Abstract
The RecQ helicases are a highly conserved family of DNA-unwinding enzymes that play key roles in protecting the genome stability in all kingdoms of life. Human RecQ homologs include RECQ1, BLM, WRN, RECQ4, and RECQ5β. Although the individual RecQ-related diseases are characterized by a variety of clinical features encompassing growth defects (Bloom Syndrome and Rothmund Thomson Syndrome) to premature aging (Werner Syndrome), all these patients have a high risk of cancer predisposition. Here, we present an overview of recent progress towards elucidating functions of RECQ1 helicase, the most abundant but poorly characterized RecQ homolog in humans. Consistent with a conserved role in genome stability maintenance, deficiency of RECQ1 results in elevated frequency of spontaneous sister chromatid exchanges, chromosomal instability, increased DNA damage and greater sensitivity to certain genotoxic stress. Delineating what aspects of RECQ1 catalytic functions contribute to the observed cellular phenotypes, and how this is regulated is critical to establish its biological functions in DNA metabolism. Recent studies have identified functional specialization of RECQ1 in DNA repair; however, identification of fundamental similarities will be just as critical in developing a unifying theme for RecQ actions, allowing the functions revealed from studying one homolog to be extrapolated and generalized to other RecQ homologs.
Keywords: Helicase, RecQ, DNA repair, DNA damage, genomic instability
Introduction
Helicases are motor proteins that utilize the energy derived from nucleotide hydrolysis to disrupt double- or multi-stranded nucleic acids during the process of DNA replication, repair, and recombination, as well as RNA transcription, maturation, and translation. Helicases are recognized by the presence of conserved signature sequence motifs; all helicases possess sequences homologous to the Walker A and B boxes that are characteristic of NTP binding and/or hydrolyzing enzymes [1]. They do not display sequence specificity for unwinding, but instead exhibit preference for the type and structure of the nucleic acid substrate. DNA helicases facilitate unwinding either passively by binding and trapping single strand DNA, or they actively destabilize paired DNA in addition to binding the released single stranded DNA [1]. Unwinding is directional, either 3’-5’ or 5’-3’ with respect to the strand on which the helicase translocates.
RecQ Helicase Family
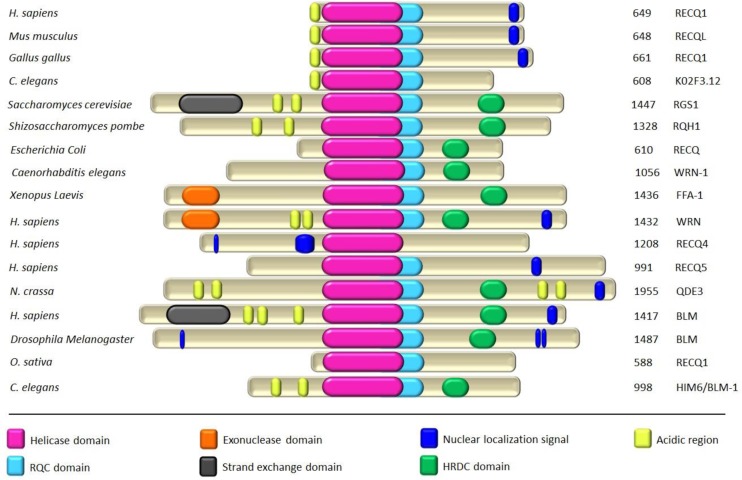
The RecQ family represents one of the most highly conserved groups of 3’-5’ DNA helicases, and is named after the prototype Escherichia coli RecQ [2–5]. A single RecQ homolog exists in bacteria and yeast whereas higher eukaryotes possess multiple markedly conserved representatives (Figure 1).
Figure 1.
Schematic representation of selected members of the RecQ-Like DNA helicases across species. Members of the RecQ family have many structural motifs that are conserved from bacteria through humans. Besides the core helicase domain, most members possess RecQ C-terminal (RQC) and Helicase and RNase D C-terminal (HRDC) domains that mediate interactions with nucleic acids and proteins. A few RecQ proteins have acidic regions that are responsible for protein-protein interactions. WRN and FFA-1 proteins are unique in that they also contain an exonuclease domain. Total number of amino acids in each protein is indicated on the right and the full color scheme is indicated. Sequence of each protein in FASTA format was used as input to an online server called MyHits (http://myhits.isb-sib.ch/) for the mapping of various motifs in each protein.
The RecQ helicase family has 5 known homologs in the human genome: RECQ1, WRN, BLM, RecQ4, and RecQ5β. Mutations in three human RecQ helicase homologs WRN, BLM and RecQ4 are related to rare genetic disorders of Werner Syndrome, Bloom Syndrome, and Rothmund-Thomson/ RAPADILINO/Baller-Gerold Syndrome, respectively, all characterized by chromosomal instability and predisposition to cancer [6, 7]. The fact that these disorders are rare attests to the critical importance of these helicases in cellular DNA metabolism. The fact that the deficiency of individual RecQ helicases is manifested as clinically distinct syndromes supports the notion that human RecQ homologs function in distinct cellular processes. A systematic analysis of the molecular interactions and cellular functions of each RecQ homolog, and the comparison of similarities and differences among them, is likely to reveal aspects of RecQ functions that are important for genome maintenance.
RECQ1 Helicase
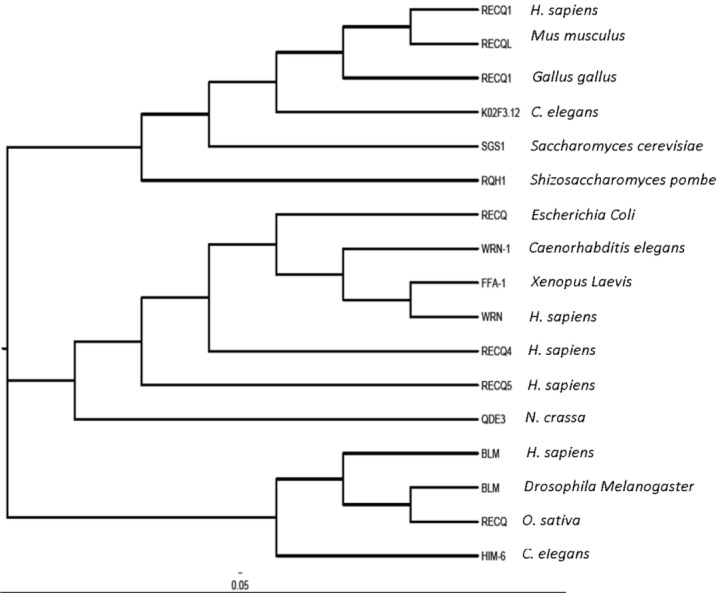
The RECQ1 (also known as RECQL or RECQL1) gene resides on chromosome 12p12 and encodes a 649 amino acid protein with a molecular mass of 73 kDa [8–10]. RECQ1 protein is smallest of the human RecQ homologs, and shares maximum homology to the prototype E. coli RecQ. RECQ1 is a DNA-stimulated ATPase and helicase [10, 11]. Phylogenetic analysis of RECQ1 with closely related proteins reveals structural divergence (Figure 2).
Figure 2.
Phylogenetic tree for the RecQ-Like proteins of DExH-Box helicase family. Phylogenetic analysis of the selected RecQ DNA helicases was performed using clustalX 2.0 and the image was generated using FigTree version 1.4.0. The branches of the tree are abbreviated with codes and detailed with genus and species name on the right.
A further insight into the anticipated biological roles of RECQ1 emanates from reviewing its structure-function relationship and molecular interactions.
Biochemical activities and substrate specificities
RECQ1 unwinds DNA with a 3’-5’ polarity [12] and needs a 3′-single strand DNA tail to unwind the substrate [11]. RECQ1 unwinds standard duplex DNA substrates such as forked duplex, 3’-overhang or 3’-flap, 5’-flap, and synthetic replication fork structures; these substrates signify model replication and repair intermediates lacking single strand character in the 3’, 5’, or both arms adjacent to the DNA duplex [11, 13–16]. Apart from conventional helicase activity, RECQ1, like BLM and WRN, also promotes branch migration of Holliday junction (HJ) and D-loops in an ATP-dependent fashion [14–16]. RECQ1 unwinds three-stranded D-loop with either a distended single stranded 3’- or 5’-tail by releasing the invading third strand from D-loop structures although a D-loop with protruding single stranded 3’-tail is a preferred substrate for unwinding [14]. In contrast to BLM helicase, RECQ1 is unable to unwind a DNA-RNA hybrid, catalyze fork regression, or displace plasmid D-loops lacking a 3’-tail but can unwind four-armed synthetic HJ structures that lacked a homologous core [14–16]. Unlike other known branch migration proteins such as BLM helicase and RAD54 both of which show no significant preference in directionality of branch migration, RECQ1 specifically catalyzes unidirectional branch migration, which may be instrumental in specific disruption of toxic, nonproductive intermediates of homologous recombination (HR) during DNA double strand break (DSB) repair in vivo [17]. However, RECQ1 is unable to use its motor ATPase to strip RAD51 from DNA during HR repair [18]. RECQ1 is also incapable of displacing streptavidin from a biotinylated oligonucleotide [19]. Consistent with a 3’-5’ directionality of RECQ1 translocation on DNA, RECQ1 helicase activity is inhibited in a strand-specific manner by an alkyl phosphotriester modification to the sugar-phosphate backbone in the predicted translocating strand [20]. Moreover, the inability of RECQ1 to unwind G-quadruplex substrates differentiates this protein from other RecQ helicases including WRN, BLM, Sgs1, or E. coli RecQ [15, 21]. Specific functions of RECQ1 in DNA metabolism are not yet clearly understood but the reported disparity in helicase substrate preference suggests functional specialization.
In addition to DNA unwinding, RECQ1 promotes annealing of complementary single strand DNA in an ATP-independent manner [14]. ATP binding induces a conformational change in RECQ1 switching it from a strand-annealing protein to a DNA unwinding activity [14]. Further studies have suggested that distinct biochemical activities of RECQ1 are dictated by different oligomeric states modulated by single strand DNA and ATP binding [22]. Overall, strand-annealing appears to be an intrinsic property of the human RecQ family since it is conserved in WRN, BLM, RecQ4 and RecQ5β [23–25].
Structural basis of biochemical functions
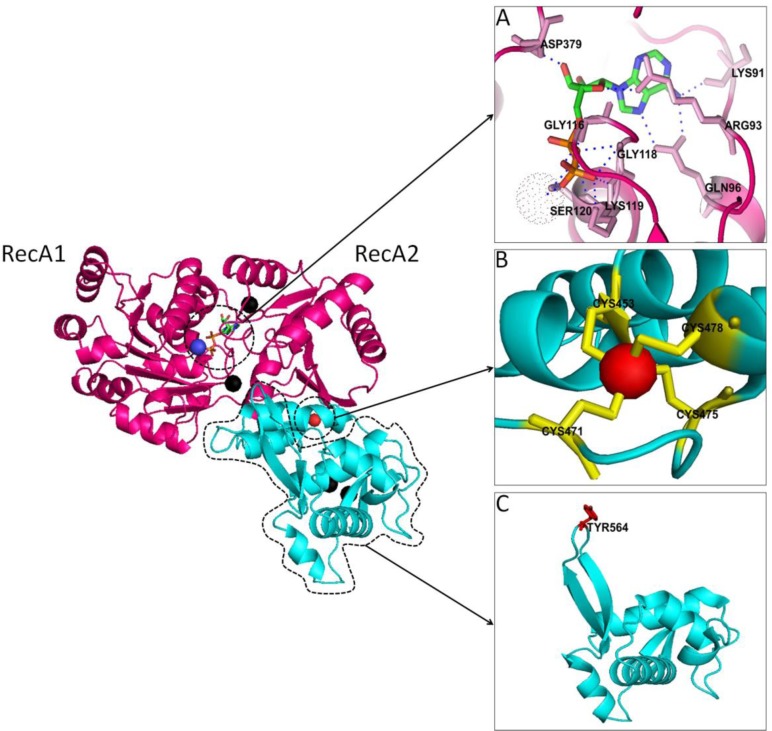
RecQ helicases share a centrally located helicase domain that couples nucleotide hydrolysis to DNA unwinding and defines the RecQ family (Figure 1). Other conserved domains include RQC (RecQ C-terminal) and HRDC ((helicase and RNaseD C-terminal), missing in RECQ1) domains, which are implicated in protein interactions and DNA binding [26–28]. The N- and C-terminal extensions in eukaryotic RecQ helicases are poorly conserved but are shown to mediate protein-protein interactions [29, 30]. Crystal structure of human RECQ1 protein lacking the first 48 and the last 33 amino acid residues (RECQ149-616) was recently reported by Gileadi's group (Figure 3) [31]. Previously it was shown that the higher oligomeric structures formed by means of N-terminus region are responsible for DNA annealing activity of RECQ1 [15, 22]; consequently, truncated RECQ149-616 failed to promote strand-annealing despite exhibiting unwinding of a forked-duplex comparable to the full-length RECQ1 [31]. Moreover, the N-terminal region of RECQ1, either direct or through the formation of higher order oligomers, also appears to be critical for the dissolution of HJs [15, 31]. Importantly, WRN protein was also shown to bind HJ and replication fork structures as an oligomer although whether this requires the N-terminal of WRN is not reported [32].
Figure 3.
Structural insight to the conserved helicase and RQC domains of human RECQ1 helicase (PDB ID: 2v1x). Cartoon representation of a single RECQ1 molecule bound to ADP; color coding of conserved structural domains is consistent with the color scheme in Figure 1. Two RecA-like domains are denoted as RecA1 and RecA2. A. The nucleotide-binding pocket. ADP forms extensive contacts with RecA1 and less with RecA2; residues in contact with ADP are shown as sticks in pink color. B. RecQ-specific Zinc-binding module in RECQ1. A single Zn2+ ion is coordinated by four Cys residues positioned on two antiparallel α-helices. C. The winged-helix (WH) domain of RECQ1. β-hairpin is shown in the upper-left corner and the important aromatic residue Tyr 564 is highlighted in red color. The figure was generated using PyMOL (http://www.pymol.org/).
RECQ1 Signature Helicase Domain
Core helicase domain of RECQ1 (amino acid residues 63-418) contains the seven signature motifs of superfamily 2 (SF2) helicases and harbors the ATP-binding pocket surrounded by highly conserved residues [3, 33]. Mutational analyses in RecQ homologs and other SF2 helicases have revealed the necessity of these amino acids for nucleotide binding as well as for functions related to nucleotide binding activity [34–36]. We and others have shown that ATP binding regulates the helicase and strand-annealing activities of RECQ1 [14, 22]. The overall fold of X-ray structure of RECQ1 bound to ADP and Mg2+ in the absence of nucleic acid was found to be comparable to other SF2 helicases having signature structure of two RecA-like domains containing the seven motifs, and retaining the general helicase fold (Figure 3) [31]. The relative orientation of the two RecA-like domains in multiple crystal forms of RECQ1 was found to be changed significantly suggesting a higher degree of flexibility between these two domains [31]. Previous studies have reported that E. coli RecQ shows slight relative rotation of the helicase domain in nucleotide-bound form compared to the unbound state and as a result of this rotation, motif I acquires an open conformation that allows the nucleotide to enter into its designated cavity [36]. Motif 0 in human RECQ1, located at the N-terminus of motif I is a RecQ-specific variant of the conserved Q motif in DEAD-box helicases and is implicated in ATP binding and hydrolysis [37]. The crystal structure of nucleotide-bound RECQ1 showed that the adenine moiety of ADP is hydrogen bonded to Gln96 within the motif 0 (Figure 3) [31]. Notably, amino acid substitution of Gln in Motif 0 of RecQ5β and BLM has been reported to reduce the ATPase and ATPase/helicase function, respectively [30, 34].
RQC Domain of RECQ1
RQC domain in RECQ1 is composed of a Zn-binding module and a helix-turn-helix fold called winged-helix (WH) domain. The Zn-binding module of RQC domain is practically identical between bacterial and human enzymes; in RQC domain of human RECQ1, a Zn2+ ion is coordinated by four Cys residues positioned on two antiparallel α-helices (Figure 3) [31]. The Zn-binding domain of E. coli RecQ [38] and human BLM [39] are important in protein/DNA binding and protein folding. Missense mutations affecting the Cys residues of the Zn-binding pocket of BLM are found in Bloom syndrome patients [39]. The WH domain, also present in transcription factors such as CAP and hRFX1, and the human DNA repair protein AGT, acts as a DNA-binding motif [31, 33, 40]. Superimposition of the WH domains from E. coli RecQ, RECQ1 and WRN show an extraordinarily conserved fold [31]. Yet, the WH of RECQ1 interacts with significantly different regions of its core helicase and Zn-binding domains as compared to those of E. coli RecQ and the human WRN protein [31]. Furthermore, a unique tyrosine residue (Tyr564) at the tip of a β-hairpin structure within the WH domain of RECQ1 is suggested to control helicase activity to unwind a simple fork duplex independent of its DNA-dependent ATPase activity (Figure 3) [31]. Vindigni's group has reported that the β-hairpin in the WH domain of RECQ1 is essential for DNA unwinding and oligomer formation [41].
Protein interactions of RECQ1
Table 1 enlists the known protein interactions of RECQ1 that are either unique or overlapping with those exhibited by other RecQ helicases. First identified protein interactions of RECQ1 were with the importin α homologs Rch1 and Qip1 [42]; Qip1 binds to the nuclear localization signal of RECQ1 (Figure 1) [43] and thus likely mediates its import to the nucleus where RECQ1 is primarily localized [44].
Table 1.
Unique and redundant protein partners of RECQ1.
| Interacting Proteins | RECQ1 | WRN | BLM | RecQ4 | RecQ5β |
|---|---|---|---|---|---|
| RPA | Yes [13] | Yes [45] | Yes [46] | Yes [47] | Yes [30] |
| RAD51 | Yes [44] | Yes [48] | Yes [49] | Yes [50] | Yes [51] |
| Top3α | Yes [52] | Yes [52] | Yes [53] | ||
| EXOI | Yes [54] | Yes [55] | Yes [56] | ||
| PARP1 | Yes [57] | Yes [58] | Yes [59] | ||
| MSH2/6 | Yes [54] | Yes [60] | Yes [61] | ||
| MLH1-PMS2 | Yes [54] | Yes [60] | Yes [62] | ||
| Ku70/80 | Yes [63] | Yes [64] | |||
| Importin alpha1/KPNA2/Rch1 | Yes [42] | ||||
| Importin alpha3/KPNA4/Qip1 | Yes [43] | ||||
| Histone H2A type 1-A | Yes [65] | ||||
| Histone H2A type 2-A | Yes [65] | ||||
| Histone H2B type 1-L | Yes [65] | ||||
| Histone 3.2 | Yes [65] |
One of the conserved physical and functional interactions of human RecQ helicases is with the single strand DNA binding protein, RPA [3]. Stimulation of WRN or BLM catalyzed DNA unwinding by RPA requires physical interaction with the helicase [66]. RPA interacts with RECQ1 and stimulates its DNA unwinding activity [13] while inhibiting strand-annealing [14]. Physical interaction of RPA heterotrimer with RECQ1 is mediated through the RPA70 subunit [13] which was also shown to be sufficient for physical and functional interaction with WRN [66]. Interaction site of RPA on RECQ1 has not yet been mapped, but RECQ1 does contain an acidic region similar to that in WRN which is reported to mediate interaction of WRN with RPA (Figure 1) [66]. RECQ1 also associates with Topoisomerase 3α [67] and RAD51 [44]; but the functional implication of these interactions remains to be elucidated [24]. Both BLM [68] and RecQ5β [51] dissociate RAD51 from DNA in vitro serving as anti-recombinase activities; whereas BLM-Topoisomerase 3α complex is exclusively important in dissolution of double HJs [69]. RECQ1 interacts physically and functionally with mismatch repair proteins (MLH1 /PMS2, MSH2/MSH6, and EXO-1) which are also involved in regulation of genetic recombination [54]. The mismatch recognition complex MSH2/MSH6 stimulates RECQ1 helicase activity whereas RECQ1 stimulates the incision activity of human EXO-1 [54]. Thus an anti-recombination function of RECQ1 may involve suppression of homeologous recombination in conjunction with mismatch repair factors that specifically bind base pair mismatches [24, 25]. In recent years, EXO-1 has emerged to be critical in helping cells deal with stalled replication forks [70–72] and interaction of RECQ1 with EXO-1 may also be important in this capacity. Another key protein partner of RECQ1 appears to be PARP-1 [57, 65]. We first identified a direct protein interaction of RECQ1 with PARP-1 and demonstrated that RECQ1-PARP-1-RPA associate in a common protein complex [57]. A conceivable biological function of this complex might be to regulate opposing activities of RECQ1 that are known to be regulated by RPA and ATP [14]. Given the ability of PARP-1 to modulate cellular ATP pools [73], interaction with PARP-1 may be important in providing an appropriate microenvironment to regulate dual activities of RECQ1 as necessary for the given cellular context. Moreover, RECQ1 and PARP-1 are also capable of interacting with the common components of mismatch repair system [54, 74] indicating a possible role in the suppression of homeologous recombination between diverged sequences. Molecular basis for the elevated sister chromatid exchanges (SCEs) in RECQ1 deficiency is not yet understood, but its interactions with mismatch repair proteins (MLH1/PMS2, MSH2/MSH6, and EXO-1) and PARP-1 may be relevant in this regard. Vindigni's lab has recently identified an exclusive role of RECQ1-PARP-1 interaction in the process of replication restart following Topoisomerase 1 (TOP1) inhibition [65]. They have demonstrated that RECQ1 preferentially catalyzes fork reversal and promotes resetting of replication forks in vitro. Following TOP1 inhibition by Camptothecin (CPT) treatment, the poly(ADP)ribosylation activity of PARP-1 inhibits fork reversal by RECQ1 in vivo [65]. Thus, RECQ1-PARP-1 complex stabilizes regressed forks until repair of the TOP1 cleavage complex is complete thus preventing premature restart of regressed forks [65]. PARP activity is not required in RECQ1-depleted cells as they cannot promote fork restoration in the absence of RECQ1 [65]; these cells likely employ HR for restart of replication following CPT treatment. Other known members of RECQ1 complex with RPA and PARP-1 are Ku70/80 [63, 65] along with certain nucleosomal histones [65]. Interestingly, RECQ1 was also found to be present with PARP-1 and Ku80 in a multi-protein complex of APLF (APTX-PNK-Like Factor) which is implicated in recruitment of non-homologous end joining (NHEJ) proteins at DSBs [75–78]. Subsequently, we have shown that RECQ1 exhibits a direct physical interaction with the Ku70/80 subunit of DNA-PK complex and modulates in vitro end joining of DSBs [63]. Collectively, these studies have provided a framework for understanding the biological roles of RECQ1 though it is still premature to propose a comprehensive model for RECQ1 in the context of its catalytic functions and protein interactions. Identification of additional constituents of the RECQ1-containing protein complexes and elucidation of how they modulate its biochemical activities will be critical to establish precise roles of RECQ1 in pathways of replication restart and DNA strand break repair.
RECQ1 in genome maintenance
Reported cellular phenotypes of RECQ1-deficiency in mouse and humans implicate unique requirement of RECQ1 in genome stability maintenance [24, 44, 79]. Despite the lack of a phenotypic defect in unstressed RECQ1 knockout mice, primary embryonic fibroblasts from RECQ1 knockout mice display chromosomal instability [79]; and increased chromosomal instability is also observed upon depletion of RECQ1 in human cells [44]. Cellular deficiency of RECQ1 is characterized by spontaneously elevated SCEs [24] which are reminiscent of recombinogenic structures proposed to arise during replication restart following fork collapse [80]. Indeed, RECQ1-deficient cells accumulate DNA damage and display increased sensitivity to DNA damaging agents that induce stalled and collapsed replication forks [44, 65, 81, 82]. Furthermore, RECQ1-deficiency in mice [79] or human cells [44] results in heightened sensitivity to ionizing radiation, which also causes oxidative DNA damage.
Our recent findings provide preliminary evidence for a unique role of RECQ1 in repair of oxidative DNA damage [57]. When cells are exposed to hydrogen peroxide (H2O2), RECQ1 is among the first RecQ proteins to arrive on chromatin and remains associated for the period of time required for the repair of strand breaks [57]. Remarkably, the chromatin localization of RECQ1 is more robust and rapid than that of WRN helicase which has been shown to function in oxidative DNA damage repair [57]. It is plausible that the activities of these RecQ proteins are assigned to dedicated pathways or sub-pathways of oxidative DNA damage repair [83, 84]. RECQ1 and its protein partners may be part of the DNA damage response by localizing to sites of oxidative lesions where they execute catalytic functions, alone or in concert. Consistent with this notion, purified recombinant RECQ1 catalyzes unwinding of duplex DNA containing oxidative base lesion such as thymine glycol, and the presence of RPA stimulates DNA unwinding by RECQ1 when the thymine glycol is positioned in the nontranslocating strand for the helicase [85]. RECQ1-depleted cells rely on PARP activity for the repair of H2O2-induced DNA damage and when RECQ1 is deficient, these lesions are possibly repaired by an alternative mechanism that involves increased activation of PARP [57]. In contrast, WRN-deficient cells fail to activate PARP in response to oxidative damage [57]. These observations suggest a novel and non-overlapping role of RECQ1 in exogenously induced-oxidative DNA damage repair via modulation of PARP-1; but a direct role of RECQ1 in specific pathway of oxidative DNA damage repair remains to be elucidated.
PARP-1 is known to bind to DNA strand breaks and subsequently synthesizes and transfers poly(ADP-ribose) polymers to itself and various nuclear proteins [86]. BLM or WRN-depleted cells exhibit constitutively hyperactivated PARP, hypersensitivity to PARP inhibitors, and are defective in HR [87]. In contrast, depletion of RECQ1 by itself does not lead to PARP hyperactivation or enhanced sensitivity to PARP inhibitor [57]. Moreover, RECQ1-depleted cells are not compromised in their ability to repair I-SceI-induced DSB by homology directed repair [57]. In addition to HR, DSBs are repaired by NHEJ mediated by Ku70/80, the DNA-PKcs protein kinase, and the complex consisting of DNA ligase IV, XRCC4 and XLF [88]. Our more recent data indicates that RECQ1 and Ku70/80 co-bind a linear DNA and the DNA binding by Ku70/80 is modulated by the presence of RECQ1 [63]. Recent models of DSB repair propose that Ku binds DNA ends first and is subsequently released through the DNA end processing activities contributed by MRN complex, CtIP, and EXO-1 [89, 90]. Ku70/80 inhibits EXO-1-mediated DSB resection in vivo [91] and DNA end resection of the forked duplex substrate in vitro [92]. The ability of RECQ1 to bind and unwind a Ku-bound forked DNA duplex relatively efficiently and its known interaction with EXO-1 suggests that RECQ1 may enable EXO-1 to overcome Ku inhibition and thereby modulate the pathway choice for DSB repair [92].
It is important to note that HR mechanisms involved in repairing classical two ended DSBs are distinct from those provoked by replication stress [93]. Notably, RECQ1, along with RecQ4, is an integral component of replication complex in unperturbed dividing cells [94]. Association of RECQ1 with replication origins during normal replication is significantly enhanced when cells encounter replication stress [82, 94]. Consistent with this, recombinant RECQ1 binds and unwinds model replication forks [14], and promotes strand exchange on stalled replication forks in vitro [81]. RECQ1-depletion results in increased sensitivity to aphidicolin, diminished checkpoint activation in response to replication stress, and chromosomal instability [82]. Chromatin immunoprecipitation experiments revealed that RECQ1 is preferentially enriched at two major fragile sites, FRA3B and FRA16D, where replication forks have stalled in vivo following aphidicolin treatment [82]. Common fragile sites are randomly distributed slow replicating genomic regions that are particularly vulnerable to replication stress and expressed as site-specific gaps or breaks on metaphase chromosomes after partial inhibition of DNA synthesis [95]. Fragile sites often coincide with chromosomal breakpoints in tumors [95, 96] and stalled replication forks at common fragile sites are believed to be a major cause of genomic instability [95, 97]. Consistent with its demonstrated catalytic functions [65, 81], recruitment of RECQ1 at fragile sites indicates that RECQ1 facilitates repair of stalled or collapsed replication forks and preserves genome integrity [82]. These results also implicate RECQ1 in mechanisms underlying common fragile site instability in cancer. Remarkably, RECQ1 is overexpressed in transformed cells [98] and in many clinical cancer samples compared to matched normal samples indicating potential target for cancer therapy [82, 99, 100]. Single nucleotide polymorphisms of RECQ1 have been associated with reduced survival in pancreatic cancer [101, 102]. RECQ1 expression is critical for the growth and proliferation of a variety of cancer cells [44, 103]; this has also been demonstrated using xenograft models [104]. It is conceivable that cancer cells are overtly dependent upon RECQ1 activities to cope with replication-induced DNA damage during rapid cell division; in normal cells, RECQ1 can act as a tumor suppressor by facilitating DNA repair and preventing mutations.
Summary and Outlook
Recent advances in structural analyses and identification of novel molecular interactions has provided a valuable foundation to explore unique and overlapping functions of RECQ1. In particular, importance of RECQ1 in repair of DNA damage in the context of replication stress has become increasingly more apparent. DNA lesions induced by replication stress occur predominantly in early replicating and actively transcribed gene clusters [105]. Given the presence of RECQ1 at replication origins, it will be insightful to investigate whether RECQ1 has a role in preventing transcription-associated genetic instability. Molecular functions of RecQ and other DNA helicases in cancer are becoming more prominent [106]. Reported overexpression of RECQ1 in a variety of clinical cancer merits systematic investigation of clinicopathological correlation. While a disease association remains to be discovered, unique requirement of RECQ1 in suppressing genomic instability proposes that a defect in RECQ1 may be linked to cancer predisposition disorders that are distinct from known RecQ-diseases. A greater understanding of the molecular and cellular functions of RECQ1 is essential to establish its role in genome maintenance and explore its translational potential.
Acknowledgements
Research in our lab is supported by the NIGMS/NIH grant 5SC1GM093999-05 (to SS). We acknowledge infrastructure support from the NIMHD/NIH under award number G12MD007597.
Competing Interests
The authors have declared that no competing interests exist.
References
- 1.Singleton MR, Dillingham MS, Wigley DB (2007) Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 76: 23–50 [DOI] [PubMed] [Google Scholar]
- 2.Hickson ID (2003) RecQ helicases: caretakers of the genome. Nat Rev Cancer 3: 169–178 [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, Doherty KM, Brosh RM, Jr. (2006) Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J 398: 319–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosh RM. Jr., Bohr VA (2007) Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res 35: 7527–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein KA, Gangloff S, Rothstein R (2010) The RecQ DNA helicases in DNA repair. Annu Rev Genet 44: 393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachrati CZ, Hickson ID (2003) RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J 374: 577–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu WK, Hickson ID (2009) RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer 9: 644–654 [DOI] [PubMed] [Google Scholar]
- 8.Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, et al. (1994) Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res 22: 4566–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puranam KL, Kennington E, Sait SN, Shows TB, Rochelle JM, et al. (1995) Chromosomal localization of the gene encoding the human DNA helicase RECQL and its mouse homologue. Genomics 26: 595–598 [DOI] [PubMed] [Google Scholar]
- 10.Puranam KL, Blackshear PJ (1994) Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem 269: 29838–29845 [PubMed] [Google Scholar]
- 11.Cui S, Klima R, Ochem A, Arosio D, Falaschi A, et al. (2003) Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J Biol Chem 278: 1424–1432 [DOI] [PubMed] [Google Scholar]
- 12.Seki M, Yanagisawa J, Kohda T, Sonoyama T, Ui M, et al. (1994) Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J Biochem 115: 523–531 [DOI] [PubMed] [Google Scholar]
- 13.Cui S, Arosio D, Doherty KM, Brosh RM. Jr., Falaschi A, et al. (2004) Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res 32: 2158–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, et al. (2005) Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem 280: 28072–28084 [DOI] [PubMed] [Google Scholar]
- 15.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, et al. (2008) The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem 283: 17766–17776 [DOI] [PubMed] [Google Scholar]
- 16.Bugreev DV, Brosh RM, Jr., Mazin AV (2008) RECQ1 possesses DNA branch migration activity. J Biol Chem 283: 20231–20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazina OM, Rossi MJ, Deakyne JS, Huang F, Mazin AV (2012) Polarity and Bypass of DNA Heterology during Branch Migration of Holliday Junctions by Human RAD54, BLM, and RECQ1 Proteins. Journal of Biological Chemistry 287: 11820–11832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bugreev DV, Mazina OM, Mazin AV (2009) Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J Biol Chem 284: 26349–26359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, et al. (2009) FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J Biol Chem 284: 7505–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suhasini AN, Sommers JA, Yu S, Wu Y, Xu T, et al. (2012) DNA Repair and Replication Fork Helicases Are Differentially Affected by Alkyl Phosphotriester Lesion. Journal of Biological Chemistry 287: 19188–19198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Shin-ya K, Brosh RM, Jr. (2008) FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol 28: 4116–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, et al. (2007) Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol 5: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohr VA (2008) Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci 33: 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S, Brosh RM, Jr. (2008) Unique and important consequences of RECQ1 deficiency in mammalian cells. Cell Cycle 7: 989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Brosh RM., Jr. (2010) Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair (Amst) 9: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JW, Kusumoto R, Doherty KM, Lin GX, Zeng W (2005) Modulation of Werner syndrome protein function by a single mutation in the conserved RecQ domain. J Biol Chem 280: 39627–39636 [DOI] [PubMed] [Google Scholar]
- 27.Huber MD, Duquette ML, Shiels JC, Maizels N (2006) A conserved G4 DNA binding domain in RecQ family helicases. J Mol Biol 358: 1071–1080 [DOI] [PubMed] [Google Scholar]
- 28.Bernstein DA, Keck JL (2005) Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure 13: 1173–1182 [DOI] [PubMed] [Google Scholar]
- 29.Wu L, Hickson ID (2006) DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet 40: 279–306 [DOI] [PubMed] [Google Scholar]
- 30.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P (2004) Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J 23: 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, et al. (2009) Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci U S A 106: 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD (2008) The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J Biol Chem 283: 24478–24483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett RJ, Keck JL (2004) Structure and function of RecQ DNA helicases. Crit Rev Biochem Mol Biol 39: 79–97 [DOI] [PubMed] [Google Scholar]
- 34.Guo RB, Rigolet P, Ren H, Zhang B, Zhang XD, et al. (2007) Structural and functional analyses of disease-causing missense mutations in Bloom syndrome protein. Nucleic Acids Res 35: 6297–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buttner K, Nehring S, Hopfner KP (2007) Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol 14: 647–652 [DOI] [PubMed] [Google Scholar]
- 36.Bernstein DA, Zittel MC, Keck JL (2003) High-resolution structure of the E.coli RecQ helicase catalytic core. EMBO J 22: 4910–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Killoran MP, Keck JL (2006) Sit down, relax and unwind: structural insights into RecQ helicase mechanisms. Nucleic Acids Res 34: 4098–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu JL, Rigolet P, Dou SX, Wang PY, Xi XG (2004) The zinc finger motif of Escherichia coli RecQ is implicated in both DNA binding and protein folding. J Biol Chem 279: 42794–42802 [DOI] [PubMed] [Google Scholar]
- 39.Guo RB, Rigolet P, Zargarian L, Fermandjian S, Xi XG (2005) Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom's syndrome helicase. Nucleic Acids Res 33: 3109–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gajiwala KS, Burley SK (2000) Winged helix proteins. Curr Opin Struct Biol 10: 110–116 [DOI] [PubMed] [Google Scholar]
- 41.Lucic B, Zhang Y, King O, Mendoza-Maldonado R, Berti M, et al. (2011) A prominent {beta}-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res 39: 1703–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seki T, Tada S, Katada T, Enomoto T (1997) Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun 234: 48–53 [DOI] [PubMed] [Google Scholar]
- 43.Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, et al. (1997) Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem 272: 26375–26381 [DOI] [PubMed] [Google Scholar]
- 44.Sharma S, Brosh RM, Jr. (2007) Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS ONE 2: e1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brosh RM, Jr., Orren DK, Nehlin JO, Ravn PH, Kenny MK, et al. (1999) Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem 274: 18341–18350 [DOI] [PubMed] [Google Scholar]
- 46.Brosh RM, Jr., Li JL, Kenny MK, Karow JK, Cooper MP, et al. (2000) Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J Biol Chem 275: 23500–23508 [DOI] [PubMed] [Google Scholar]
- 47.Rossi ML, Ghosh AK, Kulikowicz T, Croteau DL, Bohr VA (2010) Conserved helicase domain of human RecQ4 is required for strand annealing-independent DNA unwinding. DNA Repair (Amst) 9: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, et al. (2006) Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci 119: 5137–5146 [DOI] [PubMed] [Google Scholar]
- 49.Wu L, Davies SL, Levitt NC, Hickson ID (2001) Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem 276: 19375–19381 [DOI] [PubMed] [Google Scholar]
- 50.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I (2005) The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci 118: 4261–4269 [DOI] [PubMed] [Google Scholar]
- 51.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, et al. (2007) RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 21: 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson FB, Lombard DB, Neff NF, Mastrangelo MA, Dewolf W, et al. (2000) Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res 60: 1162–1167 [PubMed] [Google Scholar]
- 53.Shimamoto A, Nishikawa K, Kitao S, Furuichi Y (2000) Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res 28: 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doherty KM, Sharma S, Uzdilla LA, Wilson TM, Cui S, et al. (2005) RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination. J Biol Chem 280: 28085–28094 [DOI] [PubMed] [Google Scholar]
- 55.Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, et al. (2003) The exonucleolytic and endonucleolytic cleavage activities of human exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J Biol Chem 278: 23487–23496 [DOI] [PubMed] [Google Scholar]
- 56.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC (2008) Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A 105: 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma S, Phatak P, Stortchevoi A, Jasin M, Larocque JR (2012) RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst) 11: 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, et al. (2003) Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol 23: 8601–8613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woo LL, Futami K, Shimamoto A, Furuichi Y, Frank KM (2006) The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp Cell Res 312: 3443–3457 [DOI] [PubMed] [Google Scholar]
- 60.Saydam N, Kanagaraj R, Dietschy T, Garcia PL, Pena-Diaz J, et al. (2007) Physical and functional interactions between Werner syndrome helicase and mismatch-repair initiation factors. Nucleic Acids Res 35: 5706–5716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pedrazzi G, Bachrati CZ, Selak N, Studer I, Petkovic M, et al. (2003) The Bloom's syndrome helicase interacts directly with the human DNA mismatch repair protein hMSH6. Biol Chem 384: 1155–1164 [DOI] [PubMed] [Google Scholar]
- 62.Pedrazzi G, Perrera C, Blaser H, Kuster P, Marra G, et al. (2001) Direct association of Bloom's syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res 29: 4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parvathaneni S, Stortchevoi A, Sommers JA, Brosh RM, Jr., Sharma S (2013) Human RECQ1 Interacts with Ku70/80 and Modulates DNA End-Joining of Double-Strand Breaks. PLoS ONE 8: e62481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, et al. (2000) Ku complex interacts with and stimulates the Werner protein. Genes Dev 14: 907–912 [PMC free article] [PubMed] [Google Scholar]
- 65.Berti MChaudhuri AR, Thangavel S, Gomathinayagam S, Kenig S, et al. (2013) Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doherty KM, Sommers JA, Gray MD, Lee JW, von Kobbe C, et al. (2005) Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J Biol Chem 280: 29494–29505 [DOI] [PubMed] [Google Scholar]
- 67.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, et al. (2000) The Bloom's syndrome gene product interacts with topoisomerase III. J Biol Chem 275: 9636–9644 [DOI] [PubMed] [Google Scholar]
- 68.Bugreev DV, Yu X, Egelman EH, Mazin AV (2007) Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev 21: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bachrati CZ, Hickson ID (2009) Dissolution of double Holliday junctions by the concerted action of BLM and topoisomerase IIIalpha. Methods Mol Biol 582: 91–102 [DOI] [PubMed] [Google Scholar]
- 70.Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, et al. (2005) Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17: 153–159 [DOI] [PubMed] [Google Scholar]
- 71.Engels K, Giannattasio M, Muzi-Falconi M, Lopes M, Ferrari S (2011) 14-3-3 Proteins Regulate Exonuclease 1–Dependent Processing of Stalled Replication Forks. PLoS Genet 7: e1001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Branzei D, Foiani M (2010) Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol 11: 208–219 [DOI] [PubMed] [Google Scholar]
- 73.Martin DS, Bertino JR, Koutcher JA (2000) ATP depletion + pyrimidine depletion can markedly enhance cancer therapy: fresh insight for a new approach. Cancer Res 60: 6776–6783 [PubMed] [Google Scholar]
- 74.Liu Y, Kadyrov FA, Modrich P (2011) PARP-1 enhances the mismatch-dependence of 5’-directed excision in human mismatch repair in vitro. DNA Repair (Amst) 10: 1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bekker-Jensen S, Fugger K, Rendtlew Danielsen J, Gromova I, Sehested M, et al. (2007) Human Xip1 (C2orf13) Is a Novel Regulator of Cellular Responses to DNA Strand Breaks. Journal of Biological Chemistry 282: 19638–19643 [DOI] [PubMed] [Google Scholar]
- 76.Iles N, Rulten S, El-Khamisy SF, Caldecott KW (2007) APLF (C2orf13) Is a Novel Human Protein Involved in the Cellular Response to Chromosomal DNA Strand Breaks. Molecular and Cellular Biology 27: 3793–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanno S-i, Kuzuoka H, Sasao S, Hong Z, Lan L, et al. (2007) A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J 26: 2094–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Macrae CJ, McCulloch RD, Ylanko J, Durocher D, Koch CA (2008) APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair 7: 292–302 [DOI] [PubMed] [Google Scholar]
- 79.Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, et al. (2007) RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol 27: 1784–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Helleday T (2003) Pathways for mitotic homologous recombination in mammalian cells. Mutat Res 532: 103–115 [DOI] [PubMed] [Google Scholar]
- 81.Popuri V, Croteau DL, Brosh RM, Jr., Bohr VA (2012) RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures. Cell Cycle 11: 4252–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu X, Parvathaneni S, Hara T, Lal A, Sharma S (2013) Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Mol Cancer 12: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Svilar D, Goellner EM, Almeida KH, Sobol RW (2011) Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal 14: 2491–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Asagoshi K, Tano K, Chastain PD, 2nd, Adachi N, Sonoda E, et al. (2010) FEN1 functions in long patch base excision repair under conditions of oxidative stress in vertebrate cells. Mol Cancer Res 8: 204–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suhasini AN, Sommers JA, Mason AC, Voloshin ON, Camerini-Otero RD, et al. (2009) FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by RPA to unwind the damaged DNA substrate in a strand-specific manner. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D'Amours D, Desnoyers S, D'Silva I, Poirier GG (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342(Pt 2), 249–268 [PMC free article] [PubMed] [Google Scholar]
- 87.Gottipati P, Vischioni B, Schultz N, Solomons J, Bryant HE, et al. (2010) Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res 70: 5389–5398 [DOI] [PubMed] [Google Scholar]
- 88.Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chapman JR, Taylor Martin RG, Boulton Simon J (2012) Playing the End Game: DNA Double-Strand Break Repair Pathway Choice. Molecular Cell 47: 497–510 [DOI] [PubMed] [Google Scholar]
- 90.Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- 91.Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, et al. (2012) Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst) 11: 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun J, Lee KJ, Davis AJ, Chen DJ (2012) Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. J Biol Chem 287: 4936–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petermann E, Helleday T (2010) Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687 [DOI] [PubMed] [Google Scholar]
- 94.Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, et al. (2010) Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol 30: 1382–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Durkin SG, Glover TW (2007) Chromosome fragile sites. Annu Rev Genet 41: 169–192 [DOI] [PubMed] [Google Scholar]
- 96.Dillon LW, Burrow AA, Wang YH (2010) DNA instability at chromosomal fragile sites in cancer. Curr Genomics 11: 326–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Freudenreich CH (2007) Chromosome fragility: molecular mechanisms and cellular consequences. Front Biosci 12: 4911–4924 [DOI] [PubMed] [Google Scholar]
- 98.Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, et al. (2000) Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene 19: 4764–4772 [DOI] [PubMed] [Google Scholar]
- 99.Futami K, Ogasawara S, Goto H, Yano H, Furuichi Y (2010) RecQL1 DNA repair helicase: A potential tumor marker and therapeutic target against hepatocellular carcinoma. Int J Mol Med 25: 537–545 [DOI] [PubMed] [Google Scholar]
- 100.Sanada S, Futami K, Terada A, Yonemoto K, Ogasawara S, et al. (2013) RECQL1 DNA Repair Helicase: A Potential Therapeutic Target and a Proliferative Marker against Ovarian Cancer. PLoS ONE 8: e72820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li D, Frazier M, Evans DB, Hess KR, Crane CH, et al. (2006) Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol 24: 1720–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li D, Liu H, Jiao L, Chang DZ, Beinart G, et al. (2006) Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res 66: 3323–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Futami K, Kumagai E, Makino H, Goto H, Takagi M, et al. (2008) Induction of mitotic cell death in cancer cells by small interference RNA suppressing the expression of RecQL1 helicase. Cancer Sci 99: 71–80 [DOI] [PubMed] [Google Scholar]
- 104.Futami K, Kumagai E, Makino H, Sato A, Takagi M, et al. (2008) Anticancer activity of RecQL1 helicase siRNA in mouse xenograft models. Cancer Sci 99: 1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Barlow JH, Faryabi Robert B, Callén E, Wong N, Malhowski A, et al. (2013) Identification of Early Replicating Fragile Sites that Contribute to Genome Instability. Cell 152: 620–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brosh RM Jr(2013) DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]