Abstract
Cellulose waste biomass is the most abundant and attractive substrate for “biorefinery strategies” that are aimed to produce high-value products (e.g. solvents, fuels, building blocks) by economically and environmentally sustainable fermentation processes. However, cellulose is highly recalcitrant to biodegradation and its conversion by biotechnological strategies currently requires economically inefficient multistep industrial processes. The need for dedicated cellulase production continues to be a major constraint to cost-effective processing of cellulosic biomass.
Research efforts have been aimed at developing recombinant microorganisms with suitable characteristics for single step biomass fermentation (consolidated bioprocessing, CBP). Two paradigms have been applied for such, so far unsuccessful, attempts: a) “native cellulolytic strategies”, aimed at conferring high-value product properties to natural cellulolytic microorganisms; b) “recombinant cellulolytic strategies”, aimed to confer cellulolytic ability to microorganisms exhibiting high product yields and titers.
By starting from the description of natural enzyme systems for plant biomass degradation and natural metabolic pathways for some of the most valuable product (i.e. butanol, ethanol, and hydrogen) biosynthesis, this review describes state-of-the-art bottlenecks and solutions for the development of recombinant microbial strains for cellulosic biofuel CBP by metabolic engineering. Complexed cellulases (i.e. cellulosomes) benefit from stronger proximity effects and show enhanced synergy on insoluble substrates (i.e. crystalline cellulose) with respect to free enzymes. For this reason, special attention was held on strategies involving cellulosome/designer cellulosome-bearing recombinant microorganisms.
Keywords: metabolic engineering, butanol, ethanol, hydrogen, cellulosome, cellulase
Construction of microorganisms for consolidated bioprocessings: economic motivation and strategies
Cellulose biomass is the largest waste produced by human activities and the most attractive substrate for “biorefinery strategies” to produce high-value products (e.g. fuels, plastics, enzymes) [1, 2]. The current price for cellulosic biomass, i.e. $50/ metric ton, makes it less expensive than all other energy sources [1]. Nonetheless, plant biomass is highly recalcitrant to biodegradation. No natural microorganisms able of efficient single-step cellulosic biomass fermentation, i.e. consolidated bioprocessings (CBP), into valuable products have been isolated so far. Traditional biomass bioconversion processes require extensive feedstock pre-treatment (e.g. by steam-explosion and/or acid treatment) and the addition of exogenously produced cellulases [3, 4]. From study to study, depending on calculation methods and base-case to be compared, enzyme production impact on the whole bioconversion process cost has been widely differently estimated [5, 6]. However, the cost of added enzyme does not show a decreasing trend over time and still is a major constraint to cost-effective processing of cellulosic biomass [6]. At the low end of recent estimates, i.e. 0.50 $ per gallon ethanol, the cost of added enzymes is comparable to feedstock purchase cost thus eliminating the economic advantage of cellulosic biomass relative to corn [1, 6]. Process cost reduction can be obtained by CBP through simpler feedstock processing, lower energy inputs, higher rates and yields, although, in some cases, economic advantages of CBP might have been over-estimated or evaluated on assumptions that may be difficult to realize [4–7].
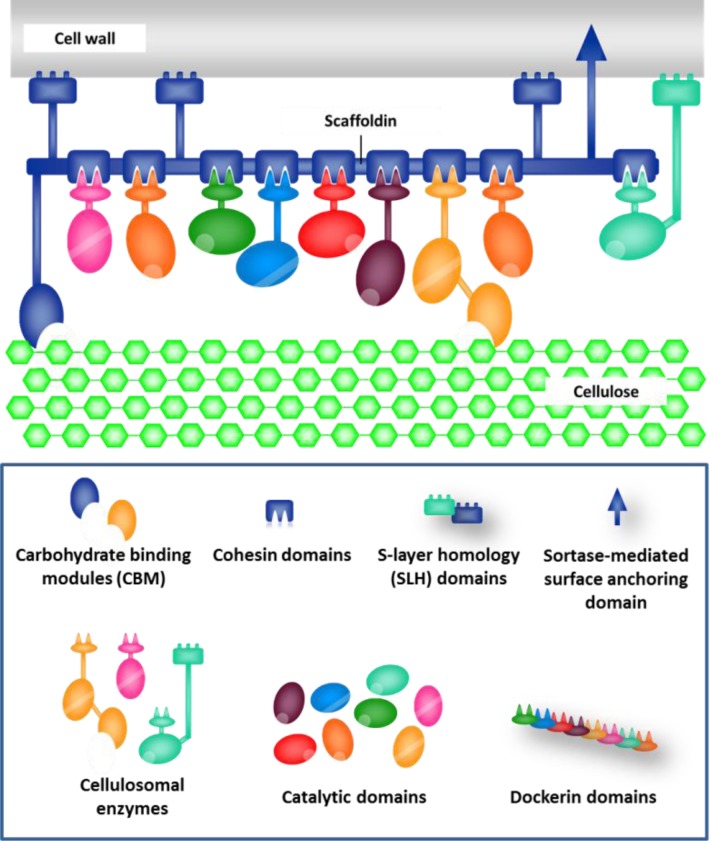
Native plant degrading microorganisms synthesize extracellular multiple enzyme systems that have different substrate specificities (e.g. cellulases, xylanases, pectinases) and catalytic mechanisms (i.e. endoglucanases, exoglucanases, processive endoglucanases and β-glucosidases) [3, 8, 9]. Enzymatic proteins can be either free or physically associated to form complexes called “cellulosomes” [3, 8]. Cellulosomes are typical of anaerobic strains (e.g. Clostridium spp. and Ruminococcus spp.) and are by far the most efficient biochemical systems for cellulose degradation [3, 8, 10–12]. Cellulosome architecture is organized by “scaffoldins”, which are able to recruit catalytic proteins by cohesin-dockerin interactions and improve complex affinity for the substrate via carbohydrate binding domains (CBMs) [3, 8] (Figure 1). In some strains, e.g. Clostridium thermocellum, Clostridium cellulovorans and Ruminococcus flavefaciens, scaffoldins also provide cell wall binding through covalent or non-covalent interactions [8].
Figure 1.
Simplistic model of a cellulosome that includes only one anchoring scaffoldin. The scaffolding protein (blue) binds the enzymatic components through cohesin-dockerin interactions, enhances the cellulosome affinity for cellulose through the CBMs, and anchors the cellulosome complex to the cell surface through either non-covalent (by means of multiple S-layer homology domains) or covalent (mediated by sortases) bonds. Apart from the catalytic domains, cellulosomal enzymes include dockerin modules and, possibly, additional domains (e.g. CBM, SLH) (modified from [60]).
Construction of recombinant microorganisms for CBP can be pursued via two alternative approaches [2–4]. Native cellulolytic strategies (NCSs) aim at introducing and/or improving high-value product biosynthetic pathways into natural cellulolytic strains. The purpose of recombinant cellulolytic strategies (RCSs) is to confer cellulolytic ability to microorganisms with valuable product formation properties and include heterologous cellulase expression. The next sections will describe limitations of each of these approaches and cutting edge solutions that were applied or could be employed in the construction of recombinant cellulolytic strains.
Strain development via native cellulolytic strategies
NCSs are currently hampered by some common limitations. Most native cellulolytic strains have been recently isolated from natural environments and are poorly characterized. Genetic manipulation tools have been set-up for relatively few of them. As far as cellulolytic fungi are concerned, most engineering efforts have been addressed to increasing cellulase production, although there is increasing interest in biofuel production engineering [6, 13–14]. C. thermocellum and C. cellulolyticum are the most established cellulosome-forming microorganisms [6, 15–17]. Furthermore, the metabolism of few strains has been investigated in depth, with C. cellulolyticum as by far the best characterized microorganism [7, 18]. Even in strains with fully sequenced genomes, many genes are annotated as hypothetical while others may have been improperly annotated since their function was deduced on the base of amino acid sequence homology only [7].
In addition, problems connected with the construction of recombinant strains for specific compound production occur. Biomass biorefinery potential for sustainable production of a large spectrum of high value products, such as building block chemicals (e.g. succinic acid, lactic acid, isoprene), higher alcohols, lipidic compounds, fine chemicals (e.g. vitamins, antibiotics), has been extensively reviewed [19–21]. Production of H2, ethanol and butanol has been targeted in this study because of their huge potential as biofuels [1, 2, 7].
Improving hydrogen production in cellulolytic microrganisms
A number of anaerobic cellulolytic bacteria, including several Clostridia (e.g. C. cellulolyticum, C. cellulovorans, C. termitidis and C. thermocellum), Ruminococci (e.g. R. albus) and the extreme thermophile Caldicellulosiruptor saccharolyticus, are able to produce H2 [7, 22]. However, H2 yields obtained by direct cellulose fermentation are usually low because of other metabolic pathways (e.g. producing acids, alcohols and ketones) which compete with proton reduction to H2 for electron donors, i.e. reduced Ferredoxin (Fdred) and/or NAD(P)H [7, 23]. H2 yields of mesophilic cellulolytic bacteria generally range from 1 to 2 mol H2/mole hexose sugar, while values close to the theoretical maximum of 4 mol H2/ mole hexose sugar can be obtained by hyper thermophiles such as C. saccharolyticus [7, 23].
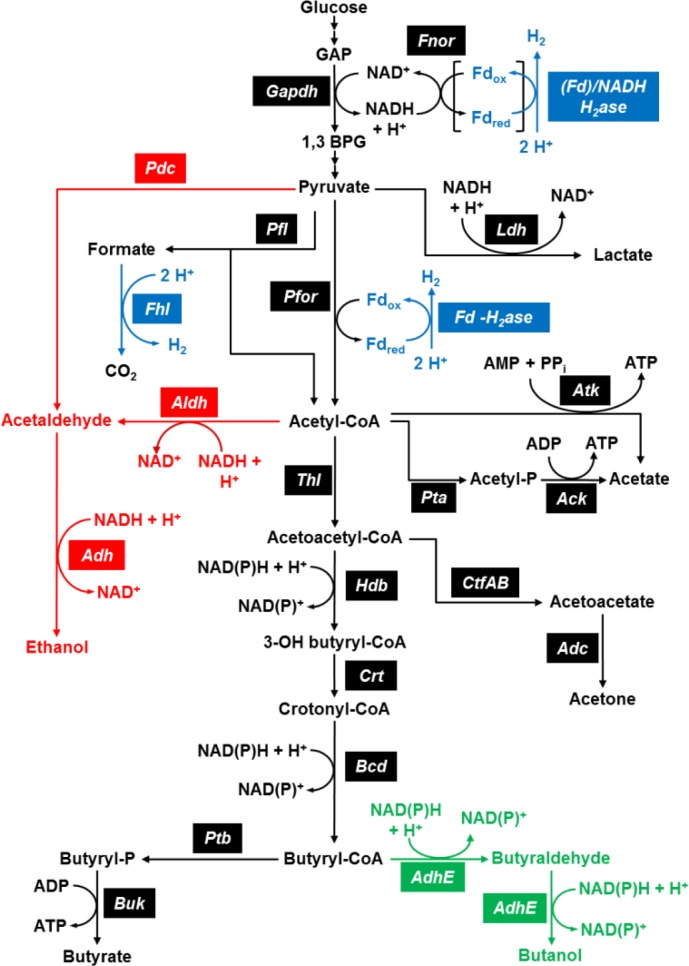
Fermentative pathways either promoting or competing with H2 biosynthesis have mostly been studied in Clostridium sp. [7, 24] and are depicted in Figure 2.
Figure 2.
Overview of Clostridium sp. central metabolic pathways. Pathways for butanol, ethanol and hydrogen production are highlighted in green, red and blue, respectively. Redox reactions involving NAD(P) or Fd and ATP generating reactions have been indicated. Glucose is catabolized through the Embden-Meyerhof route. Formate oxidation to H2 and CO2 by Formate Hydrogen Lyase (Fhl) occurs in enteric bacteria and in some species of Clostridium, although has not been observed in cellulolytic species like C. thermocellum so far [7]. Abbreviations: 1,3 BPG, 1,3 bisphosphoglycerate; Acetyl-P, acetyl phosphate; Butyryl-P, butyryl phosphate; Fd, ferredoxin; Ack, acetate kinase; Adc, acetoacetate decarboxylase; AdhE, aldehyde/alcohol dehydrogenase; Atk, acetate thiotransferase; Bcd, butyryl-CoA dehydrogenase complex; Buk, butyrate kinase; Crt, crotonase; CtfAB, acetoacetyl-CoA:acyl-CoA transferase; Fnor, ferredoxin:NAD(P)+ oxidoreductase; H2ase, hydrogenase; Hbd, 3-hydroxybutyryl-CoA dehydrogenase; Ldh, lactate dehydrogenase; Pfor, pyruvate:ferredoxin oxidoreductase; Pdc, pyruvate decarboxylase; Pta, phosphotransacetylase; Ptb, phosphotransbutyrylase; Thl, thiolase.
H2ases are regarded as the most efficient enzymes catalyzing either proton reduction (i.e. H2-evolving H2ases) or H2 oxidation (i.e. “uptake” H2ases) [24]. Construction of strains with improved H2 production has been carried out by deletion of genes encoding “uptake” H2ases and/or overexpression of H2-evolving enzymes [25–26]. Engineering more efficient H2-evolving proteins has also been proposed to increase H2 yield and/or productivity [27–28]. However, these strategies appear to have limited potential since all H2ases, although preferentially directed, are known to be reversible [7, 29].
Greater H2 yields could be obtained through repression of competing pathways (i.e. producing more reduced end-products than acetate) (Figure 2) [7, 29]. Deletion of ldhA in hemicellulolytic Thermoanaerobacterium sp. either had no effect or increased H2 production by 2-fold, depending on the strain, while repression of acetate production by ack and pta knockout reduced H2 yields by more than 25 fold [30–31]. Recent papers reported repression of lactate and/or acetate production in C. cellulolyticum and C. thermocellum by Ldh or Ack/Pta gene inactivation, respectively, but effects on H2 production were not studied [16, 32–34]. Suppression of butyrate production was recently obtained in C. acetobutylicum and Clostridium butyricum by inactivation of hdb gene encoding 3-hydroxybutyryl-CoA dehydrogenase [35–36]. However, H2 production strongly decreased in both strains. Elimination of ethanol formation alone did not increase H2 production in C. butyricum [37].
Actually, the H2 metabolic network in strict anaerobes is very complicated. A paradigm for this is the high diversity of clostridial hydrogenases and the existence of multiple forms, likely involved in different functions (e.g. redox balancing, derivation of energy from H2 oxidation, proton respiration and/or proton-gradient build-up) within one species [24]. Recently, members of the so called bifurcating hydrogenases have been identified in several clostridia, including cellulolytic strains [24, 38]. Among them, butyryl-CoA dehydrogenases/EtfAB (Bcd/EtfAB) complex from Clostridium kluyveri couples NADH-dependent exergonic reduction of crotonyl-CoA to butyryl-CoA to endergonic reduction of Fd which can be used for H2 production [38]. Discovery of such complex in clostridia provides a clue to H2 yield decrease in strains in which butyrate production was suppressed by Hdb inactivation. More detailed understanding of the metabolic networks involved in H2 production is definitely essential for successful engineering of H2 hyper producing strains.
It is worth to note that, even if 4 mol H2/mole glucose yield was attained, still H2 production from cellulose would not be competitive with other cellulosic biofuels because of lower yields and higher waste by-product disposal [27–28]. Theoretical analyses have suggested that yields close to the stoichiometry maximum of 12 moles H2/mole glucose are possible by redirecting glucose catabolism through the pentose phosphate pathway or by further acetyl-CoA oxidation through citric acid cycle [27–28]. More profound metabolic modification of natural cellulolytic strains will be necessary to assess in vivo feasibility of such strategies.
Challenges for cellulosic ethanol/butanol production by CBP
Metabolic engineering strategies aimed at developing ethanol and/or butanol hyperproducing strains face two main challenges: a) the construction of solvent tolerant strains; b) the achievement of high solvent yield, titer and productivity.
Distinct biochemical systems are generally involved in solvent resistance and biosynthesis. “Titer gap” is defined as the difference between the maximum concentration of a compound that is tolerated when it is added to a culture and the maximum concentration of that compound that is biosynthesized by a strain [6]. The development of C. thermocellum strains able to tolerate added ethanol concentrations exceeding 50 g/l has been reported. However, the maximum ethanol titer produced by this organism remains at about 25 g/l [6]. A number of engineered strains, such as Termoanaerobacterium saccharolyticum, showed solvent production titers exceeding the same solvent tolerance exhibited in exogenous addition experiments [6, 39]. The latter observations provide increasing support that with sufficient effort, stoichiometric yields of engineered products can be achieved.
Development of solvent tolerant strains
The main solvent toxicity is attributed to chaotropic effects on biological membranes [40–41]. Compound toxicity is related to its partition in an equimolar mixture of octanol and water, i.e. log P [40]. The higher is solvent polarity the lower is log P. Molecules with log P below 1 or above 4 are scarcely toxic since they are too hydrophilic to enter the membranes or too hydrophobic and therefore not bioavailable, respectively [40]. In this respect, n-butanol (log P 1) is more toxic than ethanol (log P = -0,18). Even in native solvent producers, such as C. acetobutylicum, 50% growth inhibition occur for butanol concentration as high as 7–13 g/l and metabolism ceases once solvent reaches 20 g/l [41–42]. Continuous extraction of solvents form the culture medium or two-phase (organic-aqueous) fermentation systems can be employed to overcome solvent toxicity, but they increase industrial process complexity and/or cost [39–40, 42]. The development of strains with superior tolerance features is therefore essential for sustainable production of biofuels [41].
Solvent accumulation within biological membranes increases membrane fluidity and negatively affects membrane processes, e.g. energy generation and nutrient transport [40]. Moreover, solvents may cause protein and RNA unfolding and degradation and DNA and lipid damage [40–41]. In this respect, proteins involved in cellulose hydrolysis are less affected than cells by high solvent concentration and cellulosomes appear less sensitive than free cellulases [43–44]. In response, cells induce complex stress mechanisms that include alterations in cell envelope composition, biosynthesis of heat-shock proteins and solvent active transporters (i.e. efflux pumps) and changes in cell size and shape [40–41]. However, the activation of solvent resistance systems increases cell energy expenditure. High energy costs are associated with efflux pumps and repair or re-synthesis of damaged macromolecules [40–41]. The consequences of such system activation on cell energy balance should be included in theoretical calculations of maximum solvent production yields.
Development of solvent tolerant strains has been performed by different strategies. In some cases solvent tolerant mutants also showed increased solvent production. Utilization of random mutagenesis by chemical or physical methods or by using transposable genetic elements has been reported [41]. C. beijerinckii BA101, a butanol-tolerant mutant obtained by chemical methods, showed cell inhibition at 23 g/l butanol rather than 11 g/l typical of the wild-type (WT) strain as well as improved solvent production [42]. An alternative strategy relies on overexpression of proteins involved in solvent resistance (e.g. heat-shock proteins, efflux pumps, enzymes changing membrane lipid composition) [45–47]. The overexpression of GroES and GroEL in C. acetobutylicum resulted in 85% less growth inhibition by butanol and 30% improved butanol production [45]. Global approaches, e.g. the construction of genomic and deletion libraries and the utilization transcriptomic and proteomic techniques, have been employed so as to expand the number of genes identified as involved in solvent tolerance [41]. The construction of a C. acetobutylicum genomic library led to the identification of 16 genes contributing to butanol tolerance [48]. The overexpression of one of them, i.e. CAC1869, in C. acetobutylicum resulted in 81% increase in cell density in a butanol-challenged cultures. A more straightforward approach to achieve solvent tolerant phenotype is in vivo directed evolution under selective pressure. Cellulolytic C. thermocellum strains able to tolerate ethanol concentrations as high as 80 g/l were developed by adaptation approaches [49–50]. Whole genome shuffling (WGS) can be used to improve phenotypes obtained through random mutagenesis and/or in vivo evolution [41]. Applications of WGS for improving butanol tolerance have recently been reported [51–52]. C. acetobutylicum DSM 1731 mutants able to tolerate up to 19 g/l butanol were isolated, although butanol titers obtained by batch fermentations using this strain did not exceed 15.3 g/l [52].
The same approaches could be successfully applied for developing other ethanol tolerant or butanol resistant cellulolytic strains.
Engineering efficient ethanol production in cellulolytic microorganisms
Several cellulolytic bacteria, such as Clostridium sp. (e.g. C. thermocellum and C. cellulolyticum), and fungi, such as Rhizopus, Aspergillus, Neocallimastix, and Trichoderma, can synthesize ethanol although their yields and/or titers and /or productivities are insufficient for direct utilization in CBP [6–7, 14]. Ethanol can be produced from pyruvate via two pathways (Figure 2): (i) pyruvate oxidative decarboxylation by Pfor and subsequent acetyl-CoA reduction to acetaldehyde and finally to ethanol; (ii) the pyruvate decarboxylase (Pdc) catalyzed conversion of pyruvate to acetaldehyde which is reduced by alcohol dehydrogenase (Adh) (Fig. 2). Clostridia generally employ the first pathway, however, a Pdc gene has been identified on the pSOL1 megaplasmid of C. acetobutylicum [36].
Rational metabolic engineering to increase ethanol yield and purity has been performed by two main strategies: introduce heterologous gene, and disrupt genes involved in by-product formation that compete with ethanol synthesis [34].
The first strategy, by employing the expression of Zymomonas mobilis Pdc and Adh genes, was applied to C. cellulolyticum [53]. Yet, acetate was the main end-product of the recombinant C. cellulolyticum. As compared to the wild type strain, acetate production was improved by 93% whereas final ethanol concentration was increased by 53% only [53].
Significant ethanol yield improvement was recently obtained by targeted gene disruption. The inactivation of a single gene, i.e. hdb, involved in butyrate synthesis in C. acetobutylicum, led to a strain with impressive ethanol yield (i.e. 0.38 g/g of glucose) and productivity (i.e. 0.5 g/l/h) [36]. Fed-batch cultures of the engineered C. acetobutylicum resulted in final ethanol amount of 33 g/l [36]. Ethanol titer obtained by a hdb-deficient C. butyricum was 18-fold higher that in the WT strain [35]. hdb deletion could be applied for improving ethanol production in butyrate producing cellulolytic clostridia, such as C. cellulovorans and C. thermopapyrolyticum [54].
Repression of acetate biosynthesis by inactivation of Pta and/or Ack has been suggested as a key modification for driving pyruvate flux towards ethanol [16]. However, disruption of the pta gene in C. thermocellum did not increase final ethanol amounts and led to severe growth deficiency as concerns both growth rate and final biomass [16]. Actually, acetyl-CoA conversion to acetate is a key pathway for metabolic energy production via SLP in clostridia [7, 35]. Indeed, attempts to construct pta or ack inactivated C. cellulolyticum strains were unsuccessful, so far [34].
Strategies employing Ldh disruption were more successful. A C. cellulolyticum H10 double mutant, where both Ldh paralogs, i.e. Ccel_2485 and Ccel_0137, were disrupted showed remarkable production of 0.27 g of ethanol per g of crystalline cellulose [34]. Similar ethanol yields from crystalline cellulose were obtained with a C. thermocellum strain that was constructed by both ldh and pta disruption [32]. However, maximum reported titers (5.61 g/l) remain low for this strain application to CBP [32].
Impressive results were obtained by deletion of ack, ldh and pta in the hemicellulolytic T. saccharolyticum [30]. The engineered strained showed homoethanologenic metabolism with maximum ethanol productivity and titer up to 2.2 g/l/h and 65 g/l, respectively [6, 30].
Alternative strategies for engineering butanol production in cellulolytic strains
All natural butanol producers belong to the genus Clostridium. The highest butanol amounts are synthesized by C. acetobutylicum, C. beijerinckii, C. saccharobutylicum, and C. saccharoperbutylacetonicum [39]. Development of C. acetobutylicum or C. beijerinckii strains with improved butanol production (i.e. titers up to 19 g/l) by random mutagenesis or rational metabolic engineering was reported [41, 45]. However, none of these strains can directly ferment cellulose. Few cellulolytic clostridia producing very low butanol amounts include four recently isolated strains [54–55]. By developing effective gene manipulation tools, butanol production in these microorganisms could be improved by applying strategies that were previously set up in more established butanol producers.
The expression of the clostridial butanol biosynthetic pathway in heterologous hosts, such as native cellulolytic bacteria, seems an alternative promising strategy. The whole C. acetobutylicum butanol pathway, i.e. thl, crt, bcd, etfB, etfA, bcd, and adhE genes (Figure 2), was introduced in well-established and/or solvent tolerant heterologous hosts (e.g. E. coli and Pseudomonas putida), but the highest reported butanol titers, i.e. by E. coli BUT2, were 1184 mg/l [56–57]. Inefficient or imbalanced heterologous gene expression and low catalytic efficiency of some C. acetobutilicum enzymes (i.e. thiolase and butyryl-CoA dehydrogenase), have been hypothesized as the main causes of such low butanol production [57–58]. A chimeric butanol biosynthetic pathway was constructed in E. coli by assembling genes from three different organisms [58]. The clostridial Bcd was replaced by Treponema denticola trans-enoyl-CoA reductase (Ter). Both enzymes catalyze crotonyl-CoA reduction to butyryl-CoA, but Ter reaction is more exoergonic since it does not involve concomitant Fd reduction. Anaerobic fed-batch cultures of recombinant E. coli resulted in the impressive production of 15 g/L of butanol [58].
Strategies for efficient expression of the C. acetobutylicum butanol biosynthetic pathway in other clostridium sp. hosts, such as C. tyrobutyricum, might be less complex [59]. Since C. tyrobutyricum possesses its own butyrate biosynthetic pathway, the introduction of the C. acetobutylicum acetaldehyde/alcohol dehydrogenase AdhE2 and either ptb or ack inactivation, significantly diverted carbon flux from acetate and butyrate to butanol. About 10 g/l butanol was obtained by glucose fermentation [59]. This strategy could be successfully applied to other butyrate producers, such as cellulolytic C. cellulovorans.
For a long time only the clostridial route to butanol synthesis has been known, but recently the in vivo construction of alternative pathways has been reported [19, 39]. Direct conversion of crystalline cellulose to isobutanol was performed by a modified C. cellulolyticum by the introduction of an engineered valine biosynthetic pathway [15] (Figure 3).
Figure 3.
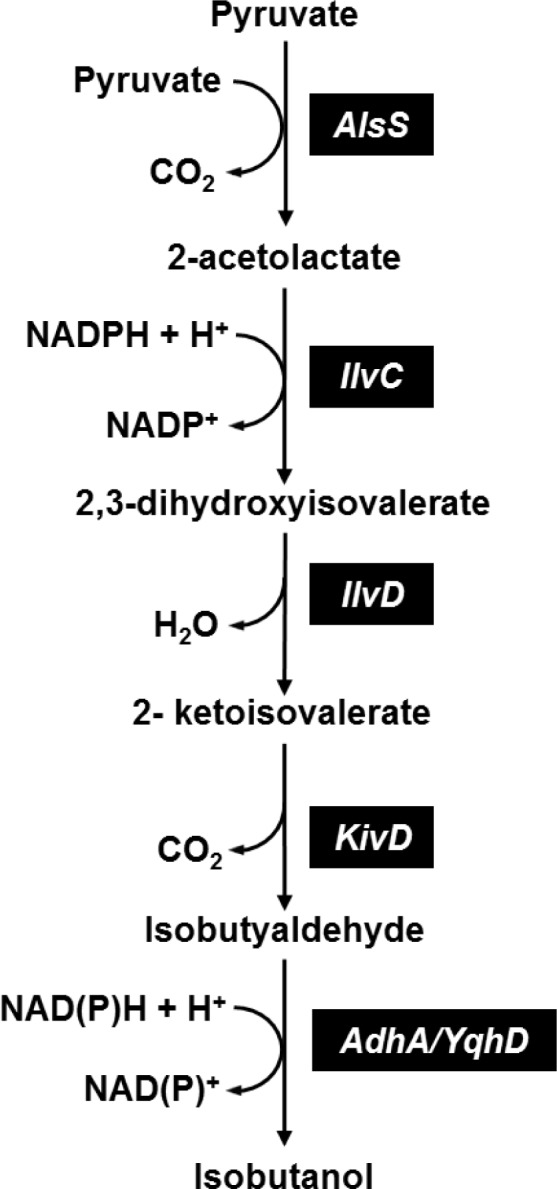
Synthetic pathway for isobutanol production in C. cellulolyticum [15]. Abbreviations: AlsS, B. subtilis a-acetolactate synthase; IlvC, E. coli acetohydroxyacid isomeroreductase; IlvD, E. coli dihydroxy acid dehydratase; KivD, L. lactis ketoacid decarboxylase; AdhA/YqhD E. coli and L. lactis alcohol dehydrogenases.
Recombinant cellulolytic strategies: efficient heterologous cellulase system expression and beyond
The secretion of efficient designer cellulase systems in heterologous hosts is among the most challenging tasks of RCSs [60]. The strategies for both i) the design of efficient artificial cellulase systems and ii) their efficient secretion in strains with product formation features will be detailed in the next sections. In-depth analyses of natural cellulolytic microorganisms metabolism provide further insights for improving recombinant strains by central metabolic pathway engineering, as described in a further section.
Design of efficient artificial cellulase systems: from nature to application
Minimal enzyme systems able to catalyze efficient cellulose hydrolysis contain at least an exoglucanase (Exg, i.e. either a cellodextrinase, EC 3.2.1.74, or a cellobiohydrolase, EC 3.2.1.91), an endoglucanase (Eng, EC 3.2.1.4) and a β-glucosidase (Bgl, EC 3.2.1.21) [3]. Yeast strains secreting a Bgl, a Exg and a Eng were able to directly ferment pretreated Whatman paper to ethanol with yields up to 94% of the theoretical maximum [61]. If RCSs involving heterologous expression of complexed cellulases (i.e. minicellulosomes or designer cellulosomes) are concerned, the additional expression of a scaffolding protein that consists of at least two cohesins is required for functional complex assembly [3].
In both free-cellulase and cellulosome biosynthesizing native organisms, optimal biomass degradation is obtained by secretion of non-equimolar ratios of different protein components with Exgs generally among the most abundant enzymatic subunits [3]. Indeed, a Saccharomyces cerevisiae strain that was engineered by introduction of two heterologous cellobiohydrolases only was able to hydrolyze up to 6 g/l of crystalline cellulose in 168 hours [62]. However, an in depth rationale able to explain and possibly predict which enzyme partners can act with the highest synergism degree is currently unavailable. Such information is essential to design optimized mixtures containing the minimal number of enzymatic subunits enabling efficient cellulose hydrolysis.
Exgs generally have a tunnel-shaped active site which retains a single glucan chain and prevents it from re-adhering to the cellulose crystal, thus enabling them to catalyze processive crystalline cellulose degradation from either the reducing or non-reducing end [9, 63]. Engs instead have cleft-shaped open active site which can cleave internal bonds of amorphous cellulose only [9, 63]. However, processive Engs, which are active on crystalline cellulose also, have also been discovered. Most processive Engs consist of a family 9 catalytic domain attached to a family 3c CBM [9, 649]. CBMs promote cellulase stable binding to cellulose, yet they allow the enzymes to diffuse along the cellulose chain. In some cases, CBM ability to disrupt non-covalent interactions between cellulose chains of crystalline cellulose has been demonstrated [63, 65].
A number of experimental observations indicate that cellulosomes are more effective than free enzymes, with special regards to insoluble (i.e. crystalline) cellulose hydrolysis, likely because the improved proximity enhances enzyme synergism [10–12]. However, in-depth understanding of the mechanisms that drive protein assembly and spatial organization in such complexes is still incomplete [8–9]. Cellulosome-biosynthesizing microorganisms adapt complex composition to the available substrate(s) and assembly non-equimolar ratios of the different subunits for optimal substrate degradation [66–68]. Cellulosome composition likely depends on both the relative amounts of available subunits and their differential affinity for cohesin domains, but with different extents depending on the microbial strain [69]. While within C. cellulolyticum and C. thermocellum cohesin-dockerin interaction seems to be non-selective or characterized by slightly different dissociation constants, up to 100-fold differences in binding affinities have been observed in C. josui and C. cellulovorans [11, 69].
Recent studies showed that linker regions that connect cohesins in scaffoldins are crucial for cellulosome plasticity and catalytic efficiency [70–71]. Linker flexibility enables cellulosome conformation to adapt to the substrate and allows glycosyl hydrolases (GHs) to work in close synergism through proximity effect [70–71]. Linker flexibility and length appear key factors mainly for very complicated and cell-bound cellulosomes, that likely need more extensible conformations [71]. Recent studies suggest that CBM3s, apart from promoting cellulosome binding to the substrate, could also induce conformational changes in the quaternary structure of cellulosomes through direct interaction with linker segments [72]. Cohesin–dockerin dual binding mode, i.e. the ability of dockerin-containing proteins to bind the cognate cohesin by two different orientations, also contribute to complex plasticity [73].
Which catalytic efficiency on native substrates can be expected for minicellulosomes with respect to natural complexes? Experimental evidences suggest an almost linear correlation between the number of cohesins that are beared by a scaffoldin and the activity on crystalline cellulose [11, 74]. The specific activity on crystalline cellulose of C. thermocellum minicellulosomes assembled by three-cohesin bearing mini-scaffoldin CipA was about 38% as compared to complexes containing the full-length CipA, which consists of 9 cohesins [11]. However, specific activity of designer cellulosomes or cellulase mixtures can be significantly increased by enzymatic components with superior activity either selected among the continuously increasing number of newly isolated GHs or developed through directed evolution or rational design [9, 12].
Strain engineering with designer cellulase systems
Cellulolytic aerobic fungi (e.g., Trichoderma reesei) usually secrete high amounts (i.e. >1 to 10 g/l) of GHs. Although cellulosome-forming microorganisms biosynthesize lower cellulase levels (i.e. 0.1 g/l), cellulase amount as high as 10 to 20% (w/w) of whole cellular proteins was estimated in C. thermocellum [12].
State-of-the-art molecular biology enables the production of large amounts of cellulases in heterologous hosts by choosing one among the several mechanisms, at either mRNA or protein level, that regulate gene expression in microorganisms [60, 62].
Heterologous cellulase gene expression under the control of constitutive transcriptional promoters appears the most appropriate for strains aimed to biorefineries, since it avoids the non negligeable supplemental cost of large amounts of specific inducers [60]. By randomized or combinatorial methods, libraries of transcriptional promoters showing strengths within a range of three orders of magnitude can be easily constructed [75]. Improvement of mRNA stability and translation efficiency can be used as further tools to increase the expression of heterologous cellulases (for extensive review refer to [60])
A more challenging task of RCSs is the coordinated expression of multiple heterologous genes that are required for efficient cellulose degradation [60, 76]. Since cellulase system optimal activity is obtained for non-equimolar ratios of the different components, the use of multiple transcriptional units under different promoter control will probably be required. As detailed understanding of mRNA processing and post-trascriptional mechanisms in microorganisms is increasing, more elegant systems, e.g. fine tuning of artificial polycistronic operons by differential RNA stability and/or translation efficiency in bacteria, will be probably available. The design of artificial multifunctional GHs and/or “covalent” cellulosomes could provide efficient cellulose hydrolysis without the need of coordinated multiple gene expression [77]. The engineering of cellulases with superior activity on native substrates could also compensate for low secretion yields.
The main concern of RCSs is to find efficient and reliable secretion methods. The products of genes coding for clostridial cellulosomal components including their original signal peptide, were efficiently secreted by C. acetobutylicum and Lactobacillus plantarum [60, 78]. In other cases, efficient cellulase secretion was promoted by the replacement of original signal peptides by either signal peptides of efficiently secreted autologous proteins or optimized synthetic sequences [79–80]. Nowadays, insufficient understanding of high complexity and specificities among different microorganisms in protein secretion mechanisms, severely limits the number of targeted approaches that can be used for improving heterologous protein secretion [60]. We are currently unable to predict if a cellulase will be secreted in high amounts in a recipient strain or it will result in saturation of membrane translocation mechanisms and cell toxicity. Nonetheless, significant progress has been achieved by trial and error approaches, as well documented by studies on C. acetobutylicum and S. cerevisiae [62, 78, 81]. First attempts to express C. cellulolyticum Cel48F and Cel9G in C. acetobutylicum were unsuccessful. C. acetobutylicum deficiency of specific chaperone(s) that maintain family 48 and 9 GHs in a competent state for translocation was hypothesized [78]. However, Cel48F/Cel9G engineering with CBM3a and X2 modules of the C. cellulolyticum CipC scaffoldin prevented toxic effects and triggered enzyme secretion in C. acetobutylicum [81]. Prior to this study, the function of X2 domains was unknown. By selecting the most efficiently secreted enzymes from a large panel of heterologous Exgs, recombinant S. cerevisiae secreting up 1 g/l of cellobiohydrolases could be engineered [62]. Secreted heterologous cellulase amount was estimated as high as 4% of total cell protein of the recombinant S. cerevisiae, demonstrating that with sufficient efforts secretion of cellulase levels which are comparable to those observed in native cellulolytic strains is possible. Selected strains from Kluyveromyces spp. and S. cerevisiae expressing a library of cellulases were able to directly convert crystalline cellulose up to 0.4-0.5 g/l of ethanol without any externally added enzyme [82]. Furthermore, cultures of such engineered strain were able to ferment crystalline cellulose to ethanol with 30% of the maximum theoretical yield, when supplemented with commercial β-glucosidase [62].
In order to avoid the hydrolysis of the heterologously expressed cellulases, utilization of protease inactivated strains, such as B. subtilis WB800 and L. lactis HtrA mutants, may be required [79, 83].
Microbial cell surface binding enhances cellulase activity [12, 79]. Higher activity of cell-bound as respect to cell-free cellulosomes is obtained by limited escape of hydrolysis products and minimal distance products must diffuse before the cellular uptake occurs [12, 79]. The effect of such improved synergism is particularly evident on crystalline cellulose as compared with amorphous substrate degradation [12].
So far, designer cellulosomes binding up to 3 catalytic subunits have been functionally displayed on the surface of engineered microbial hosts. Such minicellulosomes have covalently been linked to the cell wall of the yeast S. cerevisae by means of agglutinin/flocculin display system [68, 84]. Trifunctional-minicellulosome-displaying S. cerevisiae was able to ferment amorphous cellulose to ethanol with 62% of the theoretical yield [84]. A non-covalent surface display system for lactic acid bacteria has been developed by target protein fusion with the C-terminal cA peptidoglycan binding domain of the major autolysin AcmA from L. lactis [60]. Fragments of the scaffolding protein CipA of C. thermocellum have covalently been anchored at the surface of L. lactis by fusing them with the C-terminal anchor motif of the streptococcal M6 protein, a sortase substrate [79]. A similar strategy was used to covalently link engineered C. thermocellum scaffoldins and cellulases to the B. subtilis cell wall [83]. Higher amounts of surface displayed constructs, i.e. about 3 x 105 per cell, were estimated in engineered B. subtilis [83]. Recently, a designer cellulosome consisting of two scaffoldins, one involved in catalytic component binding and the other mediating cell-surface anchoring, was expressed in S. cerevisiae to improve complex-display level [85]. The recombinant strain was able to directly ferment crystalline cellulose to ethanol. Although the reported yields are low, as far as I know this is the first microbial strain able to biosynthesize by itself functional minicellulosomes enabling significant crystalline cellulose hydrolysis.
Lessons from natural cellulolytic microorganisms
Research on native cellulolytic strains, suggests that cellulose hydrolysis is not the only bottleneck of cellulose metabolism [18]. As compared with soluble sugar metabolizing bacteria, anaerobic cellulolytic bacteria have limited carbon consumption rates and growth capabilities, which raise some concerns about maximum productivity that could be obtained by industrial cellulose bioprocessing.
However, improved conversion efficiencies by recombinant microorganisms developed by RCSs, could be obtained by introducing cellodextrin membrane transporters [86]. The uptake of cellulose hydrolysis product by cellulolytic microorganisms mainly consists in the transport of cellodextrins with a polymerization degree up to 7 which are degraded into the cytoplasm by phosphorolytic cleavage. Both cellodextrin uptake and phosphorolytic cleavage contribute to high bioenergetic benefits of cellulose with respect to glucose or cellobiose metabolism in native organisms [18]. These activities could be engineered in heterologous hosts for optimized valuable product yields and productivity by CBP.
Metabolic flux analysis could be an essential tool to identifying further bottlenecks of cellulose catabolism in native cellulolytic microorganisms and improve recombinant strains by rational engineering of central metabolic pathways. Alternatively, evolutionary engineering strategies by continuous culture under selective pressure could be applied to optimize cellulose overall metabolism in recombinant microorganisms [60].
Conclusions and future perspectives
Both native recombinant strategies and recombinant cellulolytic strategies have made considerable progress. Outstanding results include the construction of C. cellulolyticum and C. thermocellum strains able to ferment crystalline cellulose to ethanol with yields close to 60% of the theoretical maximum and free cellulase-secreting or minicellulosome-displaying yeasts able to directly convert crystalline cellulose to ethanol [32, 34, 82, 85]. Yet, such strains are far to meet the yields, titers and productivities that are required for economically sustainable cellulose CBPs.
Rational engineering of biological systems so as to reach the high performances that are demanded by industrial processes will probably require the use of computational methods which can integrate: gene network regulation data; detailed information on in vivo enzyme catalytic parameters and metabolic fluxes; bioenergetics parameters (e.g. the energy demand of solvent tolerance mechanisms or cellulase biosynthesis, and biological reaction thermodynamics). Furthermore, this information will enable synthetic biology strategies to design new metabolic pathways for the conversion of cellulosic biomass into a virtually unlimited number of valuable products [39, 67].
Acknowledgements
Many thanks to Prof. Enrica Pessione for helpful discussion and critical reading of the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
References
- 1.Lynd LR, Laser MS, Bransby D, Dale BE, Davison B, et al. (2008) How biotech can transform biofuels. Nat Biotechnol 26: 169–172 [DOI] [PubMed] [Google Scholar]
- 2.Alper H, Stephanopoulos G (2009) Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential?. Nat Rev Microbiol 7: 715–723 [DOI] [PubMed] [Google Scholar]
- 3.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66: 506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynd LR, van Zyl WH, McBride JE, Laser M (2005) Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol 16: 577–583 [DOI] [PubMed] [Google Scholar]
- 5.Klein-Marcuschamer D, Oleskowicz-Popiel P, Simmons BA, Blanch HW (2012) The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol Bioeng 109: 1083–1087 [DOI] [PubMed] [Google Scholar]
- 6.Olson DG, McBride JE, Shaw AJ, Lynd LR (2012) Recent progress in consolidated bioprocessing. Curr Opin Biotechnol 23: 396–405 [DOI] [PubMed] [Google Scholar]
- 7.Levin DB, Carere CR, Cicek N, Sparkling R (2009) Challenges for biohydrogen production via direct lignocellulose fermentation. Int J Hydrogen Energy 34: 7390–7403 [Google Scholar]
- 8.Bayer EA, Lamed R, White BA, Flint HJ (2008) From cellulosomes to cellulosomics. Chem Rec 8: 364–377 [DOI] [PubMed] [Google Scholar]
- 9.Wilson DB (2011) Microbial diversity of cellulose hydrolysis. Curr Opin Microbiol 14: 259–263 [DOI] [PubMed] [Google Scholar]
- 10.Blanchette C, Lacayo CI, Fischer NO, Hwang M, Thelen MP (2012) Enhanced cellulose degradation using cellulase-nanosphere complexes. PLoS One 7: e42116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krauss J, Zverlov VV, Schwarz WH. (2012) In vitro reconstitution of the complete Clostridium thermocellum cellulosome and synergistic activity on crystalline cellulose. Appl Environ Microbiol 78: 4301–4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.You C, Zhang XZ, Sathitsuksanoh N, Lynd LR, Zhang YH (2012) Enhanced microbial utilization of recalcitrant cellulose by an ex vivo cellulosome-microbe complex. Appl Environ Microbiol 78: 1437–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B (2009) Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina . Biotechnol Biofuels 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Q, Singh A, Himmel ME (2009) Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr Opin Biotechnol 20: 364–371 [DOI] [PubMed] [Google Scholar]
- 15.Higashide W, Li Y, Yang Y, Liao JC (2011) Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl Environ Microbiol 77: 2727–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, et al. (2010) Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbiol 76: 6591–6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuehne SA, Heap JT, Cooksley CM, Cartman ST, Minton NP (2011) ClosTron-mediated engineering of Clostridium . Methods Mol Biol 765: 389–407 [DOI] [PubMed] [Google Scholar]
- 18.Desvaux M (2006) Unravelling carbon metabolism in anaerobic cellulolytic bacteria. Biotechnol Prog 22: 1229–1238 [DOI] [PubMed] [Google Scholar]
- 19.Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451: 86–89 [DOI] [PubMed] [Google Scholar]
- 20.Octave S, Thomas D (2009) Biorefinery: Toward an industrial metabolism. Biochimie 91: 659–664 [DOI] [PubMed] [Google Scholar]
- 21.Adkins J, Pugh S, McKenna R, Nielsen DR (2012) Engineering microbial chemical factories to produce renewable “biomonomers”. Front Microbiol. 3: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ntaikou I, Koutros E, Kornaros M (2009) Valorisation of wastepaper using the fibrolytic/hydrogen producing bacterium Ruminococcus albus . Bioresour Technol 100: 5928–5933 [DOI] [PubMed] [Google Scholar]
- 23.Oh YK, Raj SM, Jung GY, Park S (2011) Current status of the metabolic engineering of microorganisms for biohydrogen production. Bioresour Technol 102: 8357–8367 [DOI] [PubMed] [Google Scholar]
- 24.Calusinska M, Happe T, Joris B, Wilmotte A (2010) The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 156: 1575–1588 [DOI] [PubMed] [Google Scholar]
- 25.Nakayama SI, Kosaka T, Hirakawa H, Matsuura K, Yoshino S, et al. (2008) Metabolic engineering for solvent productivity by downregulation of the hydrogenase gene cluster hupCBA in Clostridium saccharoperbutylacetonicum strain N1–4. Appl Microbiol Biotechnol 78: 483–493 [DOI] [PubMed] [Google Scholar]
- 26.Jo JH, Jeon CO, Lee SY, Lee DS, Park JM (2010) Molecular characterization and homologous overexpression of [FeFe]-hydrogenase in Clostridium tyrobutyricum JM1. Int J Hydrogen Energy 35: 1065–1073 [Google Scholar]
- 27.Abo-Hashesh M, Wang R, Hallenbeck PC (2011) Metabolic engineering in dark fermentative hydrogen production; theory and practice. Bioresour Technol 102: 8414–8422 [DOI] [PubMed] [Google Scholar]
- 28.Hallenbeck PC, Abo-Hashesh M, Ghosh D (2012) Strategies for improving biological hydrogen production. Bioresour Technol 110: 1–9 [DOI] [PubMed] [Google Scholar]
- 29.Hallenbeck P.C. (2009) Fermentative hydrogen production: principles, progress, and prognosis. Int J Hydrogen Energy 34: 7379–7389 [Google Scholar]
- 30.Shaw AJ, Podkaminer KK, Desai SG, Bardsley JS, Rogers SR, et al. (2008) Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci U S A 105: 13769–13774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S., Lai C, Cai Y, Yang X, Yang S, et al. (2010) High efficiency hydrogen production from glucose/xylose by the ldh-deleted Thermoanaerobacterium strain. Bioresour Technol 101: 8718–8724 [DOI] [PubMed] [Google Scholar]
- 32.Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, et al. (2011) High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol 77: 8288–8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui GZ, Hong W, Zhang J, Li WL, Feng Y, et al. (2012) Targeted gene engineering in Clostridium cellulolyticum H10 without methylation. J Microbiol Methods 89: 201–208 [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Tschaplinski TJ, Engle NL, Hamilton CY, Rodriguez M Jr, et al. (2012) Combined inactivation of the Clostridium cellulolyticum lactate and malate dehydrogenase genes substantially increases ethanol yield from cellulose and switchgrass fermentations. Biotechnol Biofuels. 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai G, Jin B, Saint C, Monis P (2011) Genetic manipulation of butyrate formation pathways in Clostridium butyricum . J Biotechnol 155: 269–274 [DOI] [PubMed] [Google Scholar]
- 36.Lehmann D, Lütke-Eversloh T (2011) Switching Clostridium acetobutylicum to an ethanol producer by disruption of the butyrate/butanol fermentative pathway. Metab Eng 13: 464–473 [DOI] [PubMed] [Google Scholar]
- 37.Cai G, Jin B, Monis P, Saint C. 2012A genetic and metabolic approach to redirection of biochemical pathways of Clostridium butyricum for enhancing hydrogen production. Biotechnol Bioeng 10.1002/bit.24596 [DOI] [PubMed] [Google Scholar]
- 38.Buckel W, Thauer RK. 2012Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim Biophys Acta. 2012 Jul 16. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 39.Dürre P (2011) Fermentative production of butanol--the academic perspective. Curr Opin Biotechnol 22: 331–336 [DOI] [PubMed] [Google Scholar]
- 40.Heipieper HJ, Neumann G, Cornelissen S, Meinhardt F (2007) Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl Microbiol Biotechnol 74: 961–973 [DOI] [PubMed] [Google Scholar]
- 41.Nicolaou SA, Gaida SM, Papoutsakis ET (2010) A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng 12: 307–331 [DOI] [PubMed] [Google Scholar]
- 42.Huang H, Liu H, Gan YR (2010) Genetic modification of critical enzymes and involved genes in butanol biosynthesis from biomass. Biotechnol Adv 28: 651–657 [DOI] [PubMed] [Google Scholar]
- 43.Lavan LM, Van Dyk JS, Chan H, Doi RH, Pletschke BI (2009) Effect of physical conditions and chemicals on the binding of a mini-CbpA from Clostridium cellulovorans to a semi-crystalline cellulose ligand. Lett Appl Microbiol 48: 419–425 [DOI] [PubMed] [Google Scholar]
- 44.Xu C, Qin Y, Li Y, Ji Y, Huang J, et al. (2010) Factors influencing cellulosome activity in consolidated bioprocessing of cellulosic ethanol. Bioresour Technol 101: 9560–9569 [DOI] [PubMed] [Google Scholar]
- 45.Tomas CA, Welker NE, Papoutsakis ET. (2003) Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl Environ Microbial 69: 4951–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Hindorff LA, Chuang A, Monroe-Augustus M, Lyristis M, et al. (2003) Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 69: 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, et al. (2011) Engineering microbial biofuel tolerance and export using efflux pumps. Molecular Systems Biology 7: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borden JR, Papoutsakis ET (2007) Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum . Appl Environ Microbiol 73: 3061–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown SD, Guss AM, Karpinets TV, Parks JM, Smolin N, et al. (2011) Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum . Proc Natl Acad Sci U S A 108: 13752–13757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao X, Raman B, Zhu M, Mielenz JR, Brown SD, et al. (2011) Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum . Appl Microbiol Biotechnol 92: 641–652 [DOI] [PubMed] [Google Scholar]
- 51.Mao S, Luo Y, Zhang T, Li J, Bao GAH, et al. (2010) Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J Proteome Res 9: 3046–3061 [DOI] [PubMed] [Google Scholar]
- 52.Winkler J, Rehmann M, Kao KC (2010) Novel Escherichia coli hybrids with enhanced butanol tolerance. Biotechnol Lett 32: 915–920 [DOI] [PubMed] [Google Scholar]
- 53.Guedon E, Desvaux M, Petitdemange H (2002) Improvement of cellulolytic properties of Clostridium cellulolyticum by metabolic engineering. Appl Environ Microbiol 68: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendez BS, Pettinari MJ, Ivanier SE, Ramos CA, Sineriz F (1991) Clostridium thermopapyrolyticum sp. nov., a cellulolytic thermophile. Int J Syst Bacteriol 41: 281–283 [Google Scholar]
- 55.Virunanon C, Chantaroopamai S, Denduangbaripant J, Chulalaksananukul W (2008) Solventogenic-cellulolytic clostridia from 4-step-screening process in agricultural waste and cow intestinal tract. Anaerobe 14: 109–117 [DOI] [PubMed] [Google Scholar]
- 56.Inui M, Suda M, Kimura S, Yasuda K, Suzuki H, et al. (2008) Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl Microbiol Biotechnol 77: 1305–1316 [DOI] [PubMed] [Google Scholar]
- 57.Nielsen DR, Leonard E, Yoon SH, Tseng HC, Yuan C, et al. (2009) Engineering alternative butanol production platforms in heterologous bacteria. Metab Eng 11: 262–273 [DOI] [PubMed] [Google Scholar]
- 58.Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, et al. (2011) Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli . Appl Environ Microbiol 77: 2905–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu M, Zhang Y, Tang IC, Yang ST (2011) Metabolic engineering of Clostridium tyrobutyricum for n-butanol production. Metab Eng 13: 373–382 [DOI] [PubMed] [Google Scholar]
- 60.Mazzoli R, Lamberti C, Pessione E (2012) Engineering new metabolic capabilities in bacteria: lessons from recombinant cellulolytic strategies. Trends Biotechnol 30: 111–119 [DOI] [PubMed] [Google Scholar]
- 61.Khramtsov N, McDade L, Amerik A, Yu E, Divatia K, et al. (2011) Industrial yeast strain engineered to ferment ethanol from lignocellulosic biomass. Bioresour Technol 102: 8310–8313 [DOI] [PubMed] [Google Scholar]
- 62.Ilmén M, Den Haan R, Brevnova E, McBride J, Wiswall E, et al. (2011) High level secretion of cellobiohydrolases by Saccharomyces cerevisiae . Biotechnol Biofuels 4: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maki M, Leung KT, Qin W (2009) The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci 5: 500–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeon SD, Yu KO, Kim SW, Han SO (2012) The processive endoglucanase EngZ is active in crystalline cellulose degradation as a cellulosomal subunit of Clostridium cellulovorans . N Biotechnol 29: 365–371 [DOI] [PubMed] [Google Scholar]
- 65.Ciolacu D, Kovac J, Kokol V (2010) The effect of the cellulose-binding domain from Clostridium cellulovorans on the supramolecular structure of cellulose fibers. Carbohydr Res 345: 621–630 [DOI] [PubMed] [Google Scholar]
- 66.Blouzard JC, Coutinho PM, Fierobe HP, Henrissat B, Lignon S, et al. (2010) Modulation of cellulosome composition in Clostridium cellulolyticum: adaptation to the polysaccharide environment revealed by proteomic and carbohydrate-active enzyme analyses. Proteomics 10: 541–554 [DOI] [PubMed] [Google Scholar]
- 67.Cho W, Jeon SD, Shim HJ, Doi RH, Han SO (2010) Cellulosomic profiling produced by Clostridium cellulovorans during growth on different carbon sources explored by the cohesin marker. J Biotechnol 145: 233–239 [DOI] [PubMed] [Google Scholar]
- 68.Tsai SL, Goyal G, Chen W (2010) Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl Environ Microbiol 76: 7514–7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeon SD, Lee JE, Kim SJ, Kim SW, Han SO (2012) Analysis of selective, high protein-protein binding interaction of cohesin-dockerin complex using biosensing methods. Biosens Bioelectron 35: 382–389 [DOI] [PubMed] [Google Scholar]
- 70.García-Alvarez B, Melero R, Dias FM, Prates JA, Fontes CM, et al. (2011) Molecular architecture and structural transitions of a Clostridium thermocellum mini-cellulosome. J Mol Biol 407: 571–580 [DOI] [PubMed] [Google Scholar]
- 71.Molinier AL, Nouailler M, Valette O, Tardif C, Receveur-Bréchot V, et al. (2011) Synergy, structure and conformational flexibility of hybrid cellulosomes displaying various inter-cohesins linkers. J Mol Biol 405: 143–157 [DOI] [PubMed] [Google Scholar]
- 72.Yaniv O, Frolow F, Levy-Assraf M, Lamed R, Bayer EA (2012) Interactions between family 3 carbohydrate binding modules (CBMs) and cellulosomal linker peptides. Methods Enzymol 510: 247–259 [DOI] [PubMed] [Google Scholar]
- 73.Pinheiro BA, Proctor MR, Martinez-Fleites C, Prates JA, Money VA, et al. (2008) The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner. J Biol Chem 283: 18422–18430 [DOI] [PubMed] [Google Scholar]
- 74.Cha J, Matsuoka S, Chan H, Yukawa H, Inui M, et al. (2007) Effect of multiple copies of cohesins on cellulase and hemicellulase activities of Clostridium cellulovorans mini-cellulosomes. J Microbiol Biotechnol 17: 1782–1788 [PubMed] [Google Scholar]
- 75.Hammer K, Mijakovic I, Jensen PR (2006) Synthetic promoter libraries--tuning of gene expression. Trends Biotechnol 24: 53–55 [DOI] [PubMed] [Google Scholar]
- 76.Lu TK, Khalil AS, Collins JJ (2009) Next-generation synthetic gene networks. Nat Biotechnol 27: 1139–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mingardon F, Chanal A, Tardif C, Bayer EA, Fierobe HP (2007) Exploration of new geometries in cellulosome-like chimeras. Appl Environ Microbiol 73: 7138–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mingardon F, Chanal A, Tardif C, Fierobe HP (2011) The issue of secretion in heterologous expression of Clostridium cellulolyticum cellulase-encoding genes in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 77: 2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wieczorek AS, Martin VJ (2010) Engineering the cell surface display of cohesins for assembly of cellulosome-inspired enzyme complexes on Lactococcus lactis. Microb Cell Fact 9: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hyeon JE, Jeon WJ, Whang SY, Han SO (2011) Production of minicellulosomes for the enhanced hydrolysis of cellulosic substrates by recombinant Corynebacterium glutamicum. Enzyme Microb Technol 48: 371–377 [DOI] [PubMed] [Google Scholar]
- 81.Chanal A, Mingardon F, Bauzan M, Tardif C, Fierobe HP (2011) Scaffoldin modules serving as “cargo” domains to promote the secretion of heterologous cellulosomal cellulases by Clostridium acetobutylicum. Appl Environ Microbiol 77: 6277–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McBride JE, Brevnova E, Ghandi C, Mellon M, Froehlich A, et al. 2010Yeast expressing cellulases for simultaneous saccharification and fermentation using cellulose. PCT/US2009/065571
- 83.Anderson TD, Robson SA, Jiang XW, Malmirchegini GR, Fierobe HP, et al. (2011) Assembly of minicellulosomes on the surface of Bacillus subtilis . Appl Environ Microbiol 77: 4849–4858 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Wen F, Sun J, Zhao H (2010) Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl Environ Microbiol 76: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fan LH, Zhang ZJ, Yu XY, Xue YX, Tan TW (2012) Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc Natl Acad Sci U S A 109: 13260–13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Galazka JM, Tian C, Beeson WT, Martinez B, Glass NL, et al. (2010) Cellodextrin transport in yeast for improved biofuel production. Science 330: 84–86 [DOI] [PubMed] [Google Scholar]