Abstract
Corynebacterium glutamicum is well known as the amino acid-producing workhorse of fermentation industry, being used for multi-million-ton scale production of glutamate and lysine for more than 60 years. However, it is only recently that extensive research has focused on engineering it beyond the scope of amino acids. Meanwhile, a variety of corynebacterial strains allows access to alternative carbon sources and/or allows production of a wide range of industrially relevant compounds. Some of these efforts set new standards in terms of titers and productivities achieved whereas others represent a proof-of-principle. These achievements manifest the position of C. glutamicum as an important industrial microorganism with capabilities far beyond the traditional amino acid production. In this review we focus on the state of the art of metabolic engineering of C. glutamicum for utilization of alternative carbon sources, (e.g. coming from wastes and unprocessed sources), and construction of C. glutamicum strains for production of new products such as diamines, organic acids and alcohols
Keywords: amino acids, organic acids, diamines, hemicellulose, glycerol
1. Introduction
Industrial biotechnology is paving the way for a bio-based economy, which is proposed to complement and, in the long term, replace petro-based economy. Microorganisms are central to this technology and carry out the production reactions whether fermentation, whole-cell biocatalysis or enzymatic reactions. Although microbial processes have been known and used since many thousands years, it is the recombinant DNA technology and more recently the application of ‘omics’ technologies that allowed scientists to rationally engineer these microbial factories as never before.
Corynebacterium glutamicum is a Gram-positive bacterium and is referred to as the industrial workhorse for amino acid production. It was first isolated in 1950s from Japanese soil during a quest for natural L-glutamate producers [1]. Since then it has been thoroughly investigated and used as a generally-regarded-as-safe (GRAS) organism in fermentation industry for more than 50 years. Today it is used for the annual production of 2,160,000 tons of L-glutamate [2] and 1,480,000 tons of L-lysine [3]. After genetic engineering tools were developed, C. glutamicum genome was sequenced and made publically available in 2003 [4, 5] and omics technologies such as transcriptome studies evolved [6]. These milestones marked the beginning of extensive research efforts to metabolically engineer C. glutamicum, initially for L-glutamate and L-lysine production and later on for the production of a variety of products such as organic acids, alcohols or diamines. Similarly, use of alternative carbon sources, that unlike the traditional sugars sucrose, glucose and fructose, do not have competing applications in food industry, became possible by metabolic engineering.
The following pages are intended to provide the reader with a glimpse into metabolic engineering of C. glutamicum for (i) utilization of alternative carbon sources and (ii) formation of non-traditional products. Metabolic engineering approaches for L-glutamate and L-lysine production have been reviewed and monographed extensively [7–12], and therefore will not be a part of this review.
II. Metabolic engineering for carbon source utilization
II.1 Optimization of selected endogenous substrate utilization pathways
C. glutamicum can utilize many carbon sources for its growth and energy supply: monosaccharides such as glucose, fructose, and ribose; disaccharides such as sucrose, mannose, and maltose; alcohols such as inositol or ethanol and organic acids such as pyruvic acid, propionic acid, lactic acid, acetic acid, and gluconic acid, but also some amino acids such as L-glutamate and L-glutamine [13, 14]. Production processes with C. glutamicum are carried out utilizing sugars derived from starch (glucose) or molasses (sucrose and fructose) [8, 12]. Use of molasses dominates in European, South American and Asian production plants, whereas starch hydrolysates are favored in North America [12]. In C. glutamicum these sugars are substrates of the phosphotransferase system (PTS), which phosphorylates its substrate by utilizing phosphoenolpyruvate [15].
II.1.1 Glucose
The utilization of glucose via the PTS was increased by constitutive expression of the glucose specific EII subunit gene ptsG [16]. Furthermore, a PTS-independent glucose utilization pathway, which transports glucose into the cell and then phosphorylates it using ATP and/or polyphosphate dependent glucokinases [17, 18] was shown to be present in C. glutamicum. When glucose was solely utilized via this system, increased L-lysine production was observed, probably due to a higher availability of the L-lysine precursor phosphoenolpyruvate [19, 20]. This alternative pathway might be favorable for a variety of production processes [21].
II.1.2 Lactic acid
Lactic acid is not only the predominant byproduct of C. glutamicum during growth with glucose under oxygen limiting conditions, but also an interesting carbon source taking its abundance in silages into account. C. glutamicum possesses two lactate dehydrogenases for the utilization of lactic acids, one specific for L-and one for D-lactic acid oxidization to pyruvate [22, 23]. The quinone-dependent L-lactate dehydrogenase gene (lldD) is sufficiently expressed for fast growth with L-lactic acid whereas D-lactate dehydrogenase gene (dld) needs to be overexpressed for efficient D-lactic acid utilization [22–24]. Further strain optimization by overexpression of either malic enzyme or pyruvate carboxylase genes enabled L-lysine production from lactic acid [24]. The recent observation that overexpression of the lactic acid forming NAD-dependent lactate dehydrogenase gene (ldhA) rescued an lldD knockout strain [25] indicates that ldhA overexpression might prove beneficial to accelerate lactate catabolism.
II.2 Establishing access to alternative carbon sources
Several cheaply available and therefore interesting carbon sources cannot be utilized by C. glutamicum, either because the whole or part of the necessary genetic setup is missing or a sufficient level of gene expression is not present. Thus, the construction of recombinant strains towards utilization of further carbon sources is an important part of the metabolic engineering of this bacterium.
II.2.1 Starch
Glucose derived from starch hydrolysates is used in industrial amino acid production processes. In first engineering approaches towards direct utilization of starch the α-amylase gene from Streptomyces griseus was expressed in C. glutamicum and the enzyme was excreted into the medium [26]. The recombinant strain was able to grow with starch even faster as compared to glucose. However growth was only achieved when small amounts of glucose were present, and some starch degradation product remained in the medium [26]. Expression of the Streptococcus bovis α-amylase gene together with Bacillus subtilis pgsA to produce a surface displayed fusion protein, enabled C. glutamicum for the utilization of starch as sole source of carbon [27]. Starch-based production of L-glutamate [28], L-lysine [26, 27, 29], and cadaverine [30] has been reported and in the case of L-lysine, higher L-lysine production yields were reached with starch as compared to glucose [27].
II.2.2 Lactose and Galactose
Utilization of the whey sugars lactose and galactose has been engineered in C. glutamicum. Lactose utilization was achieved by heterologous expression of the lac genes from Escherichia coli [31]. As lactose was cleaved into its monomeric compounds glucose and galactose by β-galactosidase, C. glutamicum was able to utilize liberated glucose but not the galactose component of lactose [31]. Combining heterologous expression of the lactose permease and β-galactosidase genes from Lactobacillus delbrueckii with expression of Lactococcus lactis subsp. cremoris MG1363 genes for aldose-1-epimerase, galactokinase, UDP-glucose-I-P-uridylyltransferase, and UDP-galactose-4-epimerase allowed the use of both the glucose and galactose components of lactose [32]. Whey-based L-lysine production was possible, but growth from the whey components was slower as compared to that on glucose [32].
II.2.3 Arabinose
Arabinose is one of the main compounds of lignocellulosic wastes, which accrue from e.g. agricultural or forestry productions. Three enzymatic reactions are necessary for the conversion of arabinose to the pentose phosphate pathway intermediate xylulose-5-phosphate: first arabinose isomerase (AraA) converts arabinose to ribulose, second ribulokinase (AraB) phosphorylates ribulose to ribulose-5-phosphate, which finally is converted to xylulose-5-phosphate by ribulose-5-phosphate 4-epimerase (AraD) [33]. Expression of the arabinose transporter gene araE from C. glutamicum ATCC 31831 allowed utilization of at low concentrations of arabinose [34]. Arabinose has been used as a carbon source for the production of organic acids [34] as well as for the production of the amino acids L-glutamate, L-lysine, L-ornithine and L-arginine [35].
II.2.4 Xylose
As arabinose, the pentose sugar xylose is a component of lignocellulosic wastes. To funnel xylose into the pentose phosphate pathway, xylose has to be converted to xylulose and then phosphorylated to xylulose-5-phosphate. As C. glutamicum possesses a gene encoding a xylulokinase (xylB), the expression of the xylose isomerase gene (xylA) from E. coli was sufficient to enable growth from xylose as sole carbon source, but the additional expression of E. coli xylB accelerated growth of the recombinant C. glutamicum strain [36]. When optimizing arabinose utilization the arabinose transporter gene araE from C. glutamicum ATCC 31831 was expressed in the recombinant C. glutamicum xylose utilization strain, resulting in an increased xylose consumption rate and growth rate [34]. To prove its biotechnological usability, anaerobic production of succinic and lactic acid [36], as well as production of cadaverine [37] and the amino acids L-glutamate and L-lysine [38] from xylose was demonstrated.
II.2.5 Cellobiose
Cellobiose is contained in hydrolysates of cellulosic biomass. A C. glutamicum R mutant gained the ability to utilize cellobiose. This ability was due to mutations in the phosphotransferase permease subunit gene bglF, which is specific for the transport of the β-glucosides arbutin and salicin. Two mutations were published, both at the same position (317), resulting in amino acid exchanges from valine to alanine or methionine [39]. For the utilization of cellobiose also the phospho-β-glucosidase is needed (bglA). Both of these genes are present in C. glutamicum R, but are absent from others. Under oxygen deprivation conditions mixtures of cellobiose, glucose and xylose could be used for the production of lactic and succinic acid [34, 40].
II.2.6 Dicarboxylic acids
Fast utilization of the dicarboxylic acids namely succinic, fumaric, and malic acid (intermediates of the tricarboxylic acid cycle) was observed only in spontaneous mutants, which showed increased expression levels of genes encoding proton-or sodium-dependent transporters DctA and DccT, respectively [41, 42]. Increased expression was due to promoter-up mutations in the promoter regions of the genes. Overexpression of the corresponding genes dctA or dccT (dcsT in C. glutamicum R) resulted in fast growth with succinic, fumaric, and malic acid [41–43].
II.2.7 Glycerol
During the transesterfication process of oils and fats for biodiesel production, glycerol remains as a stoichiometric byproduct, with a yield of 100 kg per ton. Glycerol, previously an interesting product, also of biotechnology, became a waste material as the biodiesel industry arose. C. glutamicum, which cannot utilize glycerol naturally, was engineered for glycerol utilization by heterologous expression of the E. coli aerobic glycerol utilization genes encoding for a glycerol facilitator, glycerol kinase, and glycerol-3-phosphate dehydrogenase [44]. The plasmid borne expression of these genes enabled C. glutamicum for fast growth with glycerol as sole carbon source. When compared to glucose similar L-glutamate and L-lysine production properties were reached when using glycerol [44]. Using the same plasmid recently efficient succinic acid production from glycerol could be achieved [45].
II.2.8 Coutilization
Coutilization of substrates is a typical characteristic of C. glutamicum [14], and has been studied in detail with respect to the co-utilization of acetate and glucose [46, 47]. Coutilization is advantageous in biotechnological applications, but it is absent in other hosts e.g. E. coli [48] or B. subtilis [49], which show successive utilization of substrates, carbon catabolite repression, and diauxic growth. But, no rule without exceptions: C. glutamicum does utilize glucose before L-glutamic acid or ethanol, and acetate before ethanol [50, 51].
Moreover, when additional carbon utilization pathways are introduced into C. glutamicum coutilization is pertinent and has been shown for utilization of glycerol with glucose [44], lignocellulose compound xylose with glucose [36], arabinose with glucose [35], xylose and arabinose with glucose [38], and cellobiose together with xylose and glucose [34].
III. Metabolic engineering of C. glutamicum for product formation
This section summarizes the wide array of metabolic products for which C. glutamicum has been engineered. The majority of these products are amino acids or organic acids but diamines, alcohols and other products of industrial relevance are being continuously added to this repertoire. As mentioned earlier, metabolic engineering of L-glutamate and L-lysine will not be a part of this review because they have already been extensively reviewed [7–12]. In many cases, the strategy for product metabolic engineering has evolved over time, making use of results from ongoing research in order to overcome the bottlenecks identified and to finally get the most promising strain. All pathways and their components used for the engineering of different products discussed in this section are drawn in Figure 2 and hence a reference to the same figure will not be made while discussing each product. Readers are also recommended to refer to Table 1 of another review [52] for a summary of production parameters of different C. glutamicum production strains.
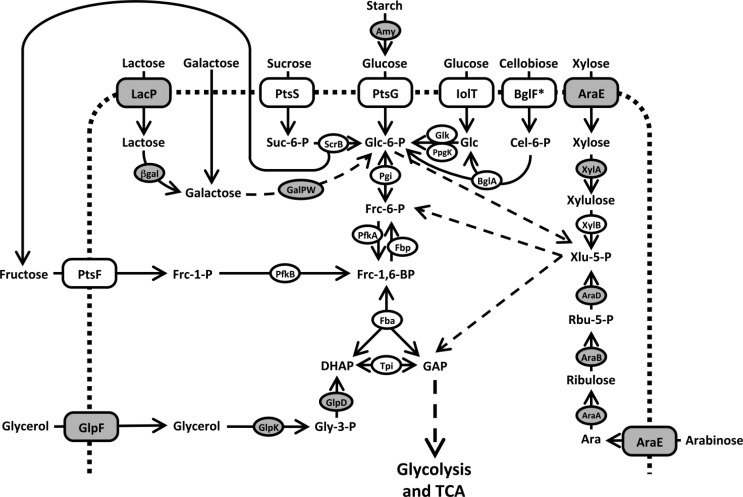
Figure 1.
Carbon source utilization in C. glutamicum. Open boxes: C. glutamicum enzymes, gray shaded boxes: enzymes from other organisms produced in C. glutamicum by heterologous expression. Abbreviations: Ara, arabinose; AraA, arabinose isomerase; AraB, ribulokinase; AraD, ribulose 5-phosphate 4-epimerase; AraE, arabinose transporter; βgal, β-galactosidase; BglA, phospho-β-glucosidase; BglF*, cellobiose specific PTS; Cel-6-P, cellobiose-6-phosphate; DHAP, dihydroxyacetonephosphate; Fba, fructose-bisphosphate aldolase; Fbp, fructose-bisphosphatase; Frc-1-,6-BP, fructose-1,6-bisphosphate; Frc-6-P, fructose-6-phosphate; Frc-1-P, fructose-1-phosphate; GalPW, galactose pathway; GAP, glyceraldehyde-3-phosphate; Glc-6-P, glucose-6-phosphate; Glc, glucose; Glk, ATP dependent glucokinase; GlpD, glycerol-3-phosphate dehydrogenase; GlpF, glycerol facilitator; GlpK, glycerol kinase; Gly-3-P, glycerol-3-phosphate; IolT, inositol transporter, also accepting glucose; LacP, lactose permease; PfkA, 6-phosphofructokinase; PfkB, 1-phosphofructokinase; PpgK, polyphosphate dependent glucokinase; PtsF, fructose specific PTS; PtsG, glucose specific PTS; PtsS, sucrose specific PTS; Rbu-5-P, ribulose-5-phosphate; ScrB, sucrose-6-phosphate hydrolase; Suc-6-P, sucrose-6-phosphate; Tpi, triosephosphate isomerase; Xlu-5-P, xylulose-5-phosphate; XylA, xylose isomerase; XylB, xylulokinase.
III.1 Amino acids
III.1.1 L-valine
L-valine is an essential amino acid with application in feed and pharmaceutical industry and has a demand of several hundred tons per year [9]. Production of L-valine by C. glutamicum has been demonstrated both aerobically and anaerobically. Blombach et al. were able to engineer an aerobic L-valine producer by starting with a first-generation strain having an inactivated pyruvate dehydrogenase complex (PDHC) and overexpressing L-valine biosynthesis pathway genes ilvBNCE. In order to channel all pyruvic acid towards L-valine production, pqo encoding pyruvate:quinone oxidoreductase and pyc encoding pyruvate carboxylase were deleted. The former enzyme converts pyruvic acid to acetic acid and the latter converts it to oxaloacetic acid. Furthermore, to increase the flux via pentose phosphate pathway in order to meet the cofactor (NADPH) demand, pgi encoding phosphoglucose isomerase was also deleted (Fig. 2). The resulting strain produced 48 g l-1 L-valine in 90 hours in a growth decoupled manner during fed-batch fermentation [53].
For anaerobic production, Hasegawa et al. engineered a C. glutamicum strain by deletion of ldh encoding lactate dehydrogenase to prevent lactic acid formation and overexpression of a modified ilvBNCD (feedback resistant acetohydroxyacid synthase (encoded by ilvN) and a modified acetohydroxy acid isomeroreductase (encoded by ilvC) with preference for NADH instead of NADPH). Additionally, to allow use of NH3 instead of glutamate as amino donor, a Lysinibacillus sphaericus leucine dehydrogenase gene (ilvE) was expressed in C. glutamicum to catalyze the final transamination step. This approach yielded a C. glutamicum capable of producing up to almost 230 g l-1 L-valine in 48 hours under oxygen deprivation conditions [54].
III.1.2 L-isoleucine
L-isoleucine belongs to the aspartate family of amino acids and finds applications in food and pharmaceutical industry. In addition to being relatively long, L-isoleucine biosynthesis pathway shares genes with L-valine and L-leucine pathways and hence is subject to tight genetic and feedback regulation at multiple steps. Accordingly, these steps have been the targets of metabolic engineering in order to overcome the regulation and allow higher carbon flux towards L-isoleucine. An L-lysine producer was selected as a starting strain for L-isoleucine production because it has a high flux through the feedback resistant aspartate kinase (lysCfbr) reaction, which is the first step towards L-isoleucine formation from L-aspartate. Additionally leuCD was mutated which reduces formation of L-leucine as a byproduct. In this genetic background, chromosomal expression of three copies of homfbr, a single copy of ilvAfbr and overexpression of ilvAfbr on a multiple copy vector yielded a strain that produced up to 21 g l-1 isoleucine via fed-batch fermentation [55]. homfbr encodes for a feedback-resistant homoserine dehydrogenase that catalyzes homoserine formation and its overexpression increases flux towards threonine, which is the precursor amino acid of L-isoleucine. ilvAfbr encodes for a feedback-resistant threonine dehydratase that catalyzes the first step in conversion of L-threonine to L-isoleucine. Upon investigation of another L-isoleucine producer it was also shown that L-isoleucine export, rather than precursor supply or enzyme activities, is the limiting factor in obtaining higher titers and therefore presents a prospective opportunity for further improvement.
III.1.3 L-serine
L-serine finds applications in pharmaceutical and cosmetic industry and is produced in the range of 300 tons per year [56]. It is degraded by C. glutamicum and holds a key position in central metabolism. Hence, mere overexpression of serine biosynthetic pathway genes serA▵197 (made insensitive to feed-back regulation by truncation of 197 amino acids at C-terminus), serC and serB was not enough for serine production. serA▵197 encodes for 3-phosphoglycerate dehydrogenase (PGDH) [57] that catalyzes the first step of serine formation by converting the glycolysis intermediate 3-phosphoglycerate to hydroxypyruvate phosphate. serC and serB encode phosphoserine aminotransferase (PSAT) and phosphoserine phosphatase (PSP) respectively that are responsible for the next two steps resulting in L-serine formation. [56, 58]. In addition to the overexpression of these L-serine biosynthesis pathway genes, a disruption of serine hydroxymethyltransferase gene (glyA) and serine dehydratase gene (sdaA), whose products are responsible for L-serine conversion to glycine and pyruvic acid respectively, was essential to form L-serine (9 g l-1) as a product [56, 58].
III.1.4 L-arginine
L-arginine is the most abundant nitrogen carrier in animals and is essential for ammonia detoxification. Its clinical application in treatment of a variety of diseases including diabetes, AIDS and heart diseases has been reported [59]. Using reverse engineering, Ikeda et al., combined mutations identified in classical producers to engineer an L-arginine and L-citrulline producer. These mutations were found to be in the arginine repressor gene argR and in the argB gene whose product catalyzes the N-acetylglutamate kinase reaction in the beginning part of the arginine biosynthesis pathway [60]. Accordingly, argR was deleted and the two amino acid substitutions observed in classical producer (A26V and M31V) were introduced in the wild type N-acetylglutamate kinase resulting in higher productivity of L-arginine and L-citrulline at the suboptimal temperature of 38°C. However, this strain could not produce L-arginine at the optimal temperature of 30°C. Later, Schneider et al., [35] starting with a similar strain carrying argR deletion and overexpressing a feedback resistant argB, created C. glutamicum ARG1 which led to production of about 7 g l-1 L-arginine at 30°C. Overexpression of the arabinose utilization operon araBAD (Fig. 1) in ARG1 allowed it to utilize arabinose or a mixture of glucose and arabinose as carbon source, resulting in production of about 8.3 g l-1 L-arginine, although complete utilization of arabinose was not possible [35].
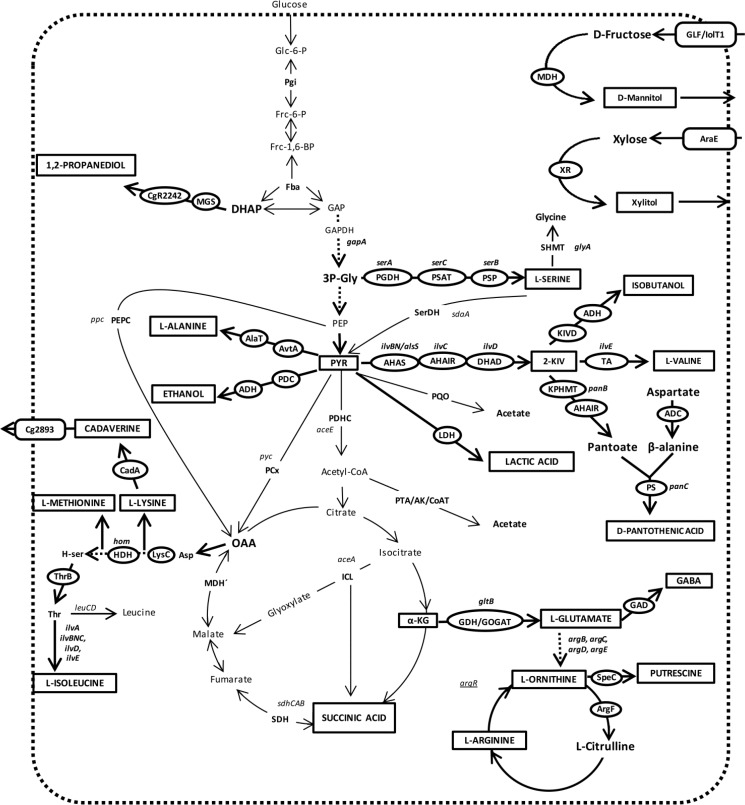
Figure 2.
Products for which C. glutamicum has been metabolically engineered. The products are depicted in rectangles and the enzymes involved in the biosynthesis pathway (bold arrows) are shown in ovals. The gene names for which the designation is not the same as enzyme name are mentioned separately next to ovals. Broken arrows represent multiple steps. argR (underlined) is a gene-encoding regulator of the arginine pathway. Abbreviations: 2-KIV, 2-Ketoisovaleric acid; 3P-Gly, 3-phosphoglycerate; α-KG, α-Ketoglutaric acid; ADC, Aspartate decarboxylase; ADH, alcohol dehydrogenase; AHAIR, acetohydroxyacid isomeroreductase; AHAS, acetohydroxyacid synthase; AK, acetate kinase; AlaT, alanine aminotransferase; AraE, arabinose transporter; ArgF, L-ornithine carbamoyltransferase; Asp, aspartate, AvtA, valine-pyruvate aminotransferase; CadA, L-lysine decarboxylase; CoAT, acetyl-CoA:CoA transferase; DHAD, dihydroxyacid dehydratase; DHAP, dihydroxyacetonephosphate; Fba, fructose-bisphosphate aldolase; Frc-6-P, fructose-6-phosphate; Frc-1,6-BP, fructose-1,6-bisphosphate; GAD, glutamate decarboxylase; GAP, glyceraldehyde-3-phosphate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GDH, glutamate dehydrogenase; Glc-6-P, glucose-6-phosphate; GLF, glucose/fructose facilitator; GOGAT, glutamate-2-oxoglutarate aminotransferase; HDH, homoserine dehydrogenase; H-ser, homoserine; ICL, isocitrate lyase; IolT1, inositol transporter; KIVD, 2-ketoacid decarboxylase; KPHMT, ketopantoate hydroxymethyltransferase; LDH, lactate dehydrogenase; LeuCD, 3-isopropylmalate dehydratase; LysC, aspartate kinase; MDH, mannitol dehydrogenase; MDH′, malate dehydrogenase; MGS, methylglyoxal synthase; OAA, Oxaloacetate; PCx, pyruvate carboxylase; PDC, pyruvate decarboxylase; PDHC, pyruvate dehydrogenase complex; PEP, phosphoenolpyruvate; PEPC, phosphoenolpyruvate carboxylase; PGDH, 3-phosphoglycerate dehydrogenase; PQO, pyruvate:quinone oxidoreductase; PSAT, phosphoserine aminotransferase; Pgi, glucose-6-phosphate isomerase; PS, pantothenate synsthase; PSP, phosphoserine phosphatase; PTA, phosphotransacetylase; PYR, pyruvate; SDH, succinate dehydrogenase; SerDH, serine dehydratase; SHMT, serine hydroxymethyltransferase; SpeC, ornithine decarboxylase; TA, transaminase; Thr, threonine; ThrB, homoserine kinase; XR, xylose reductase.
III.1.5 L-methionine
L-methionine belongs to the aspartate family of amino acids and is the third most-produced amino acid (tons per annum), next to L-glutamate and L-lysine. However, so far, its fermentative production has not been proven to be industrially feasible. This is probably because with a requirement of 7 mol ATP and 8 mol NADPH per mol L-methionine, it is the most demanding amino acid for fermentative production. By deletion of a gene producing the competing amino acid L-threonine (thrB) and by deregulating feedback inhibition of homoserine dehydrogenase (hom), Park et al, engineered a L-lysine producing C. glutamicum strain for production of 2.9 g l-1 L-methionine [61]. However, depletion of glucose in the medium led to disappearance of the produced L-methionine. Although not reflected in academic literature, commercial methionine fermentation could be a reality in the near future.
III.1.6 L-ornithine
L-ornithine is a non-proteinogenic amino acid that is an intermediate of the arginine biosynthesis pathway and might be used for treating liver diseases [62]. It was shown that deletion of argF gene encoding L-ornithine carbamoyltransferase to stop L-ornithine conversion and of argR encoding repressor of arginine biosynthesis pathway were sufficient for L-ornithine formation in C. glutamicum in milligrams per liter range [63]. Further optimization for L-arginine requirement of strains and introduction of arabinose utilization pathway (Fig. 1) resulted in L-ornithine titers of up to almost 26 g l-1 in 72 hours using glucose and arabinose as a carbon source [35].
III.1.7 γ-Aminobutyric acid
γ-Aminobutyric acid (GABA) is a non-proteinogenic amino acid that functions as the main inhibitory neurotransmitter of the mammalian nervous system and has pharmaceutical relevance for use as anti-anxiety agent and in tranquilizers, diuretics and analgesics. It can be formed from glutamate via a single decarboxylation step and hence C. glutamicum, being a natural L-glutamate producer, can serve as a competitive production platform. Using heterologous expression of Lactobacillus brevis genes for: i) glutamate decarboxylase (gadB2) to catalyze glutamate decarboxylation to GABA, ii) L-glutamate/GABA antiporter gene gadC and iii) the transcriptional regulator gene gadR, Shi and Li constructed a strain that could produce more than 2 g l-1 GABA in 72 hours [64]. Zhao et al. reported the GABA transporter in C. glutamicum responsible for its uptake and were able to show that disruption of its gene led to increased GABA production [65]. Production of GABA using C. glutamicum has the advantage that it is independent of external glutamate supply and complex media ingredients, which is the case when using lactic-acid bacteria (LAB) for its production. However the GABA production levels are higher using LAB and hence there is further need for improvement of C. glutamicum strains.
III.1.8 D-amino acids
Production of D-amino acids was also shown to be possible in C. glutamicum whereas other bacteria such as E. coli are sensitive to their presence even in low concentrations. Heterologous expression of Pseudomonas taetrolens racemase (argR) resulted in extracellular accumulation of equimolar amounts of L- and D- enantiomers of arginine, ornithine or lysine whereas D-serine concentration exceeded that of L-serine [66]. D-amino acids are used as building blocks in chemistry and hence their production via fermentation is of interest.
III.2 Diamines
III.2.1 Putrescine
Putrescine is a biogenic diamine that serves as a monomer in the production of polyamides e.g. nylons-4,6. Polyamides are produced on a multi-million-ton scale per annum predominantly from petrochemical routes and currently there is an increasing interest in their fermentative production. Putrescine can be formed via arginine-or ornithine decarboxylase pathways and it was shown that the latter results in better production levels [67]. The genetic engineering involved deletion of argF and argR in order to channel ornithine (putrescine precursor) solely towards putrescine and to relieve genetic repression of the arginine biosynthesis pathway respectively. Heterologous expression of the E. coli ornithine decarboxylase gene (speC) then led to formation of up to 6 g l-1 putrescine in 60 hours in a growth independent manner when supplemented with arginine [67]. In the next step, introduction of a plasmid addiction system expressing argF resulted in both relief of arginine auxotrophy and growth dependent putrescine production [68].
III.2.2 Cadaverine
Like putrescine, cadaverine is also a diamine, which can be used for polyamide production [69]. Mimitsuka et al. were the first to engineer C. glutamicum for cadaverine production by replacing hom with the L-lysine decarboxylase gene cadA from E. coli, resulting in production of 2.6 g l-1 cadaverine from glucose in 18 hours [70]. hom deletion functioned to stop carbon flow downstream to the formation of branched chain amino acids (L-valine, L-leucine, L-isoleucine) and diverted it instead towards cadaverine formation via lysine. In separate studies with different strains, expression of xylA and xylB that encode xylose utilization enzymes (Fig. 1) [37] and of amyA that encodes starch hydrolysis enzyme, (Fig. 1) [30] allowed cadaverine production from these substrates. Later on, overexpression of a putative permease, encoded by cg2893 [37] and deletion of a putative diaminopentane acetyltransferase, encoded by NCgl1469 (cg1722) [37] were also shown to result in higher product formation and elimination of acetylated byproduct respectively.
III.3 Organic acids
III.3.1 Succinic acid
Succinic acid can serve as a platform chemical and hence there is plentiful research going on to produce it via fermentation [45, 71–74]. The annual demand for the free acid is 15,000 tons whereas its salts are used annually in amounts up to 92,000 tons as deicers. For a feasible industrial process, the target succinic acid amount and productivity have been set at 150 g l-1 and 5 g l-1 h-1 respectively [75]. By deleting lactate dehydrogenase gene (ldhA) and overexpressing the anaplerotic pyruvate carboxylase gene (pyc), a C. glutamicum strain was made that produced 146 g l-1 succinic acid with a productivity of 3.2 g l-1 h-1 under anaerobic conditions [73]. The former modification avoided lactic acid as byproduct and the latter drives the carbon flux in to the reductive tricarboxylic acid (TCA) cycle. In a pioneering study redox-balanced anaerobic succinate production could be achieved [72]. Besides the aforementioned modifications (overexpressing a modified pycP458S instead of wild type pyc) and deletion of all known acetic acid forming genes (cat, pqo, ackA-pta), overexpression of the glyceraldehyde 3-phosphate dehydrogenase (gapA) gene led to significantly reduced acetic acid (byproduct) formation, accelerated glucose consumption and very high succinic acid titers. Heterologous production of Mycobacterium vaccae formate dehydrogenase (fdh) entailed use of formate as cosubstrate [72]. Thus, coutilization of glucose with formate allowed for redox-balanced anaerobic succinate production by this recombinant strain.
Under aerobic conditions combined overexpression of phosphoenolpyruvate carboxylase (ppc) and pyruvate carboxylase (pyc) genes – to propel anaplerosis – and deletion of acetic acid forming genes (see above) and of succinic acid oxidizing genes (sdhCAB) – to prevent succinic acid conversion – resulted in a strain producing 10.6 g l-1 succinic acid in a growth decoupled manner. This is the highest titer reported for aerobic succinic acid production in C. glutamicum [74]. Subsequent overexpression of glycerol utilization genes glpFKD (Fig. 1) allowed this strain to produce almost the same amount of succinic acid from glycerol [45].
III.3.2 Pyruvic acid
Pyruvic acid finds applications in many industries including food, cosmetic and pharmaceutical. Production costs for chemical synthesis of pyruvate of US$ 8000-9000/ton have been reported [76]. Hence, there is a lot of interest in its fermentative production. It also holds an important position in the central carbon metabolism and serves as a connecting point between glycolysis and TCA cycle. Additionally, it is a branching point for the formation of a variety of organic and amino acids. Consequently, the approach for pyruvic acid engineering focused on attenuation of these ‘branches’ to allow for its accumulation. Acetyl-CoA, acetic acid, lactic acid and alanine production were minimized by deletion of pyruvate dehydrogenase (aceE), pyruvate:quinone oxidoreductase (pqo), lactate dehydrogenase (ldh) and of alanine aminotransferase (alaT) plus valine-pyruvate aminotransferase (avtA) genes respectively. Additionally, the C-terminus of acetohydroxyacid synthase (AHAS) was deleted (▵C-T ilvN) to prevent overflow metabolism into L-valine synthesis. Titers of more than 45 g l-1 were achieved using this strain [77].
III.3.3 Lactic acid
Lactic acid is used in cosmetic and pharmaceutical industry. Its major market, however, is packaging where polymers of lactic acid are in demand. C. glutamicum wild type was shown to produce L-lactic acid under oxygen deprivation conditions and in fed-batch fermentation could produce 52 g l-1 L-lactic acid in 8 hours. In a continuous process using cell-recycling L-lactic acid production continued for 360 hours [78]. Succinic acid and acetic acid were also produced but only in lower amounts.
A drawback associated with poly L-lactic acid (PLLA) is its low melting point. One way to improve this property is production of a stereocomplex of PLLA and Poly D-lactic acid. To engineer a strain that can produce D-lactic acid a L. delbrueckii D-lactate dehydrogenase gene was expressed in C. glutamicum with a deletion of the lactate dehydrogenase (ldhA) gene. The resulting strain produced 120 g l-1 of 99.9% optically pure D-lactic acid in less than 30 hours [79].
III.3.4 α-Ketoglutaric acid
Like succinic acid, α-ketoglutaric acid is a TCA cycle intermediate and is in demand by pharmaceutical and food industry. Jo et al. engineered an α-ketoglutaric acid producer by starting with a L-glutamate producing strain as it already has a high α-ketoglutarate availability. Glutamate dehydrogenase and glutamate-2-oxoglutarate aminotransferase (GOGAT) encoding genes, gdh and gltB, respectively, were disrupted to abolish conversion of α-ketoglutaric acid to L-glutamate. In order to channel the precursor isocitrate solely to α-ketoglutaric acid, aceA encoding isocitrate lyase that converts isocitric acid into glyoxylic acid and succinic acid was also disrupted. The resulting triple mutant was able to accumulate 12.4 g l-1 α-ketoglutaric acid after 72 hours [80].
III.3.5 2-Ketoisovaleric acid
2-Ketoisovaleric acid (2-KIV) is used by pharmaceutical industry in drugs for treating kidney patients. It is a precursor of the amino acid L-valine and is formed from pyruvic acid in three reactions catalyzed by acetohydroxyacid synthase (AHAS, encoded by ilvBN), acetohydroxyacid isomeroreductase (AHAIR, encoded by ilvC) and dihydroxyacid dehydratase (DHAD encoded by ilvD). Accordingly, the engineering approach adopted by Krause et al. [81]involved overexpression of these biosynthetic pathway genes (ilvBNCD) in combination with deletion of: i) ilvE to prevent L-valine formation ii) aceE to channel pyruvate to 2-KIV formation and to allow growth decoupled production and iii) pqo to further enhance carbon flow to 2-KIV synthesis. This resulted in the growth-decoupled formation of approximately 22 g l-1 2-KIV by the engineered C. glutamicum strain [81].
III.3.6 D-Pantothenic acid
D-Pantothenic acid (Vitamin B5) can be synthesized by microorganisms and plants but not by animals and is therefore demanded as nutritional requirement for livestock and humans. Its worldwide market is reported to be 10,000 tons/year [82]. Starting from 2-ketoisovaleric acid, pantothenic acid can be synthesized in four enzymatic steps. The enzymatic reactions of the pantothenic acid biosynthesis pathway (starting from pyruvic acid) are also partly shared by the L-isoleucine and L-valine biosynthesis pathways. Its production was elucidated in a first-generation C. glutamicum production strain by overexpression of ketoisovaleric acid pathway genes (ilvBNCD) and of D-pantothenic acid biosynthesis pathway genes (panBC). To prevent L-isoleucine formation, ilvA was deleted [83]. This strain accumulated up to 1 g l-1 D-pantothenic acid. Metabolic analysis of this strain identified a high flux of the precursor 2-ketoisovaleric acid to L-valine instead of D-pantothenic acid as the main limitation for pantothenic acid production [84]. Using this information, a promoter down mutation was introduced in the first generation strain to attenuate ilvE expression and hence to stop L-valine formation. A duplication of panBC expression to increase flux into D-pantothenic acid formation was also done. This second-generation strain was able to attain titers of 1.75 g l-1 [85]. The precursor and other byproducts from associated pathways were still secreted by this strain indicating an opportunity for further metabolic engineering.
III.4 Alcohols
III.4.1 Ethanol
Ethanol fermentation for use as fuel has a well-known history and ethanol production in USA alone was more than 13 billion gallons in 2010 [86]. Ethanol production was achieved in C. glutamicum by heterologous expression of the Zymomonas mobilis pyruvate decarboxylase gene (pdc) and overexpression of an alcohol dehydrogenase gene (adhB). The former enzyme catalyzes decarboxylation of pyruvic acid to acetaldehyde, which is in turn reduced by the latter enzyme to ethanol. Ethanol production was observed with these modifications but lactic acid and succinic acid were also produced as byproducts. Disruption of the lactate dehydrogenase (ldh) and phosphoenolpyruvate carboxylase (ppc) genes relieved this problem and the resulting strain produced about 10 g l-1 ethanol under oxygen deprivation conditions [87]. Interestingly, C. glutamicum can well resist various growth inhibitors such as furans, phenols and acids that are formed during lignocellulose pretreatment under the chosen conditions, thereby opening up the prospect of ethanol production from biomass [88].
III.4.2 Isobutanol
Recently, the research interest has shifted from ethanol fermentation to production of higher alcohols for use as biofuel due to various advantages offered by the latter such as higher energy density, compatibility with existing infrastructure and lower corrosivity, vapor pressure and hygroscopicity. Isobutanol is one such higher alcohol and can be produced from 2-KIV in two steps. Because C. glutamicum has been successfully engineered for production of the isobutanol precursor 2-KIV [81] it appeared as a suitable organism for isobutanol production. Smith et al. engineered a producer strain by heterologous expression of a B. subtilis alsS, whose product (AHAS) catalyzes the conversion of pyruvic acid to acetolactate, in combination with the overexpression of ilvC and ilvD whose products in turn catalyze the next two steps towards 2-KIV formation. Heterologous expression of a Lactococcus lactis 2-ketoacid decarboxylase gene (kivd) and overexpression of native alcohol dehydrogenase gene (adhA) then converted 2-KIV to isobutanol. pyc encoding pyruvate carboxylase (PCx) and ldh were deleted to direct more carbon through the isobutanol pathway. The resultant strain produced 4.9 g l-1 isobutanol in 120 hours [89]. Blombach et al achieved more than twofold higher titers after re-engineering their 2-KIV producing strain for isobutanol biosynthesis [90]. Apart from deleting ldh, they also deleted a number of other genes to further increase pyruvic acid, 2-KIV and cofactor availability: aceE and pqo deletions served the first purpose, ilvE deletion the second and mdh deletion the third and last. However, the resultant strain grew very poorly on glucose. To overcome this drawback and to improve redox balance, E. coli pntAB operon encoding transhydrogenase was additionally expressed. This strain could then produce 13 g l-1 isobutanol under oxygen deprivation conditions [90].
III.4.3 1,2-propanediol
1,2-propanediol holds a worldwide market of more than half million tons per annum and is used in food, cosmetic and pharmaceutical industries. Production of 1,2-propanediol by C. glutamicum was observed after heterologous expression of E. coli methylglyoxal synthase gene (mgs) and overexpression of a putative aldo-keto reductase gene (cgR_2242). The former enzyme catalyzes the conversion of the glycolytic intermediate dihydroxyacetone phosphate (DHAP) to methylglyoxal, which is reduced to 1,2-propanediol by the latter enzyme. This approach resulted in a strain with a production capability of 1.8 g l-1 in 90 hours via fed-batch fermentation [91]. A high concentration of acetol, a 1,2-propanediol precursor, was also produced and presents a potential hurdle to reach higher 1,2-propanediol titers.
III.5 Sugar alcohols
III.5.1 D-Mannitol
D-Mannitol is used as an osmotic diuretic in pharmaceutical industry and as an alternative sweetener for diabetic patients in food industry and is produced using a chemical hydrogenation procedure. D-Mannitol production from D-fructose as substrate was demonstrated using resting cells of C. glutamicum [92]. The metabolic engineering steps included heterologous expression of: i) Leuconostoc pseudomesenteroides mdh, encoding a mannitol dehydrogenase, that catalyzes D-fructose to D-mannitol conversion, ii) Mycobacterium vaccae N10 fdh, encoding a formate dehydrogenase, that ensures ample cofactor supply in the presence of formate and iii) Z. mobilis glucose/fructose facilitator glf whose product enhances direct D-fructose uptake. The strain was able to form 285 g l-1 D-mannitol over a period of 96 hours using a fed-batch approach [92]. Later on, the native transporters of C. glutamicum for D-fructose were identified and it was shown that overexpression of one of these (iolT1), instead of heterologous expression of glf, also resulted in increased D-fructose uptake rate [93].
III.5.2 Xylitol
Xylitol finds relevance in food industry where it is used as a dietetic sucrose alternative because it has one third the calories of sucrose while being equally sweet and can be produced directly from xylose via a single reduction step. The metabolic engineering for creating a C. glutamicum xylitol producer included: i) heterologous expression of a C. glutamicum ATCC31831 pentose transporter gene (araE), inserted into the chromosomal DNA to improve direct xylose uptake, ii) heterologous expression of a Candida tenuis gene encoding a single-site mutated xylose reductase (XR) to catalyze xylitol formation, iii) deletion of the lactate dehydrogenase gene (ldh) to allow use of cofactor utilized in lactic acid formation reaction for xylitol production instead, and iv) deletion of phosphoenolpyruvate-dependent fructose phosphotransferase gene (ptsF) and xylulokinase gene (xylB) (Fig. 1) to prevent formation of xylitol-phosphate which is toxic for cells. The former enzyme catalyzes xylitol uptake via the phosphoenolpyruvate-dependent fructose phosphotransferase system and the latter is probably involved in its phosphorylation to yield the toxic intermediate. Indeed the deletion of these genes resulted in insensitivity to xylitol and enhanced xylitol productivity. The strain was shown to produce 166 g l-1 xylitol in 21 hours using resting cells and a fed-batch approach [94].
III.6 Cofactor regeneration
Cofactor availability is critical for biotransformation reactions as mentioned above for sugar alcohols. It has recently been shown that deletion of either 6-phosphofructokinase gene (pfkA) or glyceraldehyde 3-phosphate dehydrogenase gene (gapA) resulted in a higher NADPH per glucose yield as compared to wild-type strain, which could then be used for reductive biotransformation of the prochiral methyl acetoacetate to the chiral (R)-methyl 3-hydroxybutyrate (MHB) via expression of a Lactobacillus brevis alcohol dehydrogenase. With this approach, an almost three fold increase of MHB yield could be observed. Deletion of these genes halts the flow through glycolysis, diverts it to pentose phosphate pathway (PPP) and generates a semi-cyclic (pfkA deletion) or cyclic (gapA deletion) PPP, resulting in an increased cofactor production and availability [95].
IV. Summary and Outlook
It is evident that currently there is plentiful research going on to metabolically engineer C. glutamicum: production of more than two-third of the products discussed in this review has been made possible in the last four years only. The availability of the genome sequence and genetic engineering tools for this organism make its rapid and rational manipulation possible. So far, ‘omics’ technologies have not been extensively used in context of metabolic engineering of this bacterium, whereas they do hold the potential to elucidate production mechanisms and constraints on a systems-wide level. It can be expected that metabolic engineering of C. glutamicum will benefit from the increasing availability of gene sequences for pathway enzymes from many different organisms (not only E. coli and S. cerevisiae). In many cases, however, biochemical data rather than only a gene sequence are required to guide metabolic engineering and it is likely that more resources need to be directed towards physiological and biochemical experimental work. It is expected that pathway modeling, ideally in the context of a genome-scale metabolic model will contribute to metabolic engineering. Moreover, technical and conceptual advances to gradually control expression of not one or two, but rather tens of genes will be required to install larger synthetic pathways.
C. glutamicum is an attractive candidate for metabolic engineering as the biotechnological industry looks back at more than fifty years of safe production in the food and feed industry with this bacterium. Moreover, annual million-ton-scale production, such as of L-lysine and L-glutamate by C. glutamicum, cannot be overlooked. The examples described here demonstrate that C. glutamicum holds the potential as a general microbial production platform.
Competing Interests
The authors have declared that no competing interests exist.
References
- 1.Kinoshita S, Udaka S, Shimono M (1957) Studies on the amino acid fermentation. Production of L-glutamic acid by various microorganisms. J Gen Appl Microbiol 3: 193–205 [PubMed] [Google Scholar]
- 2.Ajinomoto (2010) Food Products Business. Available: http://www.ajinomoto.com/ir/pdf/Food-Oct2010.pdf.
- 3.Ajinomoto (2011) Feed-Use Amino Acids Business. Available: http://www.ajinomoto.com/ir/pdf/Feed-useAA-Oct2011.pdf
- 4.Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, et al. (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol 104: 5–25 [DOI] [PubMed] [Google Scholar]
- 5.Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl Microbiol Biotechnol 62: 99–109 [DOI] [PubMed] [Google Scholar]
- 6.Wendisch VF (2003) Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J Biotechnol 104: 273–285 [DOI] [PubMed] [Google Scholar]
- 7.Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104: 155–172 [DOI] [PubMed] [Google Scholar]
- 8.Kelle R, Hermann T, Bathe B (2005) L-Lysine production In Handbook of Corynebacterium glutamicum (Eggeling L, Bott M, eds), CRC Press, Boca Raton, USA, pp 465–488 [Google Scholar]
- 9.Leuchtenberger W, Huthmacher K, Drauz K (2005) Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol 69: 1–8 [DOI] [PubMed] [Google Scholar]
- 10.Shimizu H, Hirasawa T (2007) Production of Glutamate and Glutamate-Related Amino Acids: MolecularMechanism Analysis and Metabolic Engineering In Amino Acid Biosynthesis – Pathways, Regulation and Metabolic Engineering (Wendisch VF, ed), Springer, Heidelberg, Germany: 5: 1–38 [Google Scholar]
- 11.Wittmann C, Becker J (2007) The L-Lysine Story: From Metabolic Pathways to Industrial Production In Amino Acid Biosynthesis – Pathways, Regulation and Metabolic Engineering (Wendisch VF, ed), Springer, Heidelberg, Germany: 5: 39–70 [Google Scholar]
- 12.Kimura E (2005) L-Glutamate Production In Handbook of Corynebacterium glutamicum (Eggeling L, Bott M, eds), CRC Press: Boca Raton, USA: 439–464 [Google Scholar]
- 13.Arndt A, Eikmanns BJ (2008) Regulation of carbon metabolism in Corynebacterium glutamicum In Corynebacteria: genomics and molecular biology (Burkovski A, ed), Caister Academic Press, Wymondham, UK: 155–182 [Google Scholar]
- 14.Blombach B, Seibold GM (2010) Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of L-lysine production strains. Appl Microbiol Biotechnol 86: 1313–1322 [DOI] [PubMed] [Google Scholar]
- 15.Parche S, Burkovski A, Sprenger GA, Weil B, Kramer R, et al. (2001) Corynebacterium glutamicum: a dissection of the PTS. J Mol Microbiol Biotechnol 3: 423–428 [PubMed] [Google Scholar]
- 16.Krause FS, Henrich A, Blombach B, Kramer R, Eikmanns BJ, et al. (2010) Increased glucose utilization in Corynebacterium glutamicum by use of maltose, and its application for the improvement of L-valine productivity. Appl Environ Microbiol 76: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindner SN, Knebel S, Pallerla SR, Schoberth SM, Wendisch VF (2010) Cg2091 encodes a polyphosphate/ATP-dependent glucokinase of Corynebacterium glutamicum. Appl Microbiol Biotechnol 87: 703–713 [DOI] [PubMed] [Google Scholar]
- 18.Park SY, Kim HK, Yoo SK, Oh TK, Lee JK (2000) Characterization of glk, a gene coding for glucose kinase of Corynebacterium glutamicum. FEMS Microbiol Lett 188: 209–215 [DOI] [PubMed] [Google Scholar]
- 19.Lindner SN, Seibold GM, Henrich A, Kramer R, Wendisch VF (2011) Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl Environ Microbiol 77: 3571–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda M, Mizuno Y, Awane S, Hayashi M, Mitsuhashi S, et al. (2011) Identification and application of a different glucose uptake system that functions as an alternative to the phosphotransferase system in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90: 1443–1451 [DOI] [PubMed] [Google Scholar]
- 21.Lindner SN, Seibold GM, Kramer R, Wendisch VF (2011) Impact of a new glucose utilization pathway in amino acid-producing Corynebacterium glutamicum. Bioeng Bugs 2. [DOI] [PubMed] [Google Scholar]
- 22.Kato O, Youn JW, Stansen KC, Matsui D, Oikawa T, et al. (2010) Quinone-dependent D-lactate dehydrogenase Dld (Cg1027) is essential for growth of Corynebacterium glutamicum on D-lactate. BMC Microbiol 10: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, et al. (2005) Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl Environ Microbiol 71: 5920–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neuner A, Heinzle E (2011) Mixed glucose and lactate uptake by Corynebacterium glutamicum through metabolic engineering. Biotechnol J 6: 318–329 [DOI] [PubMed] [Google Scholar]
- 25.Sharkey MA, Maher MA, Guyonvarch A, Engel PC (2011) Kinetic characterisation of recombinant Corynebacterium glutamicum NAD+-dependent LDH over-expressed in E. coli and its rescue of an lldD-phenotype in C. glutamicum: the issue of reversibility re-examined. Arch Microbiol 193: 731–740 [DOI] [PubMed] [Google Scholar]
- 26.Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ (2006) Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J Biotechnol 124: 381–391 [DOI] [PubMed] [Google Scholar]
- 27.Tateno T, Fukuda H, Kondo A (2007) Production of L-Lysine from starch by Corynebacterium glutamicum displaying alpha-amylase on its cell surface. Appl Microbiol Biotechnol 74: 1213–1220 [DOI] [PubMed] [Google Scholar]
- 28.Yao W, Chu C, Deng X, Zhang Y, Liu M, et al. (2009) Display of alpha-amylase on the surface of Corynebacterium glutamicum cells by using NCgl1221 as the anchoring protein, and production of glutamate from starch. Arch Microbiol 191: 751–759 [DOI] [PubMed] [Google Scholar]
- 29.Tateno T, Fukuda H, Kondo A (2007) Direct production of L-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl Microbiol Biotechnol 77: 533–541 [DOI] [PubMed] [Google Scholar]
- 30.Tateno T, Okada Y, Tsuchidate T, Tanaka T, Fukuda H, et al. (2009) Direct production of cadaverine from soluble starch using Corynebacterium glutamicum coexpressing alpha-amylase and lysine decarboxylase. Appl Microbiol Biotechnol 82: 115–121 [DOI] [PubMed] [Google Scholar]
- 31.Brabetz W, Liebl W, Schleifer KH (1991) Studies on the utilization of lactose by Corynebacterium glutamicum, bearing the lactose operon of Escherichia coli. Arch Microbiol 155: 607–612 [DOI] [PubMed] [Google Scholar]
- 32.Barrett E, Stanton C, Zelder O, Fitzgerald G, Ross RP (2004) Heterologous expression of lactose-and galactose-utilizing pathways from lactic acid bacteria in Corynebacterium glutamicum for production of lysine in whey. Appl Environ Microbiol 70: 2861–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaguchi H, Sasaki M, Vertes AA, Inui M, Yukawa H (2008) Engineering of an L-arabinose metabolic pathway in Corynebacterium glutamicum. Appl Microbiol Biotechnol 77: 1053–1062 [DOI] [PubMed] [Google Scholar]
- 34.Sasaki M, Jojima T, Kawaguchi H, Inui M, Yukawa H (2009) Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl Microbiol Biotechnol 85: 105–115 [DOI] [PubMed] [Google Scholar]
- 35.Schneider J, Niermann K, Wendisch VF (2011) Production of the amino acids L-glutamate, L-lysine, L-ornithine and L-arginine from arabinose by recombinant Corynebacterium glutamicum. J Biotechnol 154: 191–198 [DOI] [PubMed] [Google Scholar]
- 36.Kawaguchi H, Vertes AA, Okino S, Inui M, Yukawa H (2006) Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol 72: 3418–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buschke N, Schroder H, Wittmann C (2011) Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol J 6: 306–317 [DOI] [PubMed] [Google Scholar]
- 38.Gopinath V, Meiswinkel TM, Wendisch VF, Nampoothiri KM (2011) Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl Microbiol Biotechnol 92: 985–996 [DOI] [PubMed] [Google Scholar]
- 39.Kotrba P, Inui M, Yukawa H (2003) A single V317A or V317M substitution in Enzyme II of a newly identified beta-glucoside phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiology 149: 1569–1580 [DOI] [PubMed] [Google Scholar]
- 40.Sasaki M, Jojima T, Inui M, Yukawa H (2008) Simultaneous utilization of D-cellobiose, D-glucose, and D-xylose by recombinant Corynebacterium glutamicum under oxygen-deprived conditions. Appl Microbiol Biotechnol 81: 691–699 [DOI] [PubMed] [Google Scholar]
- 41.Youn JW, Jolkver E, Kramer R, Marin K, Wendisch VF (2009) Characterization of the dicarboxylate transporter DctA in Corynebacterium glutamicum. J Bacteriol 191: 5480–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youn JW, Jolkver E, Kramer R, Marin K, Wendisch VF (2008) Identification and characterization of the dicarboxylate uptake system DccT in Corynebacterium glutamicum. J Bacteriol 190: 6458–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teramoto H, Shirai T, Inui M, Yukawa H (2008) Identification of a gene encoding a transporter essential for utilization of C4 dicarboxylates in Corynebacterium glutamicum. Appl Environ Microbiol 74: 5290–5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rittmann D, Lindner SN, Wendisch VF (2008) Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl Environ Microbiol 74: 6216–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Litsanov B, Brocker M, Bott M (2012) Glycerol as a substrate for aerobic succinate production in minimal medium with Corynebacterium glutamicum. Microb Biotechnol: Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wendisch VF, de Graaf AA, Sahm H, Eikmanns BJ (2000) Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J Bacteriol 182: 3088–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerstmeir R, Wendisch VF, Schnicke S, Ruan H, Farwick M, et al. (2003) Acetate metabolism and its regulation in Corynebacterium glutamicum. J Biotechnol 104: 99–122 [DOI] [PubMed] [Google Scholar]
- 48.Kremling A, Kremling S, Bettenbrock K (2009) Catabolite repression in Escherichia coli-a comparison of modelling approaches. Febs J 276: 594–602 [DOI] [PubMed] [Google Scholar]
- 49.Hueck CJ, Hillen W (1995) Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria?. Mol Microbiol 15: 395–401 [DOI] [PubMed] [Google Scholar]
- 50.Kramer R, Lambert C, Hoischen C, Ebbighausen H (1990) Uptake of glutamate in Corynebacterium glutamicum. 1. Kinetic properties and regulation by internal pH and potassium. Eur J Biochem 194: 929–935 [DOI] [PubMed] [Google Scholar]
- 51.Arndt A, Auchter M, Ishige T, Wendisch VF, Eikmanns BJ (2008) Ethanol Catabolism in Corynebacterium glutamicum. J Mol Microbiol Biotechnol 15: 222–233 [DOI] [PubMed] [Google Scholar]
- 52.Becker J, Wittmann C (2012) Bio-based production of chemicals, materials and fuels -Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23: 631–640 [DOI] [PubMed] [Google Scholar]
- 53.Blombach B, Schreiner ME, Bartek T, Oldiges M, Eikmanns BJ (2008) Corynebacterium glutamicum tailored for high-yield L-valine production. Appl Microbiol Biotechnol 79: 471–479 [DOI] [PubMed] [Google Scholar]
- 54.Hasegawa S, Uematsu K, Natsuma Y, Suda M, Hiraga K, et al. (2012) Improvement of the Redox Balance Increases L-Valine Production by Corynebacterium glutamicum under Oxygen Deprivation Conditions. Appl Environ Microbiol 78: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelle R, Hermann T, Weuster-Botz D, Eggeling L, Krämer R, et al. (1996) Glucose-controlled L-isoleucine fed-batch production with recombinant strains of Corynebacterium glutamicum. J Biotechnol 50: 123–136 [Google Scholar]
- 56.Peters-Wendisch P, Stolz M, Etterich H, Kennerknecht N, Sahm H, et al. (2005) Metabolic Engineering of Corynebacterium glutamicum for L-Serine Production. Appl Environ Microbiol 71: 7139–7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peters-Wendisch P, Netzer R, Eggeling L, Sahm H (2002) 3-Phosphoglycerate dehydrogenase from Corynebacterium glutamicum: the C-terminal domain is not essential for activity but is required for inhibition by L-serine. Appl Microbiol Biotechnol 60: 437–441 [DOI] [PubMed] [Google Scholar]
- 58.Stolz M, Peters-Wendisch P, Etterich H, Gerharz T, Faurie R, et al. (2007) Reduced folate supply as a key to enhanced L-serine production by Corynebacterium glutamicum. Appl Environ Microbiol 73: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Appleton J (2002) Arginine: Clinical potential of a semi-essential amino. Altern Med Rev 7: 512–522 [PubMed] [Google Scholar]
- 60.Ikeda M, Mitsuhashi S, Tanaka K, Hayashi M (2009) Reengineering of a Corynebacterium glutamicum L-arginine and L-citrulline producer. Appl Environ Microbiol 75: 1635–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park SD, Lee JY, Sim SY, Kim Y, Lee HS (2007) Characteristics of methionine production by an engineered Corynebacterium glutamicum strain. Metab Eng 9: 327–336 [DOI] [PubMed] [Google Scholar]
- 62.Soarez PC, Oliveira AC, Padovan J, Parise ER, Ferraz MB (2009) A critical analysis of studies assessing L-ornithine-L-aspartate (LOLA) in hepatic encephalopathy treatment. Arq Gastroenterol 46: 241–247 [DOI] [PubMed] [Google Scholar]
- 63.Hwang JH, Hwang GH, Cho JY (2008) Effect of increased glutamate availability on L-ornithine production in Corynebacterium glutamicum. J Microbiol Biotechnol 18: 704–710 [PubMed] [Google Scholar]
- 64.Shi F, Li Y (2011) Synthesis of gamma-aminobutyric acid by expressing Lactobacillus brevis-derived glutamate decarboxylase in the Corynebacterium glutamicum strain ATCC 13032. Biotechnology Letters 33: 2469–2474 [DOI] [PubMed] [Google Scholar]
- 65.Zhao Z, Ding JY, Ma WH, Zhou NY, Liu SJ (2012) Identification and characterization of gamma-aminobutyric acid uptake system GabPCg (NCgl0464) in Corynebacterium glutamicum. Appl Environ Microbiol 78: 2596–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stabler N, Oikawa T, Bott M, Eggeling L (2011) Corynebacterium glutamicum as a host for synthesis and export of D-Amino Acids. J Bacteriol 193: 1702–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider J, Wendisch VF (2010) Putrescine production by engineered Corynebacterium glutamicum. Applied Microbiology and Biotechnology 88: 859–868 [DOI] [PubMed] [Google Scholar]
- 68.Schneider J, Eberhardt D, Wendisch VF (2012) Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system. Appl Microbiol Biotechnol 95: 169–178 [DOI] [PubMed] [Google Scholar]
- 69.Kind S, Jeong WK, Schroder H, Zelder O, Wittmann C (2010) Identification and elimination of the competing N-acetyldiaminopentane pathway for improved production of diaminopentane by Corynebacterium glutamicum. Appl Environ Microbiol 76: 5175–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mimitsuka T, Sawai H, Hatsu M, Yamada K (2007) Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem 71: 2130–2135 [DOI] [PubMed] [Google Scholar]
- 71.Fukui K, Koseki C, Yamamoto Y, Nakamura J, Sasahara A, et al. (2011) Identification of succinate exporter in Corynebacterium glutamicum and its physiological roles under anaerobic conditions. J Biotechnol 154: 25–34 [DOI] [PubMed] [Google Scholar]
- 72.Litsanov B, Brocker M, Bott M (2012) Toward Homosuccinate Fermentation: Metabolic Engineering of Corynebacterium glutamicum for Anaerobic Production of Succinate from Glucose and Formate. Appl Environ Microbiol 78: 3325–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okino S, Noburyu R, Suda M, Jojima T, Inui M, et al. (2008) An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol 81: 459–464 [DOI] [PubMed] [Google Scholar]
- 74.Litsanov B, Kabus A, Brocker M, Bott M (2012) Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microbial Biotechnology 5: 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McKinlay JB, Vieille C, Zeikus JG (2007) Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol 76: 727–740 [DOI] [PubMed] [Google Scholar]
- 76.Li Y, Chen J, Lun SY (2001) Biotechnological production of pyruvic acid. Appl Microbiol Biotechnol 57: 451–459 [DOI] [PubMed] [Google Scholar]
- 77.Wieschalka S, Blombach B, Eikmanns BJ (2012) Engineering Corynebacterium glutamicum for the production of pyruvate. Appl Microbiol Biotechnol 94: 449–459 [DOI] [PubMed] [Google Scholar]
- 78.Okino S, Inui M, Yukawa H (2005) Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 68: 475–480 [DOI] [PubMed] [Google Scholar]
- 79.Okino S, Suda M, Fujikura K, Inui M, Yukawa H (2008) Production of D-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 78: 449–454 [DOI] [PubMed] [Google Scholar]
- 80.Jo JH, Seol HY, Lee YB, Kim MH, Hyun HH, et al. (2012) Disruption of genes for the enhanced biosynthesis of alpha-ketoglutarate in Corynebacterium glutamicum. Can J Microbiol 58: 278–286 [DOI] [PubMed] [Google Scholar]
- 81.Krause FS, Blombach B, Eikmanns BJ (2010) Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl Environ Microbiol 76: 8053–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Demain AL (2007) REVIEWS: The business of biotechnology. Industrial Biotechnology 3: 269–283 [Google Scholar]
- 83.Sahm H, Eggeling L (1999) D-Pantothenate synthesis in Corynebacterium glutamicum and use of panBC and genes encoding L-valine synthesis for D-pantothenate overproduction. Appl Environ Microbiol 65: 1973–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chassagnole C, Diano A, Letisse F, Lindley ND (2003) Metabolic network analysis during fed-batch cultivation of Corynebacterium glutamicum for pantothenic acid production: first quantitative data and analysis of by-product formation. J Biotechnol 104: 261–272 [DOI] [PubMed] [Google Scholar]
- 85.Hüser AT, Chassagnole C, Lindley ND, Merkamm M, Guyonvarch A, et al. (2005) Rational design of a Corynebacterium glutamicum pantothenate production strain and its characterization by metabolic flux analysis and genome-wide transcriptional profiling. Appl Environ Microbiol 71: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.EIA (2011) U.S. Energy Information Administration. Annual Energy Review 2010. Available: http://www.eia.gov/totalenergy/data/annual/pdf/aer.pdf
- 87.Inui M, Kawaguchi H, Murakami S, Vertes AA, Yukawa H (2004) Metabolic Engineering of Corynebacterium glutamicum for Fuel Ethanol Production under Oxygen-Deprivation Conditions. J Mol Microbiol Biotechnol 8: 243–254 [DOI] [PubMed] [Google Scholar]
- 88.Sakai S, Tsuchida Y, Nakamoto H, Okino S, Ichihashi O, et al. (2007) Effect of lignocellulose-derived inhibitors on growth of and ethanol production by growth-arrested Corynebacterium glutamicum R. Appl Environ Microbiol 73: 2349–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith KM, Cho KM, Liao JC (2010) Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol 87: 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, et al. (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77: 3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niimi S, Suzuki N, Inui M, Yukawa H (2011) Metabolic engineering of 1,2-propanediol pathways in Corynebacterium glutamicum. Appl Microbiol Biotechnol 90: 1721–1729 [DOI] [PubMed] [Google Scholar]
- 92.Bäumchen C, Bringer-Meyer S (2007) Expression of glf Z.m. increases D-mannitol formation in whole cell biotransformation with resting cells of Corynebacterium glutamicum. Appl Microbiol Biotechnol 76: 545–552 [DOI] [PubMed] [Google Scholar]
- 93.Bäumchen C, Krings E, Bringer S, Eggeling L, Sahm H (2009) Myo-inositol facilitators IolT1 and IolT2 enhance D-mannitol formation from D-fructose in Corynebacterium glutamicum. FEMS Microbiol Lett 290: 227–235 [DOI] [PubMed] [Google Scholar]
- 94.Sasaki M, Jojima T, Inui M, Yukawa H (2010) Xylitol production by recombinant Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 86: 1057–1066 [DOI] [PubMed] [Google Scholar]
- 95.Siedler S, Lindner SN, Bringer S, Wendisch VF, Bott M (2012) Reductive whole-cell biotransformation with Corynebacterium glutamicum: improvement of NADPH generation from glucose by a cyclized pentose phosphate pathway using pfkA and gapA deletion mutants. Appl Microbiol Biotechnol: Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]