Abstract
Membrane ion channels regulate key cellular functions and their activity is dependent on their 3D structure. Atomic force microscopy (AFM) images 3D structure of membrane channels placed on a solid substrate. Solid substrate prevents molecular transport through ion channels thus hindering any direct structure-function relationship analysis. Here we designed a ~70 nm nanopore to suspend a membrane, allowing fluidic access to both sides. We used these nanopores with AFM and total internal reflection fluorescence microscopy (TIRFM) for high resolution imaging and molecular transport measurement. Significantly, membranes over the nanopore were stable for repeated AFM imaging. We studied structure-activity relationship of gap junction hemichannels reconstituted in lipid bilayers. Individual hemichannels in the membrane overlying the nanopore were resolved and transport of hemichannel-permeant LY dye was visualized when the hemichannel was opened by lowering calcium in the medium. This integrated technique will allow direct structure-permeability relationship of many ion channels and receptors.
Ion channels are key transmembrane macromolecules that regulate several critical cellular functions and their activity is tightly linked to their underlying three dimensional (3D) structures. However, molecular scale single channel structure-activity relationship is poorly understood due to the lack of direct information. The best way to correlate the structure-function of biomolecules, including channels and receptors, is through direct and simultaneous study of the 3D structure-activity of isolated preparations in single molecule format. Unfortunately, there is a paucity of techniques available for simultaneous 3D molecular structure-activity study of biomolecules and almost all previous studies have been correlative studies using techniques and approaches in parallel. Such combinatorial and correlative approaches linking structure-activity of ion channels often provide incomplete information. Atomic force microscopy (AFM) is capable of high resolution biological imaging in physiological environments and can provide important mechanical and other physiological properties1. However, most studies are currently performed with lipid bilayers supported on a solid substrate that restrict the fluidity of bilayers and conformation change of proteins reconstituted in them thus limiting their function. Further, they inhibit the movement of molecules through the membrane pores. A number of studies have been performed using non-supported membranes2,3,4,5,6,7. However those studies used lipid membranes without any protein for low resolution imaging2,3,4,5,6, or 2D-protein crystals with high density protein packing and thus high membrane rigidity7. They however are not reflective of the majority of native biological systems where channels and receptors are randomly distributed in disordered clusters8.
Here, we describe the design and fabrication of a 70–90 nm diameter nanopore on a silicon support system. The bilayer membrane placed over the nanopore offers an excellent opportunity to study 3D conformations as well as ionic and molecular permeability of transmembrane ion channels and receptors. This nanopore support system allowed imaging with AFM and optical microscopy techniques. We have recently developed a waveguide based, LED powered, high performance total internal reflection microscopy (TIRFM) technique9 that has unique advantages over conventional TIRF systems, including uniform illumination across entire sample, imaging with low numerical aperture objectives for enhanced temporal resolution, fast switching times, no alignment requirements, among others. This TIRF system when integrated with an AFM allows simultaneous high resolution imaging of fluorescent molecules.
We were able to image connexin (Cx) hemichannels reconstituted in a lipid bilayer suspended over a nanopore-support. Connexin gap junctions and hemichannels play an essential role in cell growth, development, function and disease by controlling the flow of ions and signaling molecules (up to 1200 Dalton in size) through them10. The functions of these channels are regulated by variety of physiological signals including low Ca2+, pH, membrane potential, redox states and phosphorylation11,12,13,14,15,16,17,18. Currently, there is very little information available to directly relate structural heterogeneity of connexin hemichannels with their multitude of activity and biological functions.
AFM images of purified Cx43 hemichannels reconstituted in lipid bilayer, suspended over the nanopore support show low resolution and yet distinct hemichannels. Significantly, these images also indicate the membrane stability over the pores, a critical requirement for any repeatable and time lapse study of channels' conformations as a function of external perturbations. We were then able to image permeability of hemichannel permeable fluorescent dye (Lucifer yellow, LY) using the integrated TIRFM. Time lapse TIRF imaging shows no increase in fluorescence over the nanopore, when incubated in normal calcium buffer that keeps the hemichannels closed. On the other hand, an increase in the fluorescence was observed over time when the media was replaced with no calcium containing buffer that opens hemichannels. With further refinement of this integrated technique, it will be possible to obtain direct structure-permeability relationship of many ion channels and receptors.
Results
Cx43 hemichannels in the floating lipid membrane over the nanopore
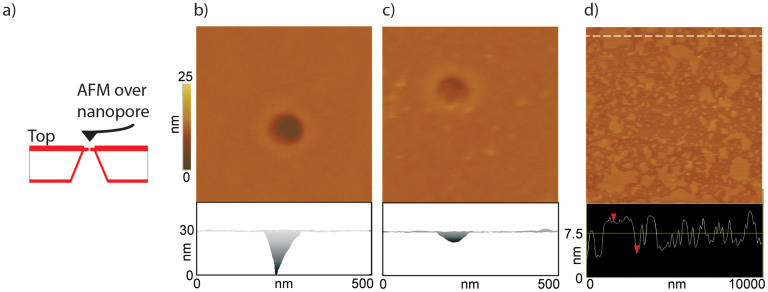
AFM images of the nanopore chip were obtained before depositing the lipids to verify its location and open/close status as several nanopores were blocked. After confirming the presence of an open pore (Fig. 2b), the hemichannels were reconstituted in situ, as explained in methods. The Cx43 reconstituted bilayer membrane was kept hydrated continuously with PBS buffer and AFM imaging was continued to ascertain that the nanopore-support is covered by the bilayer membrane. Figure 2 shows a representative example of a nanopore-support before and after deposition of Cx43 hemichannels reconstituted bilayer. The AFM image (Fig. 2b) reveals the pore size, and shape before deposition of lipids. The corresponding height profile (bottom left) across the pore shows that the pore is open. Fig. 2c shows the same pore after formation of the Cx43 reconstituted lipid bilayer. The height profile (bottom middle) obtained on the lowest section of the nanopore suggests that the pore has been covered by the lipid bilayer with reconstituted Cx43 hemichannels. We verified the applicability of our method on the chip by monitoring the formation of lipid bilayers on the chip at the periphery of the window where the nanopore was located as a function of time (Fig. 2d).
Figure 2.
Panel a: Schematic of the AFM-nanopore support imaging setup (not drawn to scale). Panels b and c: Top view and lateral view AFM images of: nanopore alone (without membrane) and the same nanopore covered with DPhPC lipid bilayer reconstituted with Cx43 hemichannels, respectively. The membrane is caved into the pore (possibly due to membrane fluidity and/or by the AFM imaging force. However, the depth of the membrane is only 5–7 nm (lateral view, bottom half of panel c) compared to the uncovered pore (lateral view, bottom half of panel b) where the depth is ~30 nm and is underestimated due to the tip shape and height. Significantly, the membrane, even though bent is stable and intact over several AFM scans. d) AFM image of the lipid bilayer patches (without any reconstituted hemichannels) on the support chip (without the nanopore) The height profile of the patches shown in the bottom half of panel d show the typical bilayer height (5–6 nm) measured by AFM. The center of the image has fewer patches, perhaps representing unfused smaller bilayer patches and/or due to the disruptive action of the AFM tip during multiple scanning in that region. A DPPC lipid was used in this experiment. All images were acquired in contact mode.
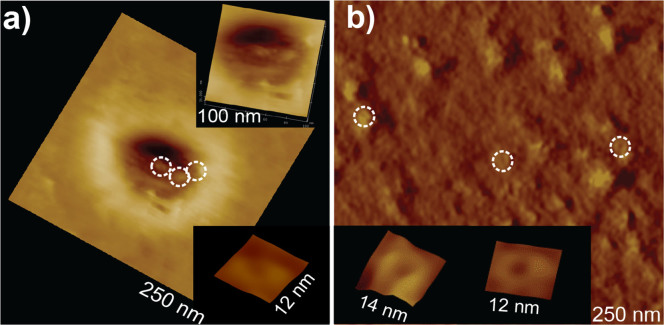
A detailed analysis of high resolution images of Cx43 reconstituted bilayer shows pore structures in areas both over the nanopore-support as well as over the solid support (Figure 3, circles). Insets in Figure 3a and b show structures of individual hemichannels. Although six subunits in each hemichannels are not clearly defined in these AFM images, the inner and outer diameters are similar to those imaged previously with AFM20. These structures over both the nanopore-support and solid support were stable during repeated multiple imaging.
Figure 3. AFM images of Cx43 hemichannels: a) covering the nanopore and b) in an area of the chip adjacent to the nanopore.
Different magnifications of the same image are shown in the insets. Height mode images are presented in a), while deflection images are shown in b) to highlight channel structures. All images were acquired in contact mode.
Dye permeability through open Cx43 hemichannels
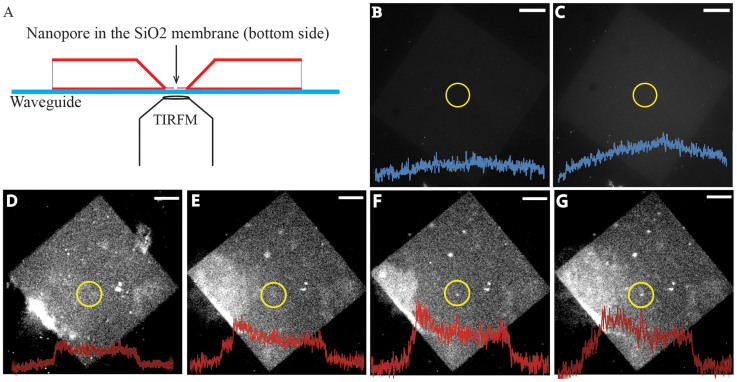
Immediately after the AFM imaging, a clean wet SF-11 coverslip was placed over the silicon chip and inverted for TIRF imaging. In the control experiments (Fig. 4B,C) where there is no Cx43 in the bilayers, showed no localized increase in the LY fluorescence over the nanopore region for up to 10 min in no calcium buffer (the membrane failed beyond this time point preventing further imaging).
Figure 4. LY dye permeability through open hemichannels.
(A): Schematic of the experimental setup for TIRF imaging. A glass coverslip was placed over the chip and inverted for TIRF imaging. (B–C): Nanopore covered by lipid bilayer without connexons (control) at 5 and 10 min in 0 mm Ca2+. (D–G): Nanopore covered with Cx43 reconstituted lipid bilayer. In normal Ca2+ levels (1.8 mM) no increase in fluorescence over the nanopore region was observed up to 15 min (D). Under nominally Ca2+ free conditions, fluorescence intensity increased with time (E,F,G at 2,10 and 20 min, respectively) suggesting hemichannel opening leading to LY permeability through the pore. Fluorescence intensity profile plots as a function of pixel position along a line segment that spans the membrane and pass through the nanopore region are overlaid over the image to show apparent increase in fluorescence. Scale bar: 5 μm.
Hemichannels remain in closed conformation in the presence of physiological Ca2+ concentration and do not allow molecular transport of fluorescent dyes. Consistently, no increase in the fluorescence was observed over the pore region for 15 min when incubated in normal Ca2+ containing buffer (Figure 4D). When the buffer was replaced with no Ca2+ buffer, the time-dependent increase in the LY fluorescence was observed over the nanopore region, consistent with the dye permeability through the open hemichannels (Figure 4E–G). The extent of dye permeability through individual hemichannels was not determined. AFM images show the presence of several hemichannels over the nanopore-support (Figure 3), however, it is unclear how many of them were open or closed.
Discussion
The best way to correlate the biophysical and molecular permeability of ion channels and receptors is by direct and simultaneous assessment of their structure and activity. Current techniques lack such capability. As a first step, we report here a combined AFM and TIRFM system that was possible with the addition of a nanopore-support platform. We adsorbed reconstituted Cx43-hemichannels lipid bilayer on the top of the nanopore-support and imaged them in PBS buffer with AFM. Using an integrated waveguide TIRF, we imaged the permeability of hemichannel-permeable LY dye when hemichannels were opened by reducing the calcium in the imaging buffer. The major advantage of this system is that the free floating bilayer over the nanopore simulates the natural biological membrane with proteins in it, placing no constraints on bilayer fluidity and protein conformation changes in response to perturbations thus offering an excellent opportunity to assess their structure-activity relationship directly. Also, it allows direct monitoring the flow of molecules through the ion-channels by any suitable technique.
The nanopore support has a pore ~70 nm in diameter. Ideally, the pore size should be small enough to include single ion-channel such that individual channel's structure-activity can be recorded. We had selected a bit larger pore for two reasons, a lack of facility to make smaller pores and more importantly, our concerns for not having any ion channels overlaying the nanopore-support with smaller pore – unlike crystalline patches of membranes with ion channels that could be obtained from crystallography or from purification from cells. Instead, we use ion channels reconstituted in random distribution and with varying concentration in bilayers with a hope that channel activity and conformational changes are not affected by cooperativity among neighboring channels possible in crystalline patches.
Our results from repeatable imaging of same structures over the nanopore-support strongly support the notion that the reconstituted bilayer retains its strengths and fluidity while imaging in PBS. The stability of the reconstituted bilayer is good enough that the channels could be imaged even when the membrane buckles around the nanopores-support rim by ~6–7 nm (Figure 2c, lateral view). Support pores without the reconstituted pores have larger depth that could be imaged with pyramidal tips.
The resolution of hemichannel structure is not high enough to decipher individual subunits of hexameric channels20,25. However, the AFM images show distinct pores and surrounding subunits, the outer and inner diameters similar to published data20,25. Since this study was designed to correlate molecular permeability with the hemichannel open and closed conformations, we did not focus on obtaining highest resolution images.
Hemichannel permeability to LY and other dyes, as a function of calcium concentration is well documented26,27. For measuring single channel LY dye permeability without the background fluorescence, we used our waveguide TIRFM system9. In our study, the thickness of the nanopore-support system (300 μm) was beyond the range for effective TIRF imaging in its upright position. After AFM imaging of hemichannels in the upright position, we put the high index glass cover slip over the reconstituted bilayer and inverted the whole system for TIRF imaging (Fig. 4A). However, successful TIRFM recording was limited as the breakage of the nanopore-support system was the most common cause that was visualized by high, uncontrolled fluorescence signal from LY dye leakage. Only 20% of experiments gave reasonable success in dye transfer study. Significantly, the structural stability of reconstituted bilayer was exceptional – dye transfer was not observed when there was no hemichannels (control, Fig 4b,c) and the hemichannels were closed (as expected in the presence of calcium in the buffer, Fig 4d) and the dye transfer began when the hemichannels were opened (after reducing the calcium in the buffer, Figure 4e–g).
In conclusion, we have developed a successful methodology to reconstitute Cx43 in-situ into the fluid bilayers over the nanopore and imaged it with AFM. We are further optimizing the setup by reducing the pore size to ~10 nm to allow for imaging of single connexons, and reducing the silicon oxide window size so that the membrane is more stable for high resolution AFM imaging. Also, depositing a small step (<50 nm) around the front window to prevent its breakage when the chip is inverted for TIRFM, while able to probe this region with evanescent field for fluorescence detection is considered. This will allow mapping true single channel conformational changes and its molecular permeability, the feat commonly unavailable.
Methods
Materials
SF-11, a high index (1.78), 25 mm diameter and #1.5 thick glass cover glass, was purchased from V.A. Optical Labs, San Anselmo, CA. Philips Luxeon K2 high power light emitting diodes (LED) of 470 nm wavelength [21 lm@1A], were purchased from Future Electronics, Canada. Thermal conductive epoxy was obtained from Epoxies, etc., (Cranston, RI). A nanopore in silicon dioxide (SiO2) membrane on a silicon chip was custom designed and fabricated at Appnano Inc., (Santa Clara, CA).
Nanopore chip
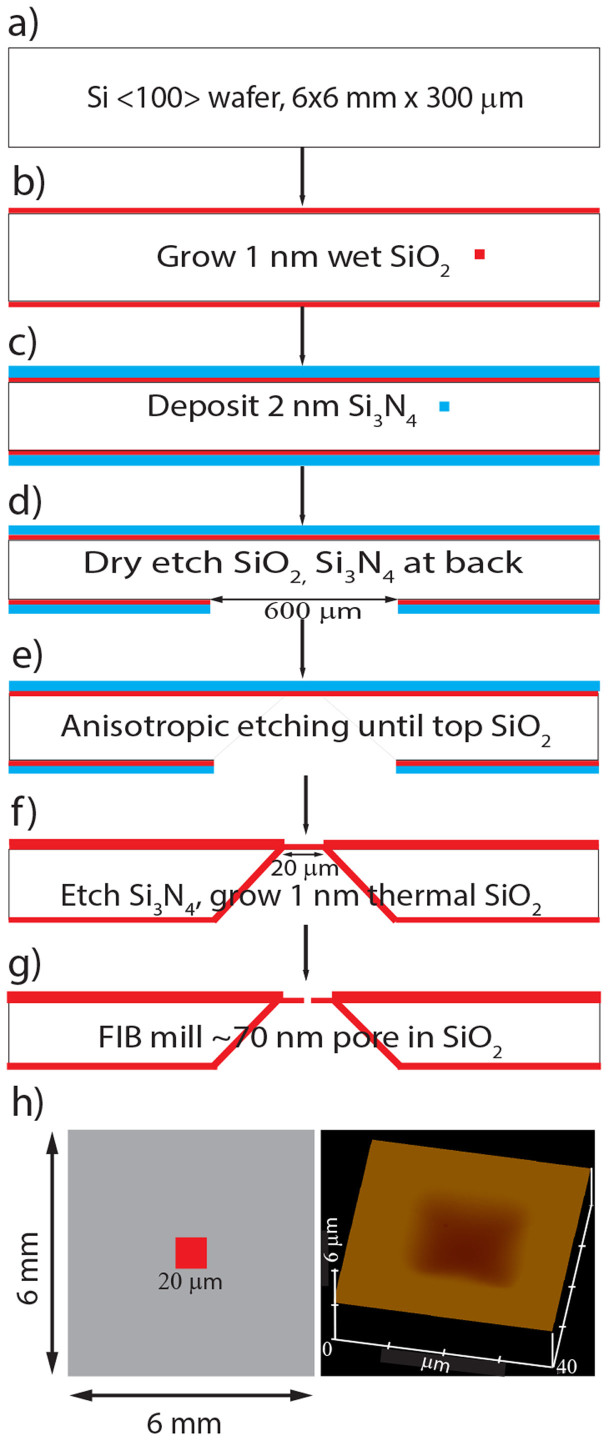
The schematic of the nanopore chip fabrication process is outlined in figure 1. A RCA clean, 300 μm thick N-type single crystal silicon chip measuring 6 x 6 mm was processed to deposit 200 nm thick SiO2 layer on top by wet thermal deposition at 1000°C. Further a 200 nm thick low stress silicon nitride layer was deposited on top of SiO2 layer by low pressure chemical vapor deposition. A window of 600 μm was defined in one of the sides “lower side” (Fig. 1d) using photo lithography process. Both silicon nitride and SiO2 layers were etched out of the window using dry etch process (plasma etching), to expose the silicon in the window region. The wafer was cleaned and put into a potassium hydroxide solution to further anisotropically etch the exposed silicon. The etching stopped at the oxide layer at front side of the wafer leaving a window of 20 μm × 20 μm (Fig. 1e). The nitride layers were etched out and 100 nm of SiO2 was deposited inside the window region to insulate the exposed silicon surface. A Nanopore of ~70 nm was then milled using focused ion beam as shown in the cross section image in the SiO2 membrane of 200 nm thickness.
Figure 1. Nanopore fabrication process outline (see text for details).
(a–g): side view, (h-left): top view of the chip, (h-right): SiO2 membrane appears depressed in AFM image due to buckling.
Cx43 hemichannel purification
Hemichannel isolation and purification was carried out as described in19,20. Briefly, Cx43 hemichannels were isolated from Marshall Cell line (BICR-M1Rk, a rat mammary fibroblastoid cell). The cells were collected and homogenized in Hanks buffer (HB) containing 20 mM HEPES (pH 7.4), 1 mM MgCl2, 1 mM dithiothreitol, and protease inhibitor cocktail (Cal-Biochem). Lysate was centrifuged at 1000 g for 5 min to remove nuclear debris. Supernatant was centrifuged at 100,000 g for 1 h. Pellets were resuspended in HB with 0.25 M sucrose, loaded on a 1.2/2.0 M sucrose gradient and centrifuged at 100,000 g for 2.5 h. The plasma membrane fraction enriched with connexons was collected between 1.2 and 2.0 M sucrose gradients. This fraction was solubilized with buffer S (20 mM HEPES, 50 mM NaCl, 50 mM octylglucoside, pH 7.4) and stirred for 30 min. The solution was centrifuged at 100,000 g for 30 min, the supernatant was incubated with an anti-peptide CT23 antibody (anti-CT 360–382 antibody, targeted to the carboxy terminus of Cx43) coupled to sepharose 4B beads for 2 h under gentle stirring. The beads were transferred onto a column and washed with 50 ml of buffer S containing phosphatidylcholine (1 mg/ml). The connexons were eluted with buffer S without octylglucoside but containing 5 μM CT23 peptide, phosphatidylcholine (1 mg/ml). The entire isolation procedure was carried at 4°C.
AFM Imaging of Cx43 hemichannels over the nanopore
The silicon chips were cleaned before AFM imaging by rinsing in acetone, isopropanol and methanol for 5 minutes each in that sequence and stored in clean DI water until imaging. We created a two-side liquid chamber by placing the chip on top of an O-ring glued to a glass cover slip attached to an AFM steel disk with the same glue (lower side of chamber). Two Ton epoxy non-reactive with water was used in order not to contaminate the buffer solution. The Multimode AFM liquid cell (Bruker) was placed on top of the sample to form the upper side of the chamber. The method used to produce Cx43 reconstituted bilayers spanned across the nanopore is a modified version of the solution-spreading method reported previously21,22,23,24. The nanopore was imaged first without the supported bilayer on a multimode AFM (Bruker, Santa Barbara, CA). For AFM imaging of Cx43 connexons, 100–500 ng of the branched 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) lipid in chloroform were applied to the nanopore and desiccated for 2 hours to allow the formation of a lipid film. Subsequently, the chip was mounted on the two-side sample chamber filled with PBS buffer solution in the lower side. The lipid film was hydrated with 20 μl of purified Cx43 connexon solution (~0.1 ng/ml) to facilitate the incorporation of Cx43 connexons into the supported lipid bilayer covering the chip and span the SiO2 membrane window harboring the ~70 nm nanopores as a free-floating bilayer across the nanopore. Imaging was performed in contact and tapping modes using Olympus cantilevers with 0.08 N/m spring constant. The force was continuously minimized to avoid sample damage.
Waveguide TIRF imaging system
Total internal reflection fluorescence microscopy (TIRFM) allows selective imaging of interfacial processes by eliminating the background signal with excellent surface sensitivity. The details of our waveguide based TIRF system are described in9. Briefly, the device consists of a high refractive index cover glass (SF-11, RI: 1.78, ø 25 mm), sandwiched between two thin flat black O-rings (4 mm rim width), placed under a stainless steel cylindrical tube. This arrangement is placed concentrically in another stainless steel dish (ø 60 mm) machined to have optical access from the bottom. Six high power LEDs of 470 nm (21 lm@1 A) mounted on a circular copper tube with thermal conductive epoxy were placed inside the outer dish. The entire setup was mounted on an Olympus IX-71 microscope equipped with 512 x 512 EMCCD (BT 2000, Diagnostic instruments) imaging system. Control experiments were carried out on a system with 1024 x 1024 EMCCD (Cascade 1K, Photometrics).
The location of nanopores in the SiO2 membrane was variable (slightly off-centered, few hundred nanometers) from chip to chip even though we intended them to be in the middle of the membrane. But we precisely located their position with AFM by imaging the entire window first and sequentially zoom into the center until we found the pore while noting the offsets and orientation of the chip. We used this spatial information to correlate the nanopore region/hemichannel activity in TIRF images. Before imaging with TIRFM, AFM was carried out on the nanopore sample to make sure the pore is covered by the bilayer and for the presence of Cx43 hemichannels in it. Immediately after this, a clean, wet SF-11 glass coverslip was placed on the chip and inverted to carry out TIRF imaging. An adhesive silicone seal was placed around the periphery of the chip to prevent any spill overs from the chip. Hemichannel permeability under normal extracellular calcium concentration (1.8 mM) was carried out by mixing Lucifer Yellow dye (0.1 mg/ml) in Ca2+ containing PBS for 15 minutes. After this, the media was replaced with no Ca2+ containing PBS buffer with LY. Any change in fluorescence over the nanopore region was monitored by time lapse imaging at frequent intervals. The gain and exposure settings were kept the same for the entire experiment. A plot of fluorescence intensities as a function of pixel positions across a line segment spanning the membrane passing over the nanopore region (plot profile) quantified the increase in fluorescence in response to low Ca2+ levels suggesting hemichannel opening leading to LY dye permeability.
Author Contributions
S.R., A.Q., D.A.C., designed TIRFM; S.R. performed TIRF experiments; F.T.A. performed AFM experiments, N.P., performed control AFM data on coverslips; R.L., F.T.A., S.R., analyzed the data; S.R. wrote the first draft and all authors reviewed the manuscript.
Supplementary Material
Supplementary Figures
References
- Lal R. & John S. A. Biological applications of atomic force microscopy. Am. J. Physiol. 266, C1-21 (1994). [DOI] [PubMed] [Google Scholar]
- Bocker M., Muschter S., Schmitt E. K., Steinem C. & Schaffer T. E. Imaging and Patterning of Pore-Suspending Membranes with Scanning Ion Conductance Microscopy. Langmuir 25, 3022–3028 (2009). [DOI] [PubMed] [Google Scholar]
- Hennesthal C. & Steinem C. Pore-spanning lipid bilayers visualized by scanning force microscopy. J. Am. Chem. Soc. 122, 8085–8086 (2000). [Google Scholar]
- Mey I. et al. Local Membrane Mechanics of Pore-Spanning Bilayers. J. Am. Chem. Soc. 131, 7031–7039 (2009). [DOI] [PubMed] [Google Scholar]
- Romer W. et al. Channel activity of a viral transmembrane peptide in micro-BLMs: Vpu (1-32) from HIV-1. J. Am. Chem. Soc. 126, 16267–16274 (2004). [DOI] [PubMed] [Google Scholar]
- Steltenkamp S. et al. Mechanical properties of pore-spanning lipid bilayers probed by atomic force microscopy. Biophys. J. 91, 217–226 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves R. P. et al. Two-chamber AFM: probing membrane proteins separating two aqueous compartments. Nat. Methods 3, 1007–1012 (2006). [DOI] [PubMed] [Google Scholar]
- Voet D. & Voet J. G. in Biochemistry (Wiley, Hoboken, NJ, 2004). [Google Scholar]
- Ramachandran S., Cohen D. A., Quist A. P. & Lal R. High performance, LED powered, waveguide based total internal reflection microscopy. Sci Rep. 3, 10.1038/srep02133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. L. Connexin channel permeability to cytoplasmic molecules. Prog. Biophys. Mol. Bio. 94, 120–143 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D. L., Ebihara L., Takemoto L. J., Swenson K. I. & Goodenough D. A. Connexin46, a novel lens gap junction protein, induces voltage-gated currents in nonjunctional plasma membrane of Xenopus oocytes. J. Cell Biol. 115, 1077–1089 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spray D. C., Harris A. L. & Bennett M. V. Gap junctional conductance is a simple and sensitive function of intracellular pH. Science 211, 712–5 (1981). [DOI] [PubMed] [Google Scholar]
- Spray D. C., Harris A. L. & Bennett M. V. Voltage dependence of junctional conductance in early amphibian embryos. Science 204, 432–4 (1979). [DOI] [PubMed] [Google Scholar]
- Retamal M. A., Cortes C. J., Reuss L., Bennett M. V. & Saez J. C. S-nitrosylation and permeation through connexin 43 hemichannels in astrocytes: induction by oxidant stress and reversal by reducing agents. P. Natl. Acad. Sci. USA. 103, 4475–80 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Xie L. H., John S. A., Subramaniam S. & Lal R. A Novel Role for Connexin Hemichannel in Oxidative Stress and Smoking-Induced Cell Injury. Plos One 2, 10.1371/journal.pone.0000712 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo R. P., Wang S. Y., John S. A., Weiss J. N. & Goldhaber J. I. Metabolic inhibition activates a non-selective current through connexin hemichannels in isolated ventricular myocytes. J. Mol. Cell. Cardiol. 32, 1859–72 (2000). [DOI] [PubMed] [Google Scholar]
- Axelsen L. N., Calloe K., Holstein-Rathlou N. H. & Nielsen M. S. Managing the complexity of communication: regulation of gap junctions by post-translational modification. Front. Pharmacol. 4, 130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras J. E., Sáez J. C., Bukauskas F. F. & Bennett M. V. L. Gating and regulation of connexin 43 (Cx43) hemichannels. P. Natl. Acad. Sci. USA. 100, 11388–11393 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Arce F. T., Ramachandran S. & Lal R. Nanomechanics of hemichannel conformations - Connexin flexibility underlying channel opening and closing. J. Biol. Chem. 281, 23207–23217 (2006). [DOI] [PubMed] [Google Scholar]
- Thimm J., Mechler A., Lin H., Rhee S. & Lal R. Calcium-dependent open/closed conformations and interfacial energy maps of reconstituted hemichannels. J. Biol. Chem. 280, 10646–10654 (2005). [DOI] [PubMed] [Google Scholar]
- Maeda N., Senden T. J. & di Meglio J. M. Micromanipulation of phospholipid bilayers by atomic force microscopy. BBA-Biomembranes 1564, 165–172 (2002). [DOI] [PubMed] [Google Scholar]
- Spangenberg T., de Mello N. F., Creczynski-Pasa T. B., Pasa A. A. & Niehus H. AFM in-situ characterization of supported phospholipid layers formed by solution spreading. Phys. Status Solidi A 201, 857–860 (2004). [Google Scholar]
- Higgins M. J. et al. Structured water layers adjacent to biological membranes. Biophys. J. 91, 2532–2542 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Song Y. H., Han X. J., Zhang B. L. & Wang E. K. Growth of cationic lipid toward bilayer, lipid membrane by solution spreading: scanning probe microscopy study. Chem. Phys. Lipids 123, 177–185 (2003). [DOI] [PubMed] [Google Scholar]
- Allen M. J., Gemel J., Beyer E. C. & Lal R. Atomic Force Microscopy of Connexin40 Gap Junction Hemichannels Reveals Calcium-dependent Three-dimensional Molecular Topography and Open-Closed Conformations of Both the Extracellular and Cytoplasmic Faces. J. Biol. Chem. 286, 22139–22146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries S. H. & Schwartz E. A. Hemi-gap-junction channels in solitary horizontal cells of the catfish retina. J. Physiol. 445, 201–30 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quist A. P., Rhee S. K., Lin H. & Lal R. Physiological role of gap-junctional hemichannels: Extracellular calcium-dependent isosmotic volume regulation. Journal of Cell Biology 148, 1063–1074 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures