Abstract
The polymeric immunoglobulin (Ig) receptor (pIgR) is an integral transmembrane glycoprotein that plays an important role in the mammalian immune response by transporting soluble polymeric Igs across mucosal epithelial cells. Single pIgR genes, which are expressed in lymphoid organs including mucosal tissues, have been identified in several teleost species. A single pigr gene has been identified on zebrafish chromosome 2 along with a large multigene family consisting of 29 pigr-like (PIGRL) genes. Full length transcripts from 10 different PIGRL genes that encode secreted and putative inhibitory membrane bound receptors have been characterized. Although PIGRL and pigr transcripts are detected in immune tissues, only PIGRL transcripts can be detected in lymphoid and myeloid cells. In contrast to pIgR which binds Igs, certain PIGRL proteins bind phospholipids. PIGRL transcript levels are increased after infection with Streptococcus iniae, suggesting a role for PIGRL genes during bacterial challenge. Transcript levels of PIGRL genes are decreased after infection with Snakehead rhabdovirus, suggesting that viral infection may suppress PIGRL function.
Keywords: Teleost, innate immunity, lipid binding, genome evolution
Introduction
The polymeric immunoglobulin (Ig) receptor (pIgR) is an integral transmembrane glycoprotein that plays an important role in the mammalian immune response by transporting soluble polymeric immunoglobulins (pIg; dimeric IgA and in some mammals, pentameric IgM) across mucosal epithelial cells, such as those lining the gastrointestinal tract. Polymeric Ig is expressed by plasma cells present in the lamina propria underlying the intestinal epithelium and is bound by pIgR on the basolateral surface of epithelial cells. pIgR transports pIgA through the epithelial cell by transcytosis. Once on the apical surface, pIgR undergoes a cleavage event in which the secretory component is released from cells either as a free form or bound to pIgA as the secretory IgA complex (SIgA) that liberates it from the plasma membrane (Asano and Komiyama 2011;Kaetzel 2005). The transport of pIg by pIgR to the intestinal lumen is essential for protecting the host from invading pathogens and maintaining homeostasis (Johansen et al. 1999).
The mammalian pIgR is encoded by a single copy gene (PIGR) that encodes five extracellular Ig domains (D1-D5), a cleavage site, a transmembrane domain and cytoplasmic tail (Asano and Komiyama 2011;Kaetzel 2005). An alternatively spliced PIGR transcript lacking the D2 and D3 domains has been reported in rabbit and cow (Deitcher and Mostov 1986;Kulseth et al. 1995). pIgR from chicken and Xenopus possesses four Ig domains; the D2 domain found in mammals is missing in these species (Braathen et al. 2007;Wieland et al. 2004). Full-length transcripts encoding a pIgR homolog, which possesses two Ig domains, have been identified in multiple fish species; however, only partial transcripts of this gene have been reported in zebrafish (Danio rerio) (Feng et al. 2009;Hamuro et al. 2007;Montgomery et al. 2011;Rombout et al. 2008;Tadiso et al. 2011;Zhang et al. 2010). Teleost (bony fish) pIgR is expressed by lymphoid organs including mucosal tissues (intestine, skin and gill) (Feng et al. 2009;Hamuro et al. 2007;Rombout et al. 2008;Tadiso et al. 2011). Teleost pIgR has been shown to bind both IgM and IgZ/IgT (Feng et al. 2009;Hamuro et al. 2007;Zhang et al. 2010).
Single pIgR-like (PIGRL) transcripts that have been identified in Atlantic salmon (Salmo salar) and common carp (Cyprinus carpio) share sequence and structural similarities with teleost pIgR, modular domain immune-type receptors (MDIRs) from clearnose skate (Raja eglanteria) and the mammalian CD300/TREM family of receptors (Cannon et al. 2006;Ribeiro et al. 2011;Tadiso et al. 2011). The carp PIGRL protein was shown to be abundantly expressed in macrophages and secreted upon immune stimulation (referred to as a soluble immune-type receptor, SITR, in: Ribeiro et al. 2011). Datamining of the zebrafish genome database previously revealed the presence of multiple PIGRL sequences on chromosome 2 (Cannon et al. 2006;Ribeiro et al. 2011;Tadiso et al. 2011). We herein report the characterization of this multi-gene family that includes the single pigr gene and 29 PIGRL genes; characterize full-length transcripts from the zebrafish pigr gene and ten PIGRL genes; demonstrate that certain PIGRL proteins bind phospholipids, examine the expression of pigr and PIGRL genes in adult tissues, during embryonic development and from leukocyte lineages; and quantify changes in pigr and PIGRL transcript levels after bacterial and viral infection.
Methods
Animals
Zebrafish were purchased from EkkWill WildLife Resources (Ruskin, FL). All experiments involving live zebrafish were performed in accordance with relevant institutional and national guidelines and regulations and were approved by the Institutional Animal Care and Use Committees of North Carolina State University, University of Maine or Wayne State University.
Bioinformatics
Genomic sequences encoding zebrafish pigr and the majority of candidate PIGRL Ig domains were identified on chromosome 2 (Zv9 scaffold 234, GenBank NW_001878710.3; Zv9 scaffold 235, GenBank NW_001878708.3; and Zv9 scaffold 3509 GenBank NW_003336263.1) with BLAST searches using skate MDIR1 (GenBank ABC86795), carp pIgR (GenBank ADB97624) and salmon pIgRL (GenBank ADM18015) sequences as queries. Additional Ig domains were identified by individually translating segments of each scaffold in silico in all six reading frames and submitting the output to SMART analyses (Letunic et al. 2012). Ig domains from two predicted PIGRL genes encode a frame shift and/or premature stop codon and have been classified as pseudogenes. Protein sequences were aligned by Clustal W (Larkin et al. 2007). Phylogenetic trees were constructed from pairwise Poisson correction distances with 2000 bootstrap replications by MEGA5 software (Tamura et al. 2011). Protein sequence domains were identified with SMART software (Letunic et al. 2009). Variability plots were generated with the Protein Variability Server (Garcia-Boronat et al. 2008). Sequences analyzed include pIgR from common carp (Cyprinus carpio, Cyca; GenBank ADB97624), fugu (Takifugu rubripes, Taru; GenBank BAF56575), orange-spotted grouper (Epinephelus coioides, Epco; GenBank ACV91878), Atlantic salmon (Salmo salar, Sasa; GenBank ACX44838), rainbow trout (Oncorhynchus mykiss, Onmy; GenBank ADB81776), human (Homo sapiens, Hosa; GenBank NP_002635), mouse (Mus musculus, Mumu; GenBank NP_035212), chicken (Gallus gallus, Gaga; GenBank NP_001038109) and Xenopus laevis (Xela; GenBank ABK62772).
Zebrafish pigr and PIGRL transcripts and genes
Rapid amplification of cDNA ends (RACE) and RT-PCR strategies were employed to obtain full-length pigr and PIGRL cDNAs. RACE-ready cDNA was prepared from RNA pooled from zebrafish kidney and spleen using the GeneRacer Kit™, Superscript™ III Reverse Transcriptase (Invitrogen) and Titanium Taq DNA polymerase (Clontech). The resulting amplicons were ligated into pGEM-T Easy (Promega) and sequenced. Experimental details are included in Electronic Supplementary Methods and Supplementary Table S1. The genomic organization of pigr and PIGRL genes was deduced by comparing cDNA sequences to zebrafish Zv9 genomic reference sequences: chromosome 2 scaffolds 234 and 235 and unplaced scaffold 3509.
Lipid binding assays
Recombinant soluble proteins of zebrafish pIgR and PIGRL D1 and D2 ectodomains fused to a human IgG Fc domain were generated by cloning various ectodomains (amplified from pooled hematopoietic tissue cDNA) into the pcDNA3-hsIgG1Fc-Avi fusion vector as described (Cannon et al. 2011;Cannon et al. 2012;Haire et al. 2012). Chimeric proteins were expressed and secreted by HEK293T cells (Haire et al. 2012). Cleared cell culture supernatant harvests were concentrated 10- to 100-fold and the hFc fusion proteins were characterized by Western analyses and quantified using the Easy-Titer Human IgG Assay kit (Thermo Scientific). The mouse CLM7-hFc protein was previously reported to bind lipids (Cannon et al. 2012;Haire et al. 2012).
Solid phase ELISA assays were conducted as described previously (Cannon et al. 2012;Haire et al. 2012). Bacterial sources and the method for phospholipid extraction have been described (Haire et al. 2012). Either 0.5 μg purified cardiolipin or 50 μl of MBTE/methanol bacterial extract was used to coat plates. Negative control wells were treated in parallel with solvent (100% methanol). Binding efficiency was determined after color development as absorbance at 450 nm. Values were corrected by subtracting the value from negative control wells. Corrected ELISA values less than 0.10 were scored as zero; 0.10–0.15 as +1; 0.15–0.20 as +2; 0.20–0.25 as +3; 0.25–0.30 as +4; >0.30 was scored as +5.
Reverse Transcriptase – Polymerase Chain Reaction (RT-PCR)
Tissues were harvested from twenty adult zebrafish. Embryos were collected by natural mating, maintained at 28 °C (Westerfield 2007) and ten embryos from each of the following time points were collected: 0, 6, 12, 24, 36, 48, 72 hours post fertilization (hpf) and 6 days post fertilization (dpf). Myeloid and lymphoid cell lineages were purified from the kidneys of ten adult zebrafish as described (Traver et al. 2003;Traver 2004). In brief, kidneys from adult zebrafish were dissected and homogenized with a 40 μm nylon-mesh filter in ice-cold PBS + 5% FBS. Propidium iodide was added to a concentration of 1 μg/ml. Myeloid and lymphoid cells were isolated from this single-cell suspension by sorting based on propidium iodide exclusion, forward scatter and side scatter with a BD FACS Aria II SORP flow cytometer (Beckton Dickinson). Cell populations were sorted twice to optimize cell purity. Total RNA was purified from zebrafish tissues, embryos and cells using TRIzol reagent (Life Technologies). Five μg RNA from each tissue, 2.0 μg RNA from each embryonic time point, or 0.5 μg RNA from each leukocyte population were reverse transcribed using Superscript™ III Reverse Transcriptase (Life Technologies) and oligo dT primers. Tissue and embryonic cDNA was diluted 1:5 and leukocyte cDNA was diluted 1:2.5 in H2O for PCR with Titanium Taq DNA polymerase (Clontech) using the gene family specific primers and cycling parameters specified in Electronic Supplementary Table S2. The resulting amplicons were cloned into pGEM-T Easy and sequenced to confirm their identity.
Streptococcus iniae infections
Adult zebrafish were anesthetized with 0.016% tricaine and either injected intramuscularly with 10 μl 1 × 107 CFU/ml S. iniae or mock injected with media (THY + P) as described (Neely et al. 2002). We have shown previously that this dose is lethal to over 90% of the fish within 4 days (Lowe et al. 2007). Zebrafish were maintained at 28 °C and euthanized for dissection at 2, 4, 8, 12 and 24 hpi. Tissues from ten zebrafish per treatment and time point were pooled for RNA extraction.
Snakehead rhabdovirus (SHRV) infections
Adult zebrafish were anesthetized as above and either injected intraperitoneally with 105 TCID50 (Tissue Culture Infectious Dose) of SHRV/ml or mock injected with PBS as described (Phelan et al. 2005;Pressley et al. 2005). Mortalities exceeded 40% under these conditions. Zebrafish were maintained at 28 °C and euthanized for dissection at 2, 4, 8, 12 and 24 hpi. Tissues from ten zebrafish per treatment and time point were pooled for RNA extraction.
Reverse Transcriptase – Quantitative Polymerase Chain Reaction (RT-qPCR)
TaqMan primer/probe sets designed to amplify and detect zebrafish pigr, pigrl1.4, pigrl2.3, pigrl3.10 and pigrl4.2 transcripts were purchased from Applied Biosystems. Reverse transcription was completed as described above. Quantitative PCRs were executed on a MyiQ Real time PCR detection system with IQ5 Optical system software (Bio-Rad). Reactions were completed in triplicate and average relative transcript levels calculated by normalizing to transcript levels of the eukaryotic translation elongation factor 1 alpha 1, like 1 (eef1a1l1) gene by the ΔΔCt method (Livak and Schmittgen 2001).
Results and Discussion
Zebrafish polymeric immunoglobulin receptor, pIgR
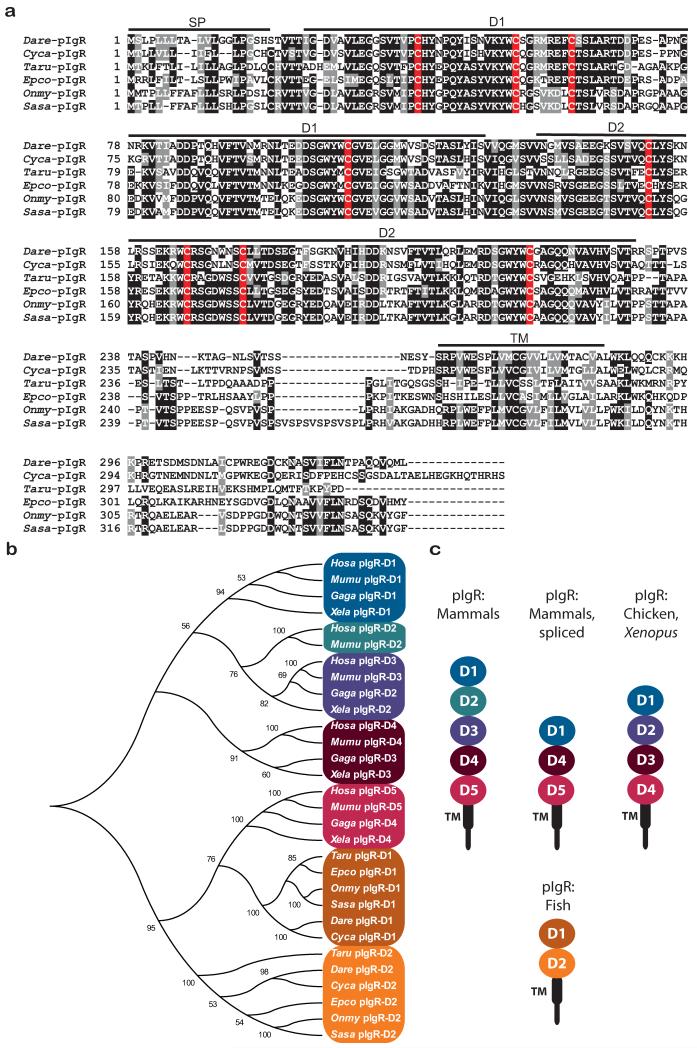
The zebrafish pigr gene maps to chromosome 2 and a partial, 5⍰ truncated cDNA sequence (GenBank EF539183) has been reported (Feng et al. 2009;Rombout et al. 2008;Tadiso et al. 2011). 5⍰ RACE and RT-PCR were employed to amplify two full-length allelic variants of zebrafish pigr that differ by three residues (GenBank KF932324 and KF932325). Alignment of pIgRs from multiple fish species at the peptide level indicates a high degree of similarity in both Ig domains (Fig. 1a). Prior sequence comparisons of teleost pIgR to mammalian pIgR led to the conclusion that the first and second Ig domains of teleost pIgR corresponds to D1 and D5 of mammalian pIgR (Feng et al. 2009;Hamuro et al. 2007;Rombout et al. 2008). Comparison of individual Ig domains from pIgR of human, mouse, chicken, Xenopus, zebrafish and multiple teleost species (Figures 1b and 1c) confirms that the second Ig domain from teleost pIgRs is most similar to D5 from tetrapod pIgR. However, the analyses presented here with an expanded gene set, indicate that the first Ig domains of teleost pIgR and tetrapod pIgR are phylogenetically distinct. It also was reported that the first Ig domain of teleost pIgR shares more similarity to D5 of mammalian pIgR than does the second Ig domain of teleost pIgR (Tadiso et al. 2011). Although the first and second Ig domains of the teleost pIgR have been referred to as D1 and D5 (suggesting similarity to mammalian D1 and D5), the domains are designated as D1 and D2 in this report.
Fig. 1. Zebrafish pIgR.
(a) Alignment of full-length pIgR proteins encoded by zebrafish (GenBank KF932324) and other teleost species. Positions that are 70% or greater identical are shaded in black and those that are structurally related are shaded in gray. Conserved cysteines are shaded red. Sequences corresponding to signal peptide (SP), D1, D2 and transmembrane (TM) domains are indicated. (b) The immunoglobulin domains from pIgR of multiple vertebrate species were aligned and a neighbor joining tree constructed. Species include: human (Homo sapiens, Hosa), mouse (Mus musculus, Mumu), chicken (Gallus gallus, Gaga), frog (Xenopus laevis, Xela), pufferfish (Takifugu rubripes, Tafu), orange-spotted grouper (Epinephelus coioides, Epco), rainbow trout (Oncorhynchus mykiss, Onmy), Atlantic salmon (Salmo salar, Sasa), zebrafish (Danio rerio, Dare) and carp (Cyprinus carpio, Cyca). (c) Structures of pIgR from various species are depicted.
It has been shown that a conserved sequence in complementarity determining region 1 (CDR1) in the D1 domain of the tetrapod pIgR is required for non-covalent Ig binding (Kaetzel 2005;Roe et al. 1999); however, a conserved equivalent of this sequence is lacking in teleost pIgR (Feng et al. 2009). The sequence differences between tetrapod and teleost pIgRs could reflect co-evolution of the receptor function with different classes of Igs. Work in other systems has demonstrated that mammalian pIgR binds IgA and IgM whereas teleost pIgR binds IgM and IgZ/IgT (Feng et al. 2009;Hamuro et al. 2007;Zhang et al. 2010).
Zebrafish pIgR-like (PIGRL) Ig domains and genes
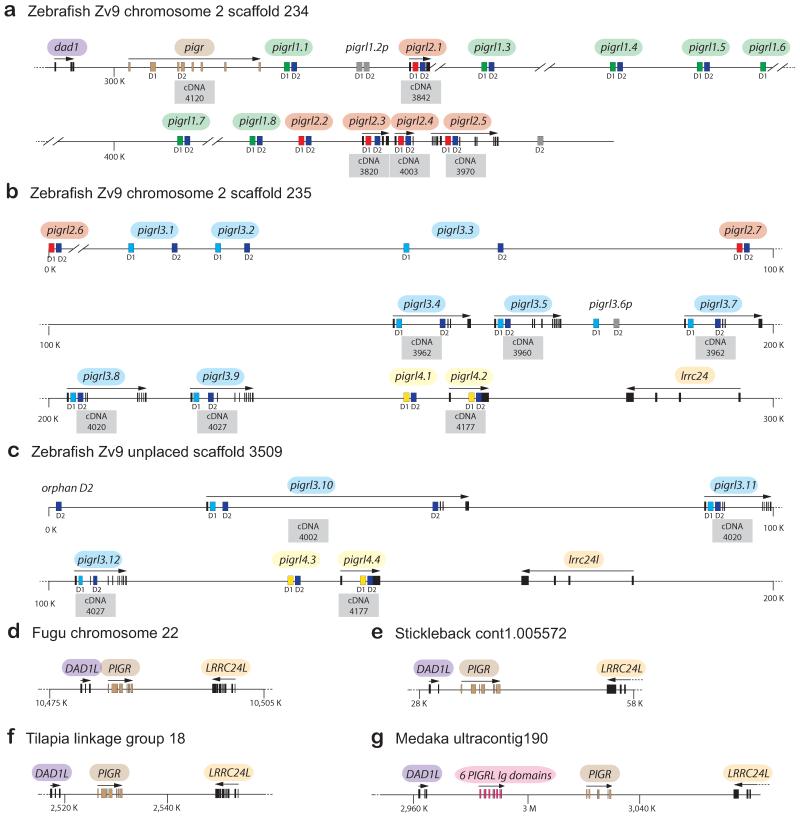
The zebrafish PIGRL sequences on chromosome 2 were identified initially through BLAST searches of the zebrafish genome with Ig domains from modular domain immune-type receptors (MDIRs) from skate, a salmon PIGRL and a carp PIGRL/SITR (Cannon et al. 2006;Ribeiro et al. 2011;Tadiso et al. 2011). All identifiable pigr and PIGRL sequences in the current version of the zebrafish genome (Zv9) can be identified in three scaffolds; 234, 235 and 3509. The zebrafish pigr gene, thirteen PIGRL genes (including one pseudogene) and the defender against cell death 1 (dad1) gene are linked tightly on scaffold 234 (Fig. 2a). An additional eighteen PIGRL genes (including one pseudogene), one “orphan” Ig domain and a leucine rich repeat containing 24 gene (lrrc24 or lrcc24-like) map to scaffolds 235 and 3509 (Figures 2b and 2c). No overlap can be identified between scaffold 234 and either scaffold 235 or scaffold 3509, suggesting that additional PIGRL genes may be present in the zebrafish genome. The high level of identity between parts of scaffold 235 (pigrl3.8, pigrl3.9, pigrl4.1, pigrl4.2, lrrc24) and scaffold 3509 (pigrl3.11, pigrl3.12, pigrl4.3, pigrl4.4, lrrc24l, respectively), coupled with the high degree of allelic polymorphism in immune-type genes in zebrafish make it likely that these scaffolds represent two different haplotypes for a single loci (Electronic Supplementary Fig. S1).
Fig. 2. Zebrafish pigr/PIGRL gene cluster.
(a-c) The zebrafish pigr gene and multiple PIGRL genes were identified on genomic scaffolds 234, 235 and 3509. The relative position of pigr and PIGRL genes along with flanking genes (dad1, lrrc24 and lrcc24-like) are shown. Exons are indicated by rectangles. Each PIGRL gene, with the exception of pigrl3.10, is predicted to encode a single D1 domain and a single D2 domain (indicated below the exons). D1 exons are color–coded to indicate different groups of D1 domains. D1 and D2 domains possessing internal frame shifts are considered psuedogenes and are shaded gray. All exons for a gene are identified with an arrow indicating relative transcriptional orientation (above the gene) and by the cDNA ID number (below the gene). (d-g) The genomic organization of DAD1-PIGR-LRRC24 loci is shown for fugu (FUGU5 chromosome 22 whole genome shotgun sequence GenBank NC_018911), three-spined stickleback (stickleback whole genome sequence cont1.0055772, GenBank AANH01005573), Nile tilapia (Oreochromis niloticus isolate 000638D3DF linkage group LG18, Orenil1.1, whole genome shotgun sequence, GenBank NC_022216) and medaka (Oryzias latipes chromosome 17 genomic scaffold, ASM31367v1 ultracontig190, whole genome shotgun sequence, GenBank NW_004088020).
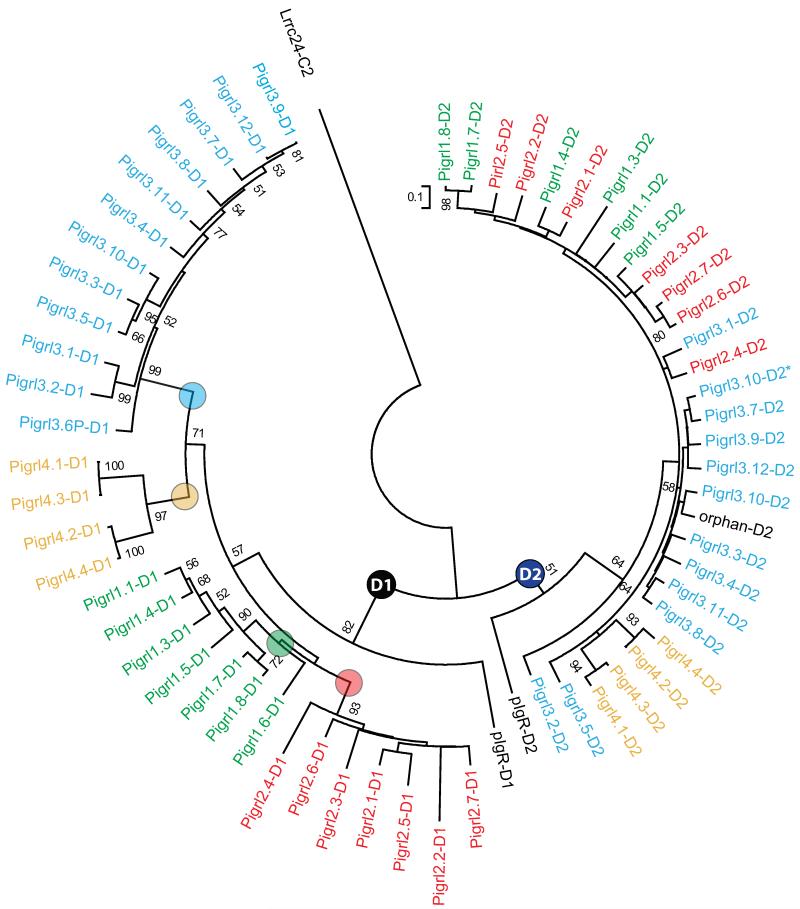
Phylogenetic comparisons indicate that all D1 and D2 domains predicted to be encoded by PIGRL genes (Electronic Supplementary Fig. S2) group in two major distributions (radiations) (Fig. 3). Whereas, the D2 domains of the PIGRLs are highly conserved between individual loci, the D1 domains distribute into four major groups named 1 through 4. As multiple genes are present in each group, genes are differentiated by an additional number (e.g. the gene symbol for PIGRL group 2, gene 1 is = pigrl2.1). The relationship between D2 domains does not consistently mirror the relationships between D1 domains, e.g., the D2 domains of Pigrl1.4 and Pigrl2.1 group together, but the D1 domains from these genes are distinctly different (Fig. 3). Recombination and exon swapping between innate immune receptor genes has been described previously (Litman et al. 2001).
Fig. 3. D1 and D2 Ig domains of PIGRL proteins.
The Ig domains identified in Fig. 2 and listed in Electronic Supplementary Fig. S2 were aligned and a neighbor joining tree constructed. The nodes defining the D1 and D2 domains are labeled. The four major groups of D1 domains are indicated by color-coded text and circles at their defining nodes. The C2-type Ig domain of zebrafish Lrrc24 is included as an out-group. Bootstrap values less than 50 are not shown. The Ig domain encoded by the second D2 domain of pigrl3.10 is indicated by an asterisk (*; see text for details).
A number of observations can be made from a phylogenetic analysis comparing Ig domains from teleost pIgRs, zebrafish PIGRLs, a salmon PIGRL, a carp PIGRL/SITR, a putative PIGRL from medaka (Oryzias latipes) and representative zebrafish novel immunoglobulin-like transcript (NILT; Stet et al. 2005), diverse immunoglobulin domain-containing protein (DICP; Haire et al. 2012) and novel immune-type receptor (NITR; Yoder 2009) proteins (Electronic Supplementary Fig. S3): 1) PIGRL proteins are distinct from NITR and DICP proteins, 2) zebrafish NILTs are more related closely to pIgR than they are to NITRs and DICPs, but are distinct from PIGRLs, 3) the previously reported carp SITR (Ribeiro et al. 2011) can be classified as a PIGRL, 4) the D2 domain of the salmon PIGRL is more related to the D2 of teleost pIgRs than it is to the D2 of zebrafish PIGRLs, and 5) the current medaka genome reveals eight Ig domains that likely represent a single pIgR as well as three PIGRL genes (discussed below).
The PIGRL gene cluster is absent in fugu, stickleback and tilapia but present in medaka
As a PIGRL transcript has been identified from salmon (Tadiso et al. 2011), we mined the salmon genome in order to determine the gene organization at the PIGR locus. From the salmon shotgun sequence database, contigs could be identified that encode DAD1, PIGR, PIGRL or LRRC24L, but none of these contigs overlap precluding any conclusion about the PIGR/PIGRL loci in this species. Although, a carp PIGRL/SITR transcript has also been identified, a carp genome sequence database is not yet available.
The genome sequence databases of other teleosts were searched in an effort to identify the genomic organization of pIgR/PIGRL loci in other species. In zebrafish the pigr gene and a number of PIGRL genes are adjacent to dad1 on scaffold 234, whereas a different set of PIGRL genes are adjacent to lrrc24 on scaffold 235 (Fig. 2a-c). In fugu pufferfish (Takifugu rubripes), stickleback (Gasterosteus aculeatus) and tilapia (Oreochromis niloticus) the PIGR gene is flanked by the DAD1L and LRRC24L genes. No genomic evidence is seen for PIGRL genes at this locus in these species (Fig. 2d-f).
A tBLASTn search of the current medaka reference genome using zebrafish Dad1, Lrrc24 and pIgR as queries identified a region of ultracontig 190 (GenBank NW_004088020.1) that encodes eight Ig domains between the DAD1L and LRRC24L genes (Fig. 2g). Although transcripts that correspond to these Ig domains have not been identified, gene prediction software predicts that the two Ig domains closest to LRRC24L are encoded by a single transcript (GenBank XM_004079122) and the remaining six Ig domains are encoded by a second transcript (GenBank XM_004079121). The former transcript encodes a membrane bound receptor with high similarity to pIgR from other fish (61% identical to grouper pIgR, E value = 1e-152) as well as D1 and D2 domains that are phylogenetically similar to pIgR from other teleosts (Electronic Supplementary Fig. S3). The additional six Ig domains encoded by the latter transcript are more similar to pIgR than to PIGRL; phylogenetic analyses indicate that these Ig domains are present in a D1-D2-D1-D2-D1-D2 configuration and are likely derived from tandem D1-D2 duplication events. Although transcripts encoding these medaka sequences are not available, the organization of the zebrafish PIGRL genes suggests that the eight homologous medaka Ig domains represent a cluster of four genes with a D1-D2 configuration, consisting of a single pIgR gene and three PIGRL genes. As mentioned above, the salmon PIGRL D1 domain is most similar to zebrafish PIGRL D1 domains whereas the D2 domain from the same protein is most similar to teleost pIgR D2 domains.
Although the DAD1-PIGR-LRRC24 gene organization appears to be well conserved in multiple teleost lineages, it may be that only certain species encode PIGRL genes. Teleosts include four major lineages: otomorphs (cyprinids [zebrafish and carp] and siluriformes [catfish]), euteleostomorphs (salmonids [salmon], tetraodontiformes [fugu], perciformes [tilapia] and ovalentariae [medaka]), osteoglossomorphs (arapaima) and elopomorphs (tarpons, ladyfishes and eels) (Betancur et al. 2013;Broughton et al. 2013). The identification of definitive PIGRL sequences from zebrafish and carp indicates that these genes are present within the otomorph lineage of teleostei. However, the presence of PIGRL sequences in some euteloestomorphs (salmon and medaka) but not others (fugu and tilapia) and the phylogenetic differences between the zebrafish, salmon and medaka PIGRL sequences (Electronic Supplementary Fig. S3) confounds efforts to model the history of the PIGRL gene cluster. The PIGRL gene clusters in zebrafish and medaka may reflect recent and independent gene expansions or a more ancient gene family that has been lost in multiple teleost lineages. The complete sequencing of additional teleost genomes will help understand the origins of these genes.
The DAD1-PIGR-LRRC24 locus is not conserved in mammals
In the human genome, PIGR, LRRC24 and DAD1 map to three different chromosomes (chromosomes 1, 8 and 14 respectively), demonstrating a lack of conserved synteny between the zebrafish and human PIGR loci. Although LRRC24 encodes an Ig domain of the C2 type, no other genes encoding Ig domains can be identified in the vicinity of this human gene. In contrast, the human, mouse and Xenopus tropicalis DAD1 gene is adjacent to the TCRα constant domain locus (TRAC), whereas chicken DAD1 is adjacent to the TCRδ locus (Electronic Supplementary Fig. S4). Although, the zebrafish TCRα constant domain (trac) and the pigr/PIGRL gene cluster both map to chromosome 2, they are predicted to be separated by approximately 15 Mbp. The human PIGR gene is flanked by FAIM3 (Fas apoptotic inhibitory molecule 3, aka FcμR and TOSO) and FCAMR (Fcα/μR and CD351); this three-gene cluster (Electronic Supplementary Fig. S4) is conserved in numerous mammalian species (Murakami et al. 2012;Sakamoto et al. 2001;Shibuya et al. 2000;Shimizu et al. 2001). Both of these mammalian receptors possess a single Ig domain that binds IgM (FAIM3) or both IgA and IgM (FCAMR). Although the PIGR gene has been identified in mammals, birds and amphibians, this cluster of Ig receptors (FAIM3, PIGR, FCAMR) may be specific to mammals as FAIM3 and FCAMR have not been identified adjacent to PIGR in chicken, zebra finch, turkey or Xenopus.
Conserved features of PIGRL Ig domains
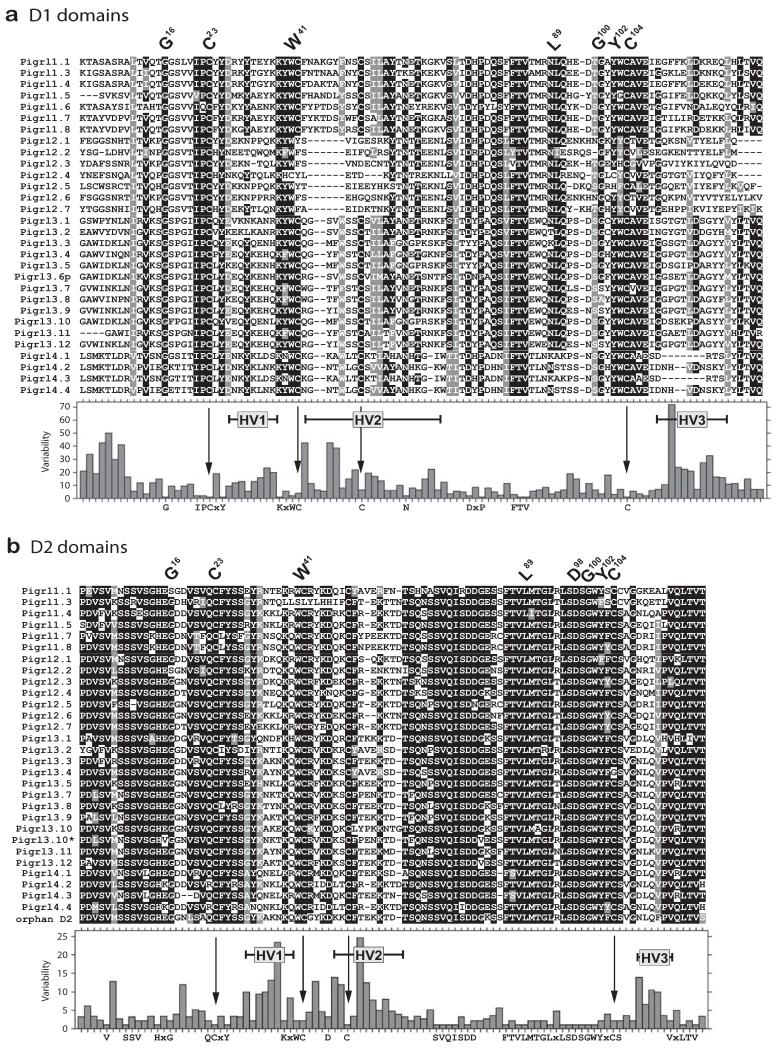
An alignment of the zebrafish PIGRL D1 and D2 domains reveals residues and extended regions that are conserved and variable (Fig. 4). Both D1 and D2 domains encode residues that are conserved with V domains (G16, C23, W41, L89, G100, Y102, C104) (Barclay et al. 1997;Barclay 2003;Williams and Barclay 1988). The D1 domains of zebrafish pIgR and PIGRLs share an IPCXY motif at the C23 position, and a FTV motif amino terminal to L89 (Figures 1 and 4). A comparison of the zebrafish PIGRL D1 and D2 domains reveals three main regions of hypervariability (HV1-HV3), which likely influence ligand recognition and binding (Fig. 4).
Fig. 4. Hypervariable regions of PIGRL Ig domains.
PIGRL Ig domains identified in Fig. 2 and listed in Electronic Supplementary Fig. S2 were organized into (a) D1 and (b) D2 domains and aligned. Positions that are 70% or greater identical are shaded in black and those that are structurally related are shaded in gray. Protein names are shown on the left. Conserved residues characteristic of immunoglobulin domains are indicated by the IMGT numbering system above the alignments (Giudicelli et al. 2006). Levels of sequence variability and hypervariable regions (HV) are indicated below the alignments. Positions of cysteines conserved in pIgR (see Fig. 1) are indicated with arrows. The Ig domain encoded by the second D2 domain of pigrl3.10 is indicated with an asterisk (*).
The D1 domains of the zebrafish pIgR, PIGRL1, PIGRL3 and PIGRL4 proteins as well as the D2 domain of pIgR and nearly all PIGRL D2 domains possess two additional cysteines (C42 and C49/C50/C52). These cysteines also are present in the D1 and D5 domains of tetrapod pIgR and the D1 and D2 domains of teleost pIgR in which their spacing is highly conserved at seven residues (CX7C). This spacing is conserved in zebrafish PIGRL3 and PIGRL4 D1 domains and in all PIGRL D2 domains. In contrast, this spacing is nine residues (CX9C) in the D1 domain of zebrafish PIGRL1 proteins. Structural models of the salmon PIGRL predict that these additional cysteines form intrachain disulfides (Tadiso et al. 2011).
PIGRL transcripts encode secreted and inhibitory forms
RACE strategies were used to clone 24 different full-length PIGRL2, PIGRL3 and PIGRL4 transcripts (GenBank KF932326 – KF932349), which likely represent polymorphic variations of 10 different genes (Fig. 5 and Electronic Supplementary Fig. S5). Only partial PIGRL1 transcripts were recovered (see below, GenBank KF932350 – KF932357). Using these full-length transcripts and the reference genome, the exon organization of multiple PIGRL genes could be defined or predicted. Twenty-one of the PIGRL transcripts encode a single D1 domain and a single D2 domain. The PIGRL3.10 gene and its three transcripts have a D1-D2-D2 exon organization that maintains the reading frame, allowing for both D2 domains to be translated (Fig. 5 and Electronic Supplementary Fig. S5). One distinguishing difference between the organization of the zebrafish pigr and PIGRL genes is that the pigr D2 domain is encoded by two exons whereas the D2 domain of all PIGRL genes is encoded by a single exon (Fig. 5 and Tadiso et al. 2011).
Fig. 5. PIGRL transcripts, exons and predicted proteins.
(a) Transcript (cDNA) numbers are listed on the left, adjacent to the gene that best matches the transcript. Exons are represented by rectangles. The position of start codons and stop codons are indicated by green and red vertical lines, respectively. D1 domain exons are color-coded as in Figures 2 and 3. Other exons are color-coded to indicate nearly identical sequences. (b) Schematic representations of zebrafish pIgR and PIGRL sequences from salmon and carp (Ribeiro et al. 2011;Tadiso et al. 2011). (c) Schematic representation of PIGRL proteins deduced from transcripts. Positioning of D1, D2, transmembrane (TM), cytoplasmic tyrosines (Y), immunoreceptor tyrosine-based inhibition motif (ITIM) and ITIM-like (itim) sequences are indicated. Transcripts that best match the gene shown are listed below the protein structure and the amino acid (aa) variation between the proteins encoded by these transcripts indicated. (d) Peptide sequences corresponding to cytoplasmic domains encoded by three PIGRL genes are listed. Variable residues encoded by polymorphic transcripts are in parentheses. Full-length protein sequences are available in Electronic Supplementary Fig. S5. Sequences have been deposited in GenBank with accession numbers KF932324 – KF932349.
Transcripts recovered from pigrl2.4, pigrl2.5 and pigrl3.5 encode type I transmembrane proteins that possess cytoplasmic immunoreceptor tyrosine-based inhibitory motif (ITIM) or ITIM-like (itim) sequences (Barrow and Trowsdale 2006;Billadeau and Leibson 2002), consistent with inhibitory function upon ligand recognition (Fig. 5d). It is possible that activating forms may be encoded by PIGRL genes for which transcripts have not yet been identified. The remaining PIGRL transcripts encode secreted proteins similar to the carp PIGRL/SITR with no identifiable signaling motifs. The role of secreted PIGRL proteins remains to be resolved; however, the carp PIGRL/SITR is secreted from macrophages after infection with a protozoan parasite (Ribeiro et al. 2011).
BLASTp searches of the human reference protein database using full-length Pigrl2.4, Pigrl3.5, Pigrl4.2 protein sequences as queries identified pIgR as the human protein with the most similarity to PIGRLs (Electronic Supplementary Table S3). Additional proteins that share similarity to these PIGRL sequences include members of the CD300, CD300-like, TREM-like and natural cytotoxicity receptors (NKp44); however, this similarity is restricted to the Ig domains. This relationship between PIGRL and human innate immune receptor families has been reported for the salmon and carp PIGRLs (Ribeiro et al. 2011;Tadiso et al. 2011).
PIGRL1 D1 domains bind bacterial lipid extracts
Based on the observations that zebrafish PIGRL Ig domains share similarity to members of the CD300 and TREM multigene families and reports that Ig domains of these mammalian receptors bind phospholipids (Cannon et al. 2012;Gasiorowski et al. 2013), the ability of select PIGRL Ig domains to bind phospholipid extracts from various bacteria was evaluated. Four PIGRL D1 domains (Pigrl1.1, Pigrl1.7, Pigrl2.6 and Pigrl3.2), two PIGRL D2 domains (Pigrl3.2 and Pigrl4.1) and the zebrafish pIgR D1 domain were amplified from cDNA and cloned into a hFc expression vector. The pIgR and Pigrl2.6 D1 domains encode sequences that are identical to the reference genome. The remaining D1 and D2 domains encode predicted proteins that differ by 1–14 residues from the reference genome (GenBank KF932318 – KF932323 and Electronic Supplementary Fig. S6). Upon transfection into cultured mammalian cells, these clones successfully produced secreted, soluble hFc fusion proteins.
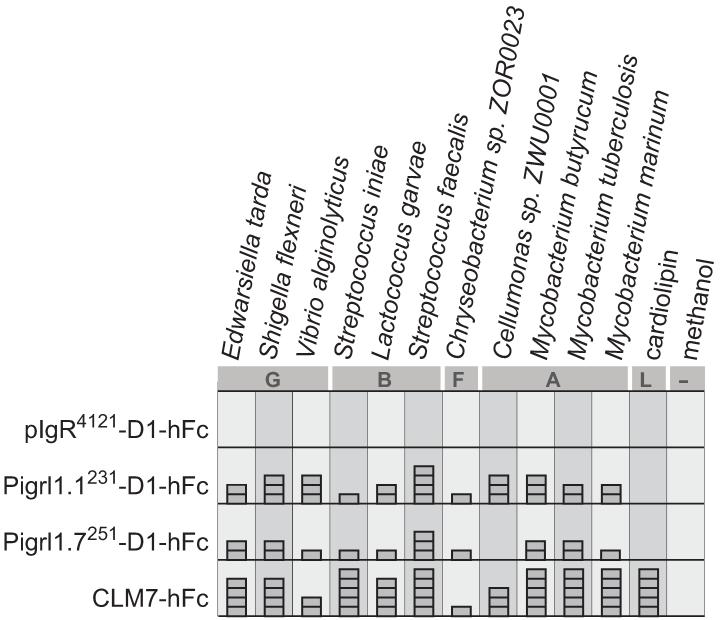
The hFc fusion proteins were used in enzyme-linked immunosorbant assays (ELISAs) in order to characterize binding to phospholipid extracts from bacteria representing the Gammaproteobacteria, Bacilli, Actinobacteria and Flavobacteria classes (Haire et al. 2012). Only the PIGRL1-D1-hFc fusion proteins were found to exhibit lipid binding (Fig. 6). The recombinant proteins Pigrl1.1-D1-hFc and Pigrl1.7-D1-hFc bound lipid extracts from all classes of bacteria. The pIgR-D1-hFc protein did not bind lipids in this assay as would be predicted based on other teleost pIgRs binding IgM and IgZ/IgT (Feng et al. 2009;Hamuro et al. 2007;Zhang et al. 2010). Lipid binding could not be detected for: Pigrl2.6-D1-hFc, Pigrl3.2-D1-hFc, Pigrl3.2-D2-hFc and Pigrl4.1-D2-hFc. The ligands for PIGRL2, PIGRL3 and PIGRL4 proteins remain to be identified. The ability of PIGRL1 D1 domains to bind lipids is shared with Ig domains from the zebrafish diverse immunoglobulin domain-containing protein (DICP) family of innate immune receptors that also share sequence and structural similarities with the CD300 and TREM families (Haire et al. 2012).
Fig. 6. PIGRL1 D1 domains bind phospholipids.
ELISA analyses of interactions of pIgR-D1-hFc and PIGRL1-D1-hFc fusion proteins (Electronic Supplementary Fig. S6) with soluble organic extracts from four different classes of bacteria; Gammaproteobacteria (G), Bacilli (B), Actinobacteria (A) and Flavobacterium (F) and the lipid (L), cardiolipin. Mouse CLM7-hFc was included as a positive control (Cannon et al. 2012). The fusion proteins pIgR-D1-hFc, Pigrl2.6-D1-hFc, Pigrl3.2-D1-hFc, Pigrl3.2-D2-hFc and Pigrl4.1-D2-hFc failed to score above background in this assay: the pIgR-D1-hFc result is shown as a negative control.
Expression analyses of zebrafish pigr and PIGRL genes
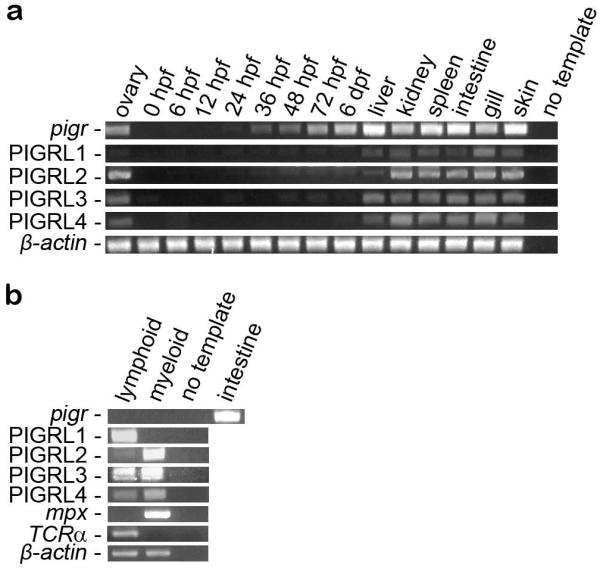
PIGR transcripts have been detected using RT-PCR in multiple lymphoid organs of fugu, carp and grouper including the kidney, spleen, gut, skin and gills (Feng et al. 2009;Rombout et al. 2008) and in the ovary or gonad of fugu and grouper (Feng et al. 2009;Hamuro et al. 2007). Similar expression profiles are observed for pigr in zebrafish (Fig. 7a). The observations that pigr is expressed in the ovary and during embryogenesis suggests that it may function in some aspect of development, possibly for transport of maternal antibodies (Wang et al. 2012).
Fig. 7. Analyses of pigr and PIGRL transcripts in normal tissues.
(a) RT-PCR analyses of pigr and PIGRL transcript levels from zebrafish ovaries, embryos and adult tissues. (b) RT-PCR analyses of pigr and PIGRL transcript levels from sorted leukocyte populations. PIGRL primers were designed to amplify multiple members within each group of genes. RT-PCR of β-actin is included as a control for cDNA quality. RT-PCR of mpx and T cell receptor α (TCRα) are included as positive controls for the myeloid and lymphoid lineages, respectively. In order to verify specificity, various amplicons were cloned and sequenced: all sequences were as expected except for two of twelve PIGRL4 amplicons were PIGRL3 sequences. PIGRL1 amplicon sequences have been deposited in GenBank (KF932350 – KF932357). hpf = hours post fertilization. dpf = days post infection.
Four sets of degenerate primers were used to evaluate the expression of zebrafish PIGRL1, PIGRL2, PIGRL3 and PIGRL4 genes (Fig. 7a). Transcripts from all PIGRL groups can be detected from liver, kidney, spleen, intestine, gill and skin as well as ovary; however, PIGRL transcripts are not abundant during embryonic development.
In order to evaluate pigr and PIGRL expression in zebrafish hematopoietic lineages, RT-PCR analyses were conducted using lymphoid and myeloid cells sorted from zebrafish kidneys (Traver 2004;Yoder et al. 2010). Although pigr transcripts were not detected in either leukocyte lineage, PIGRL gene expression was detected in both myeloid and lymphoid cells (Fig. 7b). The absence of pigr transcripts from zebrafish leukocytes is consistent with a role in transporting antibodies across mucosal epithelium. Expression of pigr in epidermal cells of the skin and intestine of fugu has been reported (Hamuro et al. 2007). PIGRL1 transcripts were detected primarily in the lymphoid lineage, whereas PIGRL2 transcripts were detected primarily from the myeloid lineage. This is consistent with the report that a carp PIGRL (aka SITR), which groups with the zebrafish PIGRL2 sequences (Electronic Supplementary Fig. S3), is expressed primarily on myeloid cells (Ribeiro et al. 2011). In contrast, PIGRL3 and PIGRL4 transcripts were expressed at comparable levels in myeloid and lymphoid cell lineages. The observation that the PIGRL genes are expressed in leukocytes, whereas pigr transcripts are not detected in these cell types, suggests that the PIGRLs are functionally distinct from pigr in zebrafish.
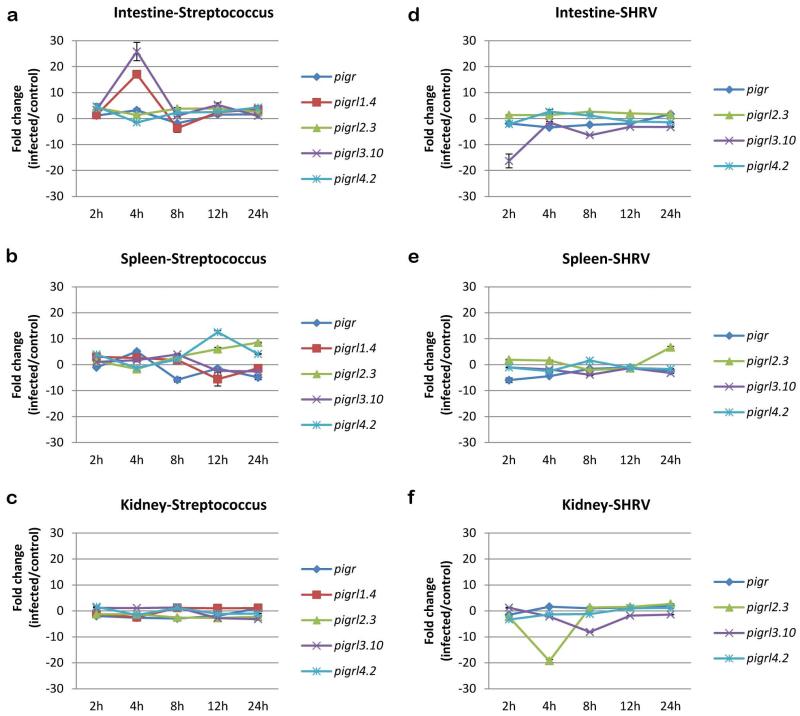
As PIGR and PIGRL transcript levels were shown to be increased after copepod infection in salmon (Tadiso et al. 2011) we employed RT-qPCR to investigate if transcript levels of zebrafish pigr, pigrl1.4, pigrl2.3, pigrl3.10 and pigrl4.2 were altered after bacterial infection (Fig. 8a-c). After infection with the fish pathogen Streptococcus iniae (Lowe et al. 2007;Neely et al. 2002) transcript levels of all five genes were altered. In the intestine of infected zebrafish, pigrl1.4 and pigrl3.10 had the largest increases in transcript level which occurred at 4 hpi (17-fold and 25-fold increases, respectively). In the spleen, pigrl4.2 (12-fold increase) and pigrl2.3 (8-fold increase) had the largest increases in transcript level which occurred at 12 and 24 hpi, respectively. No dramatic increase in pigr or PIGRL transcript levels was observed in the kidney over the 24 hr of infection. The most dramatic change in pigr transcript levels occurred at 4 hpi in the intestine (3-fold increase) and spleen (5-fold increase). Thus, the highest increase in PIGRL expression during bacterial infection was observed in the intestine which is a major site for mucosal immunity.
Fig. 8. Analyses of pigr and PIGRL transcripts after infection.
RT-qPCR was employed to quantify the relative levels of pigr and PIGRL transcripts during the initial stages of infection. Adult zebrafish were infected with either (a-c) Streptococcus iniae or (d-f) Snakehead rhabdovirus or mock infected. Expression analyses was conducted from (a, d) intestine, (b, e) spleen and (c, f) kidney. RNA samples were evaluated from 2 h to 24 h after infection. Transcript levels were normalized to eef1a1l1 by the ΔΔCt method (Livak and Schmittgen 2001). Analyses of pigrl1.4 are excluded for SHRV infections as expression was not reliably detected.
An effort was made to determine if transcript levels of pigr, pigrl2.3, pigrl3.10 and pigrl4.2 changed during infection with the viral fish pathogen Snakehead rhabdovirus (Phelan et al. 2005). In contrast to changes in transcript level after bacterial infection, the most dramatic changes after viral infection were decreased transcript levels (Fig. 8d-f). Transcript levels of pigrl3.10 in the intestine decreased by 16-fold by 2 hpi and transcript levels of pigrl2.3 in the kidney decreased by 19-fold by 4 hpi. The most dramatic change in pigr transcript levels (6-fold decrease) was observed 2 hpi in the spleen. The virally induced down-regulation of PIGRL transcripts is similar to the response of CD300a/c in human dendritic cells (DCs): TLR7 and TLR9 activation on DCs leads to down-regulation of CD300a/c transcripts (Ju et al. 2008). Collectively, these observations suggest that SHRV may suppress the zebrafish immune response by down-regulating pigr and PIGRL gene expression.
Summary
Teleost pIgR 1) is expressed in epithelial cells, 2) binds soluble immunoglobulins and 3) is well conserved across multiple species. In contrast, zebrafish PIGRL proteins 1) are expressed in leukocytes, 2) bind lipids and 3) are not well conserved between fish species. The zebrafish PIGRL gene cluster encodes both membrane-bound receptors with potential inhibitory function and secreted proteins. Some PIGRL proteins bind bacterially derived phospholipid extracts. Following bacterial infection the transcript levels of multiple PIGRL genes increase, suggesting a role in immunity. In contrast, after a viral infection the transcript levels of multiple PIGRL genes decrease possibly as a result of viral induced immune suppression. These features of the PIGRL family are reminiscent of the CD300 and TREM families of mammalian innate immune receptors.
Supplementary Material
Acknowledgements
We thank John Whitesides and Patti McDermott (Duke Human Vaccine Institute) for assistance with cell sorting; John Rawls (Duke University), Karen Guillemin and Erika Mittge (University of Oregon), and Ed Noga (North Carolina State University) for bacteria; and Barb Pryor for editorial assistance. The zebrafish PIGRL gene nomenclature has been approved by the Zebrafish Nomenclature Committee. The authors are supported the National Institutes of Health (R01 AI057559 to GWL and JAY and R01 AI23337 to GWL).
Footnotes
Electronic supplementary material
The online version of this article contains supplementary material, which is available to authorized users.
References
- Asano M, Komiyama K. Polymeric immunoglobulin receptor. J. Oral Sci. 2011;53:147–156. doi: 10.2334/josnusd.53.147. [DOI] [PubMed] [Google Scholar]
- Barclay AN. Membrane proteins with immunoglobulin-like domains—a master superfamily of interaction molecules. Semin Immunol. 2003;15:215–223. doi: 10.1016/s1044-5323(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Barclay AN, Brown MH, Law SKA, McKnight AJ, Tomlinson MG, van der Merwe PA. The Leucocyte Antigen FactsBook. Academic Press; San Diego: 1997. [Google Scholar]
- Barrow AD, Trowsdale J. You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur. J. Immunol. 2006;36:1646–1653. doi: 10.1002/eji.200636195. [DOI] [PubMed] [Google Scholar]
- Betancur R, Broughton RE, Wiley EO, Carpenter K, Lopez JA, Li C, Holcroft NI, Arcila D, Sanciangco M, Cureton Ii JC, Zhang F, Buser T, Campbell MA, Ballesteros JA, Roa-Varon A, Willis S, Borden WC, Rowley T, Reneau PC, Hough DJ, Lu G, Grande T, Arratia G, Orti G. The tree of life and a new classification of bony fishes. PLoS. Curr. 2013;5 doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billadeau DD, Leibson PJ. ITAMs versus ITIMs: striking a balance during cell regulation. J. Clin. Invest. 2002;109:161–168. doi: 10.1172/JCI14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braathen R, Hohman VS, Brandtzaeg P, Johansen FE. Secretory antibody formation: conserved binding interactions between J chain and polymeric Ig receptor from humans and amphibians. J. Immunol. 2007;178:1589–1597. doi: 10.4049/jimmunol.178.3.1589. [DOI] [PubMed] [Google Scholar]
- Broughton RE, Betancur R, Li C, Arratia G, Orti G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS. Curr. 2013;5 doi: 10.1371/currents.tol.2ca8041495ffafd0c92756e75247483e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JP, Haire RN, Mueller MG, Litman RT, Eason DD, Tinnemore D, Amemiya CT, Ota T, Litman GW. Ancient divergence of a complex family of immune-type receptor genes. Immunogenetics. 2006;58:362–373. doi: 10.1007/s00251-006-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon JP, O’Driscoll M, Litman GW. Construction, expression, and purification of chimeric protein reagents based on immunoglobulin fc regions. Methods Mol. Biol. 2011;748:51–67. doi: 10.1007/978-1-61779-139-0_4. [DOI] [PubMed] [Google Scholar]
- Cannon JP, O’Driscoll M, Litman GW. Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics. 2012;64:39–47. doi: 10.1007/s00251-011-0562-4. [DOI] [PubMed] [Google Scholar]
- Deitcher DL, Mostov KE. Alternate splicing of rabbit polymeric immunoglobulin receptor. Mol. Cell Biol. 1986;6:2712–2715. doi: 10.1128/mcb.6.7.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng LN, Lu DQ, Bei JX, Chen JL, Liu Y, Zhang Y, Liu XC, Meng ZN, Wang L, Lin HR. Molecular cloning and functional analysis of polymeric immunoglobulin receptor gene in orange-spotted grouper (Epinephelus coioides) Comp Biochem. Physiol B Biochem. Mol. Biol. 2009;154:282–289. doi: 10.1016/j.cbpb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Garcia-Boronat M, Diez-Rivero CM, Reinherz EL, Reche PA. PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res. 2008;36:W35–W41. doi: 10.1093/nar/gkn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiorowski RE, Ju X, Hart DN, Clark GJ. CD300 molecule regulation of human dendritic cell functions. Immunol Lett. 2013;149:93–100. doi: 10.1016/j.imlet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, Lefranc MP. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haire RN, Cannon JP, O’Driscoll ML, Ostrov DA, Mueller MG, Turner PM, Litman RT, Litman GW, Yoder JA. Genomic and functional characterization of the diverse immunoglobulin domain-containing protein (DICP) family. Genomics. 2012;99:282–291. doi: 10.1016/j.ygeno.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamuro K, Suetake H, Saha NR, Kikuchi K, Suzuki Y. A teleost polymeric Ig receptor exhibiting two Ig-like domains transports tetrameric IgM into the skin. J. Immunol. 2007;178:5682–5689. doi: 10.4049/jimmunol.178.9.5682. [DOI] [PubMed] [Google Scholar]
- Johansen FE, Pekna M, Norderhaug IN, Haneberg B, Hietala MA, Krajci P, Betsholtz C, Brandtzaeg P. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J. Exp. Med. 1999;190:915–922. doi: 10.1084/jem.190.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X, Zenke M, Hart DN, Clark GJ. CD300a/c regulate type I interferon and TNF-alpha secretion by human plasmacytoid dendritic cells stimulated with TLR7 and TLR9 ligands. Blood. 2008;112:1184–1194. doi: 10.1182/blood-2007-12-127951. [DOI] [PubMed] [Google Scholar]
- Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev. 2005;206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- Kulseth MA, Krajci P, Myklebost O, Rogne S. Cloning and characterization of two forms of bovine polymeric immunoglobulin receptor cDNA. DNA Cell Biol. 1995;14:251–256. doi: 10.1089/dna.1995.14.251. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Hawke NA, Yoder JA. Novel immune-type receptor genes. Immunol Rev. 2001;181:250–259. doi: 10.1034/j.1600-065x.2001.1810121.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowe BA, Miller JD, Neely MN. Analysis of the polysaccharide capsule of the systemic pathogen Streptococcus iniae and its implications in virulence. Infect. Immun. 2007;75:1255–1264. doi: 10.1128/IAI.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BC, Cortes HD, Mewes-Ares J, Verheijen K, Stafford JL. Teleost IgSF immunoregulatory receptors. Dev. Comp Immunol. 2011;35:1223–1237. doi: 10.1016/j.dci.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Narayanan S, Su S, Childs R, Krzewski K, Borrego F, Weck J, Coligan JE. Toso, a functional IgM receptor, is regulated by IL-2 in T and NK cells. J. Immunol. 2012;189:587–597. doi: 10.4049/jimmunol.1200840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely MN, Pfeifer JD, Caparon M. Streptococcus-zebrafish model of bacterial pathogenesis. Infect Immunol. 2002;70:3904–3914. doi: 10.1128/IAI.70.7.3904-3914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan PE, Pressley ME, Witten PE, Mellon MT, Blake S, Kim CH. Characterization of snakehead rhabdovirus infection in zebrafish (Danio rerio) J. Virol. 2005;79:1842–1852. doi: 10.1128/JVI.79.3.1842-1852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pressley ME, Phelan PE, III, Witten PE, Mellon MT, Kim CH. Pathogenesis and inflammatory response to Edwardsiella tarda infection in the zebrafish. Dev. Comp Immunol. 2005;29:501–513. doi: 10.1016/j.dci.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Ribeiro CM, Bird S, Raes G, Ghassabeh GH, Schijns VE, Pontes MJ, Savelkoul HF, Wiegertjes GF. A novel soluble immune-type receptor (SITR) in teleost fish: carp SITR is involved in the nitric oxide-mediated response to a protozoan parasite. PLoS. One. 2011;6:e15986. doi: 10.1371/journal.pone.0015986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe M, Norderhaug IN, Brandtzaeg P, Johansen FE. Fine specificity of ligand-binding domain 1 in the polymeric Ig receptor: importance of the CDR2-containing region for IgM interaction. J. Immunol. 1999;162:6046–6052. [PubMed] [Google Scholar]
- Rombout JH, van der Tuin SJ, Yang G, Schopman N, Mroczek A, Hermsen T, Taverne-Thiele JJ. Expression of the polymeric Immunoglobulin Receptor (pIgR) in mucosal tissues of common carp (Cyprinus carpio L.) Fish. Shellfish. Immunol. 2008;24:620–628. doi: 10.1016/j.fsi.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Sakamoto N, Shibuya K, Shimizu Y, Yotsumoto K, Miyabayashi T, Sakano S, Tsuji T, Nakayama E, Nakauchi H, Shibuya A. A novel Fc receptor for IgA and IgM is expressed on both hematopoietic and non-hematopoietic tissues. Eur. J. Immunol. 2001;31:1310–1316. doi: 10.1002/1521-4141(200105)31:5<1310::AID-IMMU1310>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Shibuya A, Sakamoto N, Shimizu Y, Shibuya K, Osawa M, Hiroyama T, Eyre HJ, Sutherland GR, Endo Y, Fujita T, Miyabayashi T, Sakano S, Tsuji T, Nakayama E, Phillips JH, Lanier LL, Nakauchi H. Fc alpha/mu receptor mediates endocytosis of IgM-coated microbes. Nat. Immunol. 2000;1:441–446. doi: 10.1038/80886. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Honda S, Yotsumoto K, Tahara-Hanaoka S, Eyre HJ, Sutherland GR, Endo Y, Shibuya K, Koyama A, Nakauchi H, Shibuya A. Fc(alpha)/mu receptor is a single gene-family member closely related to polymeric immunoglobulin receptor encoded on Chromosome 1. Immunogenetics. 2001;53:709–711. doi: 10.1007/s00251-001-0375-y. [DOI] [PubMed] [Google Scholar]
- Stet RJ, Hermsen T, Westphal AH, Jukes J, Engelsma M, Lidy Verburg-van Kemenade BM, Dortmans J, Aveiro J, Savelkoul HF. Novel immunoglobulin-like transcripts in teleost fish encode polymorphic receptors with cytoplasmic ITAM or ITIM and a new structural Ig domain similar to the natural cytotoxicity receptor NKp44. Immunogenetics. 2005;57:77–89. doi: 10.1007/s00251-005-0771-9. [DOI] [PubMed] [Google Scholar]
- Tadiso TM, Sharma A, Hordvik I. Analysis of polymeric immunoglobulin receptor- and CD300-like molecules from Atlantic salmon. Mol. Immunol. 2011;49:462–473. doi: 10.1016/j.molimm.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2004;76:127–149. doi: 10.1016/s0091-679x(04)76008-2. [DOI] [PubMed] [Google Scholar]
- Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Wang H, Ji D, Shao J, Zhang S. Maternal transfer and protective role of antibodies in zebrafish Danio rerio. Mol. Immunol. 2012;51:332–336. doi: 10.1016/j.molimm.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Guide for the Laboratory Use of Zebrafish (Danio rerio) University of Oregon Press; Eugene: 2007. [Google Scholar]
- Wieland WH, Orzaez D, Lammers A, Parmentier HK, Verstegen MW, Schots A. A functional polymeric immunoglobulin receptor in chicken (Gallus gallus) indicates ancient role of secretory IgA in mucosal immunity. Biochem. J. 2004;380:669–676. doi: 10.1042/BJ20040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily-domains for cell surface recognition. Ann. Rev. Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Yoder JA. Form, function and phylogenetics of NITRs in bony fish. Dev. Comp Immunol. 2009;33:135–144. doi: 10.1016/j.dci.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Turner PM, Wright PD, Wittamer V, Bertrand JY, Traver D, Litman GW. Developmental and tissue-specific expression of NITRs. Immunogenetics. 2010;62:117–122. doi: 10.1007/s00251-009-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.