Abstract
The evolutionary survival of Mycobacterium tuberculosis, the cause of human tuberculosis (TB), depends on its ability to invade the host, replicate, and transmit infection. At its initial peripheral infection site in the distal lung airways, M. tuberculosis infects macrophages which transport it to deeper tissues1. How mycobacteria survive in these broadly microbicidal cells is an important question. Here we show that M. tuberculosis, and its close pathogenic relative Mycobacterium marinum, preferentially recruit and infect permissive macrophages while evading microbicidal ones. This immune evasion is accomplished by using cell surface associated phthiocerol dimycoceroserate (PDIM) lipids2 to mask underlying pathogen-associated molecular patterns (PAMPs). In the absence of PDIM, these PAMPs signal a toll-like receptor (TLR)-dependent recruitment of macrophages that produce microbicidal reactive nitrogen species. Concordantly, the related phenolic glycolipids (PGL)2, promote recruitment of permissive macrophages via a host chemokine receptor 2 (CCR2)-mediated pathway. Thus, we have identified coordinated roles for PDIM, known to be essential for mycobacterial virulence3 and PGL, which (along with CCR2) is known to be associated with human TB4,5. Our findings also suggest an explanation for the longstanding observation that M. tuberculosis initiates infection in the relatively sterile environment of the lower respiratory tract, rather than in the upper respiratory tract, where resident microflora and inhaled environmental microbes may continually recruit microbicidal macrophages through TLR-dependent signaling.
Pattern recognition receptors (PRRs) such as the TLRs enable host recognition of diverse microbes through their PAMPs6. Macrophages recruited through TLR signaling pathways can eradicate organisms invading the oropharyngeal mucosa, e.g. Streptococcus pneumoniae7. In contrast, pathogenic mycobacteria appear to use macrophages and myeloid dendritic cells for transport across epithelial barriers to their infection niche1,8. Mycobacteria are replete with TLR PAMPs, such as lipoproteins and bacterial cell wall peptidoglycan, that have been shown to activate cytokine responses in cultured macrophages1. Yet in vivo studies find TLR signaling to be dispensable in the early stages of infection9, suggesting that mycobacteria have evolved mechanisms to circumvent the bactericidal consequences of TLR signaling.
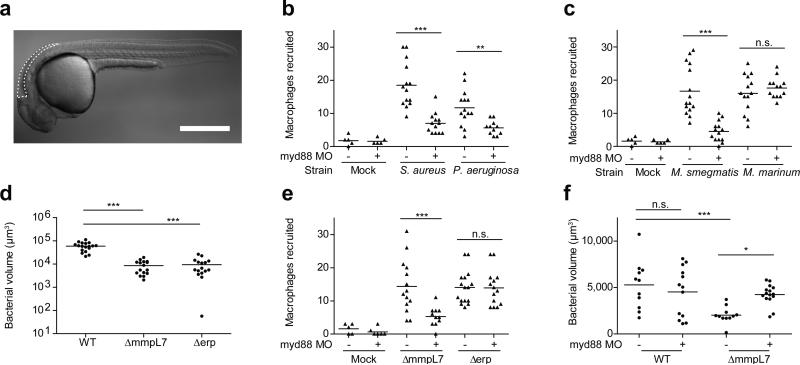
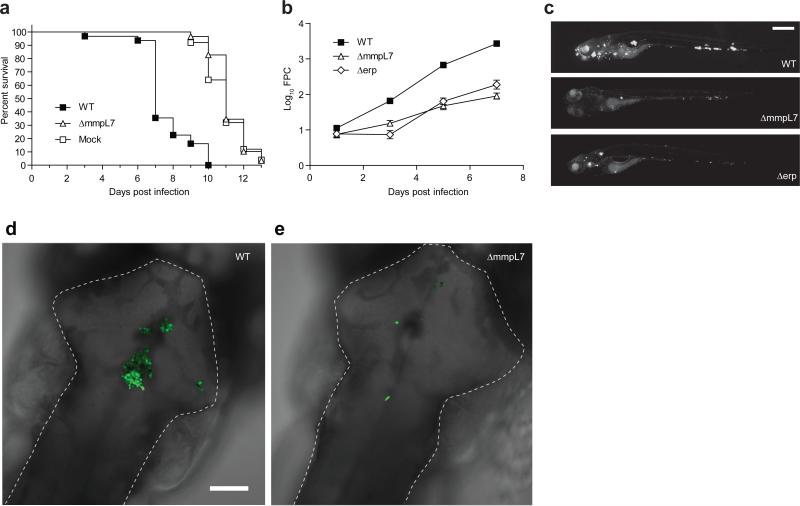
To explore these mechanisms, we used zebrafish larvae infected with M. marinum, a close genetic relative of M. tuberculosis, and the causative agent of TB in ectotherms. This model has yielded important insights into the pathogenesis and genetics of human TB10. In humans, the earliest interactions between mycobacteria and phagocytes occur at the lung epithelial surface. Such interactions can be modeled in the larva by injection of bacteria or other chemical stimuli into the hindbrain ventricle (HBV), a neuroepithelium-lined cavity to which phagocytes are recruited8 (Fig.1a). We used morpholino knockdown to create zebrafish deficient in MyD88, a common downstream adaptor molecule for TLR signaling pathways6. As expected, MyD88 morphants had decreased macrophage recruitment to Staphylococcus aureus and Pseudomonas aeruginosa, mucosal bacteria that can be commensal or pathogenic11-13 (Fig. 1b). Similarly, macrophage recruitment to the nonpathogenic Mycobacterium smegmatis was MyD88-dependent. In contrast, macrophage recruitment to M. marinum was MyD88-independent (Fig. 1c). This finding suggested that pathogenic mycobacteria have the ability to mask PAMPs that would otherwise induce TLR signaling during the initial infection phase. We hypothesized that such a factor would be a cell surface associated virulence determinant. In this light, PDIM seemed a likely candidate, particularly because it is present only in the pathogenic mycobacteria, including M. tuberculosis and M. marinum, but absent in M. smegmatis2. We created a M. marinum mutant that lacks PDIM on its surface by knocking out the PDIM transporter, encoded by the mmpL7 gene, and confirmed that it was attenuated in zebrafish larvae (Fig. 1d and Extended Data Fig. 2). If PDIM is masking PAMPs, then macrophage recruitment to ΔmmpL7 bacteria should be MyD88-dependent, and this was the case (Fig. 1e). In contrast, macrophage migration remained MyD88-independent in response to M. marinum deficient in another cell surface-associated virulence determinant, Erp (Δerp) (Fig. 1d, e and Extended Data Fig. 2)14. This result was consistent with M. smegmatis possessing a functional erp14, and suggested further that the evasion of MyD88-dependent immune detection was mediated specifically by PDIM.
Figure 1. PDIM mediated evasion of MyD88 dependent macrophage recruitment.
a, Schematic of a 2dpf zebrafish showing the hindbrain ventricle (HBV) injection site outlined with dashed white line. Scale bar = 500 μm. b-c, Mean macrophage recruitment at 3 HPI into the HBV of wild-type or MyD88-morphant (MO) fish following infection with 150 S. aureus, 200 P. aeruginosa (b), 80 M. marinum or 85 M. smegmatis (c). Representative of three separate experiments. d, Mean bacterial burdens at 3 DPI following HBV infection of wild-type fish with 80 wild-type, ΔmmpL7, or Δerp M. marinum. Representative of three separate experiments. e, Mean macrophage recruitment at 3 HPI into the HBV of wild-type or MyD88 MO fish following infection with ΔmmpL7 or Δerp M. marinum. Representative of four separate experiments. f, Mean bacterial burdens of wild-type or MyD88 MO fish at 3 DPI following HBV infection with wild-type or ΔmmpL7 M. marinum. Representative of three separate experiments. Significance testing for all panels done using one-way ANOVA, with Bonferroni's post-test for comparisons shown, *P < 0.05; ***P < 0.001.
Our model posits that pathogenic mycobacteria use PDIM to evade recruitment of MyD88-dependent macrophage populations detrimental to their survival. Therefore, we predicted that wild-type mycobacteria should be unaffected in MyD88 morphants, whereas the attenuation of ΔmmpL7 should be reversed. We found both to be the case (Fig. 1f). For these assays, ~ 80 M. marinum were injected into the HBV. However, MyD88 morphants were previously reported to be susceptible to higher M. marinum inocula delivered intravenously15. We confirmed these findings, showing that MyD88 deficiency increased susceptibility at later time points after intravenous administration of >300 CFU (Extended Data Fig. 3). It is likely that MyD88 exerts its protective responses at these later stages through mechanisms distinct from the ones we have uncovered, such as through IL-1-mediated responses9. Indeed, IL-1 expression was undetectable 3 hours following infection when we observed MyD88-dependent macrophage recruitment (data not shown) suggesting an IL-1 independent role for MyD88 in mediating recruitment towards PDIM-deficient mycobacteria.
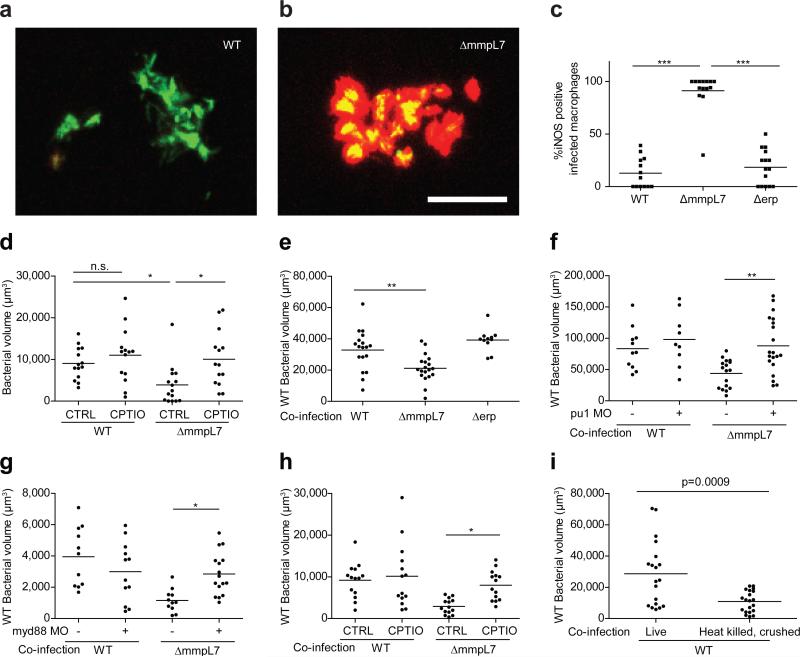
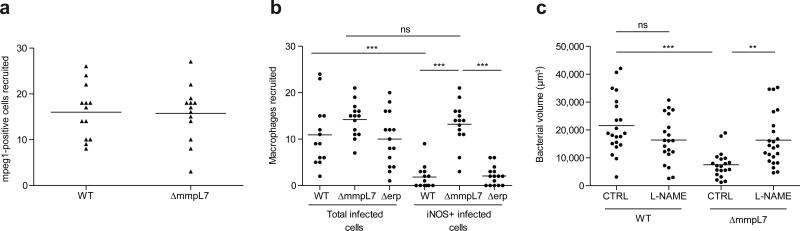
Further characterization of wild-type versus PDIM-deficient bacteria revealed that both strains recruited cells expressing the macrophage-specific marker mpeg18 (Extended Data Fig. 4a and Extended Data Videos 1, 2). We next asked whether these macrophages possessed differential microbicidal potential. We examined the expression of inducible nitric oxide synthase (iNOS) in these recruited cells because: 1) it is induced in macrophages upon TLR signaling6, and can be expressed by zebrafish16, mouse17 and human18 macrophages following mycobacterial infection and 2) mycobacteria are known to be susceptible to reactive nitrogen species (RNS) in both murine17 and human18 macrophages. We found very few iNOS-positive macrophages arriving to wild-type M. marinum, whereas the majority of those arriving in response to ΔmmpL7 bacteria were iNOS-positive (Fig. 2a-c, and Extended Data Fig. 4b). Δerp bacteria elicited very few iNOS-expressing macrophages (Fig. 2c and Extended Data Fig. 4b), further showing that this early manipulation of macrophage recruitment and/or activation is a specific characteristic of PDIM. We confirmed that RNS were the major mediators of MyD88-dependent macrophage microbicidal activity by showing that the iNOS inhibitors CPTIO and L-NAME reversed growth attenuation of the ΔmmpL7 mutant (Fig. 2d and Extended Data Fig. 4c).
Figure 2. Increased iNOS-dependent microbicidal activity of macrophages recruited to PDIM-deficient mycobacteria.
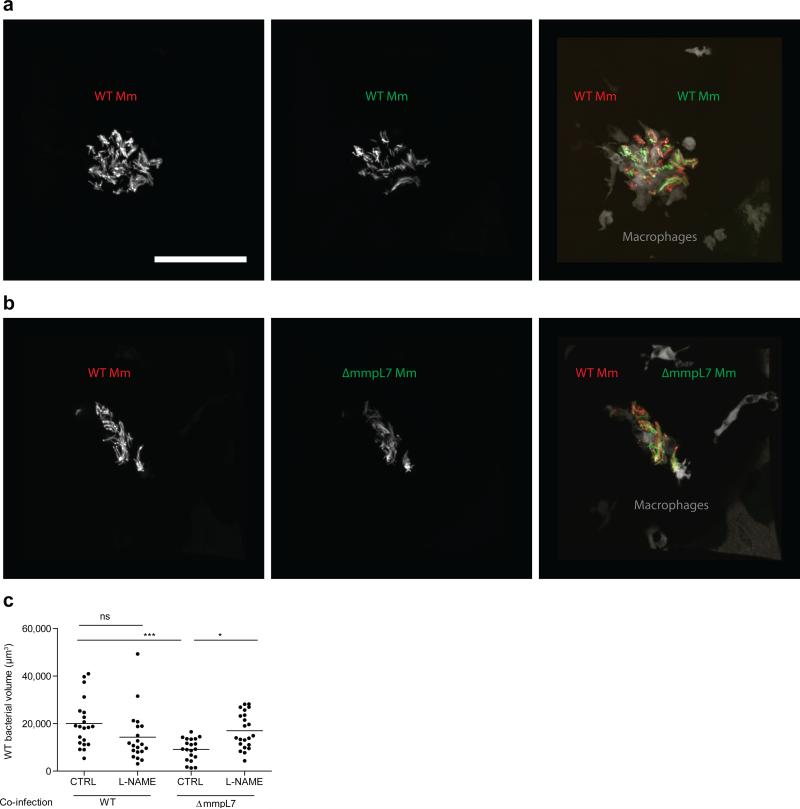
a, b, Representative images of wild-type (a) and ΔmmpL7 (b) M. marinum-infected fish from (c). N=13 (wild-type) and 14 (ΔmmpL7) larvae per group. Scale bar = 50μm. c, Percent of infected macrophages that were iNOS-positive, in the HBV at 3 DPI with 80 wild-type, ΔmmpL7, or Δerp M. marinum. Representative of three separate experiments. d, Mean bacterial burdens of 2dpf control (CTRL) or RNS scavenger (CPTIO) treated fish following HBV infection with 80 wild-type or ΔmmpL7 M. marinum. Representative of two separate experiments. e-h, Mean bacterial volume of red fluorescent wild-type M. marinum (infection inoculum 30-40) when co-infected with 30-40 green fluorescent wild-type, ΔmmpL7, or Δerp M. marinum at 3 DPI in wild-type (e), pu1 MO (f), myd88 MO (g), or CPTIO-treated (h) larvae. e, g, Co-infection of wild-type Mm and ΔmmpL7 M. marinum in wild-type or myd88 MO fish is representative of at least three separate experiments, and co-infection with Δerp is representative of two separate experiments. f, h, Representative of two separate experiments. a-h, Significance testing done using one-way ANOVA, with Bonferroni's post-test for comparisons shown, *P < 0.05; **P < 0.01; ***P < 0.001. i, Mean bacterial volume of red fluorescent wild-type M. marinum at 3 DPI (infecting inoculum 30-40) when co-infected with the volume equivalent of 30-40 heat-killed, crushed wild-type M. marinum. Representative of two separate experiments. Student's unpaired t test.
Together our findings suggested that PDIM mediates an immune evasion strategy, whereby mycobacteria evade detection by TLRs so as to avoid recruitment of iNOS-expressing, microbicidal macrophages. To test this idea, we co-infected red fluorescent wild-type bacteria with green fluorescent wild-type or ΔmmpL7 bacteria. We found that wild-type bacteria were attenuated in the presence of ΔmmpL7 bacteria, and that this attenuation transfer was specifically caused by co-infection with ΔmmpL7 and not with wild-type or Δerp bacteria (Fig. 2e and Extended Data Fig. 5a, b). Furthermore, this transfer of attenuation from ΔmmpL7 to wild-type bacteria was dependent on macrophages; no attenuation was observed when macrophages were depleted prior to infection using a morpholino against the myeloid transcription factor, PU.1 (Fig. 2f)16. Attenuation transfer was similarly dependent on MyD88 signaling, as well as on RNS production (Fig. 2g, h and Extended Data Figure 5c).
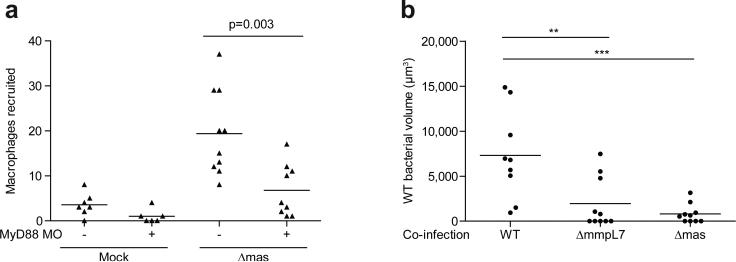
Since PDIM is not the only substrate for the MmpL7 transporter, we confirmed that the effects were due to the lack of PDIM per se by using a PDIM synthesis mutant, Δmas, showing it to both recruit macrophages in a MyD88-dependent fashion and to transfer attenuation to wild-type bacteria (Extended Data Fig. 6). Finally, to rule out the possibility that the PDIM-deficient mutants simply had increased expression of the culpable PAMP(s), we co-injected heat-killed, crushed wild-type bacteria together with live wild-type bacteria. If the culpable PAMP(s) are expressed by wild-type bacteria, then they should become exposed by crushing the bacteria and cause attenuation of the live bacteria. We found this to be the case (Fig. 2i). Altogether, these results suggest that PDIM physically masks underlying mycobacterial PAMPs, thereby preventing mycobacterial delivery into microbicidal macrophages.
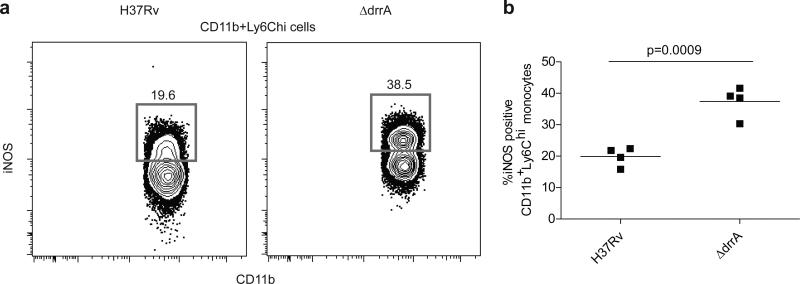
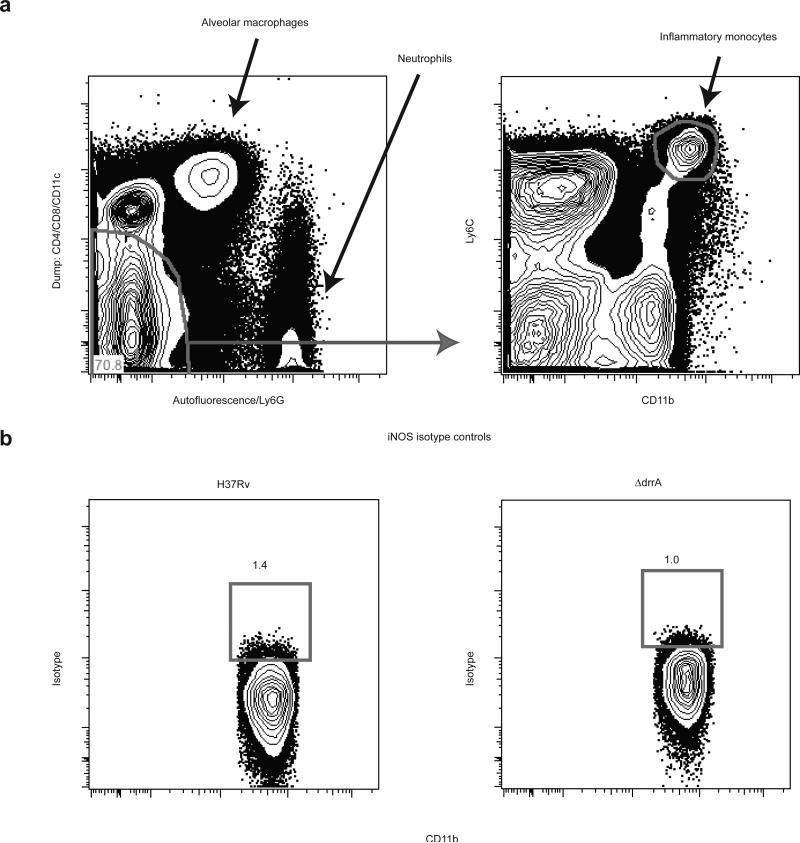
To corroborate our findings in a second model, we infected mice via aerosol with wild-type M. tuberculosis (H37Rv) or with an isogenic strain (ΔdrrA) defective for proper PDIM surface localization and virulence in mice3. At 21 DPI, we found substantially greater proportions of iNOS-producing cells among the CD11b+Ly6Chi inflammatory monocyte population in the lungs of mice infected with the ΔdrrA mutant compared to mice infected with the wild type strain (Fig. 3 and Extended Data Fig. 7). Thus PDIM-mediated evasion of TLR-dependent immune recognition is shared by M. tuberculosis in the context of the mammalian lung, consistent with its central role in avoidance of TLR-dependent anti-microbial mechanisms such as iNOS and antimicrobial peptides6.
Figure 3. Elevated frequencies of iNOS expressing inflammatory monocytes in mice infected with PDIM-deficient M. tuberculosis.
C57B6 mice were infected via the aerosol route with H37Rv or an isogenic PDIM-deficient mutant (ΔdrrA). Lung tissue was harvested on 21 DPI and iNOS (protein expression was measured via flow cytometry. Representative FACS plots (a) and graphical depiction (b) of frequencies of iNOS expressing cells within the Ly6C+CD11b+ inflammatory monocyte population. Representative of two separate experiments. Student's unpaired t test.
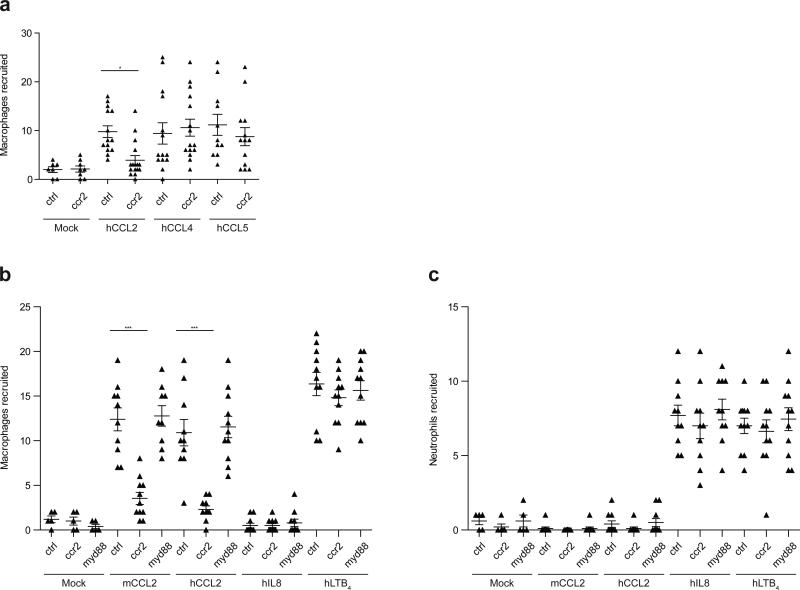
We next sought to understand the mechanism by which mycobacteria recruit the permissive macrophages that are essential for their transport into host tissues. Given our prior finding that M. marinum recruits only macrophages (and not neutrophils) to the HBV8, we considered macrophage-specific chemokines as candidates for mediating this recruitment. We investigated CCR2, which has been implicated in macrophage migration to bacterial pathogens in mice19, including macrophages that are permissive to M. tuberculosis replication after aerosol infection20. We identified the functional zebrafish CCR2 orthologue (see online methods section) and confirmed that its knockdown resulted in reduced macrophage migration in response to recombinant human chemokine ligand 2 (hCCL2) and not to the closely related human macrophage chemokines CCL4 and CCL5 (Extended Data Fig. 8a). The specificity of CCL2-mediated macrophage migration was revealed by the following findings: 1) human and mouse CCL2 induced macrophage but not neutrophil migration (Extended Data Fig. 8b, c) 2) recombinant human IL-8, a neutrophil chemokine, induced neutrophil but not macrophage migration (Extended Data Fig. 8b, c) 3) human LTB4 induced recruitment of both neutrophils and macrophages (Extended Data Fig. 8b, c), as expected16 and 4) Myd88 knockdown did not diminish CCL2-mediated macrophage migration, ruling out TLR-mediated migration in response to any endotoxin that might be contaminating the chemokine preparations (Extended Data Fig. 8b).
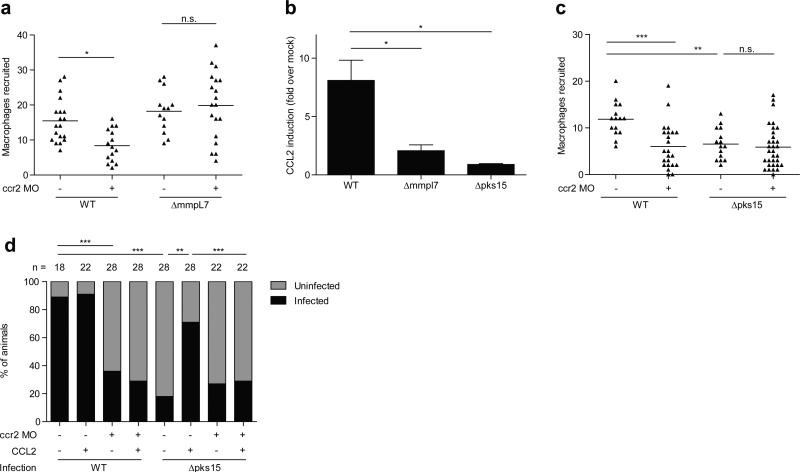
CCR2 morphants had reduced macrophage migration in response to wild-type M. marinum, confirming the role of this pathway in recruitment (Fig. 4a). Recruitment to PDIM-deficient M. marinum was unaffected showing that TLR PAMPs trigger recruitment through a CCR2-independent pathway (Fig. 4a). Accordingly, we found that M. marinum infection induced CCL2, and that CCL2 morphants also had reduced macrophage recruitment in response to infection (Fig. 4b and Extended Data Figure 9).
Figure 4. Macrophage recruitment and subsequent infectivity is mediated by mycobacterial PGL and host CCR2.
a, Mean macrophage recruitment at 3 HPI into the HBV of wild-type or CCR2 MO fish following infection with 80 wild-type or ΔmmpL7 M. marinum. Representative of three independent experiments. One-way ANOVA, with Bonferroni's post-test for comparisons shown, *P < 0.05 b, CCL2 mRNA levels (mean +/- SEM of four biological replicates) induced at 3 hours post caudal vein infection of 2 DPF larvae with 250-300 wild-type, Δpks15, or ΔmmpL7 M. marinum. One-way ANOVA with Tukey's post-test, *P < 0.05 c, Mean macrophage recruitment at 3 HPI into the HBV following infection with 80 wild-type or Δpks15 M. marinum. Representative of three separate experiments. One-way ANOVA with Bonferroni's post-test for comparisons shown, **P < 0.01; ***P < 0.001. d, Wild-type and CCR2 MO fish, with or without the addition of μg/mL CCL2, were infected in the HBV with 1-3 wild-type or Δpks15 M. marinum. Graph shows the percent of fish that were infected (black) or uninfected (gray) after five days. n=number of larvae per group. Representative of two separate experiments. Significance was evaluated using Fisher's exact test for each comparison, **P < 0.01; ***P < 0.001.
Turning to the question of which bacterial determinant induced the CCR2 pathway, we considered PGL, a molecule closely related to PDIM in both M. marinum and M. tuberculosis2. While many clinical M. tuberculosis isolates have lost PGL, its presence has been linked to increased virulence5. Moreover, among M. tuberculosis clinical isolates, PGL expression was linked to ccl2 expression in a mouse lung infection model21. Similarly, we found that PGL was required for ccl2 induction in the larva; deletion of the M. marinum pks15 locus specifically abrogates PGL, but not PDIM, production (data not shown) and resulted in loss of ccl2 induction. ΔmmpL7 bacteria, which lack surface expression of both PGL and PDIM, similarly failed to induce ccl2, highlighting that this chemokine is not induced through TLR interactions, but rather is specifically induced through PGL-mediated interactions (Fig. 4b). Furthermore, Δpks15 bacteria recruited fewer macrophages upon infection of wild-type larvae, and this reduction was similar to that seen in CCR2 morphants infected with wild-type bacteria (Fig. 4c). There was no additional reduction in recruitment when CCR2 morphants were infected with PGL-deficient bacteria, suggesting that PGL recruits macrophages solely through the CCR2 pathway (Fig. 4c).
Our findings implicate PGL in bacterial virulence and correspondingly, the CCR2 pathway in host susceptibility. Globally, a large proportion of M. tuberculosis isolates are PGL deficient due to a frameshift in pks152. However, the importance of PGL in mediating virulence and/or transmission is underscored by its presence in many of the W-Beijing strains, which are becoming rapidly enriched among M. tuberculosis isolates globally5, and have predominated in outbreaks in North America where TB is not prevalent5. Infectivity is a key requirement for transmission, and our data suggested that PGL may enhance infectivity through CCR2-mediated recruitment of permissive macrophages at the earliest stages of infection. This enhancement may be particularly relevant in the context of human infections, in which the infectious dose is thought to be as low as 1-3 bacteria22,23. To test the hypothesis that PGL enhances infectivity at low doses, we compared the ability of wild-type and PGL-deficient strains to establish infection. Confocal microscopy was used five hours after HBV injection to select those animals that had received 1-3 bacteria (Extended Data Fig. 10), and then again at 5 DPI to identify which animals were still infected. We found that 89% of the wild-type but only 18% of the Δpks15 infections were successful (Fig. 4d). Concurrent administration of recombinant CCL2 restored the infectivity of Δpks15 bacteria, provided the CCR2 pathway was intact (Fig. 4d). Correspondingly, we found that wild-type bacteria had a lower infectivity rate in CCR2 morphants (Fig. 4d). Consistent with our finding that PGL recruits macrophages solely through CCR2, there was no further decrease in infectivity in CCR2 morphants infected with the PGL mutant (Fig. 4d). Finally, the infectivity of wild-type bacteria in MyD88 morphants was undiminished (90% for wild-type vs. 83% for morphants), consistent with our finding that TLR signaling is not involved in macrophage recruitment to wild-type bacteria.
These findings highlight the interdependency between bacterial PGL and host CCR2 signaling in driving bacterial infectivity under the low inoculum conditions relevant to human infection. Prior investigations into the role of PGL and CCR2 may have failed to reveal these mechanisms because those studies used higher inocula24 and, in the study of CCR2 signaling, a PGL-deficient strain25. Indeed, our finding that CCR2 signaling is a host susceptibility factor is reinforced by human studies showing an association between the high expression of CCL2 and TB susceptibility4. Furthermore, the association appears to be stronger in East Asian populations26, where clinical isolates are enriched for the predominantly PGL-expressing, W-Beijing strains27. In light of our findings, we propose that the enrichment of PGL expression among these strains is influencing this association, as the CCR2 pathway would be most relevant in the context of bacterial PGL stimulation.
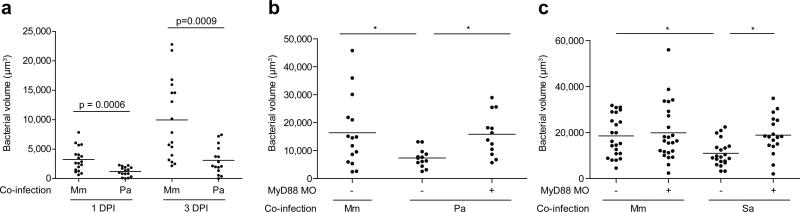
Finally our data suggested an explanation for why M. tuberculosis must reach the alveolar surfaces of the distal lung in order to initiate infection23 (Extended Data Fig. 1). It is well established that TB results from inhalation of small aerosol droplets containing ~1-3 bacteria, which are capable of reaching the alveolar surfaces of the distal lung; in contrast, large droplets harboring ~104 bacteria are trapped in the upper bronchial passages and are far less successful at establishing infection22,23. These observations have led to the idea that the alveolar surfaces of the distal lung offer a more favorable environment for mycobacterial proliferation. We hypothesized that commensal microbes from the oropharyngeal surfaces as well as inhaled environmental organisms, might lead to continual TLR signaling in the upper respiratory tract that would then override the mycobacterial PDIM-dependent immune evasion strategies we identified. In contrast, the lower respiratory tract, which is relatively sterile28, would favor recruitment of Mycobacterium-permissive macrophages. To test this hypothesis we co-infected animals with M. marinum together with bacterial colonizers of the pharynx that induce TLR signaling - either S. aureus, a common Gram-positive colonizer of the nasopharynx in both adults and children12 or the Gram-negative bacterium P. aeruginosa, also reported to colonize the pharynx of asymptomatic adults and children11. Co-infection with P. aeruginosa resulted in the attenuation of wild-type mycobacteria even by 1 DPI, and continuing into 3 DPI (Fig. 5a). Mycobacterial growth was attenuated despite rapid clearing of P. aeruginosa: 56% of the animals had cleared the co-infected P. aeruginosa by 1 DPI, and 76% by 3 DPI, with only a few residual bacteria in the remaining animals. Thus, it was not the physical presence of, but rather the detrimental immunological milieu induced by P. aeruginosa that was responsible for the attenuation of M. marinum. Consistent with our hypothesis, we found that the detrimental effect of P. aeruginosa on mycobacterial survival was MyD88-dependent (Fig.5b). S. aureus co-infection also had a MyD88-dependent detrimental effect on M. marinum survival (Fig. 5c)
Figure 5. MyD88-dependent macrophage recruitment elicited by other bacterial pathogens and commensals attenuates pathogenic mycobacteria.
a, Mean bacterial volume of red fluorescent M. marinum (infecting inoculum 30-40) following co-infection with either 30-40 green fluorescent M. marinum or 300 P. aeruginosa at 1 and 3 DPI. Representative of three separate experiments. Significance assessed using Student's t test. b-c, Mean bacterial volume of 30-40 red fluorescent wild-type M. marinum (infecting inoculum 30-40) following co-infection with either 30-40 green fluorescent wild-type M. marinum or 300 P. aeruginosa (b) or 300 S. aureus (c) at 3 DPI, in wild-type or MyD88 MO larvae. Significance tested by one-way ANOVA with Bonferroni's post–test for comparisons shown, *P < 0.05.
Our prior work identified strategies by which intracellular mycobacteria manipulate host pathways after having traversed epithelial barriers; these involve a bacterial protein secretion system that expands the bacterial niche through macrophage recruitment to the nascent granuloma10. We now describe what may be the first contact between mycobacteria and their hosts, and the manner in which mycobacteria manipulate recruitment, and potentially influence the differentiation or activation state, of the first responding macrophages so as to gain access to their preferred niche (Extended Data Fig. 1). The choreographed entry involves two related mycobacterial lipids acting in concert to avoid one host pathway while inducing another. Our findings link PDIM, recognized as an absolutely essential mycobacterial virulence factor, to the evasion of TLR detection and thus explain the dispensability of TLR-mediated immunity in protection against M. tuberculosis infection in both human and animal studies9,29. In contrast, PGL is dispensable for virulence, being variably present among clinical isolates. Yet, its presence in the ancestral M. cannetti strains as well as in M. marinum, the closest genetic relative of the M. tuberculosis complex2, suggests its integral role in the evolution of mycobacterial pathogenicity. TB is an ancient disease and the enhanced infectivity conferred by PGL may have been essential for most of its history before human crowding, with its greatly increased opportunities for transmission, made it dispensable30.
Our findings suggest that commensal flora may play a central role in choreographing mycobacterial entry. Not only must pathogenic mycobacteria possess a physical barrier to prevent host TLR-mediated detection, but they must also evade TLR signaling initiated by other organisms, by entering through the distal lung (Extended Data Fig. 1). Our work may also explain the paradox that smaller M. tuberculosis droplets are more infectious than larger ones. However the requirement placed on mycobacteria to gain entry through the distal lung makes TB less contagious than most other respiratory infections, thus assigning a protective role to the commensal flora. Conversely, the persistence of human TB for over 70,000 years30 attests to the effectiveness of the mycobacterial evolutionary survival kit (masking lipid, recruiting lipid and small infection droplets) to simultaneously evade and manipulate the host and its commensal flora.
Online Methods
Bacterial Strains and Methods
M. marinum strain M (ATCC BAA-535) and the Δerp mutant have been described32. The ΔmmpL7 and Δpks15 mutants were generated as described in the following section. Fluorescently labeled bacterial strains were generated by transformation with the pTEC15 or pTEC27 plasmids (deposited with Addgene, plasmids 30174 and 30182 respectively), resulting in msp12-driven expression of the wasabi or tdTomato fluorescent proteins, respectively. Mycobacteria were grown at 33°C in Middlebrook 7H9 broth or on 7H10 agar (both by Difco) supplemented with 0.5% bovine serum albumin, 0.005% oleic acid, 0.2% glucose, 0.2% glycerol, 0.085% sodium chloride and 0.05% Tween-80 (broth culture only). 50 μg/mL hygromycin was added as appropriate. For sucrose counter-selection, 7H10 agar was supplemented with 10% sucrose. Single-cell suspensions of bacteria were prepared as described33. To prepare heat-killed crushed M. marinum, bacteria were incubated at 80°C for 20 minutes and then homogenized in a Biospec Bead Beater together with 0.1 mm silica spheres for 1 minute. The P. aeruginosa PAO1 fluorescent strain used in this study has been described34. The S. aureus Newman strain expressing pOS1-SdrC-mCherry #391 was a gift from Dr. Juliane Bubeck Wardenburg.
Targeted Deletion of mmpL7 and pks15
A 2638 bp PstI fragment containing part of the mmpL7 (MMAR_1764) ORF was cloned into a pBluescript-derived vector, pBSXKpn.2 (CLC, unpublished). A 1124 bp KpnI fragment internal to mmpL7 was then excised and replaced with the aph cassette, conferring kanamycin resistance. The sucrose counter-selectable marker, sacB, and an additional marker hygA, conferring hygromycin resistance, were then added to create pJENK7.1::Hyg. This construct was transformed into the wild-type reference strain, M, and kanamycin-resistant colonies were selected. Subsequent screening for sucrose-sensitivity identified merodiploids that were verified by southern blotting. One such merodiploid was then grown in liquid culture for ten days and plated on sucrose-containing medium. Sucrose- and kanamycin-resistant, hygromycin-sensitive colonies were then verified by southern blotting to identify the ΔmmpL7 mutant, KT15. This strain was verified to be deficient in surface localization of PDIM (data not shown), and exhibited colony morphology defects previously reported for M. marinum PDIM mutants35. The pks15 (MMAR_1762) locus was deleted as follows. Flanking regions upstream and downstream of pks15 were amplified by PCR using primers 5′pks15F (5′CCGCTCGAGGGTGCGATGCGTGGTATC3′), 5′pks15R.2 and (5′CGACTAGTTCAGTTGCTCCTGTTCATG3′), 3′pks15F, and (5′GGAGCAACTGAACTAGTACCATCCGACACCGACTG3′) and 3′pks15R.2 (5′CCGTCTAGAGTGGTGCTGTTCGGCGTC3′), respectively. These fragments were sequentially inserted, directly adjacent to each other into pBluescriptSK+::SacBHyg.1 (CLC, unpublished), a pBluescript derivative which contains sacB and hygA external to the MCS. The resulting construct, pPKS15KO, bears an unmarked deletion of the pks15 ORF, and was used to transform strain M, and hygromycin resistant colonies were selected. Putative merodiploids were verified by Southern blotting and then counter-selected on sucrose as above, to produce the sucrose-resistant, hygromycin-sensitive isolate (KT21) which was then verified by Southern blotting. Additional verification by thin layer chromatography determined that PGL was absent, while PDIM production was retained (data not shown), consistent with deletion of pks15 in M.tuberculosis5.
Zebrafish Husbandry and Infections
Wild-type AB zebrafish were maintained as described36. Larvae (of undetermined sex given the early developmental stages used) were infected at 36–48 hours post-fertilization (hpf) via caudal vein or hindbrain ventricle injection using thawed single-cell suspensions of known titer33,36. Number of animals to be used for each experiment was guided by past results with other bacterial mutants and/or zebrafish morphants. Larvae were randomly allotted to the different experimental conditions. Zebrafish husbandry and all experiments performed on them were in compliance with Institutional Animal Care and Use Committee approved protocols.
Microscopy and Image-Based Quantification of Infection Level
Wide-field microscopy was performed using a Nikon Eclipse Ti-E equipped with a C-HGFIE 130W mercury light source, Chroma FITC (41001) filter, and 2x/0.10 Plan Apochromat objective. Fluorescence images were captured with a CoolSNAP HQ2 Monochrome Camera (Photometrics) using NIS-Elements (version 3.22). Quantification of fluorescent M. marinum infection using images of individual embryos using Fluorescent Pixel Count (FPC) was performed as previously described33. For confocal imaging, larvae were imbedded in 1.5% agarose (low melting point)37. A series of z-stack images with a 2 μm step size was generated through the infected HBV, using the galvo scanner (laser scanner) of the Nikon A1 confocal microscope with a 20x Plan Apo 0.75 NA objective. Bacterial burdens were determined by using the 3D surface-rendering feature of Imaris (Bitplane Scientific Software)8.
Hindbrain Assays
Macrophage recruitment assays were performed as previously described36. For determination of hindbrain ventricle infection burdens, 1 and 3 DPI larvae were mounted in 1.5% agarose and confocal z-stacks of 2 μm were obtained.
iNOS Staining
Antibody staining of larvae was performed as described31. Larvae were then imaged using confocal microscopy and the number of infected macrophages that were positive for iNOS staining was determined for each larva.
iNOS Scavengers
Fish were treated as previously described38. CPTIO or L-NAME (Sigma) were used at a final concentration of 500 μM and 1 mM, respectively in 0.1% DMSO in fish water. Fish were incubated immediately following infection and fresh inhibitor was added every 24 hours until bacterial burden was determined.
Morpholinos
Morpholinos described in Table S1 were injected at the 1-4 cell stage as previously described16.
RT-PCR to verify efficacy of Myd88 MO
RNA was extracted from pools of 15-40 embryos using TRIzol reagent (Life Technologies), treated with Turbo DNA-Free Kit (Life Technologies) and cDNA synthesized with PrimeScript (Takara). Primers used for PCR were as follows: actin FWD 5′-ACCTGACAGACTACCTGATG, REV 5′-TGAAGGTGGTCTCATGGATAC, myd88 FWD 5′-ATGGCATCAAAGTTAAGTATAGACC, REV 5′-AGGGCAGTGAGAGTGCTTTG.
Identification of candidate CCR2 orthologue in zebrafish
BLAST searches of the zebrafish genome (www.ensembl.org) identified two closely-related CCR-like genes on Chromosome 16, ENSDARG00000079829 and ENSDARG00000062999. In BLAST comparisons to the human genome ENSDARG00000079829 was found to have the highest homology to human CCR2 (E value 8.8e-112), while ENSDARG00000062999 was most highly homologous to human CCR4 (E value 2e-90). In addition, annotation of the zebrafish genome from NCBI annotates ENSDARG00000079829 as a CCR-2-like gene. We confirmed expression of the mRNA and identified the short 5′ upstream exon ATGTCGGCGACACAAAACAGTA via 5′-RACE39.
Identification of zebrafish CCL2 orthologue
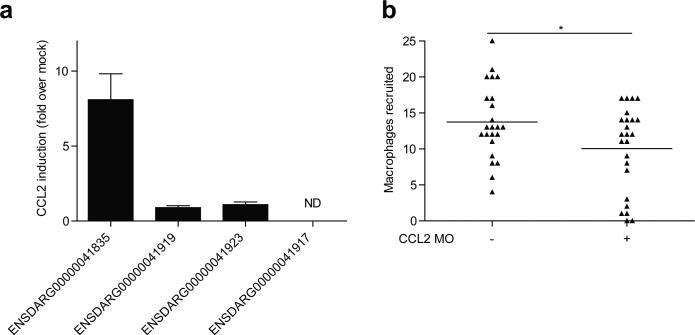
Protein sequences of human and mouse CCL2 were used to interrogate the zebrafish genome by BLAST. Expression levels of the four most closely-related zebrafish proteins was then examined at 3 HPI to identify the likely functional orthologue (Extended Data Fig. 9a). Of the four candidates, only ENSDARG00000041835 was significantly induced at 3 HPI. Knockdown of ENSDARG00000041835 resulted in a decrease in macrophage recruitment into the HBV at 3 HPI (Extended Data Fig. 9b).
Quantitative Real-time PCR (qRT-PCR)
cDNA was synthesized from pools of 20-40 larvae as previously described31. Quantification of ccl2 RNA levels were determined using SYBR green and the following primer pair; 5′GTCTGGTGCTCTTCGCTTTC3′ and 5′TGCAGAGAAGATGCGTCGTA3′.
Infectivity Assay
2 DPF larvae were infected via the HBV36 with an average of 0.8 bacteria per injection. Fish harboring 1-3 bacteria were then identified at 5 HPI by confocal microscopy. These infected fish were then evaluated at 5 DPI and were scored as infected or uninfected, based on the presence or absence of fluorescent bacteria.
Mice, aerosol infections, and flow cytometry
C57BL/6 mice were purchased from Jackson Laboratories. All mice were housed under specific pathogen free conditions at Seattle Biomedical Research Institute, and all experiments were performed in compliance with the respective Institutional Animal Care and Use Committee approved protocols. Ten-week old female mice were randomized to the different experimental groups. The number of mice to be used to adequately power the experiment was guided by the results of the corresponding zebrafish experiments. A stock of M. tuberculosis strain H37Rv or the isogenic PDIM-deficient ΔdrrA strain was sonicated before use and mice were infected in an aerosol infection chamber (Glas-Col) with approximately 200 colony forming units (CFUs) of H37Rv or 1000 CFUs of ΔdrrA to achieve similar bacterial burdens at 21 days post infection. The infectious dose in each experiment was determined by plating lung tissue of two mice from each group. Colonies on 7H10 agar plates were counted after 21 days of incubation at 37°C. Lung tissue was perfused with 5 ml of PBS administered through the right ventricle of the heart, finely chopped using a gentleMACS Octo Dissociator (Miltenyi Biotec) and incubated at 37°C for 30 minutes in HEPES buffer containing Liberase Blendzyme 3 (Roche Applied Science). Following digestion, single cell suspensions were prepared by passing tissue through a cell strainer. Single cells suspensions were then stained for flow cytometric analysis. Lung single cell suspensions were surface stained at 4°C for 20 minutes in the presence of Fc block (24G2) with the following antibodies from eBioscience: PE-Cy7 labeled anti-CD4 (GK1.5, eBioscience), anti-CD8α (53-6.7, eBioscience), anti-CD11c (N418), and FITC-labeled anti-Ly6G (1A8) to exclude T cells, DCs, and neutrophils. Alveolar macrophages were excluded based on their high CD11c expression and autofluorescence. PerCPCy5.5 labeled anti-Ly6C (HK1.4) and APC-efluor 780 labeled anti-CD11b (M1/70) were used to identify CD11b+Ly6CHI monocytes. Intracellular staining was done following fixation and permeabilization following manufacturer recommendations (eBioscience). Cells were fixed and permeabilized using eBioscience's Fix/Perm buffer for 1 hour at 4°C, followed by staining for iNOS with anti-NOS2 Alexa Fluor 405 (C-11, Santa Cruz Biotechnology) or mouse IgG1 isotype control for 30 minutes at 4°C. Samples were analyzed on an LSR-II (BD Biosciences) and FlowJo Software (Treestar).
Statistics
Statistical analyses were performed using Prism 5.01 (GraphPad). For data sets requiring log10 transformation prior to ANOVA, embryos with no detectable fluorescence above background were assigned a value of 0.9, with 1 being the limit of detection, prior to log10 transformation. Post-test P values are as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
Supplementary Material
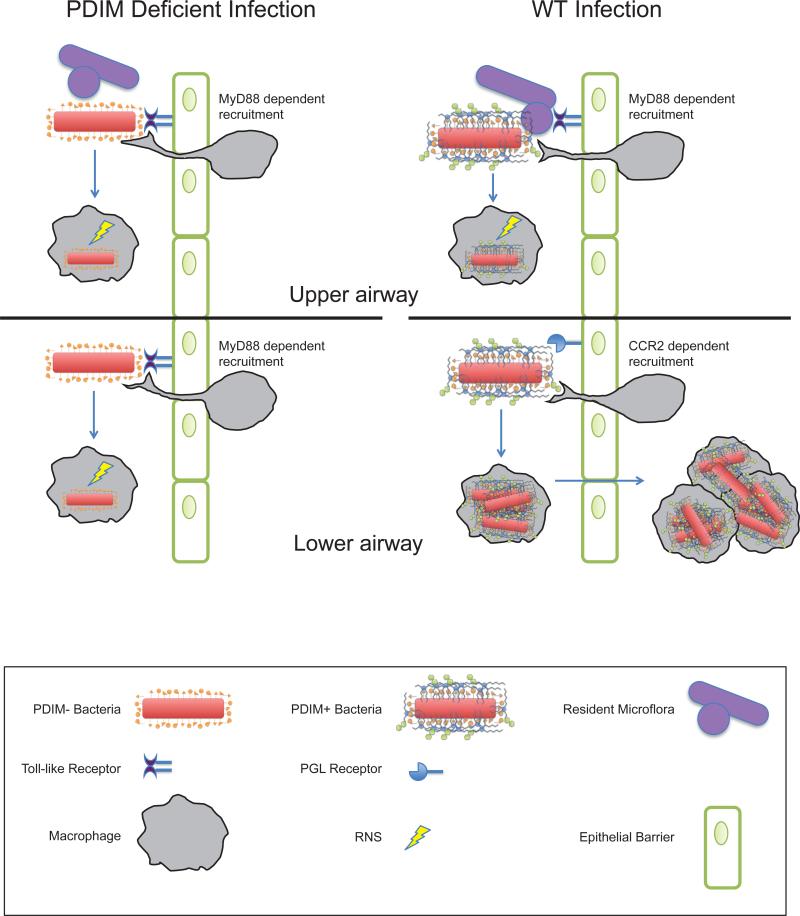
Extended Data Fig. 1.
Coordinate use of PDIM-mediated immune evasion and PGL-mediated recruitment by pathogenic mycobacteria. Model for infection with wild-type (WT) and PDIM-deficient mycobacteria are shown in the context of the relatively sterile lower airway versus the upper airway, with its higher levels of resident microflora and inhaled environmental organisms.
Extended Data Fig. 2.
ΔmmpL7 bacteria are attenuated in zebrafish larvae. a, Kaplan-Meier graph showing daily survival of larvae infected via caudal vein injection with medium (mock), 29 wild-type or 70 ΔmmpL7 M. marinum. N=25 (mock), 31 (wild-type), or 29 (ΔmmpL7) larvae per group. Mean time to Death (days): Mock (11), wild-type (7.6) and ΔmmpL7 (11.2). Survival was compared by log-rank test: wild-type vs. mock and wild-type vs. ΔmmpL7, p<0.0001; mock vs. ΔmmpL7, p=0.5601. b, c, Larvae were infected via caudal vein injection 1 DPF with 550 wild-type, 650 ΔmmpL7, or 700Δerp, fluorescent M. marinum. b, Infection burdens were measured by Fluorescent Pixel Count (FPC, mean +/- SEM). c, Representative images at 7 DPI. N= 29 (wild-type and ΔmmpL7) or 30 (Δerp) larvae per group. Scale bar = 500 μm. At 3, 5 and 7 DPI, Log10 FPC was compared by ANOVA, with Dunnett's post-test. ***, p<0.001. d,e, Representative images from wild-type (d) and ΔmmpL7 (e) M. marinum HBV infections quantified in Figure 1d. N=18 (wild-type) or 16 (ΔmmpL7) larvae per group. HBVs are outlined with a dashed white line. Scale bar = 100μm.
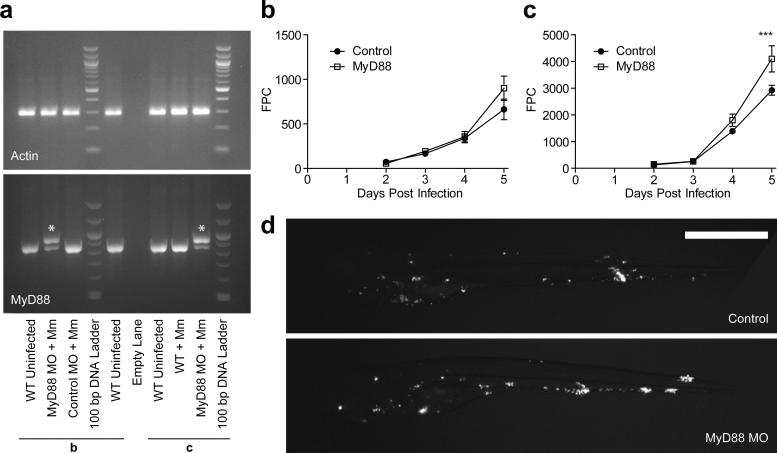
Extended Data Fig. 3.
Knockdown of MyD88 results in a late, dose dependent hypersusceptibility to M. marinum systemic infection. a, RT-PCR for actin (upper panel) and myd88 (lower panel) demonstrating that that the majority of myd88 transcripts at 7 DPF are abnormal in MyD88 morphants. Lanes marked ‘b’ and ‘c’ correspond to morphants from the same experiments depicted in panels b and c respectively. The abnormal larger transcript (indicated by *) results from the inclusion of intron 2 in the final transcript, incorporating a premature stop codon that truncates the protein prior to the TIR (Toll/Interleukin Receptor) domain. (b, c) Caudal vein infection of MyD88 morphants with 141 (b) or 325 (c) CFU M. marinum/larva. Bacterial burden was assessed by FPC, values plotted represent the mean +/- SEM. Time points were compared by one-way ANOVA and Bonferroni's post-tests, *** p<0.001. d, Representative images of larvae at 5 DPI from experiment in (c), N=30 control, 15 Myd88 MO.
Extended Data Fig. 4.
Characteristics of macrophages recruited to wild-type and PDIM-deficient bacteria. a,. Mean mpeg1 positive macrophages recruited at 3 HPI into the HBV of wild-type fish following infection with 80 wild-type or ΔmmpL7 M. marinum. b, Data from Figure 2C expressed as mean numbers of total infected macrophages and iNOS expressing infected macrophages following HBV infection with 80 wild-type, ΔmmpL7, or Δerp M. marinum. c, bacterial burdens after L-NAME treatment. Mean bacterial burdens of 2 DPF control (CTRL) or iNOS inhibitor (L-NAME) treated fish following HBV infection with 80 wild-type or ΔmmpL7 M. marinum.
Extended Data Fig. 5.
Wild-type bacterial burdens after co-infection with wild-type or ΔmmpL7 bacteria. Representative images from the HBV co-infections quantified in Figure 2e. a-b, Red fluorescent wild-type (WT) M. marinum co-infected with green fluorescent wild-type (a) or ΔmmpL7 (b) M. marinum. N=18 (wild-type) and 19 (ΔmmpL7) larvae per group. Scale bar = 50 μm. c, Wild-type bacterial burdens after co-infection with wild-type or ΔmmpL7 M. marinum with and without L-NAME treatment. Significance tested by one-way ANOVA with Bonferroni's post–test for comparisons shown.
Extended Data Fig. 6.
MyD88-dependent macrophage recruitment occurs in response to PDIM deficiency rather than due to a loss of another MmpL7 exported product. a, Mean macrophage recruitment at 3 HPI into the HBV of wild-type or MyD88 MO larvae following infection with 80 Δmas M. marinum. Student's unpaired t test. b, Mean surviving bacterial volume of red fluorescent wild-type M. marinum (initial infection dose of 30-40 CFU) when co-infected with 30-40 green fluorescent wild-type, ΔmmpL7 or Δmas M. marinum at 3 DPI. Representative of two separate experiments. Significance tested by one-way ANOVA with Tukey's post-test.
Extended Data Fig. 7.
Gating Strategy and Isotype Controls for iNOS Staining of Mouse Lung. a, Representative gating strategy for isolation of inflammatory monocytes. A dump channel containing anti-CD4, CD8, and CD11c was plotted against a channel exhibiting autofluorescence and also containing anti-Ly6G. Using these markers, T cell, dendritic cell, alveolar macrophage, and neutrophil cell populations were excluded from the double negative gate. Inflammatory monocytes were identified within the double negative population by their co-expression of Ly6C and CD11b. These cells were then evaluated for intracellular iNOS expression, N=4 per group (Figure 3a,b) or b, with isotype control antibodies, N=4 per group.
Extended Data Fig. 8.
Specificity of CCL2-mediated macrophage recruitment in wild-type and CCR2 morphant larvae a, Mean macrophage recruitment at 3 HPI into the HBV of control (ctrl), or CCR2 MO (ccr2) larvae following injection of vehicle control (“mock”; 0.1% BSA in PBS), human CCL2 (hCCL2), human CCL4 (hCCL4), or human CCL5 (hCCL5) b-c, Mean macrophage (b) and neutrophil (c) recruitment at 3HPI into the HBV of control (ctrl), CCR2 MO (ccr2), or MyD88 MO (myd88) larvae following injection of vehicle control (mock), murine CCL2 (mCCL2), human CCL2 (hCCL2), human interleukin-8 (hIL-8), or human leukotriene B4 (hLTB4). Representative of three separate experiments. Significance assessed by one-way ANOVA with Bonferroni's post-test for the comparisons shown, *P < 0.05; ***P < 0.001.
Extended Data Fig. 9.
Identification of zebrafish CCL2 orthologue. a, mRNA levels of potential CCL2 orthologues (mean +/- SEM of four biological replicates) induced at 3 hours post caudal vein infection of 2dpf larvae with 250-300 wild-type M. marinum. These assays were performed on the same cDNA pools as the data presented in Figure 4b. b, Mean macrophage recruitment at 3 HPI into the HBV of wild-type or CCL2 MO fish following infection with 80 M. marinum. Representative of two separate experiments.
Extended Data Fig. 10.
Infectivity Assay. a-b, Representative 5 HPI images from Figure 4d following HBV infection with (a) one or (b) three M. marinum. Scale bar = 100 μm. Ns for fish represented in (a) and (b) (i.e. those found to be infected with one to three bacteria) are presented in Figure 4d (18, 22, 28, 28, 28, 28, 22, 22 for the respective conditions as specified in the figure). c, Mean bacterial burdens 5 HPI HBV infection with 1-3 wild-type (WT), ΔmmpL7, or Δpks15 M. marinum.
Acknowledgments
We thank S. Falkow and P. Edelstein for sharing their knowledge and insights, P. Donald for discussions about human infectivity in TB, B. Cormack for manuscript review and editing, J. Bubeck-Wardenberg for the fluorescent S. aureus strain, K. Hicks for initial MyD88 experiments, T.-Y. Chen, B. Moody, P. Manzanillo and J. Cox for help with lipid analyses, and J. Cameron for fish facility management. Supported by an NSF predoctoral fellowship to CJC, a Senior Research Training Fellowship from the American Lung Association and the National Institutes of Health Training Grant “Training Clinical and Basic Immunologists” to RPL, a National Institutes of Health “Academic Pediatric Infectious Disease” Training Grant award to REH, an American Cancer Society Postdoctoral Fellowship and National Institutes of Health Bacterial Pathogenesis Training Grant Award to DMT, and National Institutes of Health grants to KBU and LR. DMT is a recipient of the National Institutes of Health Director's New Innovator Award and LR is a recipient of the National Institutes of Health Director's Pioneer Award.
Footnotes
The authors declare no competing financial interests.
Author Contribution
CJC, CLC, KKT, DMT, REH and LR conceived and designed M. marinum-zebrafish experiments and analyzed data, CJC, CLC, KKT, DMT and REH performed these experiments, RPL and KBU designed the M. tuberculosis-mouse experiments and analyzed the data, RPL performed the mouse experiments, CJC, CLC and LR wrote the paper, CJC, RPL and KKT prepared the figures and all authors edited the paper.
Supplementary Information
References
- 1.Philips JA, Ernst JD. Tuberculosis Pathogenesis and Immunity. Annual Review of Pathology: Mechanisms of Disease. 2012;7:353–384. doi: 10.1146/annurev-pathol-011811-132458. [DOI] [PubMed] [Google Scholar]
- 2.Onwueme KC, Vos CJ, Zurita J, Ferreras JA, Quadri LEN. The dimycocerosate ester polyketide virulence factors of mycobacteria. Progress in Lipid Research. 2005;44:259–302. doi: 10.1016/j.plipres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Murry JP, Pandey AK, Sassetti CM, Rubin EJ. Phthiocerol dimycocerosate transport is required for resisting interferon-gamma-independent immunity. The Journal of Infectious Diseases. 2009;200:774–782. doi: 10.1086/605128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores-Villanueva PO, et al. A functional promoter polymorphism in monocyte chemoattractant protein-1 is associated with increased susceptibility to pulmonary tuberculosis. The Journal of experimental medicine. 2005;202:1649–1658. doi: 10.1084/jem.20050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed MB, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;449:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 7.Weiser JN. The pneumococcus: why a commensal misbehaves. Journal of Molecular Medicine. 2009;88:97–102. doi: 10.1007/s00109-009-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C-T, et al. Neutrophils Exert Protection in the Early Tuberculous Granuloma by Oxidative Killing of Mycobacteria Phagocytosed from Infected Macrophages. Cell Host & Microbe. 2012;12:301–312. doi: 10.1016/j.chom.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer-Barber KD, et al. Cutting Edge: Caspase-1 Independent IL-1 Production Is Critical for Host Resistance to Mycobacterium tuberculosis and Does Not Require TLR Signaling In Vivo. The Journal of Immunology. 2010;184:3326–3330. doi: 10.4049/jimmunol.0904189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramakrishnan L. Revisiting the role of the granuloma in tuberculosis. Nature Reviews Immunology. 2012;12:352–366. doi: 10.1038/nri3211. [DOI] [PubMed] [Google Scholar]
- 11.Rosenthal S, Tager IB. Prevalence of gram-negative rods in the normal pharyngeal flora. Annals of internal medicine. 1975;83:355–357. doi: 10.7326/0003-4819-83-3-355. [DOI] [PubMed] [Google Scholar]
- 12.Wertheim HFL, et al. The role of nasal carriage in Staphylococcus aureus infections. The Lancet Infectious Diseases. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 13.Eddens T, Kolls JK. Host defenses against bacterial lower respiratory tract infection. Current Opinion in Immunology. 2012;24:424–430. doi: 10.1016/j.coi.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Mendonça-Lima L, et al. The allele encoding the mycobacterial Erp protein affects lung disease in mice. Cellular Microbiology. 2003;5:65–73. doi: 10.1046/j.1462-5822.2003.00237.x. [DOI] [PubMed] [Google Scholar]
- 15.van der Vaart M, van Soest JJ, Spaink HP, Meijer AH. Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Disease Models & Mechanisms. 2013;6:841–854. doi: 10.1242/dmm.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobin DM, et al. The lta4h Locus Modulates Susceptibility to Mycobacterial Infection in Zebrafish and Humans. Cell. 2010;140:717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. The Journal of experimental medicine. 1992;175 doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kröncke KD, Fehsel K, Kolb-Bachofen V. Inducible nitric oxide synthase in human diseases. Clinical and experimental immunology. 1998;113:147–156. doi: 10.1046/j.1365-2249.1998.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-Mediated Defense Against Microbial Pathogens. Annual Review of Immunology. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonelli LRV, et al. Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. Journal of Clinical Investigation. 2010;120 doi: 10.1172/JCI40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ordway D, et al. The hypervirulent Mycobacterium tuberculosis strain HN878 induces a potent TH1 response followed by rapid down-regulation. Journal of immunology. 2007;179:522–531. doi: 10.4049/jimmunol.179.1.522. [DOI] [PubMed] [Google Scholar]
- 22.Bates JH, Potts WE, Lewis M. Epidemiology of Primary Tuberculosis in an Industrial School. The New England journal of medicine. 1965;272:714–717. doi: 10.1056/NEJM196504082721403. [DOI] [PubMed] [Google Scholar]
- 23.Wells WF, Ratcliffe HL, Grumb C. On the mechanics of droplet nuclei infection; quantitative experimental air-borne tuberculosis in rabbits. American journal of hygiene. 1948;47:11–28. doi: 10.1093/oxfordjournals.aje.a119179. [DOI] [PubMed] [Google Scholar]
- 24.Sinsimer D, et al. The Phenolic Glycolipid of Mycobacterium tuberculosis Differentially Modulates the Early Host Cytokine Response but Does Not in Itself Confer Hypervirulence. Infection and Immunity. 2008;76:3027–3036. doi: 10.1128/IAI.01663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott HM, Flynn JL. Mycobacterium tuberculosis in chemokine receptor 2-deficient mice: influence of dose on disease progression. Infection and Immunity. 2002;70:5946–5954. doi: 10.1128/IAI.70.11.5946-5954.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng WX, et al. CCL2−2518 (A/G) polymorphisms and tuberculosis susceptibility: a meta-analysis. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2012;16:150–156. doi: 10.5588/ijtld.11.0205. [DOI] [PubMed] [Google Scholar]
- 27.Gagneux S. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlson ES, et al. Topographical Continuity of Bacterial Populations in the Healthy Human Respiratory Tract. American Journal of Respiratory and Critical Care Medicine. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Bernuth H, Picard C, Puel A, Casanova J-L. Experimental and natural infections in MyD88- and IRAK-4-deficient mice and humans. European Journal of Immunology. 2012;42:3126–3135. doi: 10.1002/eji.201242683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clay H, et al. Dichotomous Role of the Macrophage in Early Mycobacterium marinum Infection of the Zebrafish. Cell Host & Microbe. 2007;2:29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosma CL, Klein K, Kim R, Beery D, Ramakrishnan L. Mycobacterium marinum Erp Is a Virulence Determinant Required for Cell Wall Integrity and Intracellular Survival. Infection and Immunity. 2006;74:3125–3133. doi: 10.1128/IAI.02061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takaki K, Cosma CL, Troll MA, Ramakrishnan L. An in vivo platform for rapid high-throughput antitubercular drug discovery. Cell reports. 2012;2:175–184. doi: 10.1016/j.celrep.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brannon MK, et al. Pseudomonas aeruginosaType III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cellular Microbiology. 2009;11:755–768. doi: 10.1111/j.1462-5822.2009.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, et al. Both Phthiocerol Dimycocerosates and Phenolic Glycolipids Are Required for Virulence of Mycobacterium marinum. Infection and Immunity. 2012;80:1381–1389. doi: 10.1128/IAI.06370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takaki K, Davis JM, Winglee K, Ramakrishnan L. Evaluation of the pathogenesis and treatment of Mycobacterium marinum infection in zebrafish. Nature Protocols. 2013;8:1114–1124. doi: 10.1038/nprot.2013.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis JM, Ramakrishnan L. The Role of the Granuloma in Expansion and Dissemination of Early Tuberculous Infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lepiller S, et al. Imaging of nitric oxide in a living vertebrate using a diaminofluorescein probe. Free Radical Biology and Medicine. 2007;43:619–627. doi: 10.1016/j.freeradbiomed.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 39.Maruyama IN, Rakow TL, Maruyama HI. cRACE: a simple method for identification of the 5′ end of mRNAs. Nucleic Acids Research. 1995;23:3796–3797. doi: 10.1093/nar/23.18.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.