Abstract
Aims:
To study the prevalence of extended spectrum β-lactamase (ESBL) producing Enterobacteriaceae and coresistance to other commonly used antibiotics from the Bhopal region of Central India.
Settings and Design:
A prospective study was conducted from September 2011 to August 2012 in Microbiology Department of our tertiary health care center.
Materials and Methods:
A total of 1044 Enterobacteriaceae isolates were recovered from various specimens. ESBL production was detected by using Clinical Laboratory Standard Institute (CLSI) that described the phenotypic confirmatory test along with routine antibiotic susceptibility testing.
Statistical Analysis:
Two-tailed Z-test.
Results:
Escherichia coli was the most common isolate (65.32%). ESBL production was confirmed in 504 (48.27%) isolates. The isolates of E. coli (50.14%) were the most common ESBL producers. Maximum ESBL isolates were obtained from urine samples (52.28%) and male patients (52.54%). Sensitivity to imipenem was 100% followed by piperacillin–tazobactam (89.28%), meropenem (87.5%), and amikacin (83.92%). Significant resistance was detected against trimethoprim–sulfomethoxazole, fluoroquinolones, and gentamicin.
Conclusion:
This is the only study conducted from Central India and shows high prevalence of ESBL production among Enterobacteriaceae. Imipenem seems to be more sensitive than meropenem. Piperacillin–tazobactam combination was found to be the best among the β-lactam–β-lactamase inhibitor combinations. Prevalence of ESBL producers were more in males than females.
Keywords: Antibiotic coresistance, Enterobacteriaceae, extended spectrum β-lactamase
INTRODUCTION
Beta-lactam antimicrobial agents are the most commonly used treatment of bacterial infections.[1] Productions of β-lactamases are reported to be the leading cause of resistance to these antimicrobial agents, especially by gram-negative bacteria.[1,2] These enzymes are numerous and they mutate continuously in response to heavy pressure of antibiotic use and have lead to the development of extended spectrum β-lactamases (ESBL).[3] In recent years, there has been an increased incidence and prevalence of ESBLs among family Enterobacteriaceae.[3]
ESBL are placed under Bush's functional class 2be.[4] They are plasmid-mediated enzymes and are derived from point mutation of TEM on SHV β-lactamases that are widely distributed among the Enterobacteriaceae.[3,5] In recent years, several new ESBLs of the non-TEM and the non-SHV types emerged, such as the enzymes of the CTX-M, PER, VEB, and the GES lineages.[6] ESBL inactivate β-lactam antibiotics containing the oxyimino group such as oxyimino-cephalosporin and oxyimino-monobactam.[5] They have no effect on cephamycins and carbapenems and are commonly inhibited by β-lactamase inhibitors such as clavulinic acid, sulbactam, and Tazobactam.[7,8]
Plasmid coding for ESBL enzymes may carry coresistance genes for other non-β-lactam antibiotics.[9] Therefore, it is common for organisms expressing an ESBL to express coresistance to other antibiotics, such as aminoglycosides, trimethoprim, sulfomethoxazole, and tetracycline, leading to treatment failure. Being plasmid mediated, these enzymes spread fast among various bacteria. Hence, reliable detection of ESBL production by the clinical microbiology laboratory is essential.
Although various studies have been conducted on the ESBL-producing strains of Enterobacteriaceae in different regions of India like Dibrugarh,[10] Bangalore,[11] Pondicherry,[12] Bijapur,[13] New Delhi,[14] Hyderabad,[15] Mumbai,[16] no published data are available on the prevalence of ESBL production in the Bhopal region of Madhya Pradesh in Central India.
Therefore, the study was conducted at a tertiary care hospital, Bhopal, to know the prevalence of ESBL among members of the family Enterobacteriaceae isolated in various clinical specimens and to know associated coresistance for other commonly used antimicrobial agents.
MATERIALS AND METHODS
A prospective study was conducted over a period of 1 year (September 2011 to August 2012) at a tertiary care teaching hospital.
Bacterial isolates
A total of 1044 consecutive, nonrepetitive clinical isolates of Enterobacteriaceae isolated from various clinical samples such as urine (656), pus/wound swabs (231), sputum (49), tracheal aspirate (39), blood (40), vaginal swab (19), and ascitic fluid (10) were included in the study. Samples were processed and isolates were identified by standard laboratory methods.[17]
Antimicrobial agents susceptibility testing
Antimicrobial susceptibility was determined by the Kirby-Bauer disc diffusion method[18] according to Clinical Laboratory Standard Institute (CLSI) guidelines.[19] Antibiotic discs were obtained from HiMedia, Mumbai.
The antibiotics used for antibiogram determination of collected strains were ampicillin (10 μg), cefuroxime (30 μg), ceftriaxone (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg), cefoxitin (30 μg), aztreonam (30 μg), amoxicillin–clavulinic acid (20/10 μg), ampicillin–sulbactam (10/10 μg), piperacillin–tazobactam (100/10 μg), gentamicin (10 μg), amikacin (30 μg), ciprofloxacin (5 μg), imipenem (10 μg), meropenem (10 μg), trimethoprim–sulfamethoxazole (1.25/23.75 μg). Norfloxacin (10 μg) and nitrofurantoin (300 μg) were tested against isolates from urine samples only.
Detection of ESBL
ESBL production was detected by using the CLSI described phenotypic confirmatory test along with routine antibiotic susceptibility testing. Cefotaxime and ceftazidime discs alone and in combination with clavulinic acid were used. A >5 mm increase in zone was considered as confirmation of ESBL production.[19]
Although CLSI described phenotypic confirmatory test is applicable for Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis, an attempt was made to look for ESBL production among the other members of Enterobacteriaceae.
ESBL-producing Enterobacteriaceae isolates which were found to be resistant to cefoxitin (zone diameter < 18 mm)[19] were not considered for the study. This is to exclude associated Amp-C type of β-lactamases.[20,21]
Throughout the study, K. pneumoniae ATCC 700603 and E. coli ATCC 25922 (HiMedia Laboratories, Mumbai) were used as positive and negative controls, respectively, for ESBL production.
RESULTS
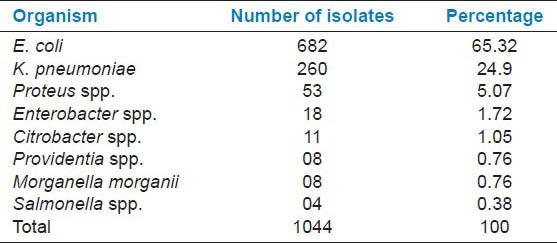
In 1 year study period, a total of 1044 Enterobacteriaceae isolates were obtained. Table 1 shows different isolates of family Enterobacteriaceae isolated from clinical samples. E. coli was the most common (65.32%) isolate followed by K. pneumoniae (24.9%) and others.
Table 1.
Isolates of family Enterobacteriaceae from clinical samples
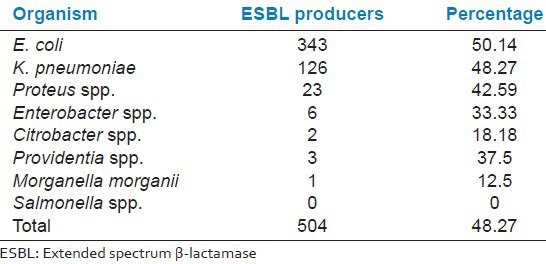
A total of 504 of these isolates produced ESBL. The detection rate of ESBL in our study was 48.27%. E. coli was the most common ESBL producing Enterobacteriaceae (50.14%) followed by K. pneumoniae (48.27%) and others [Table 2].
Table 2.
ESBL producers among different isolates of family Enterobacteriaceae
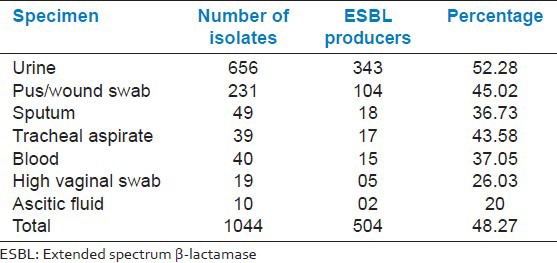
ESBL producing strains were obtained from various specimens as shown in Table 3. The majority of ESBL isolates were from urine samples (52.28%).
Table 3.
Specimen wise distribution of ESBL producers
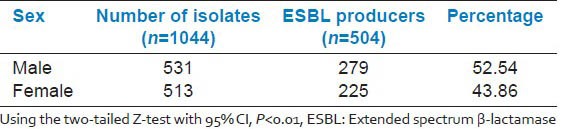
Sex wise distribution of ESBL producers as shown in Table 4 revealed that the prevalence was more among the males as compared to females.
Table 4.
Sex wise distribution of ESBL producers
The antibiotic susceptibility pattern of ESBL isolates to other antibiotics is shown in Figure 1. All the isolates were sensitive to imipenem. The majority of ESBL producers (>80%) were sensitive to meropenem, piperacillin-tazobactam, and amikacin. Only 50% of ESBL isolates were sensitive to gentamicin. Occurrence of coresistance among ESBL isolates was observed. Poor sensitivity (<15%) was found against trimethoprim–sulfamethoxazole, amoxicillin–clavulinic acid, ampicillin–sulbactam, and ciprofloxacin. The susceptibility of ESBL-producing urinary isolates to nitrofurantoin and nalidixic acid was 82.5% and 15.16%, respectively.
Figure 1.
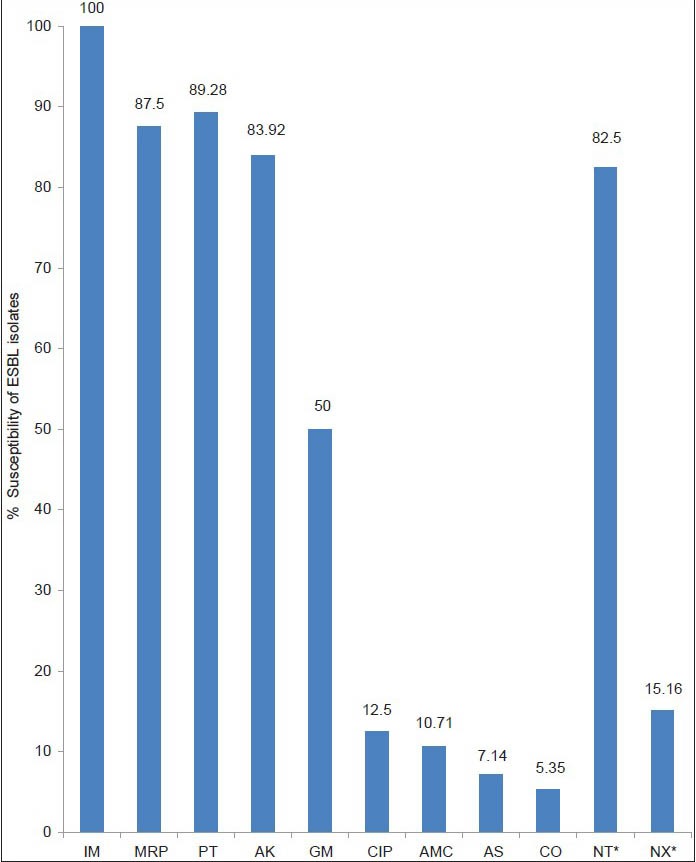
Susceptibility of the ESBL isolates to various antibiotics. IM: imipenem, MRP: meropenem, PT: piperacillin– tazobactam, AK: amikacin, GM: gentamicin, CIP: ciprofloxacin, AMC: amoxicillin–clavulinic acid, AS: ampicillin–sulbactam, CO: trimethoprim–sulfamethoxazole, NT: nitrofurantoin, NX: norfloxacin. Asterisk indicates antibiotics tested against urinary isolates only
DISCUSSION
The members of the family Enterobacteriaceae are among the most important bacterial human pathogens accounting for the majority of bacteria isolated from clinical samples.[22] The β-lactam group of drugs including extended spectrum cephalosporins are commonly used for treatment of such infections. In recent years, bacterial resistance to these drugs has increased dramatically with ESBL contributing to this increase.[3,5] These enzymes are plasmid coded which may also carry coresistance genes for other commonly used non-β-lactam antibiotics and thus limiting the number of useful drugs against these bacteria.[9] To make problems worse, plasmid-mediated ESBL enzymes spread fast among various bacteria resulting into a number of nosocomial outbreaks.[23,24,25] Hence, reliable and accurate detection of ESBL in a microbiology laboratory is a must. In a recent study conducted by Gavin et al.,[26] it was found that the majority of physicians changed therapy after a report of an ESBL-producing pathogen from microbiology laboratory highlighting the importance of ESBL detection. Since no data on ESBL prevalence in our area were available, a study was conducted in our institute to look for prevalence of ESBL among members of the family Enterobacteriaceae and antimicrobial susceptibility pattern of such isolates.
In one-year study period, a total of 1044 Enterobacteriaceae isolates were analysed. The majority were E. coli (65.82%) followed by K. pneumoniae (24.9%), Proteus spp. (5.07%), and others Table 1. Metri et al.[13] from Bijapur have also reported E. coli and K. pneumoniae as the most common Enterobacteriaceae which were prevalent in their clinical samples, and this was well comparable to the reports from our study. Rudresh et al.[11] from Bangalore too reported the prevalence of 40.2% E. coli and 33.1% Klebsiella spp.
No countrywide study has been conducted so far for detection of the prevalence of ESBL production in India. Individual studies were done in different parts of the country, which showed various prevalence rates.
The prevalence of ESBL-producing organisms in this study was found to be 48.27% which was higher than that which was reported by other studies done in Hyderabad (19.8%),[15] Dibrugarh (24.56%),[10] and Bijapur (32.1%).[13] The prevalence was lower when compared with the studies which were done in Mumbai (53%),[16] Bangalore (62.3%),[11] and Pondicherry (66.7%).[12] The wide variation in the prevalence is probably due to the variation in the risk factors and in the extent of antibiotic use.
A report from Pondicherry, India, showed that ESBL production was 81% in E. coli and 74% in K. pneumonia.[12] In a similar study by Rudresh et al.,[11] 40.2% of the E. coli and 33.1% of K. pneumoniae isolates were reported to be ESBL producers. This study also reveals similar findings with E. coli as the major ESBL producer (50.14%) followed by K. pneumoniae (48.27%) Table 2. Although Salmonella spp. is known to produce ESBLs,[15,27] none of the Salmonella spp. in our study showed ESBL production. This could be due to the few isolates (4) obtained during the study period.
In our study, ESBL isolates were obtained from different clinical specimens [Table 3]. The majority of ESBL isolates were from urine samples. A study from North Karnataka, India, revealed similar findings.[13] Umadevi et al.[12] also found a maximum number of ESBL-producing isolates from the urine sample of patients.
Shah et al.[28] studied the relation of ESBL-producing Enterobacteriaceae with respect to age and gender and reported more ESBL-positive isolates in males (65.33%) than females (34.67%). Nibedita Das et al.[10] also found a slight male preponderance for ESBL production among the study subjects. Similar findings were observed in the present study. 52.54% of ESBL isolates were obtained from males while 43.46% from females [Table 4]. After applying the two-tailed Z-test with 95% CI, this difference was found to be statistically significant (P < 0.01).
In this study, all ESBL isolates were found to be sensitive to imipenem and more than 80% were susceptible to piperacillin–tazobactam (89.28%), meropenem (87.5%), and amikacin (83.92%) [Figure 1].
Although belonging to the same carbapenem group, imipenem showed better in vitro activity against ESBL-producing isolates than meropenem in our study. This result correlates well with the findings in other studies.[29,30] This is likely to be due to overuse of meropenem in our health care setting leading to heavy selection pressure and development of resistance among Enterobacteriaceae isolates. Hence we need to keep in mind that carbapenems are antimicrobials that should be kept in reserve.[29] They should be used only in life-threatening infections and in outbreak situations. This approach intends to preserve the therapeutic value of these precious drugs.
In our study, when sensitivity of the β-lactam–β-lactamase inhibitor combination was compared, it was observed that sensitivity of piperacillin–tazobactam was far better than amoxicillin–clavulinic acid and ampicillin–sulbactum [Figure 1]. Wong-Beringer[31] and al Zahrani et al.[32] have also reported good sensitivity of piperacillin–tazobactam against ESBL isolates. Bano et al.[33] conducted a study in patients with bacteremia due to ESBL-producing E. coli and suggested that piperacillin–tazobactam and amoxicillin–clavulinic acid are suitable alternatives to carbapenems for treating such patients if active in vitro and would be particularly useful as definitive therapy. According to Peterson,[34] piperacillin–tazobactam is clinically reliable for the treatment of serious infections caused by susceptible strains of ESBL-producing E. coli and Klebsiella spp. Our study also highlights the same results.
Antibiotic coresistance among ESBL isolates have been noted as a serious problem even at our tertiary health center. In our study, only 50% ESBL-producing organisms were sensitive to gentamicin, 12.5% to ciprofloxacin, and 5.35% to trimethoprim–sulfamethoxazole. Norfloxacin and nitrofurantoin were tested only for urinary isolates. ESBL isolated from urine samples showed good in vitro activity against nitrofurantoin (82.5% sensitive), but only 15.16% were sensitive to norfloxacin. Thus, low sensitivity of ESBL-producing Enterobacteriaceae has been observed for gentamicin, fluoroquinolones, and trimethoprim–sulfamethoxazole. Rudresh et al.[11] reported a similar susceptibility pattern for ESBL isolates with 46.9% isolates sensitive to gentamicin followed by ciprofloxacin (29.5%) and trimethoprim–sulfomethoxazole (23.4%). Nibebita Das et al.[10] in their study have showed 30.96% sensitivity toward fluoroquinolones and 2.39% toward trimethoprim–sulfamethoxazole. Ullah et al.[35] have also observed coresistance of ESBL isolated to different antibiotics. This may be due to occurrence of genes encoding resistance to aminoglycoside, trimethoprim–Sulfomethoxazole, and quinolones on the same plasmid that encodes for ESBL production.[5,9] Martínez-Martínez and colleagues[36] have performed an analysis of mechanism of quinolone resistance in K. pneumoniae isolates of clinical origin and found that porin loss was observed only in those K. pneumoniae strains producing an ESBL. A significant number of these porin deficient strains also showed active efflux of quinolones.
CONCLUSIONS
High prevalence of ESBL production among the members of Enterobacteriaceae is a matter of concern. This study showed that the phenotypic confirmatory test can reliably detect ESBL production. Instead of screening and confirming ESBL production, direct phenotypic confirmatory test along with routine antibiotic susceptibility testing can help to report ESBL production within 48 hours. This protocol can be followed on a routine basis and for all Enterobacteriaceae isolates to save time. This will help clinicians in selecting and prescribing proper antibiotics for treatment of such infections. Carbapenems still remains most effective drug against ESBL-producing organisms. Imipenem seems to be superior to meropenem. There is a need to “reserve” these precious drugs and should be used only in life-threatening infections. Their overuse or misuse can pose great problem.[29] If piperacillin–tazobactam or amikacin is found to be sensitive in vitro, either of these drugs can be used as alternatives to carbapenems. The situation may vary from region to region, so institutional antibiograms or local patterns of susceptibility are necessary and this helps for preparation of antibiotic policy of individual institute. An appropriate and judicious antibiotic use may lead to withdrawal of the selective pressure and there is possibility that the resistance bacteria will no longer have survival advantages.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Kotra LP, Samama J, Mobashery S. Beta-lactamases and their resistance to beta-lactam antibiotics. In: Lewis K, Salyers AA, Taber HW, Wax RG, editors. Bacterial resistance to antimicrobials. New York: Marcel Decker; 2002. pp. 123–60. [Google Scholar]
- 2.Samaha –Kfoury JN, Araj GF. Recent development in β-lactamases and extended spectrum β – lactamases. BMJ. 2003;327:1209–13. doi: 10.1136/bmj.327.7425.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodford PA. Extended spectrum β –lactamases in the 21st century. Characterization, epidemiology and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–51. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for β lactamases and its correlation with molecular structures. Antimicrob Agents Chemother. 1995;39:1211–33. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paterson DL, Bonomo RA. Extended spectrum β – lactamases; a clinical update. Clin Microbiol Rev. 2005;18:657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luzzaro F, Mezzatesta M, Mugnaioli C, Perilli M, Stefani S, Amicosante G, et al. Extended – spectrum β –lactamases among enterobacteria of Medical interest: Report of the second Italian Nationwide survey. J Clin Microbiol. 2006;44:1659–64. doi: 10.1128/JCM.44.5.1659-1664.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby GA. Extended spectrum beta-lactamases and other enzymes providing resistance to oxyimino-beta lactams. Infect Dis Clin North Am. 1997;11:875–87. doi: 10.1016/s0891-5520(05)70395-0. [DOI] [PubMed] [Google Scholar]
- 8.Thomson KS, Prevan AM, Lagrange PH. Novel plasmid mediated beta-lactamases in Enterobacteriaceae: Emerging problems for new beta-lactam antibiotics. Curr Clin Topics Infect Dis. 1996;16:151–63. [PubMed] [Google Scholar]
- 9.Jacoby GA, Sutton L. Properties of plasmids responsible for production of extended spectrum beta lactamases. Antimicrob Agents Chemother. 1991;35:164–9. doi: 10.1128/aac.35.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das N, Borthakur AK. Antibiotic coresistance among extended-spectrum beta lactamase- producing urinary isolates in a tertiary medical center: A prospective study. Chron Young Sci. 2012;1:53–6. [Google Scholar]
- 11.Rudresh SM, Nagarathnamma T. Extended spectrum β-lactamase producing Enterobacteriaceae and antibiotic co- resistance. Indian J Med Res. 2011;133:116–8. [PMC free article] [PubMed] [Google Scholar]
- 12.Umadevi S, Kandhakumari G, Joseph NM, Kumar S, Easow JM, Stephen S, et al. Prevalence and antimicrobial susceptibility pattern of ESBL producing Gram Negative Bacilli. J Clin Diagn Res. 2011;5:236–9. [Google Scholar]
- 13.Metri Basavaraj C, Jyothi P, Peerapur Basavaraj V. The prevalence of ESBL among Enterobacteriaceae in a tertiary care hospital of North Karnataka, India. J Clin Diagn Res. 2011;5:470–5. [Google Scholar]
- 14.Khan MK, Thukral SS, Gaind R. Evaluation of a modified double disc synergy test for detection of extended spectrum β-lactamases in AmpC β-lactamase producing proteus mirabilis. Indian J Med Microbiol. 2008;26:58–61. doi: 10.4103/0255-0857.38860. [DOI] [PubMed] [Google Scholar]
- 15.Kumar MS, Lakshmi V, Rajagopalan R. Occurrence of extended spectrum beta-lactamases among Enterobacteriaceae spp. isolated at a tertiary care institute. Indian J Med Microbiol. 2006;24:208–11. [PubMed] [Google Scholar]
- 16.Rodrigues C, Joshi P, Jani SH, Alphonse M, Radhakrishnan R, Mehta A. Detection of β-Lactamases in nosocomial Gram negative clinical isolates. Indian J Med Microbiol. 2004;22:247–250. [PubMed] [Google Scholar]
- 17.Win WC, Allen SD, Janda WM, Koneman EW, Procop GW, Schreckenberger PC, Woods G, editors. 6th ed. Philadelphia: Lippincott Williams and Wilkins; Enterobacteriaceae; 2006. Colour atlas and text book of diagnostic microbiology; pp. 211–302. [Google Scholar]
- 18.Bauer AW, Kirby WM, Sherris JC, Jurck M. Antibiotic susceptibility testing by a Standardized Single disc diffusion method. Am J Clin Pathol. 1996;45:493–6. [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standard Institute. Performance Standards for Antimicrobial Susceptibility Testing; 21st Informational Supplement. CLSI document. 2011 Jan;31(No. 1):48–9. Supplemental Table 2A-S1, M100-S21. [Google Scholar]
- 20.Mohamudha PR, Harish BN, Parija SC. Amp C beta lactamases among gram negative clinical isolates from a tertiary hospital, South India. Braz J Microbiol. 2010;41:596–602. doi: 10.1590/S1517-83822010000300009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polsfuss S, Bloemberg GV, Giger J, Meyer V, Bottger EC, Hambach M. Practical approach for reliable detection of AmpC beta-lactamases producing Enterobacteriaceae. J Clin Microbiol. 2011;49:2798–803. doi: 10.1128/JCM.00404-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisentein BI, Zaleznik DF. Enterobacteriaceae. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and practice of infectious diseases. 5th ed. Philadelphia, Pa: Churchill Living Stone; 2000. pp. 2294–310. [Google Scholar]
- 23.Komatsu M, Ikeda N, Aihara M, Nakamachi Y, Kinoshita S, Yamasaki K, et al. Hospital outbreak of MEN-1-derived extended spectrum beta-lactamase-producing Klebsiella pneumoniae. J Infect Chemother. 2001;7:94–101. doi: 10.1007/s101560100015. [DOI] [PubMed] [Google Scholar]
- 24.Rice LB, Willey SH, Papanicolaou GA, Medeiros AA, Eliopoulos GM, Moellering RC, Jr, et al. Outbreak of ceftazidime resistance caused by extended-spectrum beta-lactamases at a Massachusetts chronic – care facility. Antimicrob Agent Chemother. 1990;34:2193–9. doi: 10.1128/aac.34.11.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiener J, Quinn JP, Bradford PA, Goering RV, Nathan C, Bush K, et al. Multiple antibiotic resistant Klebsiella and Escherichia coli in nursing homes. JAMA. 1999;281:517–23. doi: 10.1001/jama.281.6.517. [DOI] [PubMed] [Google Scholar]
- 26.Gavin PJ, Bolden JR, Jr, Peterson LR, Thomson RB., Jr Does identification of an extended spectrum beta-lactamase (ESBL) producing organism by the microbiology laboratory influence patient management? Infect Dis Clin Pract. 2006;14:81–3. [Google Scholar]
- 27.Gotale M, Manthalkar P, Kandle S, Yamul V, Jahagirdhar V. Co-relation of extended spectrum β-lactamase production with cephalosporins resistance in Gram negative bacilli. Indian J Pathol Microbiol. 2004;47:82–4. [PubMed] [Google Scholar]
- 28.Shah AA, Hasan F, Ahmed S, Hameed A. Extended Spectrum beta-lactamase in Enterobacteriaceae: Related to age and gender. New Microbiol. 2002;25:363–6. [PubMed] [Google Scholar]
- 29.Gupta E, Mohanty S, Sood S, Dhawan B, Das BK, Kapil A. Emerging resistance to carbapenems in a tertiary care hospital in north India. Indian J Med Res. 2006;124:95–8. [PubMed] [Google Scholar]
- 30.Moczygemba LR, Frei CR, Burgess DS. Pharmacodynamic modeling of carbapenems and fluoroquinolones against bacteria that produce extended-spectrum beta-lactamases. Clin Ther. 2004;26:1800–7. doi: 10.1016/j.clinthera.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Wong-Beinger A. Therapeutic challenges associated with extended-spectrum Beta-lactamases producing Escherichia coli and Klebsiella pneumoniae. Pharmacotherapy. 2001;21:583–92. doi: 10.1592/phco.21.6.583.34537. [DOI] [PubMed] [Google Scholar]
- 32.Al- Zahrani AJ, Akhtar N. Susceptibility patterns of Extended Spectrum β- lactamase (ESBL) –producing Escherichia coli and Klebsiella pneumoniae isolated in a teaching hospital. Pakistan J Med Res. 2005;44:64–7. [Google Scholar]
- 33.Bano JR, Navarro MD, Retamar P, Picon E, Pascual A. β-lactam/βlactam inhibitor combinations for the treatment of bacteremia due to Extended –Spectrum β-lactamase-producing Escherichia coli: A post Hoc analysis of prospective cohort. Clin Infect Dis. 2012;54:167–74. doi: 10.1093/cid/cir790. [DOI] [PubMed] [Google Scholar]
- 34.Peterson LR. Antibiotic policy and prescribing strategies for therapy of extended-spectrum β-lactamase-producing Enterobacteriaceae: The role of piperacillin-tazobactam. Clin Microbiol Infect. 2008;14:181–4. doi: 10.1111/j.1469-0691.2007.01864.x. [DOI] [PubMed] [Google Scholar]
- 35.Ullah F, Malik SA, Ahmed J. Antibiotic susceptibility pattern and ESBL prevalence in nosocomial Escherichia coli from urinary tract infections in Pakistan. Afr J Biotechnol. 2009;8:3921–6. [Google Scholar]
- 36.Martínez-Martínez L, Pascual A, Conejo Mdel C, García I, Joyanes P, Doménech-Sánchez A, et al. Energy-dependent accumulation of norfloxacin and porin expression in clinical isolates of Klebsiella pneumoniae and relationship to extended-spectrum beta-lactamase production. Antimicrob Agents Chemother. 2002;46:3926–32. doi: 10.1128/AAC.46.12.3926-3932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


