Abstract
BACKGROUND
Genotype imputation substantially increases available markers for analysis in genome-wide association studies (GWAS) by leveraging linkage disequilibrium from a reference panel. We sought to (i) investigate the performance of imputation from the August 2010 release of the 1000 Genomes Project (1000GP) in an existing GWAS of prostate cancer, (ii) look for novel associations with prostate cancer risk, (iii) fine-map known prostate cancer susceptibility regions using an approximate Bayesian framework and stepwise regression, and (iv) compare power and efficiency of imputation and de novo sequencing.
METHODS
We used 2,782 aggressive prostate cancer cases and 4,458 controls from the NCI Breast and Prostate Cancer Cohort Consortium aggressive prostate cancer GWAS to infer 5.8 million well-imputed autosomal single nucleotide polymorphisms.
RESULTS
Imputation quality, as measured by correlation between imputed and true allele counts, was higher among common variants than rare variants. We found no novel prostate cancer associations among a subset of 1.2 million well-imputed low-frequency variants. At a genome-wide sequencing cost of $2,500, imputation from SNP arrays is a more powerful strategy than sequencing for detecting disease associations of SNPs with minor allele frequencies above 1%.
CONCLUSIONS
1000GP imputation provided dense coverage of previously-identified prostate cancer susceptibility regions, highlighting its potential as an inexpensive first-pass approach to fine-mapping in regions such as 5p15 and 8q24. Our study shows 1000GP imputation can accurately identify low-frequency variants and stresses the importance of large sample size when studying these variants.
Keywords: rare variants, association, fine mapping
INTRODUCTION
Prostate cancer is one of the most common chronic diseases afflicting the US and European aging male population [1,2]. So far, more than 45 independent common germline variants have been robustly associated with prostate cancer risk [3,4]. Since current commercial single nucleotide polymorphism (SNP) microarrays are primarily designed to tag genetic variants with minor allele frequencies (MAF) greater than 5%, the current signals have only explored a fraction of the potential genetic architecture of prostate cancer. New dense SNP microarrays can begin to investigate the component of the genetic architecture due to variants with MAF between 2 and 5%, but at a substantial financial cost. However, the emergence of high performance imputation programs can be applied to previously scanned data sets to search for less common variants associated with complex diseases, such as prostate cancer.
Recently, data from the 1000 Genomes Project (1000GP) have become publicly available [5]. The 1000GP is a large-scale sequencing consortium designed to survey common and uncommon human genome variation (to a standard threshold of MAF> 0.5%) by combining high and low-coverage sequencing projects. The overall goal of the 1000GP is to characterize 95% of currently accessible common variation with MAF greater than 1%, as well as to catalogue all coding functional alleles with MAF > 0.1%. Using data from the 1000GP as a reference panel, it is possible to impute over 11.5 million autosomal single nucleotide polymorphisms (SNPs) by utilizing existing SNP microarray data.
Fine mapping studies have attempted to localize signals from prostate cancer susceptibility loci by using custom genotyping panels, HapMap imputation, and earlier versions of 1000GP imputation to improve the coverage of variants around genomic regions of interest [6–9]. These studies were successful in localizing association signals, identifying statistically independent markers, and suggesting variants for functional analysis. While these studies indicated that imputation can be a useful tool for fine mapping, each of these studies only focused on a particular susceptibility locus of interest. GWAS leveraging 1000GP data have begun to appear [10–15], but to our knowledge, imputation of a prostate cancer GWAS based on the 1000GP reference panel has yet to be published as a full analysis across the entire genome, which would enable an ‘agnostic’ investigation of potential new SNPs associated with prostate cancer. Moreover, the approach can also be useful for exploring fine mapping of established loci.
Our study aimed to use the 1000GP sequencing data to impute loci in the NCI Breast and Prostate Cancer Cohort Consortium (BPC3) aggressive prostate cancer genome-wide association study (GWAS). Our goals were to (i) evaluate how well the 1000GP reference panel could impute new loci in our existing GWAS, (ii) search for novel imputed SNPs associated with prostate cancer risk at genome-wide significance levels (5×10−8), (iii) further fine map existing prostate cancer loci using the higher SNP density of the 1000GP imputation, and (iv) determine optimally powered approaches to investigate disease associations with low-frequency genetic variants.
MATERIALS AND METHODS
Genotyping data for our study originated from the BPC3, a collection of 7 prospective cohort studies aimed at investigating hormone-related gene variants and environmental factors for prostate cancer [16]. The aggressive prostate cancer GWAS includes 2,782 aggressive prostate cancer cases, defined as either having extra prostatic extension (stage C/D) or high histological grade (Gleason score >7), and 4,458 controls of European background. Individuals were genotyped on one or more Illumina Infinium Human SNP arrays, resulting in a total of 569,767 SNPs that passed quality control filtering [17]. Participating subjects provided informed consent, and the institutional review boards from each participating center approved the study protocol.
We used the August 4, 2010 release of the 1000GP as the reference panel for imputation. The European continental group contains autosomal sequences for 283 individuals of European ancestry. Called SNPs were mapped using the Genome Reference Consortium Human 37 assembly (GRCh37) and dbSNP version 129. The 283 reference individuals represent 90 Utah residents with Northern and Western European ancestry from the Centre d'Etude du Polymorphisme Humain collection (CEU), 92 Italians from Toscani, Italy (TSI), 43 British from England and Scotland (GBR), 36 Finnish in Finland (FIN), 17 Mexicans from Los Angeles, California (MXL), and 5 Puerto Ricans in Puerto Rico (PUR). The release is a four-way merged set that combines data from the Broad Institute, Boston College, University of Michigan, and the National Center for Biotechnology Information and was downloaded from the MACH website (www.sph.umich.edu/csg/abecasis/MACH/download/1000G-2010-08.html) on February 3, 2011.
Our study employed a two-stage imputation approach that was applied separately to each of the seven BPC3 cohorts. The first stage consisted of phasing the genotyped data of each individual. In this step, alleles from each genetic locus are assigned to their respective chromosomal strands of DNA. We carried out phasing in our dataset using the expectation-maximization algorithm in version 3.3.1 of BEAGLE [18].
In the second stage, expected allelic counts were inferred for each imputed locus from the phased data, resulting in a continuous allelic dosage between 0 and 2; this dosage was included in subsequent logistic-regression analyses testing for association between each SNP and aggressive prostate cancer. The second-stage imputation used the European continental group from the 1,000GP as the reference panel. The minimac program, an efficient implementation of the MaCH algorithm [19] designed to work on phased genotypes and very large reference panels, was used for imputing in this second stage. Imputation R2 values were calculated for all imputed SNPs; these estimate the level of success of combining local linkage disequilibrium from genotyped BPC3 markers and the reference panel to infer probabilistic genotypes at each imputed locus. The imputation R2 compares the observed variance of the genotype scores with the expected variance of the genotype scores if they were observed without error and can be interpreted as an estimate of the squared correlation between the true genotype and the imputed genotype [19].
Association analyses for each genetic locus were carried out using logistic regression as implemented in ProbABEL version 0.1-3 [20]. ProbABEL factors in the uncertainty from imputed SNPs and carries out a one degree of freedom likelihood ratio test comparing whether the effect of each germline variant is significantly different from the null. Additionally, we included covariates for the first principal components to adjust for potential population stratification bias. We calculated principal components using the GLU struct.pca program on a set of population informative SNPs [17,21]. These analyses were conducted separately for each cohort; locus effect estimates and standard errors from each cohort were then imported into METAL (2009-02-03 release) for meta-analysis [22]. For each marker, only cohorts with imputation R2 greater than 0.80 were included in the final meta-analysis.
We used complementary approaches to fine-mapping 32 known prostate cancer susceptibility regions within the imputed 1000GP data set. First, to identify a set of SNPs that is highly likely to contain the causal variant in a region (assuming it is contained in the 1000GP reference panel), we conducted an approximate Bayesian analysis. This approach estimates the posterior probability that a given SNP is a causal variant assuming there is only one causal SNP in the region, that it has been either genotyped or imputed, and that each SNP in the region is equally likely a priori to be the causal variant. The estimate is a simple ratio of the likelihood from the logistic regression for a particular SNP and the sum across all likelihoods for individual SNPs in the region [23–25]. Posterior probabilities were estimated for all SNPs in windows spanning 1cM upstream and downstream from the most highly associated published SNP in the region. Once these posteriors are estimated, the highest posterior density set is defined as the smallest set of SNPs such that the total posterior density (summed over all SNPs in the set) is above 80%. This approximate Bayesian approach can help guide the selection of candidate SNPs for further downstream functional and bioinformatics analyses.
In addition, stepwise regression models were used to screen for potentially novel, statistically independent signals in published genome-wide significant regions (p < 5 × 10−8). Multilocus models were iteratively used that conditioned on the top-ranked variants to look for independently associated variants. With each consecutive round, the previously most significant variant within a region was included as an additional covariate in the model and additional signals were assessed for statistical significance. Signals were considered statistically independent if they had a false discovery rate [26] less than 5% (accounting for the number of SNPs within a 2 cM window). This approach can determine whether there are statistically independent markers in a region. However, the parsimonious, independent set of markers chosen via this procedure need not contain any of the causal variants, even if these are typed or imputed and analyzed (each marker may be a proxy for many other markers in strong linkage disequilibrium, any one of which may be a causal variant).
Power calculations were also conducted to compare differences in power between two designs aimed at detecting associations between low-frequency or rare variants and disease; imputation using the 1000GP reference panel into a set of samples to be genotyped, or de novo sequencing of large numbers of samples. A range of minor allele frequencies from 0.005 to 0.50 and effect estimates with relative risks ranging from 1.1 to 2.0 were investigated across varying sample sizes using a one degree of freedom genotype trend test [27]. When calculating sequencing power, we ignored any potential sequencing error and assumed all 1000GP loci could be perfectly measured. For imputation power, we took into account imputation error by factoring in the imputation R2 distribution as a function of minor allele frequency [28,29]. The minor-allele-frequency-specific imputation R2 distributions were calculated by averaging the empirical distribution of R2 across the seven cohorts in the BPC3. All power calculations assume a 1:1 case:control ratio and an alpha level of 5×10−8. Additional calculations were carried out to compare the cost to achieve 80% power when using either 1000GP imputation or whole-genome sequencing approaches. A ratio of the cost to genotype plus imputation over the cost to sequence was used as a metric to compare cost effectiveness. A ratio greater than one indicates that under the specified parameters it is more cost effective to perform whole-genome sequencing, whereas if the ratio is less than one 1000GP imputation is favored. Several cost scenarios were considered for whole genome sequencing to estimate the effects of sequencing price on overall cost-effectiveness.
RESULTS
The August 2010 release of the 1000GP was used to impute 11,572,501 autosomal loci based on SNP genotype data on 2,782 aggressive prostate cancer cases and 4,458 controls spread across the 7 BPC3 cohorts. Genotyped SNPs with empirical R2 and leave-one-out R2 estimates greater than 0.80 had a concordance greater than 99.8%, indicating that the imputation R2 is an adequate estimate for the quality of imputed SNPs. The results of the imputation R2 are presented in Table 1 for each cohort. While the overall number of subjects in each cohort ranges from 418 to 2,161, there were minor differences in the percentage of SNPs that reached the same R2 threshold across cohorts. We selected an R2 cutoff value of 0.80 for all subsequent association analyses, resulting in a total of 5.8 million loci being included into our association analyses.
Table 1.
| n | R2 ≥ 0 | R2 > 0.3 | R2 > 0.5 | R2 > 0.8 | R2 > 0.9 | |
|---|---|---|---|---|---|---|
| ATBC | 1,490 | 11,572,501 (1.00) | 7,632,434 (0.66) | 7,015,561 (0.61) | 5,957,745 (0.51) | 5,180,557 (0.45) |
| CPSII | 1,258 | 11,572,501 (1.00) | 7,649,969 (0.66) | 6,922,561 (0.60) | 5,713,739 (0.49) | 4,834,759 (0.42) |
| EPIC | 857 | 11,572,501 (1.00) | 7,715,156 (0.67) | 7,041,970 (0.61) | 5,931,981 (0.51) | 5,114,239 (0.44) |
| HPFS | 418 | 11,572,501 (1.00) | 7,504,336 (0.65) | 6,885,596 (0.59) | 5,802,486 (0.50) | 4,970,764 (0.43) |
| MEC | 503 | 11,572,501 (1.00) | 7,615,594 (0.66) | 6,967,897 (0.60) | 5,843,127 (0.50) | 4,983,357 (0.43) |
| PHS | 553 | 11,572,501 (1.00) | 7,611,015 (0.66) | 6,968,790 (0.60) | 5,868,505 (0.51) | 5,035,674 (0.44) |
| PLCO | 2,161 | 11,572,501 (1.00) | 7,810,713 (0.67) | 7,076,176 (0.61) | 5,895,120 (0.51) | 5,039,260 (0.44) |
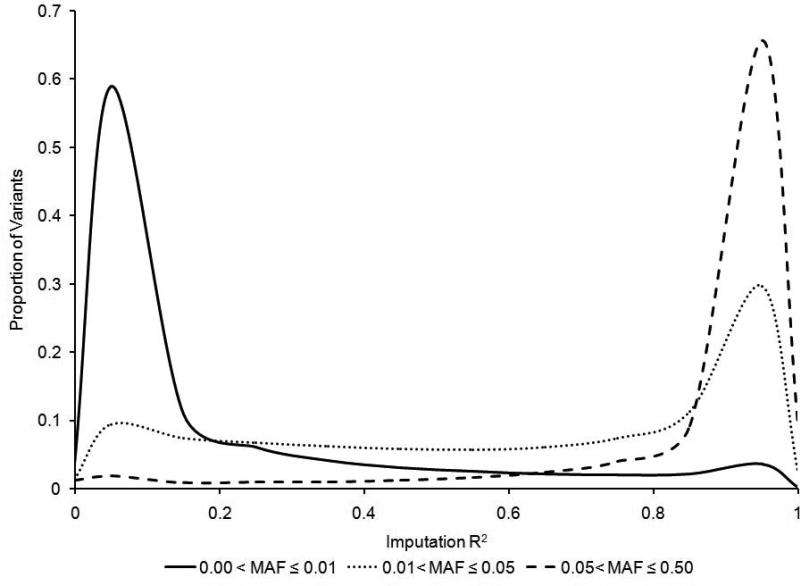
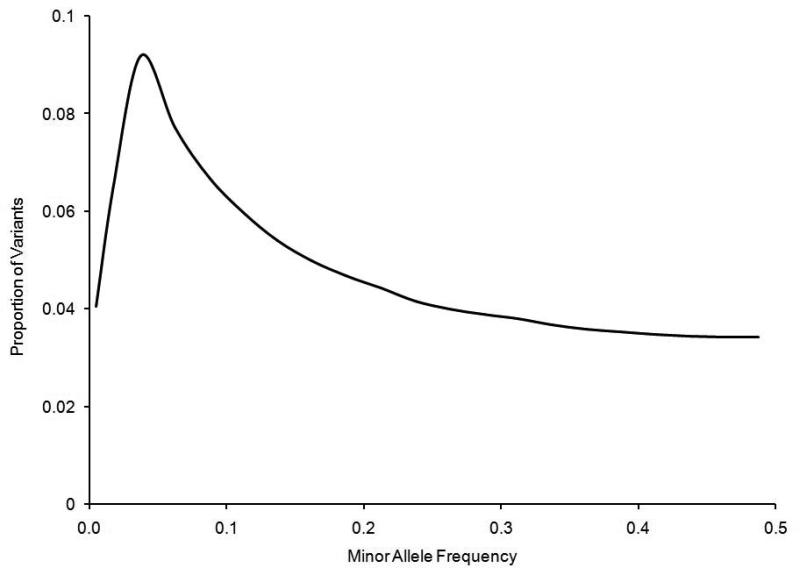
Figure 1 shows the relationship between the MAF of a SNP and the ability of the 1000GP reference set to impute the locus. In general, as the MAF increased, the average imputation R2 increased. For rare variants (MAF≤0.01), the 1000GP reference panel imputed 237,399 (6%) of these variants with an R2 value greater than 0.80. For low frequency variants (0.01<MAF≤0.05), the number of variants with an R2 greater than 0.80 was 915,708 (43%). For common variants (MAF>0.05), the 1000GP reference panel was used to impute 4,705,850 (85%) of the SNPs with R2 values greater than 0.80. Even though it is more difficult to impute rare variants, the data indicate that the distribution of well-imputed SNPs (R2≥0.8) as a function of minor allele frequency was skewed toward lower-frequency variants (Figure 2). This is a result of the large number of rare variants in the 1000GP panel (34% of 1000GP reference panel); even though a smaller proportion of rare variants could be imputed well, the absolute number of well-imputed rare variants was larger than the number of well-imputed common variants. We also observed that a greater proportion of variants were removed from the analysis when we filtered for high imputation R2 (>80%) than would be expected if the distribution of 1000GP variants was more uniformly distributed over MAF.
FIGURE 1.
Imputation R2 distribution for rare (MAF ≤ 0.01, solid line), low-frequency (0.01 < MAF ≤ 0.05, dotted line), and common (MAF > 0.05, dashed line) variants.
FIGURE 2.
Minor allele frequency distribution for the 5,858,958 well-imputed (imputation R2≥0.8) autosomal SNPs from the August 2010 release of the 1000GP data. The maximum frequency is at a MAF of 0.04.
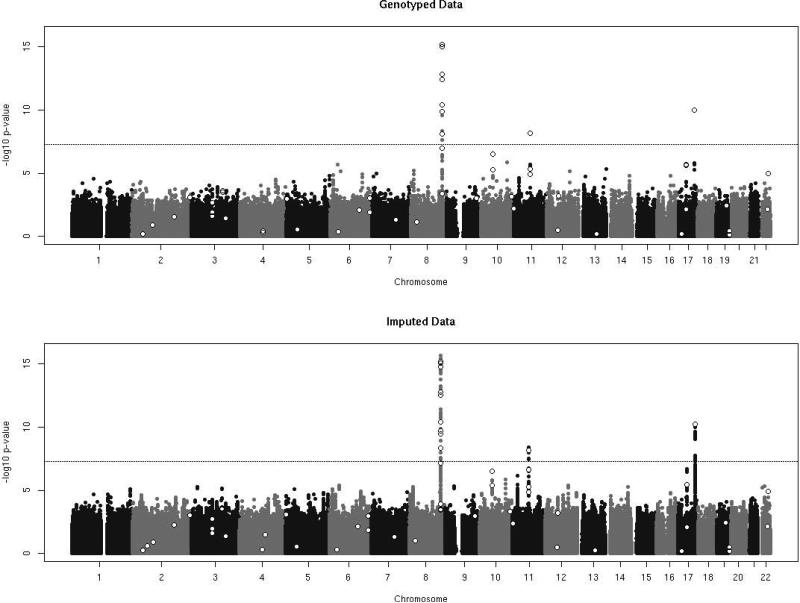
Association results from the genotyped analysis and the imputed analysis were combined by meta-analysis and plotted in Figure 3. The Manhattan plots for the genotyped data and imputed data highlight qualitative differences in marker density between the two association analyses. Although no novel loci were found to be associated with prostate cancer risk at genome-wide significance levels (p<5×10−8), additional imputed variants in LD with previously published loci were observed with p-values of comparable magnitude. These results provide a higher resolution association analysis of known prostate cancer regions.
FIGURE 3.
Manhattan plots of the genotyped data (569,767 SNPs) and the well-imputed data with R2 greater than 0.80 (5,858,958 SNPs). Open white points represent previously published loci with genome-wide significant associations with prostate cancer risk.
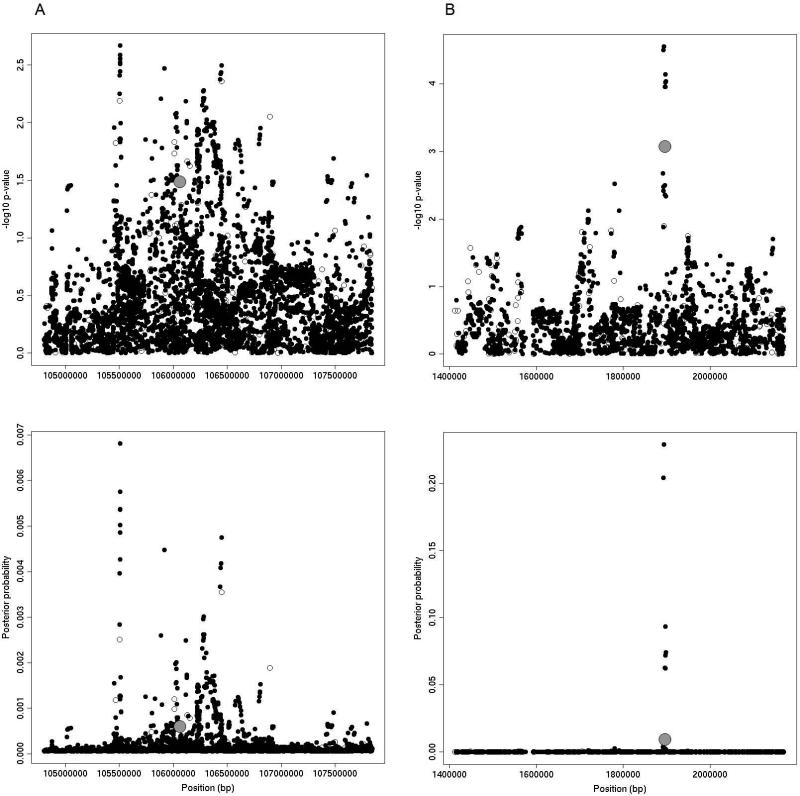
When applying a Bayesian framework to these regions, we found instances where 1000GP imputation was of little assistance for highlighting variants for future functional study (Figure 4a) and also instances where 1000GP imputation was successful in aiding selection of variants for further investigation (Figure 4b). For example, Figure 4a shows the TET2 region on chromosome 4q24, which is within a 2cM window flanking rs7679673 and includes 6,122 SNPs. The minimum p-value in our dataset was 2.15×10−3 for rs2905651, which was an imputed variant. After applying the approximate posterior Bayesian framework to this region, many variants had posterior probabilities that remained close to their prior probabilities, with 3,162 loci (52%) needing to be carried forward for further study to reach a posterior probability sum greater than 0.80. The lack of high posterior probabilities underscores the large sample sizes needed to narrow the list of potential causal variants using an association approach [25]. In this particular case, although this region has been shown to be robustly associated with prostate cancer, the evidence for association in our data set remains relatively weak. In contrast, for the IRX4 region on chromosome 5p15 (Figure 4b), the approximate posterior Bayesian framework is useful in selecting variants for further analysis. This region is a 2,413 SNP window that spans 2cM around SNP rs12653946. Again, an imputed SNP (rs34695572) is the most significantly associated variant (p-value= 8.51×10−4). In this region, only 8 SNPs (<1% of the total SNPs in the window) were required to reach a posterior probability sum greater than 0.80.
FIGURE 4.
Results from 2cm windows around TET2 (A) and IRX4 (B) regions on chromosome 4q24 and chromosome 5p15, respectively. The top panels show –log10 association p-values for each tested variant. The bottom panels display posterior probabilities from the approximate posterior Bayesian approach. Genotyped variants are indicated by open circles, imputed variants are solid circles, and previously published loci are shown in gray.
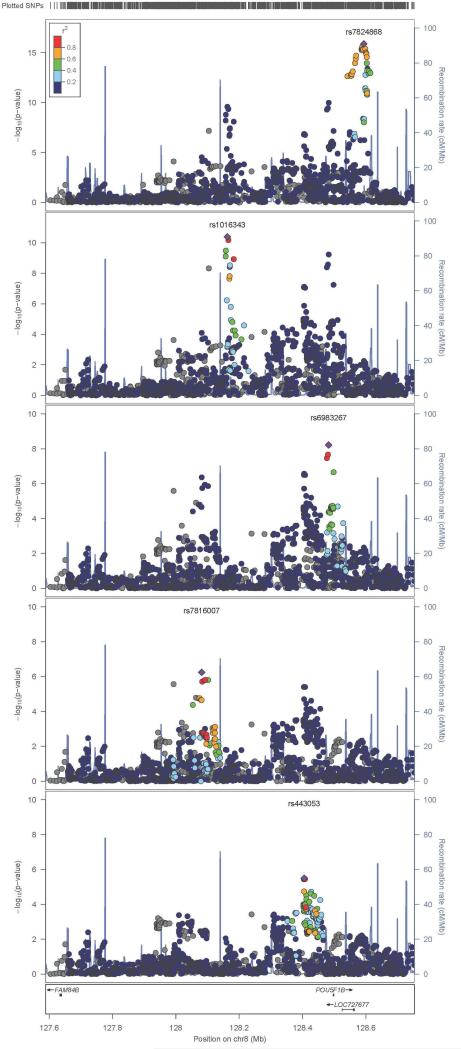
The stepwise regression models used to screen for potentially novel, statistically independent signals within published genome-wide association regions were unable to find variants that had not previously been reported. The models, however, did closely replicate known multilocus associations that have previously been published at prostate cancer susceptibility regions. For example, Figure 5 highlights five statistically independent signals located in the 8q24 region associated with prostate cancer risk. Despite the fact that the 1000GP imputation included 8 times the variants in the region than available from genotyping alone, including 15 rare and 298 low-frequency variants, the added information from imputation did not identify new independent variants in the 8q24 region associated with prostate cancer.
FIGURE 5.
LocusZoom plots of well-imputed variants from the 8q24 locus. Each panel shows statistically independent regions at the locus in order of statistical significance in our data. Color coding indicates the local pairwise linkage disequilibrium with our most significant variant in the region. Displayed –log10(p-values) are for overall association statistics with prostate cancer risk.
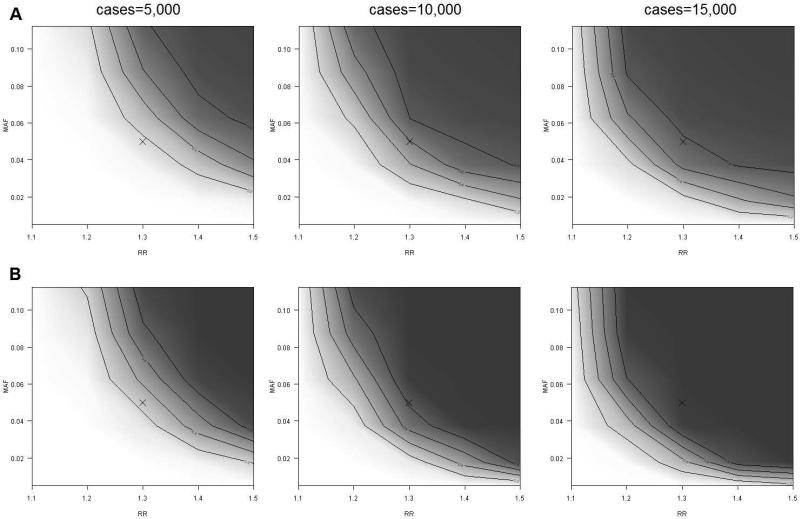
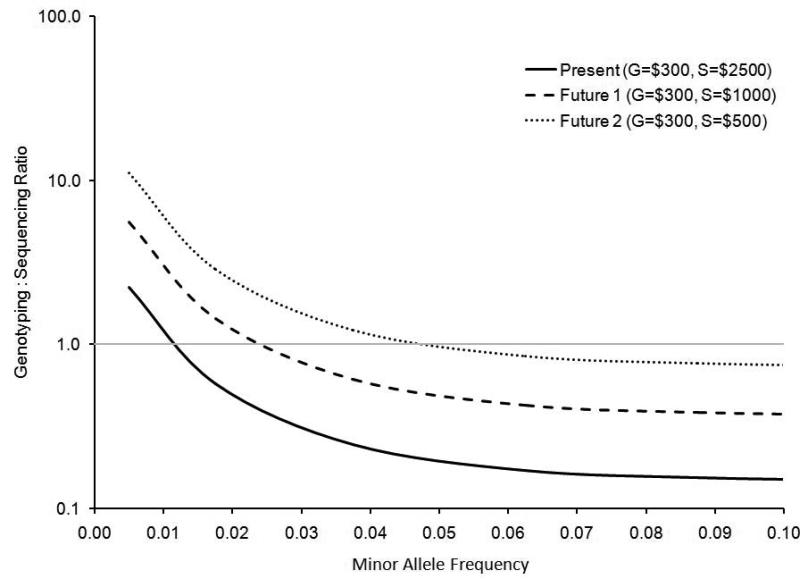
Results from power calculations comparing a hypothetical whole-genome sequencing study to a hypothetical study that used standard array based genotyping and 1000GP imputation are displayed in Figure 6. At all minor allele frequency and relative risk levels sequencing has improved power over imputation. This difference is most notable at lower minor allele frequencies and relative risks primarily due to the lower imputation R2 values when imputing lower MAF variants with the August 2010 release. The cost-effectiveness to obtain 80% power is also compared for sequencing and imputing (Figure 7). Results are displayed for 3 hypothetical pricing schemes that represent current approximate prices and two future pricing scenarios. As the cost to sequence decreases, the MAF at which it becomes more effective to sequence increases, however, genotyping plus 1000GP imputation remains most cost effective for investigating common variants [30].
FIGURE 6.
Estimated power to detect a variant for a range of relative risks and minor allele frequencies. Power was estimated for 1000GP imputation (A) and sequencing (B). Darker regions indicate areas of high power.
FIGURE 7.
Comparison of the cost effectiveness of achieving 80% power by performing genotyping plus imputation to the cost effectiveness of sequencing. Above calculations are for a hypothetical disease with prevalence 10%, per allele relative risk of 1.3, and alpha of 5×10−8. A genotyping to sequencing ratio greater than one indicates a scenario where it is more cost effective to sequence. Three scenarios of relative pricing are considered. The genotyping to sequencing ratio is plotted on a logarithmic scale.
DISCUSSION
Our study demonstrated that the 1000GP reference panel can be used to successfully impute over 5.8 million autosomal loci, of which 1.2 million have estimated MAF < 5%, based on existing GWAS data using the first generation of commercial HumanHap Illumina SNP microarrays. GWAS sample size and Illumina HumanHap array type had little effect on the ability to impute loci; the MAF of the imputed variant was the greatest determinant of successful imputation. We observed no new loci associated with prostate cancer risk below the threshold of genome-wide significance, but the 1000GP imputation did provide on average 10 times the number of variants in a region of interest resulting in a more dense resolution of variants around known associated regions with utility for fine mapping. Additionally, results from simulated power analyses showed that imputation is currently more cost effective than sequencing for SNPs with MAFs of 1-5% across a study of this size.
The ability of the 1000GP reference panel to impute over an order of magnitude more SNPs from existing genotype data provides a powerful tool for utilizing existing GWAS data to explore common and uncommon variants. For common SNPs alone (MAF 5-50%), the 1000GP panel was successful at imputing 5,560,973 variants from the 569,767 available genotyped SNPs. An additional 6,011,528 low-frequency (MAF 1-5%) and rare (MAF <1%) variants were imputed from the 1000GP reference panel, allowing for the investigation of disease associations with lower-frequency variants that had previously been infeasible. However, a majority of these low-frequency variants had low imputation R2 values, with only 1,826,551 (30%) having R2 values greater than 0.50 and 1,153,108 (19%) having R2 values greater than 0.80. This highlights the difficulty of imputing lower-frequency variants with the August 2010 European continental reference panel of 283 individuals and our sample size of 2,782 cases and 4,458 controls.
The newest release of the 1000GP (Phase I v3, March 2012) has 379 European samples and about 39.7 million markers, of which approximately 1.4 million are short indels and large deletions. To compare the performance of the added haplotypes and increased number of markers with the August 2010 build, we imputed markers for chromosome 20 using both reference panels in a separate CGEMS GWAS of 1,145 breast cancer cases and 1,142 controls. Results indicate that for an imputation R2 threshold of 0.80 approximately 20% more common variants (MAF>0.05) and 50% more low-frequency variants (0.01<MAF<0.05) would be available for analysis. The major gain was with rare variants (MAF<0.01), where up to 4 times as many well-imputed variants were available. The gains from adding information on 1.4 million short insertions and deletions have yet to be evaluated [31]. Correlations of imputed SNP dosages with imputation R2>0.3 were high between the two releases, with a correlation coefficient of 0.82. While advances are being made in 1000GP reference panels for imputation, current imputation of low-frequency and rare variants remains far from the quality achieved from sequencing, but does allow investigators to utilize existing GWAS data to investigate disease associations with a subset of well-imputed low-frequency variants.
In our meta-analysis of 2,782 aggressive prostate cancer cases and 4,458 controls, the 1000GP imputed data did not elucidate any novel loci associated with prostate cancer risk. The three genetic loci that were genome-wide significant (p < 5×10−8) were within an intergenic region on 8q24.21, a region near TPCN2 on chromosome 11q13.3, and an intergenic region on 17q24.3; all of which have been robustly replicated and reported elsewhere [8,9,32–36]. The 1000GP imputed data, however, was useful in more densely mapping variants at known prostate cancer associated loci. For some loci, using the 1000GP imputed data and utilizing an approximate Bayesian framework, we were able to select a subset of highly probable potential causal variants for further analysis. In addition, stepwise regression models were able to successfully replicate the known multi-locus associations at complex regions such as 8q24 as well as 11q13 (results not included) [8,37] found using more dense genotyping technologies in fine mapping studies in larger sample sizes, but were unable to find novel independent variants within these regions.
Our power to detect a single specific association of a low-frequency variant and prostate cancer risk is small due to the requirement of a larger sample size for lower MAFs with comparable small effects and the issues of imputation accuracy of low MAF SNPs. For example, based on our sample size and the ability of the 1000GP European reference panel to impute a variant with MAF of 0.02, we would have <1%, 2%, and 16% power to detect a variant with a relative risk of 1.3, 1.5, and 1.7, respectively. However, our power to detect at least one locus when there is more than one directly associated variant within the tested MAF and risk range substantially increases. For example, under the assumption there are 10 causal loci with a MAF of 0.02 and a relative risk of 1.7 either in the 1000GP reference panel or highly correlated with a variant in the 1000GP panel, our power to detect at least one locus is greater than 80 percent. Additionally, if there were 85 associated loci with a MAF of 0.02 and relative risk of 1.5, we would have over 80 percent power to detect at least one locus. Under these hypothetical assumptions, we can estimate an upper bound as to what the expected contribution of low-frequency variants may have on the genetic architecture of prostate cancer. Our failure to find any new such loci indicate that low-frequency variants associated with prostate cancer may have more subtle effects and may be fewer in number than the assumptions made in the above power calculations; for example, their effect sizes may be less than a relative risk of 1.7 and there may be fewer than 85 such loci that contribute directly to prostate cancer risk.
Larger studies will be needed to better assess the role that low-frequency and rare variants have in the genetic architecture of prostate cancer. Figure 5 gives insight into the sample size increase needed to be well-powered to detect some of the effects of low-frequency variants. Compared to our sample size of 2,782 cases and 4,458 controls, such sample sizes have improved power, but may still require alternative analytical techniques such as “burden of rare variant” tests [38–43] in order to provide a comprehensive look into how lower-frequency variants contribute to disease risk. Additionally, simulations based on the imputation R2 values from 1000GP imputation in our study can help maximize the cost effectiveness of future studies by guiding investigators to the optimal genotyping approach to obtain desired power levels. Currently, imputation is more cost effective when identifying associations at MAFs of 1-5%. As the 1000GP reference panel increases in size, this cost advantage could improve imputation for SNPs with MAF < 1%, but this is unlikely because of the substantial problem of the large fraction of SNPs with MAF < 1% that appear to be private to specific ethnicities [5]; this last fact indicates that it will be difficult to use reference panels for low MAF SNPs for the near future. However, this cost advantage for imputation will be reversed as the cost of sequencing decreases.
CONCLUSIONS
We have demonstrated that substantially more common and uncommon genetic variants can be imputed from existing GWAS datasets by using the August 2010 1000GP reference panel. While our imputed dataset was unable to find any novel loci associated with prostate cancer risk, we were able to demonstrate how an approximate Bayesian framework could select a highly probable subset of markers for additional analysis from an associated region of densely imputed SNPs. As our search continues to find additional prostate cancer risk loci at ever decreasing minor allele frequencies, it will become increasingly important to compile samples from larger, and thus better powered, consortia capable of detecting the contribution these low-frequency variants may have on prostate cancer risk.
ACKNOWLEDGMENTS
Special thanks to the participants and study staff from each study NCI BPC3 study. In particular, we would like to acknowledge the contributions from Demetrius Albanes (Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland), Gerald L. Andriole (Division of Urologic Surgery, School of Medicine, Washington University, St. Louis, Missouri), Sonja I. Berndt (Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, Maryland), H. Bas Bueno-de-Mesquita (National Institute for Public Health and the Environment (RIVM), Bilthoven, The Netherlands; Department of Gastroenterology and Hepatology, University Medical Centre Utrecht (UMCU), Utrecht, The Netherlands), Brian Henderson (Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California), Mattias Johansson (International Agency for Research on Cancer (IARC), Lyon, France; Department of Surgical and Perioperative Sciences, Urology and Andrology, Umeå University, Umeå, Sweden), Loic Le Marchand University of Hawaii Cancer Center, Honolulu, Hawaii), Sara Lindstrom (Program in Molecular and Genetic Epidemiology, Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts), J. Ramon Quiros (Public Health Directorate, Asturias, Spain), Susan M. Gapstur (Epidemiology Research Program, American Cancer Society, Atlanta, Georgia), J. Michael Gaziano (Division of Aging, Brigham and Women's Hospital, Boston, Massachusetts; Massachusetts Veterans Epidemiology Research and Information Center, VA Boston Healthcare System, Boston, Massachusetts), Edward Giovannucci (Departments of Nutrition and Epidemiology, Harvard School of Public Health, Boston, Massachusetts; Channing Laboratory, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts), Salvatore Panico (Department of Clinical and Experimental Medicine, Federico II University, Naples, Italy), Frederick Schumacher (Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California), Meir J. Stampfer (Departments of Nutrition and Epidemiology, Harvard School of Public Health, Boston, Massachusetts; Channing Laboratory, Department of Medicine, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts), Anne Tjonneland (Institute of Cancer Epidemiology, Danish Cancer Society), Ruth Travis (Cancer Epidemiology Unit, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, United Kingdom), Dimitrios Trichopoulos (Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts; Hellenic Health Foundation; Bureau of Epidemiologic Research of the Academy of Athens) Jarmo Virtamo (Department of Chronic Disease Prevention, National Institute for Health and Welfare, Helsinki, Finland), Walter C. Willett (Departments of Nutrition and Epidemiology, Harvard School of Public Health, Boston, Massachusetts), and Meredith Yeager (Core Genotype Facility, National Cancer Institute, National Institutes of Health, Gaithersburg, Maryland).
GRANT ACKNOWLEDGMENT
This work was supported by the US National Institutes of Health, National Cancer Institute (cooperative agreements U01-CA98233-07 to David J. Hunter, U01-CA98710-06 to Susan M. Gapstur, U01-CA98216-06 to Elio Riboli and Rudolf Kaaks, and U01-CA98758-07 to Brian E. Henderson, and Intramural Research Program of NIH/National Cancer Institute, Division of Cancer Epidemiology and Genetics) as well as support from T32-CA09001 and T32-GM074897.
Footnotes
DISCLOSURE STATEMENT
The authors report no conflicts of interest.
REFERENCES
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer Statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA: A Cancer Journal for Clinicians. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Lortet-Tieulent J, Ferlay J, Forman D, Auvinen A. Prostate cancer incidence and mortality trends in 37 European countries: an overview. European Journal of Cancer. 2010;46(17):3040–52. doi: 10.1016/j.ejca.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Kote-Jarai Z, Olama AAA, Giles GG, Severi G, Schleutker J, Weischer M, Campa D, Riboli E, Key T, Gronberg H, Hunter DJ, Kraft P, Thun MJ, Ingles S, Chanock S, Albanes D, Hayes RB, Neal DE, Hamdy FC, Donovan JL, Pharoah P, Schumacher F, Henderson BE, Stanford JL, Ostrander EA, Sorensen KD, Dörk T, Andriole G, Dickinson JL, Cybulski C, Lubinski J, Spurdle A, Clements JA, Chambers S, Aitken J, Gardiner RF, Thibodeau SN, Schaid D, John EM, Maier C, Vogel W, Cooney KA, Park JY, Cannon-Albright L, Brenner H, Habuchi T, Zhang H-W, Lu Y-J, Kaneva R, Muir K, Benlloch S, Leongamornlert DA, Saunders EJ, Tymrakiewicz M, Mahmud N, Guy M, O'Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English DR, Wahlfors T, Tammela TLJ, Klarskov P, Nordestgaard BG, Røder MA, Tybjærg-Hansen A, Bojesen SE, Travis R, Canzian F, Kaaks R, Wiklund F, Aly M, Lindstrom S, Diver WR, Gapstur S, Stern MC, Corral R, Virtamo J, Cox A, Haiman CA, Le Marchand L, Fitzgerald L, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Meyer A, Serth J, Yeager M, Berndt SI, Marthick JR, Patterson B, Wokolorczyk D, Batra J, Lose F, McDonnell SK, Joshi AD, Shahabi A, Rinckleb AE, Ray A, Sellers TA, Lin H-Y, Stephenson RA, Farnham J, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao G-W, Slavov C, Mitev V, Easton DF, Eeles RA. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nature Genetics. 2011;43(8) doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Human Genetics. 2011;130(1):59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- 5.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lou H, Yeager M, Li H, Bosquet JG, Hayes RB, Orr N, Yu K, Hutchinson A, Jacobs KB, Kraft P, Wacholder S, Chatterjee N, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Ma J, Gaziano JM, Stampfer M, Schumacher FR, Giovannucci E, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Anderson SK, Tucker M, Hoover RN, Fraumeni JF, Thomas G, Hunter DJ, Dean M, Chanock SJ. Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. Proceedings of the National Academy of Sciences. 2009;106(19):7933–8. doi: 10.1073/pnas.0902104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prokunina-Olsson L, Fu Y-P, Tang W, Jacobs KB, Hayes RB, Kraft P, Berndt SI, Wacholder S, Yu K, Hutchinson A, Spencer Feigelson H, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson BE, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Travis R, Kaaks R, Isaacs WB, Isaacs SD, Grönberg H, Wiklund F, Xu J, Vatten LJ, Hveem K, Kumle M, Tucker M, Hoover RN, Fraumeni JF, Hunter DJ, Thomas G, Chatterjee N, Chanock SJ, Yeager M. Refining the prostate cancer genetic association within the JAZF1 gene on chromosome 7p15.2. Cancer Epidemiology, Biomarkers & Prevention. 2010;19(5):1349–55. doi: 10.1158/1055-9965.EPI-09-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung CC, Ciampa J, Yeager M, Jacobs KB, Berndt SI, Hayes RB, Gonzalez-Bosquet J, Kraft P, Wacholder S, Orr N, Yu K, Hutchinson A, Boland J, Chen Q, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson BE, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs WB, Isaacs SD, Grönberg H, Wiklund F, Xu J, Vatten LJ, Hveem K, Njolstad I, Gerhard DS, Tucker M, Hoover RN, Fraumeni JF, Hunter DJ, Thomas G, Chatterjee N, Chanock SJ. Fine mapping of a region of chromosome 11q13 reveals multiple independent loci associated with risk of prostate cancer. Human Molecular Genetics. 2011;20(14):2869–78. doi: 10.1093/hmg/ddr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berndt SI, Sampson J, Yeager M, Jacobs KB, Wang Z, Hutchinson A, Chung C, Orr N, Wacholder S, Chatterjee N, Yu K, Kraft P, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman C, Henderson B, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Travis RC, Kaaks R, Isaacs W, Isaacs S, Wiley KE, Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, Vatten LJ, Hveem K, Njølstad I, Gerhard DS, Tucker M, Hayes RB, Hoover RN, Fraumeni JF, Hunter DJ, Thomas G, Chanock SJ. Large-scale fine mapping of the HNF1B locus and prostate cancer risk. Human Molecular Genetics. 2011;20(16):3322–9. doi: 10.1093/hmg/ddr213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies RW, Wells G a, Stewart AFR, Erdmann J, Shah SH, Ferguson JF, Hall AS, Anand SS, Burnett MS, Epstein SE, Dandona S, Chen L, Nahrstaedt J, Loley C, König IR, Krauss WE, Granger CB, Engert JC, Hengstenberg C, Wichmann H-E, Schreiber S, Tang WHW, Ellis SG, Rader DJ, Hazen SL, Reilly MP, Samani NJ, Schunkert H, Roberts R, McPherson R. A genome wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circulation. Cardiovascular Genetics. 2012 doi: 10.1161/CIRCGENETICS.111.961243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho MH, Castaldi PJ, Wan ES, Siedlinski M, Hersh CP, Demeo DL, Himes BE, Sylvia JS, Klanderman BJ, Ziniti JP, Lange C, Litonjua AA, Sparrow D, Regan EA, Make BJ, Hokanson JE, Murray T, Hetmanski JB, Pillai SG, Kong X, Anderson WH, Tal-Singer R, Lomas DA, Coxson HO, Edwards LD, MacNee W, Vestbo J, Yates JC, Agusti A, Calverley PM, Celli B, Crim C, Rennard S, Wouters E, Bakke P, Gulsvik A, Crapo JD, Beaty TH, Silverman EK. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Human Molecular Genetics. 2012;21(4):947–57. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meschia JF, Singleton A, Nalls M a, Rich SS, Sharma P, Ferrucci L, Matarin M, Hernandez DG, Pearce K, Brott TG, Brown RD, Hardy J, Worrall BB. Genomic risk profiling of ischemic stroke: results of an international genome-wide association meta-analysis. PloS one. 2011;6(9):e23161. doi: 10.1371/journal.pone.0023161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung YJ, Wang L, Rankinen T, Bouchard C, Rao DC. Performance of genotype imputations using data from the 1000 genomes project. Human Heredity. 2012;73(1):18–25. doi: 10.1159/000334084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. European Journal of Human Genetics. 2012:1–5. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thye T, Owusu-Dabo E, Vannberg FO, van Crevel R, Curtis J, Sahiratmadja E, Balabanova Y, Ehmen C, Muntau B, Ruge G, Sievertsen J, Gyapong J, Nikolayevskyy V, Hill PC, Sirugo G, Drobniewski F, van de Vosse E, Newport M, Alisjahbana B, Nejentsev S, Ottenhoff THM, Hill AVS, Horstmann RD, Meyer CG. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nature Genetics. 2012;44(3):257–9. doi: 10.1038/ng.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter DJ, Riboli E, Haiman CA, Albanes D, Altshuler D, Chanock SJ, Haynes RB, Henderson BE, Kaaks R, Stram DO, Thomas G, Thun MJ, Blanché H, Buring JE, Burtt NP, Calle EE, Cann H, Canzian F, Chen YC, Colditz GA, Cox DG, Dunning AM, Feigelson HS, Freedman ML, Gaziano JM, Giovannucci E, Hankinson SE, Hirschhorn JN, Hoover RN, Key T, Kolonel LN, Kraft P, Le Marchand L, Liu S, Ma J, Melnick S, Pharaoh P, Pike MC, Rodriguez C, Setiawan VW, Stampfer MJ, Trapido E, Travis R, Virtamo J, Wacholder S, Willett WC. A candidate gene approach to searching for low penetrance breast and prostate cancer genes. Nature Reviews Cancer. 2005;5(12):977–85. doi: 10.1038/nrc1754. [DOI] [PubMed] [Google Scholar]
- 17.Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, Stevens VL, Chen C, Mondul AM, Travis RC, Stram DO, Eeles RA, Easton DF, Giles G, Hopper JL, Neal DE, Hamdy FC, Donovan JL, Muir K, Al Olama AA, Kote-Jarai Z, Guy M, Severi G, Grönberg H, Isaacs WB, Karlsson R, Wiklund F, Xu J, Allen NE, Andriole GL, Barricarte A, Boeing H, Bas Bueno-de-Mesquita H, Crawford ED, Diver WR, Gonzalez CA, Gaziano JM, Giovannucci EL, Johansson M, Le Marchand L, Ma J, Sieri S, Stattin P, Stampfer MJ, Tjonneland A, Vineis P, Virtamo J, Vogel U, Weinstein SJ, Yeager M, Thun MJ, Kolonel LN, Henderson BE, Albanes D, Hayes RB, Spencer Feigelson H, Riboli E, Hunter DJ, Chanock SJ, Haiman CA, Kraft P. Genome-wide association study identifies new prostate cancer susceptibility loci. Human Molecular Genetics. 2011:1–9. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. American Journal of Human Genetics. 2007;81(5):1084–97. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiology. 2010;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu K, Wang Z, Li Q, Wacholder S, Hunter DJ, Hoover RN, Chanock S, Thomas G. Population substructure and control selection in genome-wide association studies. PloS One. 2008;3(7):e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzo Bermejo J, Garcia Perez A, Brandt A, Hemminki K, Matthews AG. Comparison of six statistics of genetic association regarding their ability to discriminate between causal variants and genetically linked markers. Human Heredity. 2011;72(2):142–52. doi: 10.1159/000332006. [DOI] [PubMed] [Google Scholar]
- 24.Gu F, Monsees G, Kraft P. Exhaustive screens for disease susceptibility loci incorporating statistical interaction of genotypes: a comparison of likelihood-ratio-based and Akaike and Bayesian information criteria-based methods. BMC Proceedings. 2007;1(Suppl 1):S25. doi: 10.1186/1753-6561-1-s1-s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Udler MS, Tyrer J, Easton DF. Evaluating the power to discriminate between highly correlated SNPs in genetic association studies. Genetic Epidemiology. 2010;34(5):463–8. doi: 10.1002/gepi.20504. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57(1):289–300. [Google Scholar]
- 27.Sasieni PD. From genotypes to genes: doubling the sample size. Biometrics. 1997;53(4):1253–61. [PubMed] [Google Scholar]
- 28.Jorgenson E, Witte JS. Coverage and power in genomewide association studies. American Journal of Human Genetics. 2006;78(5):884–8. doi: 10.1086/503751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraft P, Cox DG. Study designs for genome-wide association studies. Advances in Genetics. 2008;60(07):465–504. doi: 10.1016/S0065-2660(07)00417-8. [DOI] [PubMed] [Google Scholar]
- 30.Sampson J, Jacobs K, Yeager M, Chanock S, Chatterjee N. Efficient study design for next generation sequencing. Genetic Epidemiology. 2011;277:269–77. doi: 10.1002/gepi.20575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu JT, Wang Y, Gibbs R a, Yu F. Characterizing linkage disequilibrium and evaluating imputation power of human genomic insertion-deletion polymorphisms. Genome Biology. 2012;13(2):R15. doi: 10.1186/gb-2012-13-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier J-B, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson J-E, Bälter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nature Genetics. 2006;38(6):652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 33.Zheng SL, Stevens VL, Wiklund F, Isaacs SD, Sun J, Smith S, Pruett K, Wiley KE, Kim S-T, Zhu Y, Zhang Z, Hsu F-C, Turner AR, Johansson J-E, Liu W, Kim JW, Chang B-L, Duggan D, Carpten J, Rodriguez C, Isaacs W, Grönberg H, Xu J. Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiology, Biomarkers & Prevention. 2009;18(6):1815–20. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So W-Y, Tong PCY, Ng MCY, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CNA, Rotimi C, Chan JCN, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nature Genetics. 2007;39(8):977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 35.Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, Yu K, Hutchinson A, Wang Z, Amundadottir L, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson B, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs W, Isaacs S, Wiley KE, Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, Vatten LJ, Hveem K, Kumle M, Tucker M, Gerhard DS, Hoover RN, Fraumeni JF, Hunter DJ, Thomas G, Chanock SJ. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nature Genetics. 2009;41(10):1055–7. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, Morrison J, Maranian M, Pooley KA, Luben R, Eccles D, Evans DG, Fletcher O, Johnson N, dos Santos Silva I, Peto J, Stratton MR, Rahman N, Jacobs K, Prentice R, Anderson GL, Rajkovic A, Curb JD, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Diver WR, Bojesen S, Nordestgaard BG, Flyger H, Dörk T, Schürmann P, Hillemanns P, Karstens JH, Bogdanova NV, Antonenkova NN, Zalutsky IV, Bermisheva M, Fedorova S, Khusnutdinova E, Kang D, Yoo K-Y, Noh DY, Ahn S-H, Devilee P, van Asperen CJ, Tollenaar RAEM, Seynaeve C, Garcia-Closas M, Lissowska J, Brinton L, Peplonska B, Nevanlinna H, Heikkinen T, Aittomäki K, Blomqvist C, Hopper JL, Southey MC, Smith L, Spurdle AB, Schmidt MK, Broeks A, van Hien RR, Cornelissen S, Milne RL, Ribas G, González-Neira A, Benitez J, Schmutzler RK, Burwinkel B, Bartram CR, Meindl A, Brauch H, Justenhoven C, Hamann U, Chang-Claude J, Hein R, Wang-Gohrke S, Lindblom A, Margolin S, Mannermaa A, Kosma V-M, Kataja V, Olson JE, Wang X, Fredericksen Z, Giles GG, Severi G, Baglietto L, English DR, Hankinson SE, Cox DG, Kraft P, Vatten LJ, Hveem K, Kumle M, Sigurdson A, Doody M, Bhatti P, Alexander BH, Hooning MJ, van den Ouweland AMW, Oldenburg RA, Schutte M, Hall P, Czene K, Liu J, Li Y, Cox A, Elliott G, Brock I, Reed MWR, Shen C-Y, Yu J-C, Hsu G-C, Chen S-T, Anton-Culver H, Ziogas A, Andrulis IL, Knight JA, Beesley J, Goode EL, Couch F, Chenevix-Trench G, Hoover RN, Ponder BAJ, Hunter DJ, Pharoah PDP, Dunning AM, Chanock SJ, Easton DF. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nature Genetics. 2009;41(5):585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert D a, Tymrakiewicz M, Jhavar S, Saunders E, Hopper JL, Southey MC, Muir KR, English DR, Dearnaley DP, Ardern-Jones AT, Hall AL, O'Brien LT, Wilkinson R a, Sawyer E, Lophatananon A, Horwich A, Huddart R a, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper C, Donovan JL, Hamdy FC, Neal DE, Eeles R a, Easton DF. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nature Genetics. 2009;41(10):1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 38.Lin D-Y, Tang Z-Z. A general framework for detecting disease associations with rare variants in sequencing studies. American Journal of Human Genetics. 2011;89(3):354–67. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases : application to analysis of sequence data. Journal of Human Genetics. 2008:311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genetics. 2009;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price AL, Kryukov GV, de Bakker PIW, Purcell SM, Staples J, Wei L-J, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. American Journal of Human Genetics. 2010;86(6):832–8. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neale BM, Rivas M a, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS Genetics. 2011;7(3):e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. American Journal of Human Genetics. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]