich), and Protease Inhibitor Mixture (Promega). For click chemistry (biotinylation), lysate containing 1 mg of total protein was adjusted to a reaction volume of 1 ml containing 0.1 mm biotin-azide (Invitrogen), 1 mm neutralized TCEP (Thermo Scientific), 0.2 mm TBTA (Sigma-Aldrich) dissolved in dimethyl sulfoxide/tert-butanol (20%/80%) and 1 mm CuSO4 (final reagent volume adjusted with PBS), and incubated for 2 h at room temperature in the dark. Proteins were then precipitated with methanol:chloroform:water (4:1:3) and collected by centrifugation at 15,000 × g for 1 min, the aqueous phase was removed by aspiration retaining the protein pellet at the interface, and 400 μl of methanol was added prior to a second centrifugation at 15,000 × g for 2 min. The resultant pellet was dissolved in 1% SDS in PBS, and the protein concentration was determined. For Western blot analysis following biotinylation, 80 μg of protein was resolved by SDS-PAGE and transferred onto PVDF membrane blocked with 5% nonfat milk in PBS, 0.05% Tween 20 (PBST). Membranes were washed 2 × 5 min with PBST and incubated with streptavidin-horseradish peroxidase (Sigma, 1:1000 in PBST) for 1 h at room temperature. The membrane was then washed 5 × 5 min with PBST, and biotin-avidin complexes were visualized using Enhanced Chemiluminescence reagent (Amersham Biosciences). To verify that 17-ODYA is incorporated through a thioester linkage, Western blots were incubated with 720 mm neutralized NH2OH for 72 h at room temperature prior to incubation with streptavidin. Identification of RyR1 was on the basis of Western blotting of lysates run in parallel, and quantification of results was carried out by normalizing with respect to sample content of RyR1.
RESULTS
RyR1 Is Modified by S-Palmitoylation
We employed the acyl-RAC method (which we developed (8)) to probe for S-palmitoylated proteins in SR vesicles, a subcellular fraction highly enriched for SR that also contains some transverse tubule (T-tubule) membrane (4). In this method (8, 9), free thiols are blocked, treatment with neutral hydroxylamine selectively cleaves thioester-linked palmitate from modified Cys, and proteins are coupled to thiopropyl-Sepharose resin through the free thiols created by hydroxylamine treatment. Subsequently eluted proteins may be analyzed intact, or peptides may be eluted following proteolytic digestion for mass spectrometric identification of specific palmitate-modified Cys.
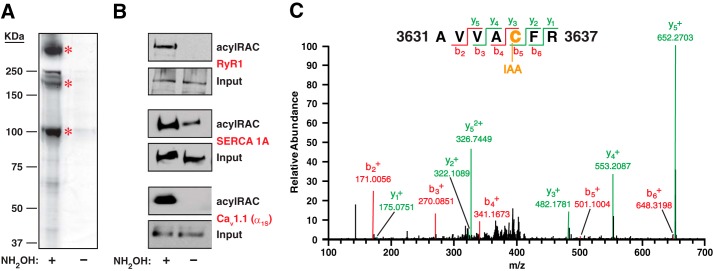
A number of proteins were pulled down by acyl-RAC from solubilized SR vesicles in a hydroxylamine-dependent fashion, as revealed by silver staining following SDS-PAGE of resin eluates (Fig. 1A). The most prominent substrates were identified as RyR1, the sarco/endoplasmic reticulum Ca2+-ATPase SERCA 1A (ATP2A1) and the α1S subunit of CaV1.1 (L-type Ca2+ channel, dihydropyridine receptor (DHPR)) on the basis of their characteristic position (Mr) and by Western blotting (Fig. 1, A and B). LC-MS/MS of peptides eluted from the immobilizing resin following trypsin digest confirmed the identification of RyR1 and SERCA 1A (Figs. 1C, Fig. 2, and Table 1). We did not identify peptides from the α1S subunit of CaV1.1 by LC-MS/MS under the conditions employed. To further control for possible false-positive mass spectrometric identification of palmitoylated Cys residues, eluates from resin incubated with solubilized SR vesicles that had not been treated with hydroxylamine were analyzed in parallel by LC-MS/MS in every experiment, and Cys-containing peptides were never identified in the absence of hydroxylamine.
FIGURE 1.
S-Palmitoylated proteins of skeletal muscle SR vesicles. A, analysis of rabbit SR vesicles by acyl-RAC followed by silver staining reveals pulldown of a number of proteins in a hydroxylamine (NH2OH)-dependent fashion, identifying them as substrates of S-palmitoylation. Asterisks indicate, from top to bottom, RyR1, the α-subunit of the L-type Ca2+ channel CaV1.1 (the lowest of three closely spaced bands) and SERCA 1A. B, Western blotting confirms the identification of RyR1, SERCA 1A, and the α-subunit of CaV1.1. C, a representative peptide spectrum produced by LC-MS/MS following acyl-RAC of SR vesicles and trypsin digest is shown. Cys-3635 (previously identified as the principal site of physiological S-nitrosylation of RyR1 (2)) was among the Cys residues identified as sites of S-palmitoylation.
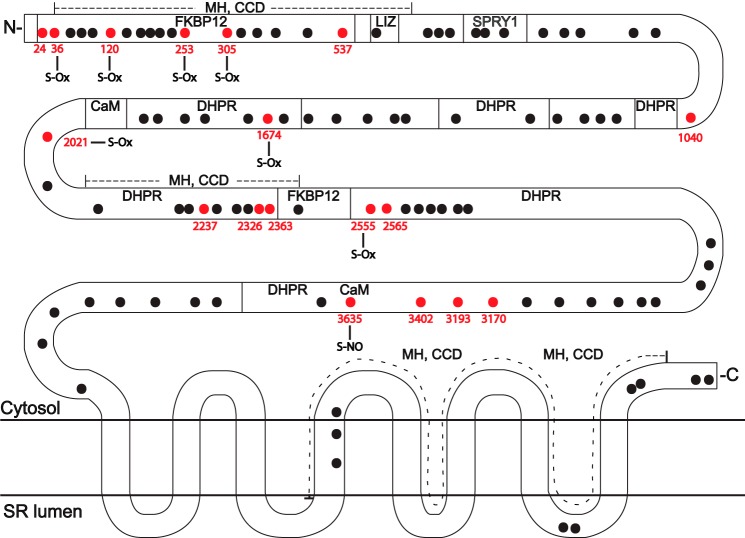
FIGURE 2.
Localization of S-palmitoylated Cys within rabbit RyR1 (shown in red), as identified by acyl-RAC. Previously identified sites of physiological S-oxidation (S-Ox) (5) and S-nitrosylation (S-NO) (2) are indicated. Transmembrane domains, regions implicated in the interaction of RyR1 with the L-type Ca2+ channel CaV1.1 (dihydropyridine receptor, DHPR), FK506-binding protein 12 (FKB12), and calmodulin (CaM), leucine/isoleucine zipper (LIZ) motifs, and the SPRY1 domain, as well as hot spot regions implicated in malignant hypothermia/central core disease (MH, CCD), are delineated on the basis of the human RyR1 sequence (reviewed recently in Ref. 39).
TABLE 1.
Conservation in RyR2 and RyR3 of Cys residues identified as sites of palmitoylation in RyR1
X = conserved Cys.
| RyR1 | RyR2 | RyR3 |
|---|---|---|
| 24 | X | X |
| 36 | X | X |
| 120 | X | X |
| 253 | ||
| 305 | ||
| 537 | X | X |
| 1040 | ||
| 1674 | X | X |
| 2021 | X | X |
| 2237 | X | X |
| 2326 | ||
| 2363 | X | X |
| 2555 | X | X |
| 2565 | X | X |
| 3170 | X | |
| 3193 | X | X |
| 3402 | X | |
| 3635 | X | X |
Identification of S-Palmitoylated Cys Residues
Acyl-RAC coupled to LC-MS/MS identified 18 palmitoylated Cys residues within RyR1 (seen in at least three of five separate experiments) (Fig. 2 and Table 1). This represents a minimum number of palmitoylated Cys residues because theoretical analysis indicates that trypsin digest provides coverage by LC-MS/MS of no more than 76% of Cys residues within RyR1 (i.e. 76 of 100 Cys residues found in one or more peptides of ∼800–3000 Da). Palmitoylated Cys residues were distributed throughout the N-terminal cytoplasmic domain of RyR1, but appeared to be grouped in three principal clusters (Fig. 2). The most N-terminal cluster comprised 6 of 18 Cys residues between and including RyR1 residues 24–537, which were localized in functional domains previously identified as those mediating the interaction of RyR1 with FK506-binding protein 12 (FKBP12) and as “hot spot” domains implicated in malignant hyperthermia and central core disease. A second cluster comprised 6 of 9 Cys residues between and including RyR1 residues 2237–2565, which were localized to functional domains previously identified as participating in the interaction of RyR1 with Cav1.1 and to hot spot domains for malignant hyperthermia and central core disease. The third cluster comprised 4 of 4 Cys residues between and including RyR1 residues 3170–3635, which were localized to a CaV1.1-interacting domain as well as the domain implicated in the interaction of RyR1 with calmodulin. All palmitoylated Cys residues identified in rabbit RyR1 are conserved in mammalian RyR1s, including human RyR1, with the exception of Cys-305. In addition, 12 of 18 and 14 of 18 palmitoylated Cys residues identified in RyR1 are conserved in RyR2 and RyR3, respectively (Table 1). Notably, 7 of 18 palmitoylated Cys residues have been identified previously as subject to physiological oxygen-coupled S-oxidation (5), including 5 of 6 Cys within the N-terminal cluster of palmitoylated Cys residues (Fig. 2). The principal site of physiological S-nitrosylation (Cys-3635) (2) was also identified as a site of S-palmitoylation (Figs. 1C and 2).
In addition, mass spectrometric analysis identified 8 palmitoylated Cys (seen in at least two of four separate experiments) located throughout the cytoplasmic domain of SERCA 1A (Cys-12, Cys-344, Cys-349, Cys-364, Cys-471, Cys-636, Cys-674, and Cys-675); trypsin digest coupled with LC-MS/MS as employed would in theory yield coverage of 18 of 24 Cys residues within SERCA 1A. Six of the 8 Cys residues identified as palmitoylated in SERCA 1A (ATP2A1) are conserved in SERCA 2 (ATP2A2) and SERCA 3 (ATP2A3); the exceptions are Cys-12 and Cys-674. Thus, the principal proteins that govern SR Ca2+ flux are subject to S-palmitoylation, and there exists the potential for extensive cross-talk between S-palmitoylation and alternative Cys-based post-translational modifications of RyR1.
Functional Effects of Diminished S-Palmitoylation
To assess a possible role for S-palmitoylation in regulation of RyR1 activity, we compared the activity of RyR1 in native SR vesicles with the activity in vesicles treated with neutral hydroxylamine to remove thioester-linked palmitate. Treatment of SR vesicles with hydroxylamine (0.5 m, 2 h) reduced RyR1 activity by ∼62% as assessed by assay of [3H]ryanodine binding (Fig. 3A). Ryanodine binds to RyR1 in the active state in which the Ca2+ pore is open, and the [3H]ryanodine binding assay therefore provides a direct indication of channel open probability (po). To verify that treatment with hydroxylamine depalmitoylated RyR1, we further purified RyR1 from CHAPS-solubilized untreated and hydroxylamine-treated SR vesicles in which free thiols had been labeled with the fluorescent thiol reporter MBB, employing density gradient centrifugation followed by SDS-PAGE of RyR1-enriched gradient fractions to complete the purification. The band corresponding to RyR1 was excised and subjected to trypsin digest followed by assay of MBB fluorescence. The average hydroxylamine-treated to untreated ratio of MBB fluorescence was 1.45 (±0.22 S.D.) (Fig. 3B). We (4) and others (21) have previously reported that RyR1 possesses an average of approximately 35 free thiols/RyR1 monomer under similar conditions (room air) as assessed by MBB assay. Thus, the 45% increase in MBB fluorescence following hydroxylamine treatment (measured within the linear range of the MBB assay) represents a gain of 16 thiols/RyR1 monomer on average, consistent with our identification of 18 Cys residues within RyR1 that are subject to S-palmitoylation. Our analysis does not allow us to deduce the stoichiometry of S-palmitoylation of individual Cys residues, and it should be noted that the magnitude of the increase in free thiols following treatment with hydroxylamine will reflect the fact that, as noted above, 7 of the Cys we identified as S-palmitoylated were identified previously as subject to (hydroxylamine-insensitive) substoichiometric S-oxidation under similar conditions (5) and that an additional palmitoylated Cys residue (Cys-3635) is subject to activity- and pO2-dependent S-nitrosylation (2) (we detect S-nitrosylation of RyR1 in SR vesicles under basal conditions by SNO-RAC (Ref. 8 and data not shown).
FIGURE 3.
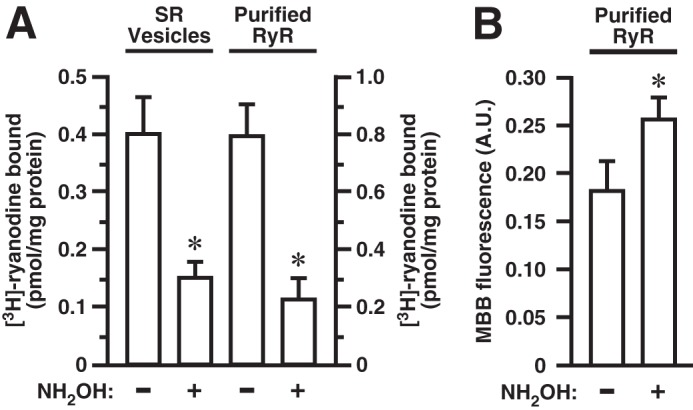
Suppression of RyR1 activity by depalmitoylation with hydroxylamine (NH2OH). A, at left, depalmitoylation by treatment with hydroxylamine diminishes RyR1 activity in SR vesicles as assessed by [3H]ryanodine binding (n = 3 separate experiments; *, p < 0.05). At right, hydroxylamine treatment diminishes the activity of RyR1 purified from untreated SR vesicles versus SR vesicles treated with hydroxylamine prior to assay (n = 4 separate experiments; *, p < 0.05). B, assay of free thiols by MBB fluorescence in RyR1 purified from untreated or hydroxylamine-treated SR vesicles demonstrates hydroxylamine-dependent generation of free thiols, indicative of depalmitoylation (n = 4 separate experiments; *, p < 0.05). In A and B, error bars indicate S.E.
In addition, we assayed RyR1 activity by [3H]ryanodine binding after further purification of RyR1 from untreated and hydroxylamine-treated SR vesicles by the well established method of density gradient centrifugation following CHAPS solubilization. Hydroxylamine treatment decreased the activity of RyR1 in RyR1-enriched gradient fractions by ∼72% (Fig. 3A), which is closely comparable with the decrease observed in intact SR vesicles. These findings are consistent with the conclusion that depalmitoylation of RyR1 suppresses RyR1 activity at least in part through a direct effect on channel configuration.
We next examined in cultured myofibers (derived from the flexor digitorum brevis muscle of mice) the effects of inhibiting S-palmitoylation with 2-BP on stimulus-coupled intracellular Ca2+ release through RyR1, induced by electrical depolarization (pacing at 1 Hz) and assessed by Fluo 3-AM fluorescence. If palmitate turns over on one or more Cys residues within RyR1 (and/or other SR proteins) under basal conditions, then incubation with 2-BP, which would prevent palmitoylation, would result in a time-dependent decrease in palmitoylation state that would reach a plateau after an interval determined by the half-life of palmitate turnover. We found that incubation of myofibers with 2-BP (1 μm) prior to assessment of intracellular Ca2+ release decreased the magnitude of release by ∼44% and that the suppressive effect of 2-BP increased with increasing incubation interval and plateaued within 2 h (Fig. 4, A and B). Metabolic labeling (2 h) with the ω-alkynyl-palmitate analog 17-ODYA followed by bio-orthogonal click chemistry with biotin-azide (18–20) demonstrated directly turnover of palmitate on RyR1 under basal conditions, which was greatly diminished by co-incubation with 2-BP (1 μm) (Fig. 4C). Finally, we verified that, as reported previously (4), depolarization-induced Ca2+ release in cultured myofibers as assessed by Fluo 3-AM fluorescence was largely dependent upon RyR1 because it was rapidly eliminated by incubation with ryanodine, to selectively inhibit RyR1 (Fig. 4D). Thus, our results indicate that depalmitoylation, likely of RyR1, substantially diminishes stimulus-coupled Ca2+ release through RyR1.
FIGURE 4.
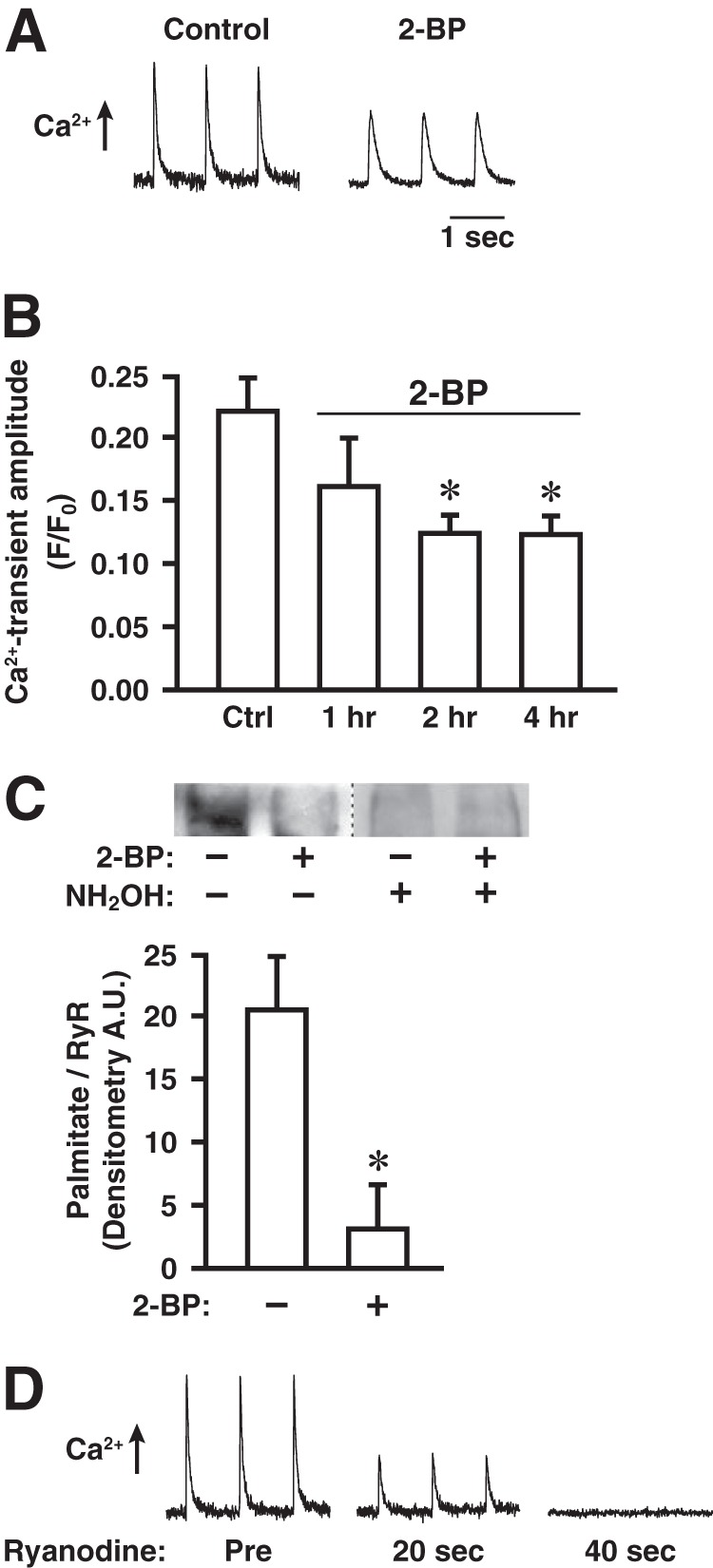
Treatment with 2-BP diminishes electrically induced Ca2+ release through RyR1 and turnover of palmitate on RyR1 in cultured myofibers. A, representative records of electrically induced (pacing at 1 Hz) intracellular Ca2+ release in a control myofiber and a myofiber treated with 2-BP (1 μm, 2 h), as assessed by Fluo 3-AM fluorescence. B, effects on electrically induced Ca2+ release of treatment with 2-BP (1 μm) for the indicated times (n = 4 separate experiments, 8–10 control and 8–10 2-BP-treated myofibers imaged per experiment; *, p < 0.05 with respect to control). Error bars indicate S.E. C, top, metabolic labeling with the ω-alkynyl-palmitate analog 17-octadecynoic acid (2 h) followed by detection of incorporated analog by bio-orthogonal click chemistry with biotin-azide demonstrating dynamic palmitoylation of at least some sites within RyR1 under basal conditions, which is greatly diminished by co-incubation with 2-BP (1 μm). Hydroxylamine (NH2OH)-sensitivity verifies that labeling represents thioester-linked palmitoylation. Bottom, quantification of the degree of RyR1 palmitoylation (normalized for RyR1 content of samples) demonstrating a decrease of approximately 85% in analog incorporation into RyR1 by co-incubation with 2-BP (*, p = 0.01; n = 4 separate experiments). Error bars indicate S.E. D, electrically induced Ca2+ release rapidly suppressed by inhibiting RyR1 with ryanodine (1 μm). Tracings shown are representative of three separate experiments.
DISCUSSION
The continuously expanding list of mammalian proteins shown to be modified by S-palmitoylation now exceeds several hundred substrates. Although the nicotinic cholinergic receptor localized to the skeletal muscle sarcolemma was identified early on as a substrate for S-palmitoylation (22), S-palmitoylation of skeletal muscle membrane proteins localized to internal, SR, and T-tubule membranes has not been reported previously. Our identification of RyR1, SERCA 1A (localized to the SR), and the α1S subunit of CaV1.1 (localized to T-tubules) as substrates for S-palmitoylation indicates that S-palmitoylation modifies the principal governors of Ca2+ release that mediate excitation-contraction coupling in skeletal muscle.
RyR1, SERCA 1A, and the α1S subunit of CaV1.1 are all previously unidentified substrates of S-palmitoylation. RyR1 represents the first example of S-palmitoylation of an intracellular Ca2+-release channel; the α1S subunit of CaV1.1, the prototypical voltage-gated Ca2+ channel, represents the first example of S-palmitoylation of a pore-forming α-subunit of CaV channels; SERCA 1A, the prototype of the family of ion-transporting P-type ATPases, represents the first verified example of S-palmitoylation of a member of the Ca2+-ATPase subfamily (although large scale proteomic screening has identified ATP2A2 and ATP2A3 as candidate S-palmitoylated substrates (23)). It may be anticipated that S-palmitoylation of additional isoforms of RyR, CaV α-subunits, and SERCA will regulate Ca2+ flux in multiple cell types in addition to skeletal muscle.
We identified 18 sites of S-palmitoylation within RyR1, which are clustered in multiple functional domains including those implicated in the activity-governing protein-protein interactions of RyR1 with CaV1.1, calmodulin, and FKBP12, as well as in hot spot regions containing sites of mutations implicated in malignant hyperthermia and central core disease. There is precedent for multisite S-palmitoylation of both voltage- and ligand-gated channels (localized to the plasma membrane) that results in site-specific effects on channel function, in the cases respectively of the large conductance Ca2+-activated potassium (BK) channels that contain the stress-axis-regulated exon (STREX) splice variant (24–26) and the GluN2A/B subunits of the neuronal N-methyl-d-aspartate glutamate receptor/ion channel (27). However, modification of a single protein by as many as 18 palmitoyl groups is unprecedented. The direct interaction between RyR1 and CaV1.1 is the principal determinant of RyR1 activation, whereas the interaction of RyR1 with FKBP12 (calstabin1) is thought to stabilize the closed conformation of the channel (28). It has recently been reported (29) that FKBP12, a cis-trans-prolyl isomerase, binds the small G protein H-Ras, dependent upon H-Ras palmitoylation, and promotes depalmitoylation that is dependent upon a Pro residue proximal to the S-palmitoylated Cys residues within H-Ras (179PGCMSC184). It is of interest that, although Pro residues are not found proximal to S-palmitoylated Cys in the N-terminal cluster of S-palmitoylated Cys that are localized to a predicted FKBP-binding domain within RyR1, Pro residues are found proximal to the two S-palmitoylated Cys that are located adjacent to the predicted C-terminal FKBP-binding domain (2325PCG2327 and 2361PECF2364; see Fig. 2). Thus, although speculative, altered palmitoylation of RyRs might play a role in the multiple striated muscle pathophysiologies that are reported to be associated with altered FKBP12-RyR binding (30).
S-Palmitoylation of various ion channels has been shown to control surface expression, localization to membrane subdomains, internalization and intracellular trafficking, as well as protein-protein interactions (31, 32). These functions have been demonstrated in some cases to reflect the regulation by S-palmitoylation of other post-translational modifications of ion channels, in particular phosphorylation and ubiquitination, which may be exerted through effects of S-palmitoylation on channel subunit configuration (31, 32). However, in addition to possible allosteric regulation, signaling cross-talk between S-palmitoylation and other Cys-based post-translational modification may directly reflect the mutual exclusivity of alternative forms of modification of Cys thiols (33). Our findings indicate substantial overlap between Cys residues identified as sites of S-palmitoylation and those shown previously to be subject to physiological S-oxidation and S-nitrosylation (1, 2, 5). It is possible that a given Cys residue may be preferentially subject to only one modification, but it seems more likely that modification of at least some sites by S-palmitoylation will be antiphasic to modification by S-oxidation or S-nitrosylation, and thus that the state of RyR1 S-palmitoylation will reflect a balance between S-palmitoylation and alternative, redox-based modifications. In addition, to the extent that they occur, interactions between S-palmitoylation and alternative, Cys-based RyR1 modifications might reflect allosteric regulation, as in the case of gating of S-nitrosylation by S-oxidation (1).
Our finding that depalmitoylation of RyR1 substantially suppresses basal channel activity as assessed by [3H]ryanodine binding in both intact SR vesicles and isolated RyR1 suggests that S-palmitoylation of at least some sites within RyR1 influences channel configuration. There are precedents for effects of S-palmitoylation on the gating and activation/inactivation of ion channels, although correlated effects on structural conformation have not been established directly (34–38). Our finding that inhibiting palmitoylation with 2-BP in intact myofibers results in depalmitoylation of at least some Cys residues within RyR1 and also substantially diminishes voltage-triggered Ca2+ release through RyR1 is consistent with the results of chemical depalmitoylation; however, the effects of 2-BP may also reflect depalmitoylation of other proteins that play a role in Ca2+ mobilization through RyR1 (e.g. SERCA 1A and CaV1.1). We conclude that S-palmitoylation is a significant regulator of Ca2+ release from SR stores in skeletal muscle that mediates excitation-contraction coupling and may be expected to play a similar role in RyR-mediated Ca2+ release in the multiple, additional cell types in which one or more forms of RyR operate.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL0591130 and R01 AR018687.
- RyR
- ryanodine receptor/Ca2+-release channel
- acyl-RAC
- resin-assisted capture of S-acylated proteins
- 2-BP
- 2-bromopalmitate
- LC-MS/MS
- liquid chromatography and tandem mass spectrometry
- MBB
- monobromobimane
- 17-ODYA
- 17-octadecynoic acid
- SERCA
- sarco/endoplasmic reticulum Ca2+-ATPase
- SR
- sarcoplasmic reticulum
- T-tubule
- transverse tubule
- TBTA
- Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine
- TCEP
- Tris(2-carboxyethyl)phosphine hydrochloride.
REFERENCES
- 1. Eu J. P., Sun J., Xu L., Stamler J. S., Meissner G. (2000) The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell 102, 499–509 [DOI] [PubMed] [Google Scholar]
- 2. Sun J., Xin C., Eu J. P., Stamler J. S., Meissner G. (2001) Cysteine 3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc. Natl. Acad. Sci. U.S.A. 98, 11158–11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun J., Xu L., Eu J. P., Stamler J. S., Meissner G. (2003) Nitric oxide, NOC-12, and S-nitrosoglutathione modulate the skeletal muscle calcium release channel/ryanodine receptor by different mechanisms: an allosteric function for O2 in S-nitrosylation of the channel. J. Biol. Chem. 278, 8184–8189 [DOI] [PubMed] [Google Scholar]
- 4. Sun Q. A., Hess D. T., Nogueira L., Yong S., Bowles D. E., Eu J., Laurita K. R., Meissner G., Stamler J. S. (2011) Oxygen-coupled redox regulation of the skeletal muscle ryanodine receptor-Ca2+ release channel by NADPH oxidase 4. Proc. Natl. Acad. Sci. U.S.A. 108, 16098–16103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun Q. A., Wang B., Miyagi M., Hess D. T., Stamler J. S. (2013) Oxygen-coupled redox regulation of the skeletal muscle ryanodine receptor/Ca2+-release channel (RyR1): sites and nature of oxidative modification. J. Biol. Chem. 288, 22961–22971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson K., Cohn A. H., Meissner G. (1994) High-affinity [3H]PN200–110 and [3H]ryanodine binding to rabbit and frog skeletal muscle. Am. J. Physiol. 266, C462–466 [DOI] [PubMed] [Google Scholar]
- 7. Lai F. A., Erickson H., Block B. A., Meissner G. (1987) Evidence for a junctional feet-ryanodine receptor complex from sarcoplasmic reticulum. Biochem. Biophys. Res. Commun. 143, 704–709 [DOI] [PubMed] [Google Scholar]
- 8. Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M. A., Stamler J. S. (2009) Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forrester M. T., Hess D. T., Thompson J. W., Hultman R., Moseley M. A., Stamler J. S., Casey P. J. (2011) Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 52, 393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 11. Xu T., Venable J. D., Park S. K., Cociorva D., Lu B., Liao L., Wohlschlegel J., Hewel J., Yates J. R. (2006) ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cell. Proteomics 5, S174 [Google Scholar]
- 12. Tabb D. L., McDonald W. H., Yates J. R., 3rd. (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cociorva D., L. Tabb D., Yates J. R. (2007) Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics Chapter 13, Unit 13.4 [DOI] [PubMed] [Google Scholar]
- 14. Xu H., Freitas M. A. (2007) A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics 8, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai F. A., Erickson H. P., Rousseau E., Liu Q. Y., Meissner G. (1988) Purification and reconstitution of the calcium release channel from skeletal muscle. Nature 331, 315–319 [DOI] [PubMed] [Google Scholar]
- 16. Shefer G., Yablonka-Reuveni Z. (2005) Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol. Biol. 290, 281–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kosower N. S., Kosower E. M. (1987) Thiol labeling with bromobimanes. Methods Enzymol. 143, 76–84 [DOI] [PubMed] [Google Scholar]
- 18. Hannoush R. N., Arenas-Ramirez N. (2009) Imaging the lipidome: ω-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem. Biol. 4, 581–587 [DOI] [PubMed] [Google Scholar]
- 19. Martin B. R., Cravatt B. F. (2009) Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yap M. C., Kostiuk M. A., Martin D. D., Perinpanayagam M. A., Hak P. G., Siddam A., Majjigapu J. R., Rajaiah G., Keller B. O., Prescher J. A., Wu P., Bertozzi C. R., Falck J. R., Berthiaume L. G. (2010) Rapid and selective detection of fatty acylated proteins using ω-alkynyl-fatty acids and click chemistry. J. Lipid Res. 51, 1566–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petrotchenko E. V., Yamaguchi N., Pasek D. A., Borchers C. H., Meissner G. (2011) Mass spectrometric analysis and mutagenesis predict involvement of multiple cysteines in redox regulation of the skeletal muscle ryanodine receptor ion channel complex. Res. Rep. Biol. 2011, 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olson E. N., Glaser L., Merlie J. P. (1984) α and β subunits of the nicotinic acetylcholine receptor contain covalently bound lipid. J. Biol. Chem. 259, 5364–5367 [PubMed] [Google Scholar]
- 23. Dowal L., Yang W., Freeman M. R., Steen H., Flaumenhaft R. (2011) Proteomic analysis of palmitoylated platelet proteins. Blood 118, e62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tian L., Jeffries O., McClafferty H., Molyvdas A., Rowe I. C., Saleem F., Chen L., Greaves J., Chamberlain L. H., Knaus H. G., Ruth P., Shipston M. J. (2008) Palmitoylation gates phosphorylation-dependent regulation of BK potassium channels. Proc. Natl. Acad. Sci. U.S.A. 105, 21006–21011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jeffries O., Geiger N., Rowe I. C., Tian L., McClafferty H., Chen L., Bi D., Knaus H. G., Ruth P., Shipston M. J. (2010) Palmitoylation of the S0-S1 linker regulates cell surface expression of voltage- and calcium-activated potassium (BK) channels. J. Biol. Chem. 285, 33307–33314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou X., Wulfsen I., Korth M., McClafferty H., Lukowski R., Shipston M. J., Ruth P., Dobrev D., Wieland T. (2012) Palmitoylation and membrane association of the stress axis regulated insert (STREX) controls BK channel regulation by protein kinase C. J. Biol. Chem. 287, 32161–32171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hayashi T., Thomas G. M., Huganir R. L. (2009) Dual palmitoylation of NR2 subunits regulates NMDA receptor trafficking. Neuron 64, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zalk R., Lehnart S. E., Marks A. R. (2007) Modulation of the ryanodine receptor and intracellular calcium. Annu. Rev. Biochem. 76, 367–385 [DOI] [PubMed] [Google Scholar]
- 29. Ahearn I. M., Tsai F. D., Court H., Zhou M., Jennings B. C., Ahmed M., Fehrenbacher N., Linder M. E., Philips M. R. (2011) FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol. Cell 41, 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Betzenhauser M.J., Marks A.R. (2010) Ryanodine receptor channelopathies. Pflugers Arch. 460, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shipston M. J. (2011) Ion channel regulation by protein palmitoylation. J. Biol. Chem. 286, 8709–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blaskovic S., Blanc M., van der Goot F. G. (2013) What does S-palmitoylation do to membrane proteins? FEBS J. 280, 2766–2774 [DOI] [PubMed] [Google Scholar]
- 33. Ho G. P., Selvakumar B., Mukai J., Hester L. D., Wang Y., Gogos J. A., Snyder S. H. (2011) S-Nitrosylation and S-palmitoylation reciprocally regulate synaptic targeting of PSD-95. Neuron 71, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gubitosi-Klug R. A., Mancuso D. J., Gross R. W. (2005) The human KV1.1 channel is palmitoylated, modulating voltage sensing: identification of a palmitoylation consensus sequence. Proc. Natl. Acad. Sci. U.S.A. 102, 5964–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mueller G. M., Maarouf A. B., Kinlough C. L., Sheng N., Kashlan O. B., Okumura S., Luthy S., Kleyman T. R., Hughey R. P. (2010) Cys palmitoylation of the β subunit modulates gating of the epithelial sodium channel. J. Biol. Chem. 285, 30453–30462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stephens G. J., Page K. M., Bogdanov Y., Dolphin A. C. (2000) The α1B Ca2+ channel amino terminus contributes determinants for β subunit-mediated voltage-dependent inactivation properties. J. Physiol. 525, 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qin N., Platano D., Olcese R., Costantin J. L., Stefani E., Birnbaumer L. (1998) Unique regulatory properties of the type 2a Ca2+ channel β subunit caused by palmitoylation. Proc. Natl. Acad. Sci. U.S.A. 95, 4690–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bosmans F., Milescu M., Swartz K. J. (2011) Palmitoylation influences the function and pharmacology of sodium channels. Proc. Natl. Acad. Sci. U.S.A. 108, 20213–20218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hwang J. H., Zorzato F., Clarke N. F., Treves S. (2012) Mapping domains and mutations on the skeletal muscle ryanodine receptor channel. Trends Mol. Med. 18, 644–657 [DOI] [PubMed] [Google Scholar]