Abstract
We use atomistic molecular dynamics simulations to reveal the binding mechanisms of therapeutic agents in PEG-ylated micellar nanocarriers (SSM). In our experiments, SSM in buffer solutions can solubilize either ≈ 11 small bexarotene molecules or ≈ 6 (2 in low ionic strength buffer) human vasoactive intestinal peptide (VIP) molecules. Free energy calculations reveal that molecules of the poorly water soluble drug bexarotene can reside at the micellar ionic interface of the PEG corona, with their polar ends pointing out. Alternatively, they can reside in the alkane core center, where several bexarotene molecules can self-stabilize by forming a cluster held together by a network of hydrogen bonds. We also show that highly charged molecules, such as VIP, can be stabilized at the SSM ionic interface by Coulombic coupling between their positively charged residues and the negatively charged phosphate head-groups of the lipids. The obtained results illustrate that atomistic simulations can reveal drug solubilization character in nanocarriers and be used in efficient optimization of novel nanomedicines.
Introduction
Many therapeutic molecules exhibit a limited aqueous solubility, a reduced in vivo stability, and major side effects due to non-specific delivery [1, 2]. These limitations can be addressed in modern nanomedicines, where drug molecules are encapsulated within various nanocarriers [3–6]. In particular, lipid-based micellar nanocarriers can solubilize hydrophobic drugs with high loading capacity, protect them from degradation, improve their availability at disease sites, and suppress their side activity [7].
Today’s predictive strategies anticipate the drug solubility in lipid nanocarriers from their bulk partition coefficients (water/octanol) [8–10]. These indirect strategies fail to provide the information on the exact location of the drugs within the nanocarrier. However, the drug location in a nanocarrier can determine its stability, protection, loading efficiency and release rate, which are all crucial for the therapeutic effects of the drug [11, 12]. Drugs that are buried in the nanocarrier interior and more strongly attached to it tend to be more stable and better protected against degradation and inactivation [7]. The number of drug molecules which can be loaded in a nanocarrier is determined by the effective volume of their preferred location. The release rate (transport) of a drug from the nanocarrier is determined by its solvation free energy and local diffusion constants in the carrier [13]. In order to develop new nanomedicines and optimize their pharmaceutical properties and physiological effects, we need to understand the stabilization mechanisms of therapeutic agents within the nanocarriers [8].
In principle, the drug-nanocarrier complexation could be described by precise atomistic simulations. In the past, a typical use of molecular dynamics (MD) simulations was to predict free energies of solvation for short alkanes in the cores of small surfactant micelles [14] or the permeation of small solutes through lipid membranes [15, 16]. In our recent large-scale atomistic MD simulations, phospholipid-based sterically stabilized micelles (SSM) were explored in water and saline solutions [17]. In Fig. 1 (b–d) we show atomistic models of SSM in pure water and aqueous solutions of different ionic strength. Our previous experiments have shown that such SSM can solubilize different hydrophobic and amphiphilic drug molecules [18, 19] and peptides [20–24]. However, the stabilization mechanisms and the exact location and organization of these drugs within SSM are not known.
FIG. 1.
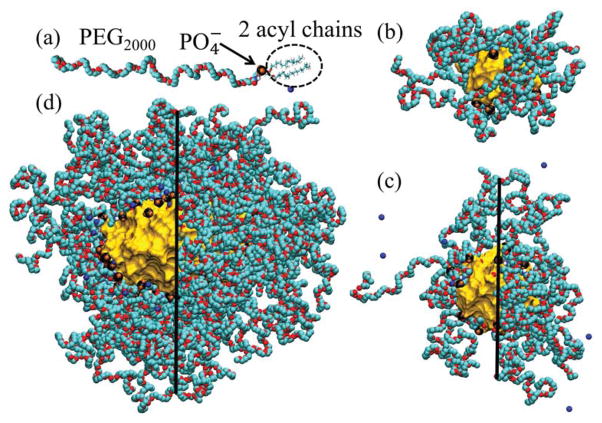
(a) DSPE-PEG2000 monomer with Na+ counterion. (b) Equilibrated SSM-10 in water. The characteristic layers in this system are alkane core (rcore ≈ 0 – 1.2 nm, shown as a gold surface), ionic interface (rint ≈ 1.2 – 1.8 nm; the negatively charged groups are shown as black and orange clusters), PEG corona (rPEG ≈ 1.8–3.0 nm, shown as turquoise and red chains), and aqueous ionic solution (raq > 3.0 nm, not shown for clarity). (c) Equilibrated SSM-20 in water, modeled after the SSM observed by DLS experiments in PB buffer (low ionic strength solution), with rcore ≈ 0 – 1.7 nm, rint ≈ 1.7 – 2.3 nm, rPEG ≈ 2.3 – 3.8 nm, and raq > 3.8 nm. (d) Equilibrated SSM-90 in 0.16 M NaCl solution, with rcore ≈ 0 – 2.4 nm (along minor axis), rint ≈ 2.4 – 3.0 nm, rPEG ≈ 3.0 – 7.5 nm, and raq > 7.5 nm. In SSM shown in (c–d), groups and PEG chains are removed for clarity on the left sides of the images.
In this work we use precise atomistic MD simulations combined with free energy calculations to model the stabilization of a small water-insoluble drug bexarotene (BEX) and a human vasoactive intestinal peptide (VIP) inside SSM, as observed in our experiments. The aim of the simulations is to identify the equilibrated locations and binding energies of the drug molecules within the SSM nanocarriers (Fig. 1 (b–d)).
Materials and Methods
Experimental Characterization of SSM
Blank and bexarotene-containing SSM nanocarriers (SSM-BEX) were prepared in HEPES buffer from PEG-ylated phospholipid 1,2- Distearoyl-sn-glycero-3-phosphatidylethanolamine-N- [methoxy(poly ethyleneglycol)-2000] sodium salt (DSPE-PEG2000). Several solutions with the bexarotene concentration in the range of 0 – 60 μg/mL and 1 mM DSPE-PEG2000 concentration were prepared by the film-rehydration method, described extensively in [5, 19]. These solutions were tested to determine the maximum solubilization capacity of SSM for bexarotene. Micelle dispersions were analyzed by dynamic light scattering (DLS) for the particle distributions and by HPLC for the drug content.
The VIP-SSM complexes were prepared in standard phosphate buffered saline (PBS, pH= 7.4) and phosphate buffer solutions (PB, 1 mM, pH=7.4). The prepared solutions had constant VIP concentration cVIP = 4.0 μM and variable DSPE-PEG2000 concentration in the range clip = 0 – 0.4 mM. The VIP-SSM assemblies were characterized by DLS and fluorescence spectroscopy. Intrinsic fluorescence of tyrosine residues in VIP was used (the excitation wavelength λex = 277 nm, the emission wavelength λem = 304 nm). Materials, methods and our experimental procedures are described in detail in the Supporting Information.
MD Simulations
In Fig. 1 (a), (b), (c) and (d) we show a single DSPE-PEG2000 monomer, the free SSM-10 (SSM with the aggregation number Nagg = 10, modeled in water), SSM-20 (Nagg=20, water), and SSM-90 (Nagg=90, 0.16 M NaCl), respectively. The free and drug filled micelles were modeled by atomistic classical MD simulations using the NAMD package [25] and CHARMM force field (CHARMM27 and C35r revision for ethers, TIP3P water model) [26, 27]. The prepared free SSMs were equilibrated for 5 – 10 ns in the NpT ensemble at p = 1 bar and T = 300 K, and the electrostatics were evaluated with the PME method [28].
In the BEX-SSM simulations, we used SSM-10 (Fig. 1 (b)) and SSM-90 (Fig. 1 (d)). The choice of SSM-10 is based on the comparison of SSM sizes in water obtained by DLS measurements and our previous MD simulations [17]; the comparison was validated by showing that the diameters of simulated SSM were in agreement with diameters of SSM in buffer obtained from DLS measurements. The SSM-90 in buffer is based on the SANS measurements of SSM in 10 mM HEPES buffered saline (pH = 7.4) at the clip = 5 mM lipid concentration [29, 30]. The ionic strength of the simulated NaCl solution matches the ionic strength of the experimental buffer solution. Since SANS measurements showed that SSM-90 is nearly spherical (slightly oblate) [29], DSPE-PEG2000 monomers were initially spherically distributed in all the simulations of empty SSMs. SSM-10 in water remained spherical in all equilibration and free energy simulations. SSM-90 became oblate during the first few nanoseconds of equilibration, and remained oblate during the equilibration and multiple free energy simulations. We used the adaptive biasing force (ABF) method [31–33] to calculate the solvation Gibbs free energy (ΔG) profiles of bexarotene inside SSM-10 and SSM-90. For comparison, we evaluated ΔG profiles of bexarotene at the interfaces of octane with aqueous solutions.
In the VIP-SSM simulations, we prepared the SSM-20 (Fig. 1 (c)) and SSM-90 (Fig. 1 (d)). The aggregation numbers (Nagg=20, 90) were chosen to match the experimental SSM sizes observed in PB and PBS solutions. The SSM-20 was solvated in water (the ionic strength of the PB solution with the λD = 9.7 nm Debye length is so weak that there is a negligible number of ions in the simulation box). The SSM-90 was solvated in 0.16 M NaCl solution. The SSMs had VIP molecules placed in PEG palisade regions. In SSM-20, VIP was used in its molecular form, representing a therapeutic agent. In SSM-90, VIP was tethered to a DSPE-PEG3400 monomer as a targeting agent (tail end of the PEG chain), whose alkane chains were inserted into the SSM core. We equilibrated the VIP-SSM systems for t ≈ 30 ns. Further information about the computational methods is given in the Supporting Information.
Results and Discussion
Solubilization of Bexarotene in SSM
The SSM is formed by DSPE-PEG2000 copolymers, containing two acyl chains, a negatively charged phosphate group and a long PEG chain, as shown in Fig. 1 (a). First, we study experimentally bexarotene solubilization in SSM by dynamic light scattering. We find the maximum solubilization of bexarotene (without formation of precipitate) by monitoring its saturation concentration, cbex, in DLS experiments. After a certain number of drug molecules occupies (saturates) SSM, the drugs start to precipitate in the form of secondary particle species. The secondary particle species were previously investigated for other drugs by DLS and transmission electron microscopy (TEM) [18, 34]. Here, we monitor the sizes of SSM and the secondary particle species by DLS only.
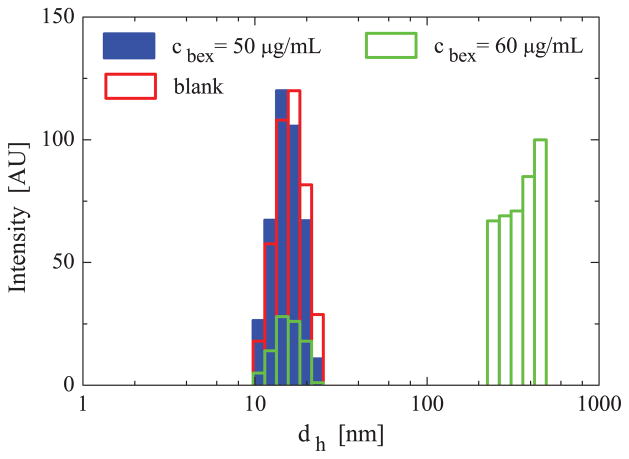
In Fig. 2, we show the particle size distributions obtained by DLS for SSM dispersions (clip = 1 mM) containing bexarotene in HEPES buffered saline, as we vary the concentration of bexarotene, cbex. The DLS measurements show similar SSM particle size distributions, dh ≈ 15 nm, for dispersions at the bexarotene concentration cbex ≤ 50 μg/mL. At cbex = 60 μg/mL, in addition to the SSM diameter of dh ≈ 15 nm, particles with an average hydrodynamic diameter of dh ≈ 500 nm were also observed. Previous DLS and TEM experiments revealed particles of similar sizes in solutions of SSM and other drug molecules (camptothecin, paclitaxel), spherical SSM (dh ≈ 15 nm) were observed at low drug concentrations, while larger particles (dh ≈ 300 – 500 nm) appeared when drug concentrations exceeded the solubility limit in SSM [18, 34].
FIG. 2.
Experimental intensity-weighted particle size distributions obtained by DLS in HEPES buffered saline with the DSPE-PEG2000 concentration of clip = 1 mM and the bexarotene concentrations of cbex = 0, 50, 60 μg/mL.
Since the intrinsic solubility of bexarotene in HEPES buffered saline is 0.56 μg/ml (determined by HPLC), the data in Fig. 2 indicates that the maximum solubility of bexarotene in SSMs is cbex = 50–60 μg/ml, at the DSPE-PEG2000 concentration of clip = 1 mM. Based on the intrinsic solubility of bexarotene in HEPES buffered saline and the fact that ≈ 90 lipid monomers form one micelle [29], we determined that on average Nbex ≈ 11 bexarotene molecules are encapsulated within SSM.
Simulations of SSM-BEX Nanomedicines
Since drugs can be solvated in different regions of the SSM interior (Fig. 1), it is good to understand the structure of the SSM regions. The central (nonpolar) SSM core region is formed by densely aggregated alkane chains of various levels of interconnection and fluidity. The core is surrounded by covalently attached groups and their free hydrated Na+ counterions, positioned at a certain distance from groups (depending on the ionic strength of the solution). The ionic interface ( groups) is surrounded by the PEG corona, which is immersed in water (SSM-10) or 0.16 M NaCl solution (SSM-90). In the corona, the Na+ counterions are often solvated in oxygen atom pockets formed by PEG chains [17, 35]. Density distributions in SSM, analyzed in [17], show that SSM regions change gradually along the radial coordinate r. On the other hand, radial distributions of ions in SSM show strong gradients around the ionic interface, observed at 1 nm < r < 3 nm for SSM-10 in water and at 1.8 nm < r < 5 nm for SSM-90 in 0.16 M NaCl [17]. The micelle interior is heterogeneous and fluctuating. While in SSM-10, the ionic interface and the alkane core are highly exposed to water, in SSM-90, PEG forms a relatively homogeneous layer, which can spread and expose the SSM core to the bulk aqueous solution [17].
Solvation of bexarotene at the SSM-core interface
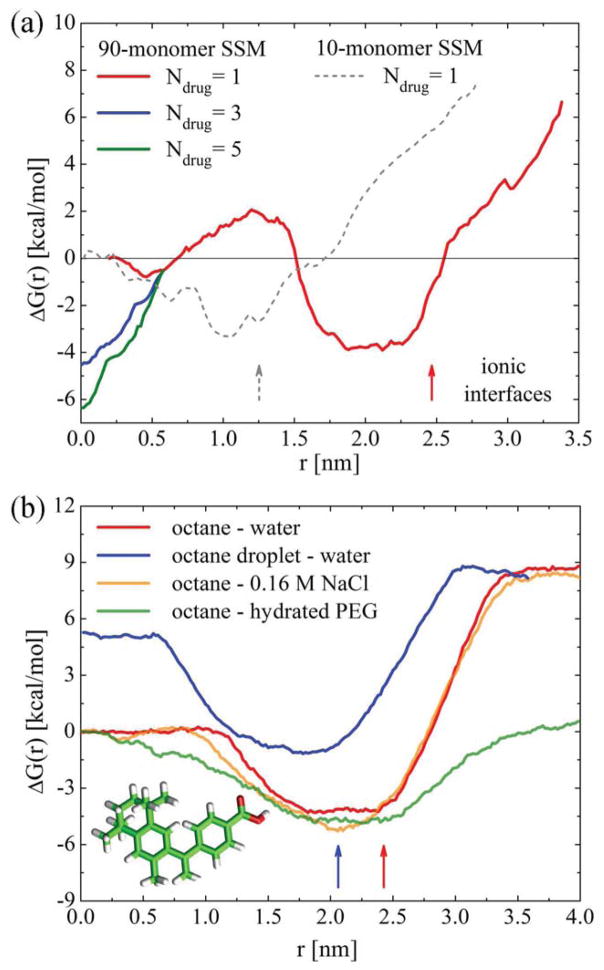
First, we examine the possibility of solvation of bexarotene at the hydrophobic/philic SSM-core interface, which might be a less favorable place of residence for drugs that are poorly soluble in water [7]. In Fig. 3 (a) , we show the Gibbs free energy profiles, ΔG(r), for a single bexarotene molecule along the radial coordinate r in SSM-10 and SSM-90. In SSM-10, ΔG(r) has a single minimum, while in SSM-90 two minima of different depths are observed. In both SSMs, the global minimum forms in a region located between the alkane cores and the ionic interfaces, at r ≈ 0.8 – 1.2 nm (SSM-10) and r ≈ 1.7 – 2.5 nm (SSM-90). The barriers for transfer of bexarotene from these minima into the alkane cores of both SSMs are ΔΔG(r) ≈ 4 kcal/mol, whereas the barriers for transfer into the aqueous PEG regions are ΔΔG(r) ≈ 10 kcal/mol. In these interfacial minima, bexarotene has its polar -COOH group oriented towards the aqueous region, while its body is immersed in the alkane region (Fig. 4 (c)).
FIG. 3.
(a) Free energy profiles of bexarotene in SSM-10 (water) and SSM-90 (0.16 M NaCl). Whole solvated micelles loaded with drugs were present in the free energy calculations; different numbers of drugs are considered in the SSM-90 core. The vertical arrows show the positions of ionic interfaces in the two SSMs. (b) Free energy profiles of bexarotene across the interfaces of octane with pure water, 0.16M NaCl, and hydrated PEG. Interfaces in these planar systems are located at r ≈ 2.5 nm (red arrow). The free energy in a hydrated octane nanodroplet of the radius 2.1 nm is also shown. The interface in this system is marked by the blue arrow.
FIG. 4.
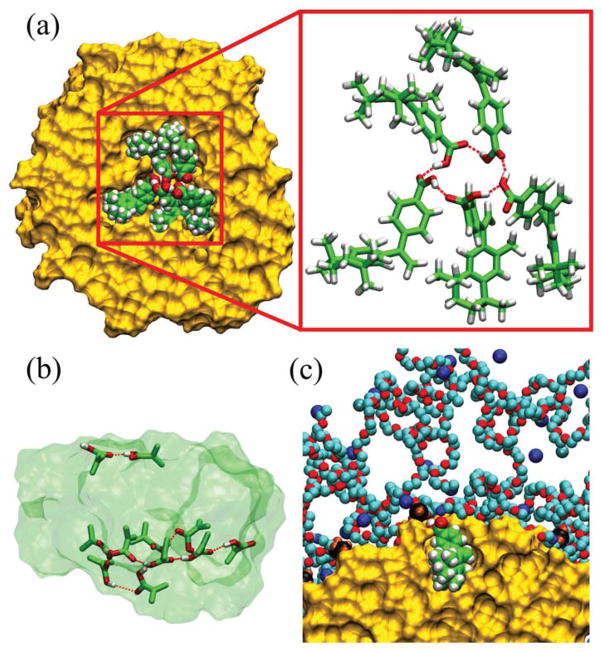
(a) A snapshot of the cluster of 5 bexarotene molecules formed inside the SSM core, after 11 ns of equilibration. A hydrogen bond network between -COOH groups is highlighted. (b) The cluster of 11 bexarotene molecules forms 2 hydrogen bond networks in the SSM core after 17 ns of equilibration (transparent background shows the whole 11-drug cluster, and the atoms shown in the stick representation participate in the H-bond network). (c) A solvated bexarotene molecule orients its non-polar part to the alkane core and its -COOH group towards the ionic interface (shown in SSM-90).
The results in Fig. 3 (a) can be compared with ΔG(r) profiles for bexarotene at flat interfaces of two bulk liquids with properties similar to the SSM regions. However, note that flat interfacial systems contain only some of the elements which contribute to the complexity of the SSM environment (curvature, strong gradients in ion concentration [17]). In Fig. 3 (b), we show ΔG(r) at octane interfaced with pure water, 0.16 M NaCl solution, and ≈ 70 w/w % hydrated PEG. The curves are aligned in the regions of the same solvent and geometry (bulk). In all the cases, bexarotene has energy minima at the interfaces. In aqueous solutions, the minima are shifted towards octane and the barriers are asymmetrical, as in SSM in Fig. 3 (a), with ΔΔG(r) ≈ 12.5 kcal/mol on the aqueous side and ΔΔG(r) ≈ 4.5 kcal/mol on the octane side. The barrier for bexarotene transfer into 70 w/w % PEG in water is significantly reduced, ΔΔG(r) ≈ 5 kcal/mol. However, since PEG in SSM has a smaller density than in Fig. 3 (b) (≈ 25 – 50 w/w % for r = 3– 4 nm in SSM-90 [17]), the barrier reduction in SSM-10 and SSM-90 was not observed.
It is also of interest to consider the intermediate case between the flat alkane-water interface and the SSM. In Fig. 3 (b), we show the free energy profile for bexarotene solvated in a hydrated octane nanodroplet with a radius of r = 2.1 nm. Here, the energy curves are aligned in the bulk water solvent. The data reveal an increase of ΔG(r) ≈ 5 kcal/mol in the core beyond the value observed in the bulk octane. This Gibbs energy growth inside octane nanodroplets can be attributed to the Laplace pressure (surface tension) [36], which is largely reduced in SSM formed by the amphiphilic monomers. To our knowledge, this Laplace pressure effect was not quantified in nanodroplets with a molecular structure.
Solvation of bexarotene in the SSM core
Next, we discuss bexarotene solvation in the SSM core, which is considered in micellar nanomedicines as the dominant residing region for poorly water-soluble drug molecules [7]. Figure 3 (a) shows that a single bexarotene already has a shallow minimum in the SSM-90 core, which is not seen in SSM-10 (no significant bulk region exist within r < 1.0 nm) and the bulk systems. Even though this minimum in the SSM-90 core is shallower than at the core interface, potential cooperative effects between multiple drugs present in the core might increase the chance for the drugs to stay there.
In our previous simulations [17], we observed that a cavity with diameter d ≈ 1 nm, present initially in the SSM-90 core, disappeared after the first 1–2 ns of simulations. The core became oblate as the cavity closed up and the alkane tail ends formed an interface-like region. The cavity has the potential to host drug molecules, since the molecular solvation free energies can be reduced over there. The core center is analogous to the middle of a lipid bilayer, where the energy required to form a cavity is decreased, due to the lower density of the alkane tails [15]. This energy reduction could potentially explain the local minimum in ΔG(r) seen to form for a single bexarotene in the SSM-90 core (Fig. 3 (a)). To further investigate the capacity of the cavity to host drugs, we try to accommodate 3 and 5 bexarotene molecules in the initial cavity within the SSM-90 core. Although these drug numbers might be realistic, due to relatively large steric forces within the SSM-90 core, we still need to examine if 11 bexarotene molecules observed in the experimental micelles (Nbex=11) are likely to occupy the SSM core as a cluster.
During the equilibration of systems with multiple drugs, the cavity in the SSM core partially collapses and the drugs reorient into a configuration with inwards pointing -COOH groups, thus forming a molecular cluster held together by a hydrogen bond network (analogue of a small inverse micelle). In Fig. 4 (a), we show the arrangement of 5 bexarotene molecules and the hydrogen bond network formed by them within the alkyl core, after t ≈ 11 ns of equilibration. To examine if bexarotene prefers to remain within the cluster of drug molecules formed in the SSM core rather than at its ionic interface, we calculate ΔG(r) for one of these bexarotene molecules as a function of its distance from the SSM center (drug cluster). In Fig. 3 (a), we show that ΔG(r) develops a deeper minimum in the core center in the presence of multiple bexarotene molecules. Its depth increases with the number of drugs present in the core and for 5 drugs it surpasses the local minimum at the ionic interface. The developed minima in the core can explain the large loading capacity of SSM-90.
Next, we briefly examine whether all the observed (Nbex ≈ 11) bexarotene molecules can possibly fit in the SSM-90 core. In Fig. 4 (b), we show 11 bexarotene molecules equilibrated for t ≈ 17 ns in the SSM-90 core. Nine molecules formed one H-bond network (not anymore a ring), while the remaining two molecules separated from this cluster when their -COOH groups associate with each other. The formation of two hydrogen bond centers indicates potential crowding in the SSM-90 core in the presence of 11 bexarotene molecules. Therefore, the 11 bexarotene drugs are potentially located both in the core and at the ionic interface of SSM-90, where the local minima in free energy are seen to form.
Solubilization of VIP in SSM
In the next example of nanomedicine modeling, we study the solubilization of VIP in SSM. The stabilization of VIP-SSM complexes is based on different principles, since the VIP peptide is chemically very different from bexarotene. Fluorescence measurements of VIP tyrosine residues were used to find the average number of VIP molecules solubilized in SSM in PB and PBS solutions containing DSPE-PEG2000 lipids (clip = 0 – 0.4 mM). In the absence of DSPE-PEG2000, the fluorescence of hydrated VIP molecules is quenched due to their aggregation [37]. On the other hand, when VIP is associated with SSM in its monomeric form, its fluorescence can be observed (increased from its background value). We increased the ratio of DSPE-PEG2000 to VIP in PB and PBS solutions of a constant VIP concentration (cVIP = 4 μM) by changing the DSPE-PEG2000 concentration, and tracked the ratio at which the VIP fluorescence saturated. When this occurred, all the present VIP should have been complexed with SSM, assuming that no aggregated VIP molecules were in the solutions.
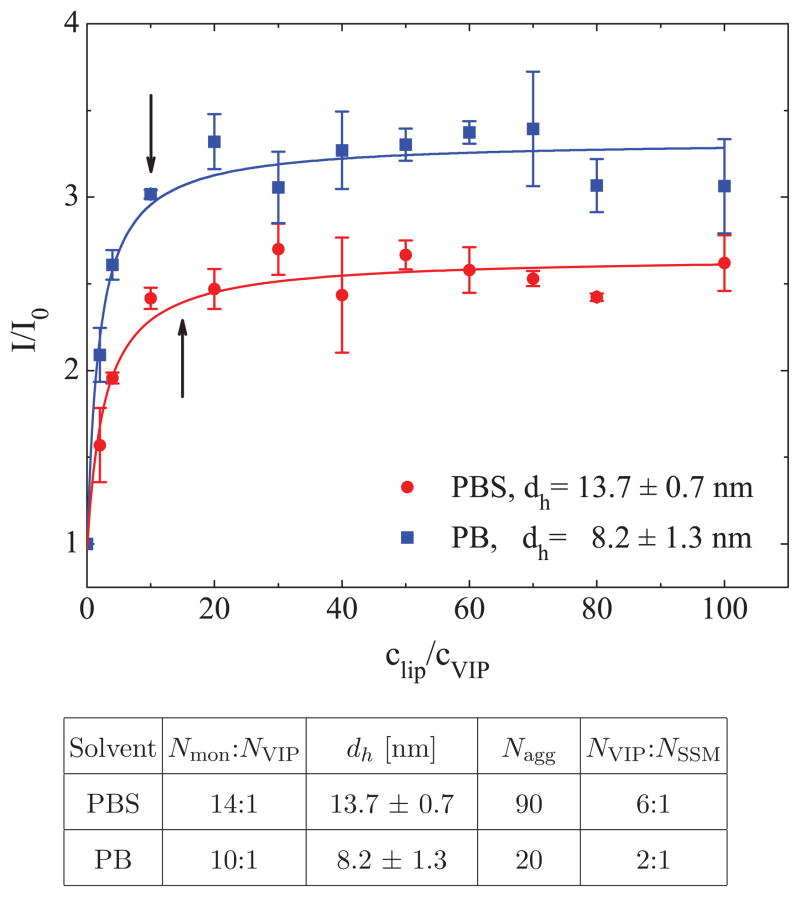
In Fig. 5 (top), we show the ratiometric intensity curves of the PB and PBS solutions containing DSPE-PEG2000 (varying concentration) and VIP (cVIP = 4 μM). The results show that there are ≈ 10 and ≈ 14 DSPE-PEG2000 monomers per each VIP in the PB and PBS solutions, respectively. In the table in Fig. 5 (bottom), we present the DLS hydrodynamic diameters of SSM (dh) in PB and PBS solutions, and inferred SSM aggregation numbers, Nagg (according to the estimate method validated in [17]). The data indicate that SSM in PB solutions have Nagg ≈ 20 (Fig. 1 (b)) and can complex with ≈ 2 VIP molecules, whereas SSM in PBS solutions have Nagg ≈ 90 (Fig. 1 (c)) and can complex with ≈ 6 VIP molecules. The finding that SSM in PBS are significantly larger than those in PB solution is in agreement with our previous finding that SSM size increases with the ionic strength of the solvent [17].
FIG. 5.
(top) The normalized VIP fluorescence intensity, I/I0, as a function of the clip/cVIP ratio, where I and I0 are fluorescence intensities observed for VIP in solutions with and without DSPE-PEG2000 monomers, respectively. A steady-state approximation (arrows) is used to obtain the Nmon:NVIP saturation molar ratio, shown in the table (bottom). The VIP concentration is cVIP = 4 μM, while the concentration of DSPE-PEG2000 is varied. The table also shows dh, determined by DLS, and estimated SSM aggregation numbers, Nagg [17], which are used to determine the VIP loading efficiency (Neff = NVIP/NSSM).
Simulations of VIP-SSM Nanomedicines
The experimental results shown in Fig. 5 indicate that individual SSMs in PB and PBS solutions can stabilize several VIP molecules. In order to reveal the interaction mechanisms leading to VIP stabilization, we simulate the VIP-SSM complexes in solvents of low (water) and high (0.16 M NaCl) ionic strengths. Our model of the VIP peptide has a net +3 charge, with 2 clusters of positively charged residues (Arg-Leu-Arg-Lys and Lys-Lys), two well separated negatively charged residues (two Asp), and the charged C and N termini. While in this work, VIP is stabilized within the SSM in its free form, in our previous experiments, the incorporated VIP was conjugated to a DSPE-PEG3400 into the SSM [38]. To examine the two representative experimental systems, we simulate binding of the free VIP to SSM-20 (in low ionic strength solvent) and binding of the conjugated VIP to SSM-90 (in high ionic strength solvent).
Complexation of VIP with SSM-20
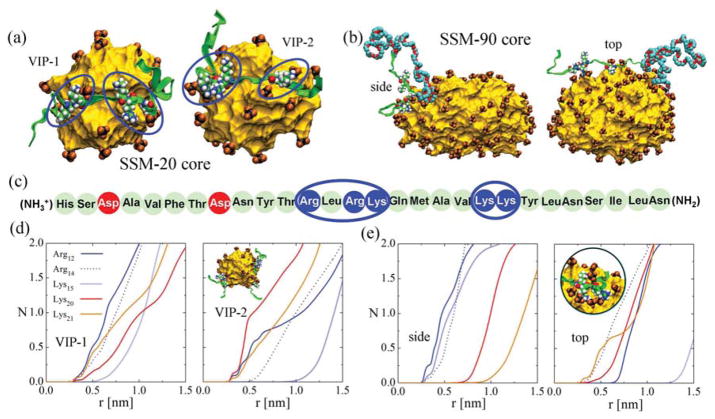
First, we model the complexation of 2 VIP molecules with SSM-20 in water, based on the experimental data for the PB buffer (low ionic strength), shown in Fig. 5. Initially, the two VIP molecules are placed on the opposite sides of SSM-20, within 0.7 nm of its core edge, and the whole system is equilibrated for t ≈ 30 ns. After the first ≈ 10 ns, both VIP molecules become closely coordinated to the groups positioned at the surface of the alkane core, as shown in Fig. 6 (a). The groups migrate primarily towards the two clusters of positively charged residues, and then redistribute more homogeneously on the alkane core surface. The coordination of groups with the positive residues occurs due to their strong Coulombic coupling, which is poorly screened in water (Debye length in 1mM PB solution is λD ≈ 9.7 nm).
FIG. 6.
(a) MD snapshots of VIP-1 and VIP-2 complexed on the opposite sides of SSM-20 (inset (d)). The alkyl core, shown as a yellow surface (PEG corona not shown for clarity), is surrounded by groups (black, orange), which coordinate with two positively charged regions on each of the VIP peptides. VIP molecules are shown as green ribbons, the residues of the positively charged regions are shown in atomic detail. The VIP molecule used in experiments has an aminated His end and an amidated Asn end. In our modeling, the His and Asn ends were C- and N-termini, respectively. (b) SSM-90 core complexed with DSPE-PEG3400-conjugated VIP viewed from the side and from the top. The PEG3400 chain is shown in teal (C-atoms) and red (O-atoms) colors. (c) VIP sequence. Neutral, negatively charged and positively charged residues are marked in light green, red and blue, respectively. (d) The average number N of groups at the distance r from each positively charged VIP-1 and VIP-2 residues (center of mass of the atoms carrying the charge) for SSM-20. The averaging is performed over the last 17 ns of the 30 ns simulations. (e) The same as in (d) for SSM-90. The averaging is performed over the last 7 ns of the 30 ns simulation. (inset) Detail of Arg and Lys coordination to the groups (VIP on the side of the SSM).
In Fig. 6 (d), we show the average number of groups at the distance r from the positive residues of VIP, calculated in VMD [39]. These results reveal that among the 5 positive residues on each VIP, there are at least 3 of them which are almost fully coordinated by counterions (≈ 1 group within r = 0.7 nm of the residue center of mass). The fact that VIP remains in the proximity of the SSM interface during the simulations suggest that Coulombic coupling is the primary VIP-SSM stabilization mechanism.
Complexation of VIP with SSM-90
Besides being a highly potent drug nanocarrier, SSM can also serve as a base for targeting agents. Such targeting can be achieved by grafting the distal end of the PEG moiety of the phospholipid with VIP [38]. Here, we model VIP complexation with SSM-90 in a 0.16 M NaCl solution, where VIP is conjugated to the tail end of the PEG chain on one DSPE-PEG3400 monomer (up to 5 VIP molecules conjugated to the distal end of a PEG-ylated lipid could complex with one micelle [38]). The long PEG chain allows easy accommodation of VIP in the SSM environment. Since the SSM-90 core is oblate [17], we separately model VIP loading along its major (top) and minor (side) axes. The VIP segment of the conjugated monomer is initially placed within 0.7 nm of the micelle core edge and the system is equilibrated for ≈ 30 ns.
During the equilibration of both VIP orientations in SSM-90, the micelle core rearranges to better coordinate with VIP. After the initial readjustment, the groups redistribute more homogeneously and reshape the micelle, as seen in Fig. 6 (b). In the side binding, the core slightly elongates along the major SSM axis. In the top binding, the core stretches along its minor axis and becomes more spherical.
In SSM-90, the VIP is again tightly coordinated by the groups on the surface of the alkane core, despite a higher ionic strength and a more sterically protected environment. In Fig. 6 (e), we show the average number of groups at the distance r from each positively charged VIP residue. Again, most of the VIP residues are surrounded (screened) by the groups at a short distance (r < 1 nm), where the top configuration seems to better accommodate VIP positive residues to the groups. Note that the coupling ratio of several positive residues and the groups is not 1 : 1, but there are pockets of several groups coordinating and stabilizing the residues (inset of Fig. 6 (e), showing VIP stabilized on the side of the SSM-90 core). The large attraction and favorable coordination ratio maintain the VIP position at the ionic interface within the PEG corona. Note that VIP folding within SSM was also experimentally observed [20, 38]. Unfortunately, modeling of VIP folding is currently beyond our simulation times. However, it is very likely that the large negatively charged interfacial area of the SSM will play a significant role in stabilizing the oppositely charged VIP, even if partially folded.
Conclusion
In summary, we examined the solvation of two representative therapeutic molecules, bexarotene and VIP, in PEG-ylated phospholipid SSM nanocarriers. Experimentally, we found that ≈ 11 bexarotene drug molecules are solvated in SSM in HEPES buffered saline, while ≈ 2 and ≈ 6 VIP molecules are solvated in SSMs in the PB and PBS buffers, respectively. Our large-scale atomistic MD simulations and free energy calculations of the BEX-SSM complexes revealed that a single bexarotene molecule preferentially solvates at the ionic interface regions of SSM-10 and SSM-90. Even though a single bexarotene molecule is less stable in the SSM-90 core than at its ionic/alkane interface, we found that the core center can potentially host multiple bexarotene molecules which can help to stabilize each other. We also found that the VIP-SSM stabilization originates in close coordination of VIP positive residues with negatively charged groups capping the micelle core.
The observed stabilization of bexarotene and VIP in the SSM nanocarriers relies on the compatibility of the drug with the polymer building blocks of the SSM. Our simulations have shown that both hydrophobic and Coulombic interactions between the drug molecules and the phospholipid polymers can stabilize the drug molecules in the SSM. Tuning these and other interactions between the drug and the polymer blocks of the nanocarrier can be used to localize the drug in a specific region of the nanocarrier. Our studies clearly illustrate that precise atomistic simulations can bring essential understanding of drug-nanocarrier complexes, which can impact the discovery of future nanomedicines.
Supplementary Material
Acknowledgments
This study was supported in part by grants from the National Institutes of Health (R01AG024026, R01CA12797). We acknowledge the support from the UIC Dean Scholar Award (LV), UIC College of Pharmacy Van Doren Scholarship (AB), UIC Herbert E. Paaren Summer and Academic Year Research Scholarships (AM and AT). The simulations were partly performed at the NERSC, NCSA, and BlueGene (ANL) supercomputers, as well as local GPU servers.
Contributor Information
Petr Král, Email: pkral@uic.edu.
Hayat Onyuksel, Email: hayat@uic.edu.
References
- 1.Brayden DJ. Controlled release technologies for drug delivery. Drug Discov Today. 2003;8:976–978. doi: 10.1016/s1359-6446(03)02874-5. [DOI] [PubMed] [Google Scholar]
- 2.Neslihan Gursoy R, Benita S. Self-emulsifying drug delivery systems (SEDDS) for improved oral delivery of lipophilic drugs. Biomed Pharmacother. 2004;58:173–182. doi: 10.1016/j.biopha.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat Nano. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 4.Duncan R. The dawning era of polymer. Nat RevDrug Discov. 2003;2:347. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 5.Koo OM, Rubinstein I, Onyuksel H. Role of nanotechnology in targeted drug delivery and imaging: a concise review. Nanomedicine. 2005;1:193–212. doi: 10.1016/j.nano.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Torchilin V. Micellar nanocarriers: Pharmaceutical perspectives. Pharm Res. 2007;24:1– 16. doi: 10.1007/s11095-006-9132-0. [DOI] [PubMed] [Google Scholar]
- 7.Lim SB, Banerjee A, Önyüksel H. Improvement of drug safety by the use of lipid-based nanocarriers. J Control Release. 2012;163:34–45. doi: 10.1016/j.jconrel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Rane SS, Anderson BD. What determines drug solubility in lipid vehicles: Is it predictable? Adv Drug Deliv Rev. 2008;60:638–656. doi: 10.1016/j.addr.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliver Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 10.Avdeef A, Testa B. Physicochemical profiling in drug research: a brief survey of the state-of-the-art of experimental techniques. Cell Mol Life Sci. 2002;59:1681–1689. doi: 10.1007/PL00012496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torchilin VP. Structure and design of polymeric surfactant-based drug delivery systems. J Control Release. 2001;73:137–172. doi: 10.1016/s0168-3659(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 12.Allen C, Maysinger D, Eisenberg A. Nano-engineering block copolymer aggregates for drug delivery. Colloids and Surfaces B: Biointerfaces. 1999;16:3–27. [Google Scholar]
- 13.Xiang TX, Anderson BD. Liposomal drug transport: A molecular perspective from molecular dynamics simulations in lipid bilayers. Adv Drug Deliv Rev. 2006;58:1357–1378. doi: 10.1016/j.addr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Yoshii N, Iwahashi K, Okazaki S. A molecular dynamics study of free energy of micelle formation for sodium dodecyl sulfate in water and its size distribution. J Chem Phys. 2006;124:184901–6. doi: 10.1063/1.2179074. [DOI] [PubMed] [Google Scholar]
- 15.Marrink SJ, Berendsen HJC. Simulation of water transport through a lipid membrane. J Phys Chem. 1994;98:4155–4168. [Google Scholar]
- 16.Bemporad D, Luttmann C, Essex J. Behaviour of small solutes and large drugs in a lipid bilayer from computer simulations. BBA-Biomembranes. 2005;1718:1–21. doi: 10.1016/j.bbamem.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Vuković L, Khatib FA, Drake SP, Madriaga A, Brandenburg KS, Král P, Onyuksel H. Structure and dynamics of highly PEG-ylated sterically stabilized micelles in aqueous media. J Am Chem Soc. 2011;133:13481–13488. doi: 10.1021/ja204043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo OM, Rubinstein I, Onyuksel H. Camptothecin in sterically stabilized phospholipid micelles: A novel nanomedicine. Nanomedicine. 2005;1:77–84. doi: 10.1016/j.nano.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Onyuksel H, Mohanty PS, Rubinstein I. VIP-grafted sterically stabilized phospholipid nanomicellar 17-allylamino-17-demethoxy geldanamycin: A novel targeted nanomedicine for breast cancer. Int J Pharm. 2009;365:157–161. doi: 10.1016/j.ijpharm.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sethi V, Rubinstein I, Kuzmis A, Kastrissios H, Artwohl J, Onyuksel H. Novel, bio-compatible, and disease modifying VIP nanomedicine for rheumatoid arthritis. Mol Pharm. 2013;10:728–738. doi: 10.1021/mp300539f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzmis A, Lim SB, Desai E, Jeon E, Lee BS, Rubinstein I, Önyüksel H. Micellar nanomedicine of human neuropeptide Y. Nanomedicine. 2011;7:464–471. doi: 10.1016/j.nano.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim S, Rubinstein I, Sadikot R, Artwohl J, Önyüksel H. A Novel peptide nanomedicine against acute lung injury: GLP-1 in phospholipid micelles. Pharm Res. 2011;28:662–672. doi: 10.1007/s11095-010-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee A, Onyuksel H. Human pancreatic polypeptide in a phospholipid-based micellar formulation. Pharm Res. 2012;29:1698–1711. doi: 10.1007/s11095-012-0718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee A, Onyuksel H. Peptide delivery using phospholipid micelles. WIREs Nanomed Nanobiotechnol. 2012;4:562–574. doi: 10.1002/wnan.1185. [DOI] [PubMed] [Google Scholar]
- 25.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular and modeling dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Lee H, Venable RM, MacKerell AD, Jr, Pastor RW. Molecular dynamics studies of polyethylene oxide and polyethylene glycol: Hydrodynamic radius and shape anisotropy. Biophys J. 2008;95:1590–1599. doi: 10.1529/biophysj.108.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darden T, York D, Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 29.Ashok B, Arleth L, Hjelm RP, Rubinstein I, Önyüksel H. In vitro characterization of PEGylated phospholipid micelles for improved drug solubilization: Effects of PEG chain length and PC incorporation. J Pharm Sci. 2004;93:2476–2487. doi: 10.1002/jps.20150. [DOI] [PubMed] [Google Scholar]
- 30.Arleth L, Ashok B, Onyuksel H, Thiyagarajan P, Jacob J, Hjelm RP. Detailed structure of hairy mixed micelles formed by phosphatidylcholine and PEGylated phospholipids in aqueous media. Langmuir. 2005;21:3279–3290. doi: 10.1021/la047588y. [DOI] [PubMed] [Google Scholar]
- 31.Darve E, Pohorille A. Calculating free energies using average force. J Chem Phys. 2001;115:9169–9183. [Google Scholar]
- 32.Darve E, Rodriguez-Gomez D, Pohorille A. Adaptive biasing force method for scalar and vector free energy calculations. J Chem Phys. 2008;128:144120–13. doi: 10.1063/1.2829861. [DOI] [PubMed] [Google Scholar]
- 33.Henin J, Chipot C. Overcoming free energy barriers using unconstrained molecular dynamics simulations. J Chem Phys. 2004;121:2904–2914. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]
- 34.Krishnadas A, Rubinstein I, Onyuksel H. Sterically stabilized phospholipid mixed micelles: in vitro evaluation as a novel carrier for water-insoluble drugs. Pharm Res. 2003;20:297–302. doi: 10.1023/a:1022243709003. [DOI] [PubMed] [Google Scholar]
- 35.Pearson RM, Patra N, Hsu H-j, Uddin S, Král P, Hong S. Positively charged dendron micelles display negligible cellular interactions. ACS Macro Lett. 2012;2:77–81. doi: 10.1021/mz300533w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menger FM. Laplace pressure inside micelles. J Phys Chem. 1979;83:893–893. [Google Scholar]
- 37.Önyüksel H, Bodalia B, Sethi V, Dagar S, Rubinstein I. Surface-active properties of vasoactive intestinal peptide. Peptides. 2000;21:419–423. doi: 10.1016/s0196-9781(00)00161-3. [DOI] [PubMed] [Google Scholar]
- 38.Dagar A, Kuzmis A, Rubinstein I, Sekosan M, Onyuksel H. VIP-targeted cytotoxic nanomedicine for breast cancer. Drug Deliv and Transl Res. 2012;2:454–462. doi: 10.1007/s13346-012-0107-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.