Summary
Neural plasticity requires protein synthesis, however the identity of newly synthesized proteins generated in response to plasticity-inducing stimuli remains unclear. We used in vivo bio-orthogonal non-canonical amino acid tagging (BONCAT) with the methionine analog, azidohomoalanine (AHA) combined with multidimensional protein identification technique (MudPIT) to identify proteins that are synthesized in tadpole brain over 24 h. We induced conditioning-dependent plasticity of visual avoidance behavior. Induction of behavioral plasticity required NMDA and Ca2+-permeable AMPA receptors, αCaMKII and rapid protein synthesis. Combining BONCAT with Western blots revealed that proteins including αCaMKII, MEK1, CPEB, and GAD65 are synthesized during conditioning. Acute synthesis of CPEB during conditioning is required for behavioral plasticity as well as conditioning-induced synaptic and structural plasticity in the tectal circuit. We outline a signaling pathway regulating protein synthesis-dependent behavioral plasticity in intact animals, identify newly synthesized proteins induced by visual experience and demonstrate a requirement for acute synthesis of CPEB in plasticity.
Introduction
Synaptic plasticity is thought to be a cellular substrate for experience-dependent behavioral plasticity. Calcium influx through NMDAR and Ca2+-permeable AMPAR drives rapid changes in synaptic efficacy (Liu and Zukin, 2007; Malenka, 2003) and triggers activity-dependent gene transcription and protein synthesis (Chen et al., 2012a; Nedivi, 1999; Sutton and Schuman, 2006; West and Greenberg, 2011). Activity-regulated protein translation by mRNA-binding proteins provides a mechanism to coordinate expression of a cohort of transcripts (Keene and Tenenbaum, 2002). Studies in hippocampal neuron cultures (Atkins et al., 2004; Wu et al., 1998) and mammalian visual cortex (Wells et al., 2001) suggest that a cascade of NMDAR activation, calcium influx and αCaMKII activation result in CPEB phosphorylation and relief of translational inhibition. Although CPEB has been shown to play a role in synaptic plasticity across phyla (Berger-Sweeney et al., 2006; Bestman and Cline, 2008; Dziembowska et al., 2012; Keleman et al., 2007; Oruganty-Das et al., 2012; Richter, 2010; Si et al., 2003; Wells et al., 2001), evidence that it is required for behavioral plasticity in vertebrates is limited (Berger-Sweeney et al., 2006). In the Xenopus visual system, NMDAR, CaMKII and CPEB regulate synaptic strength, experience-dependent structural plasticity and tectal cell visual responses (Bestman and Cline, 2008; Rajan et al., 1999; Sin et al., 2002; Wu et al., 1996; Wu and Cline, 1998). Recent work has shown that tadpoles exhibit an innate visual avoidance behavior, in which animals avoid an approaching visual stimulus (Dong et al., 2009; Shen et al., 2011), however it is unclear whether the visual avoidance behavior shows experience-dependent plasticity or what cellular mechanisms govern the behavioral plasticity.
Bio-orthogonal metabolic labeling and click chemistry have advanced the study of proteins (Best, 2009; Ngo and Tirrell, 2011; Speers and Cravatt, 2004). Azidohomoalanine (AHA) is a non-canonical amino acid (ncAA) methionine analog that is incorporated into newly synthesized proteins in place of methionine. AHA’s highly reactive azide group does not react with functional groups in cells, but efficiently reacts with biotin-alkyne using copper-catalyzed azide-alkyne cycloaddition (CuAAC) in a click chemistry reaction. Furthermore, the small size of the reactive group does not interfere with protein function and is not toxic to cells or animals (Beatty and Tirrell, 2008; Best, 2009; Dieterich et al., 2010; Dieterich et al., 2006; Hinz et al., 2012; Melemedjian et al., 2010; Ngo and Tirrell, 2011; Yang et al., 2010). Because almost all proteins have at least one methionine (97.9% of Xenopus transcripts in RefSeq begin with methionine), this method can provide an accurate report of newly synthesized proteins. AHA-biotin labeled proteins have been detected after AHA exposure in cultured neurons and non-neuronal cells (Beatty and Tirrell, 2008; Choi et al., 2012; Dieterich et al., 2010; Dieterich et al., 2006; Dziembowska et al., 2012; Melemedjian et al., 2010), and in zebrafish larvae (Hinz et al., 2013; Hinz et al., 2012) using Western blots or fluorescence (FUNCAT) to detect AHA-labeled proteins, however direct detection of AHA-biotin-modified peptides by MudPIT has been challenging.
Here we demonstrate that visual conditioning (VC) induces protein synthesis-dependent plasticity of visual avoidance behavior. Using BONCAT and MudPIT, we identify ~1000 proteins in the tadpole brain that are synthesized over 24 h. We also use BONCAT with Western blots to identify proteins that are induced in response to VC, including CPEB. Finally, we demonstrate that acute synthesis of CPEB during VC is required for behavioral plasticity and the underlying synaptic and structural plasticity in the tectum. In contrast to the prevailing model that protein synthesis is required for late maintenance phases of plasticity, our data suggest that protein synthesis is required earlier, during induction of plasticity and identifies key players in early protein synthesis-dependent plasticity.
Results
Experience-dependent plasticity of visual avoidance behavior
Tadpoles escape from an approaching object by changing their swim trajectory when an object approaches the animal at approximately right angles to the eye (Figure 1A). We determined the avoidance index (AI, % of avoidance responses per 10 trials) in response to 0.4 cm stimuli (Dong et al., 2009; Shen et al., 2011) over 4–24 hours by measuring AI during 1-minute test periods with half an hour intervals between tests. We did not observe any habituation of AI when animals were tested over 7 hours (Figure 1B), indicating our method is suitable for studies of behavioral plasticity over this time frame. Since tadpoles are not pre-screened for high performance of avoidance behavior (Shen et al., 2011), the average AI for control animals is ~40%, providing the opportunity to detect changes in AI.
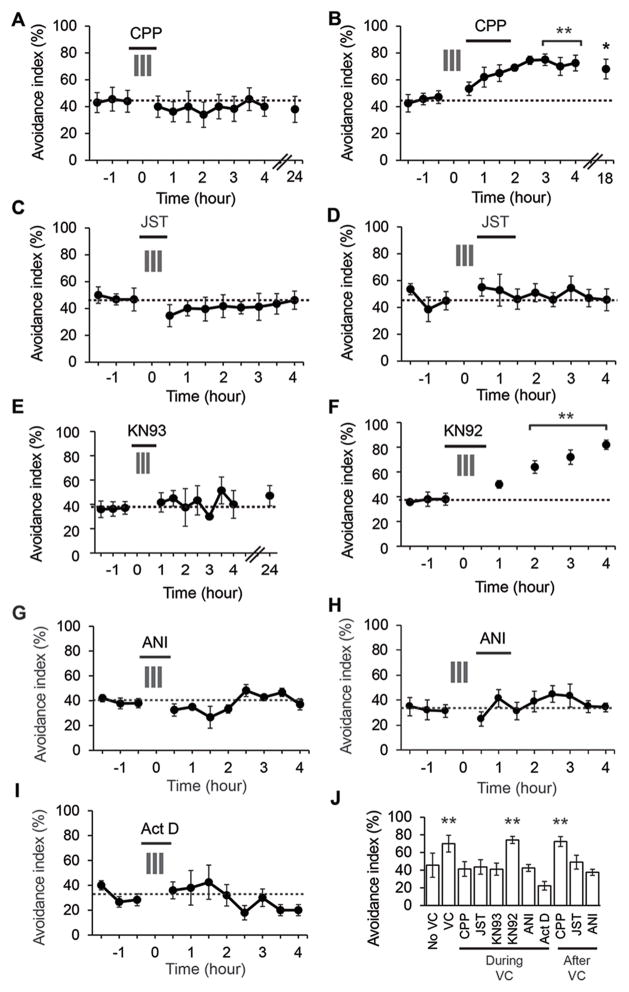
Figure 1.
Conditioning-dependent plasticity of visual avoidance behavior A. Diagram of visual avoidance behavior. When a stimulus approaches a freely swimming tadpole, the animal rapidly changes its swim trajectory. B. The avoidance index (AI) remains constant over 7 hours and is unchanged after 24 hours. Dotted line is the average AI over the first three tests. C. VC for 30 minutes during the period marked with the grey bars significantly enhanced AI for at least 24 h. D, E. AI significantly increased compared to baseline when tested up to 24 h after 2 h or 4 h of VC. **P<0.01, *P<0.05. N= 6–12 animals per group. See also Figure S1.
We tested whether VC affects visual avoidance behavior by exposing freely swimming animals to a stimulus composed of bars moving at 0.3 Hz in 4 directions in pseudorandom order and testing visual avoidance. AI was measured three times at 30-minute intervals to establish a baseline before tadpoles were exposed to VC. Exposure to 30 minutes of conditioning consisting of 3 five-minute episodes of moving bars with 5-minute intervals between episodes significantly increased AI for 24 hours (Figure 1C). Exposure to 2 or 4 hours of continuous VC significantly improved AI when tested 30 minutes or 1 day after conditioning (Figure 1D, E). VC did not significantly affect responses to other stimuli (Figure S1). We used the 30-minute VC protocol in the following experiments to investigate mechanisms underlying rapid induction of visual avoidance plasticity.
NMDAR, Ca2+-permeable AMPAR and CaMKII are required for plasticity of visual avoidance behavior
Exposing animals to (±)-3-(2-carboxypiperazin-4-yl)-propyl-1-phosphonic acid (CPP, 25 μM), a potent and selective NMDA receptor antagonist, during but not after VC blocked plasticity of avoidance behavior (Figure 2A, B). Exposing tadpoles to Joro Spider Toxin (JST, 500 nM), a specific Ca2+-permeable GluAR blocker, during or just after VC blocked behavioral plasticity (Figure 2C). Exposure to JST without VC does not affect AI (Figure S2). Exposing tadpoles to the CaMKII inhibitor KN93 (5 μM) during VC blocked behavioral plasticity (Figure 2E), while the inactive analog, KN92 (5 μM) did not (Figure 2F).
Figure 2.
Behavioral plasticity requires calcium-dependent signaling, gene transcription and protein synthesis. A,B. CPP (25 μM) during but not after VC blocked visual avoidance plasticity. C, D. Joro Spider Toxin (JST, 500 nM) during or after VC blocks behavioral plasticity. E, F. KN93 (5 μM) but not KN92 (5 μM) blocks behavioral plasticity. G,H. Anisomycin (ANI, 25 μM) during or after VC blocks plasticity. I. Actinomycin D (Act D, 25 μM) during VC blocks behavioral plasticity. J. AI (mean ± SEM) from 3–4 hours after VC. N=8–16 animals per group. **P<0.01, *P<0.05. See also Figure S2.
Visual avoidance plasticity requires protein synthesis during conditioning
Protein synthesis is thought to be necessary for late stages of learning and memory (Agranoff and Klinger, 1964; Chen et al., 2012a; Flexner et al., 1963; Sutton and Schuman, 2006). Exposing animals to anisomycin (ANI, 25 μM), a protein synthesis inhibitor, during or immediately after VC, blocked behavioral plasticity (Figure 2G, H). Furthermore, exposing animals to actinomycin D (Act D, 25 μM), a transcriptional inhibitor, during VC, also blocked behavioral plasticity (Figure 2I). Exposing tadpoles to ANI or Act D for 8 hour does not affect baseline AI (Figure S2), indicating that the short-term drug exposures used here do not alter the animal’s health or sensorimotor responses. Summary plots of AI measured 3–4 h after VC show that behavioral plasticity depends on NMDAR receptor, Ca2+-permeable AMPAR and CaMKII, as well as gene transcription and protein translation (Figure 2J).
Acute labeling of newly synthesized proteins in vivo
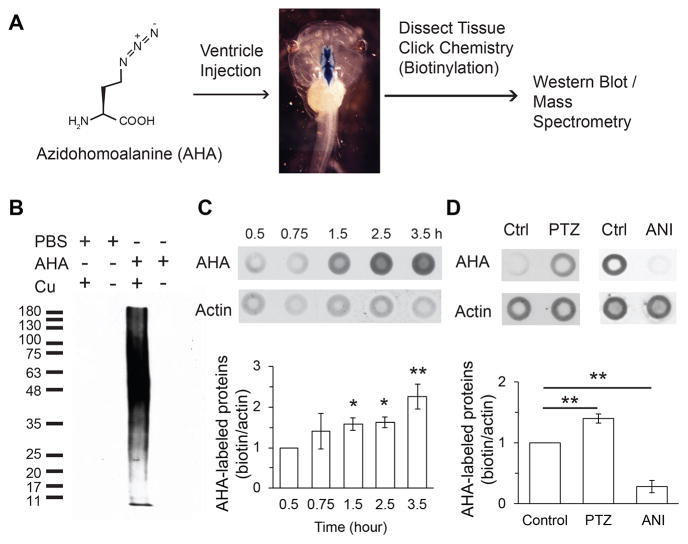
We used BONCAT with AHA incorporation to label newly synthesized CNS protein in intact tadpoles (Figure 3A). Tecta harvested 2 hours after ventricular AHA injection show extensive labeling of biotinylated proteins over a wide range of molecular weights (Figure 3B). Newly synthesized proteins labeled by BONCAT can be detected within 30 min of AHA injection and increase over 3.5 hours (Figure 3C).
Figure 3.
Rapid labeling of newly synthesized proteins in vivo
A. Diagram of AHA labeling of newly synthesized proteins and detection by Western blot or MudPIT. B. Newly synthesized proteins detected 2 h after ventricular AHA injection. PBS injection or click chemistry without copper produce minimal labeling. C,D. Quantification of AHA labeling. Top: Dot blots of AHA-biotin label and C4-actin, for normalization. Bottom. Relative changes in AHA labeling. Whole brains were dissected 3 h after AHA injection. C. AHA-labeling increases over 3.5 hours after ventricular AHA injection. D. ANI (25 μM in rearing solution) significantly decreases AHA labeling and PTZ (15 mM in rearing solution) significantly increases AHA labeling compared to controls. Relative intensity of biotin labeling: PTZ =1.40 ± 0.07; ANI = 0.28 ± 0.10. N=3–5. Ventricular injection of ANI or cycloheximide also decreased AHA labeling compared to PBS-injected controls (Figure S3). **P<0.01, *P<0.05.
To test whether AHA incorporation can be used to detect changes in protein synthesis in vivo, we exposed tadpoles to ANI and to the GABA receptor antagonist, pentylenetetrazol (PTZ). PTZ induces seizure (Hewapathirane et al., 2008), which increases gene expression and protein synthesis (Hinz et al., 2012; Worley et al., 1990). ANI significantly decreased AHA incorporation and PTZ significantly increased AHA labeling compared to untreated tadpoles (Figure 3D).
MudPIT analysis of AHA-biotin labeled newly synthesized proteins
We used MudPIT to analyze newly synthesized proteins in tadpole brains. Stage 47/48 tadpoles received 2 ventricular AHA injections over 24 h and brains were dissected 1–2 h after the 2nd injection. We optimized the CuAAC click chemistry reaction and established methods to enrich mass spectrometry samples for peptides containing biotinylated AHA. We identified proteins based on direct detection of peptides containing biotinylated AHA by measuring the molecular mass shifts of biotin-modified peptides compared to unmodified peptides to eliminate false positive calls. From the brains of 1200–1500 AHA-injected tadpoles, we detected 992 newly synthesized proteins from 1617 AHA-biotin-modified peptides. AHA-biotin labeled proteins were categorized according to the protein’s annotated cellular localization using STRAP (Software Tool for Researching Annotations of Proteins) (Bhatia et al., 2009)(Figure 4A and Table S1). The newly synthesized proteins were annotated to a wide variety of cellular compartments including the nucleus, cytoplasm and cytoskeleton. We searched our dataset for proteins that are functionally related to neural circuit development and plasticity, and present them according to functional categories including translation regulation, protein kinases and phosphatases, receptors, channels, membrane trafficking and cell adhesion (Figure 4B and Table S2). Key players in synaptic plasticity were AHA labeled, including protein phosphatases and protein kinases, such as CaMKII, MAPK, PI3K and PKA, synaptic proteins, including SNAREs, Homer, oligophrenin and AMPA and NMDA receptors, and translational regulators, such as translation initiation and elongation factors, the mRNA binding proteins, CPEB, staufen, and pumilio, and growth factors, including BDNF. These data indicate that in vivo BONCAT can be used to identify newly synthesized proteins based on direct detection of AHA-biotin modified peptides by MudPIT. The data demonstrate the breadth and diversity of the newly synthesized proteome generated over 24 h in the intact brain.
Figure 4.
Mass spectrometric analysis of AHA incorporation in vivo identifies newly synthesized CNS proteins. (A) Annotation of AHA-biotin labeled proteins according to sub-cellular compartments and organelles using STRAP. Proteins in each compartment are listed in Table S1. (B) Annotation of AHA-biotin labeled proteins according to function in neurons. Proteins are listed in Table S2.
Identification of newly synthesized proteins induced by VC
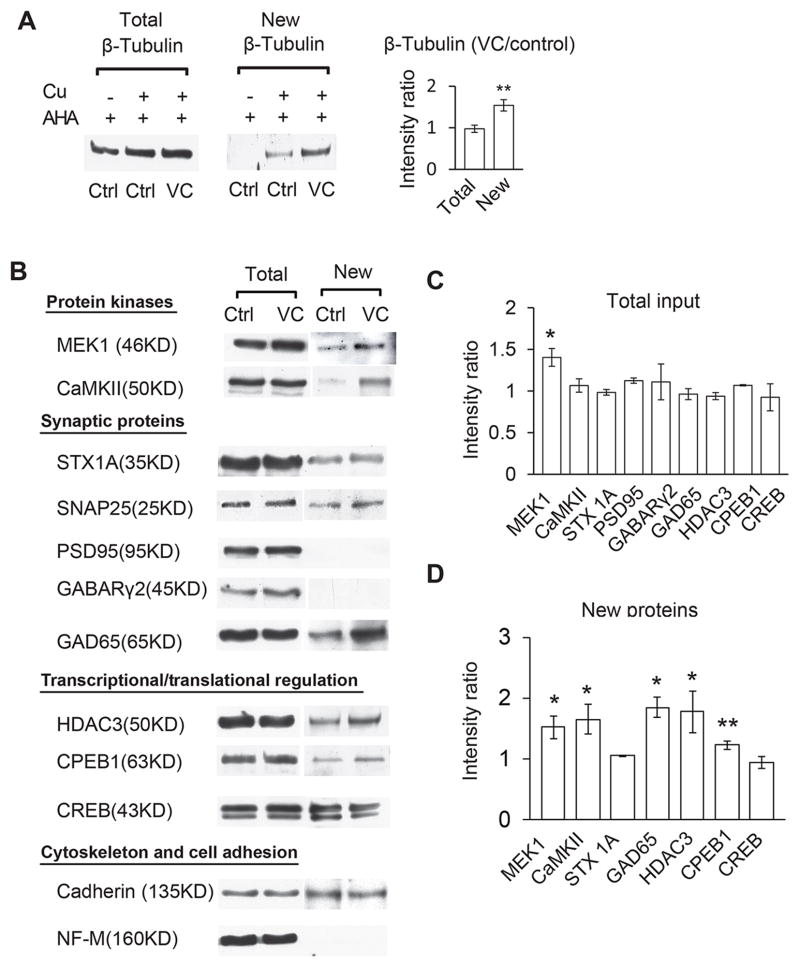
We exposed AHA-injected animals to VC for 30 minutes, and after 2 hours in ambient light, optic tecta were dissected. Ventricular injection of 500 mM AHA does not affect visual avoidance behavior or behavioral plasticity (Figure S4), confirming studies in Zebrafish (Hinz et al., 2012), indicating that AHA can be used to assess protein synthesis in vivo without interfering with neural circuit function or behavior. Proteins for which antibodies are available were analyzed as shown in Figure 5A. First we estimated protein levels in an aliquot of the tectal homogenate by measuring β-tubulin by Western blot. The remainder of the sample was processed for click chemistry and used to assess newly synthesized AHA-biotin tagged β-tubulin. The same blots from the click chemistry samples were stripped and labeled with anti-biotin antibody to reveal the newly synthesized AHA-biotin labeled β-tubulin (Figure 5A). Comparable amounts of β-tubulin were detected in samples from control and VC tadpoles (Figure 5A, left panel), but more newly synthesized AHA-biotin labeled β-tubulin was detected after VC (Figure 5A, middle and right panels). These data indicate that BONCAT can dramatically increase the sensitivity to detect changes in protein synthesis.
Figure 5.
Identification of proteins synthesized in response to visual conditioning A. Analysis of AHA-labeled β-tubulin relative to total β-tubulin in tecta from control and VC tadpoles. Left panel: Total β-tubulin in control and VC samples is comparable (VC/control = 0.98 ± 0.09). Middle and right panels: VC increases AHA-biotin labeled β-tubulin (VC/control=1.5±0.2; N=3). AHA-biotin labeled β-tubulin is not detected when copper is omitted from the reaction. B. VC increases protein synthesis. Protein candidates were measured in the total input and as biotinylated proteins from control and VC animals. C,D. Quantification of data in B. Relative labeling intensities (VC/control) of total input (C) or AHA-biotin (D). N=3 independent experiments for each candidate. * P<0.05, ** P<0.01.
Analysis of AHA-biotin labeling in candidate proteins showed that VC rapidly increased the synthesis of some proteins, including the protein kinases MEK1 and CaMKII, the histone deacetylase, HDAC3, the translational regulator, CPEB1, and the GABA synthetic protein, GAD65. AHA-labeling in other proteins was not changed by VC, including synaptic proteins, syntaxin 1A (STX1A) and SNAP25, the transcription factor, CREB, and the cell adhesion molecule, Cadherin. Finally, AHA incorporation was not detected in several candidates we tested, NF-M, PSD95, and γ2 subunit of GABAA receptors (Figure 5B-D).
Visual avoidance behavior is regulated by CPEB signaling
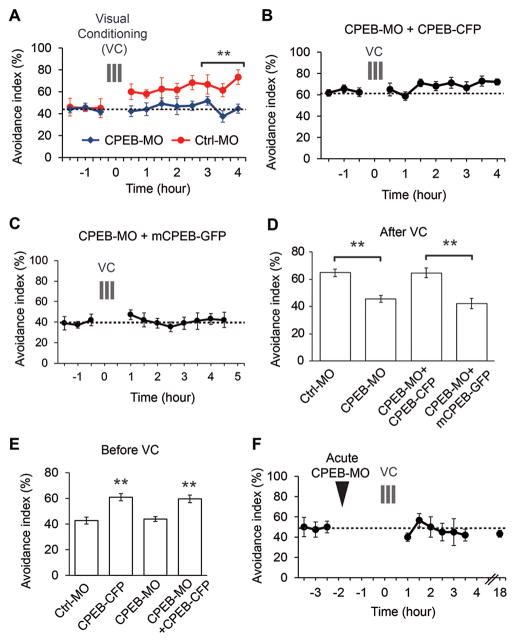
We electroporated tadpoles with translation-blocking morpholinos against CPEB (CPEB-MO), which knock down CPEB and decrease CPEB function (Bestman and Cline, 2008) and subjected them to VC 2 days later. Electroporation distributes morpholinos throughout the tectum (Bestman and Cline, 2013). CPEB-MO or control morpholino (Ctrl-MO) did not affect baseline AI, but CPEB-MO specifically blocked VC-induced visual avoidance plasticity (Figure 6A,D,E). Co-electroporating CPEB-MO and morpholino-insensitive Xenopus CPEB-CFP increased baseline AI, which was not affected by CPEB-MO (Figure 6B, D,E), suggesting that overexpression of CPEB occluded the effect of VC on behavior. By contrast, co-electroporating mutant CPEB, which lacks the phosphorylation sites required to activate CPEB (Wu et al., 1998), does not affect baseline AI and blocks the VC-induced behavioral plasticity (Figure 6C-E). Data presented in Figure 5 indicate that VC acutely increases CPEB synthesis. Electroporating CPEB-MO immediately before VC blocked behavioral plasticity (Figure 6F), suggesting that newly synthesized CPEB induced during VC is required for plasticity of visuo-motor behavior. Consistent with this, CPEB-MO blocks VC-induced CaMKII phosphorylation and decreases VC-induced AHA labeling (Figure S5).
Figure 6.
CPEB is required for plasticity of avoidance behavior.
A. CPEB-MO (blue) blocks VC-induced visual avoidance behavior seen with control morpholino (Ctrl-MO, red). Morpholinos were electroporated 2 days before VC and behavioral testing. B. Co-electroporation of CPEB-MO and full length Xenopus CPEB-CFP increases baseline AI and occludes VC-induced plasticity. C. Co-electroporation of mutant Xenopus CPEB-GFP, lacking CaMKII phosphorylation sites with CPEB-MO fails to rescue the decrease in VC-induced plasticity seen with CPEB MO alone. D. AI (mean ±SEM) 3–4 h after VC. E. Baseline AI (mean ±SEM) before VC. F. CPEB-MO electroporated 2 h before VC blocks behavioral plasticity for at least 18 h. N=8–16 animals per group. **P<0.01. See also Figure S5.
VC-dependent retinotectal circuit plasticity is blocked by acute CPEB knockdown
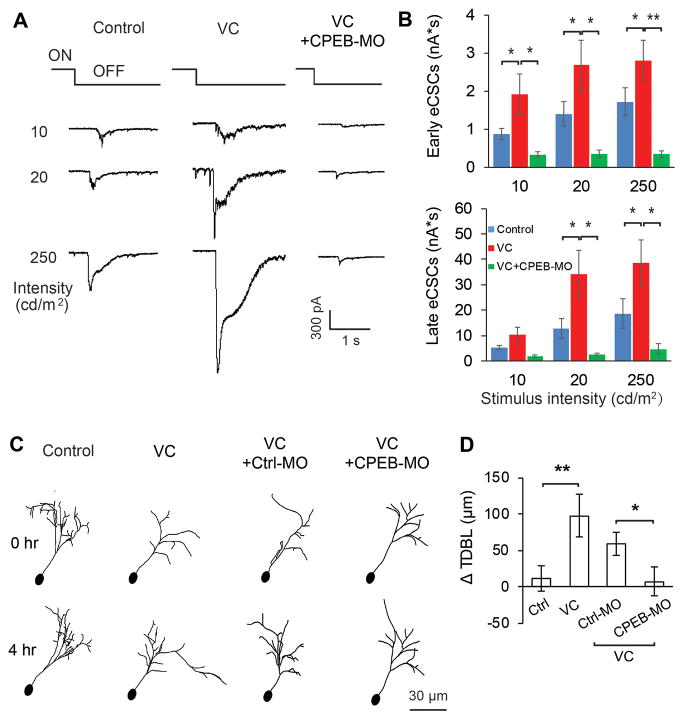
We recorded visually-evoked excitatory compound synaptic currents (CSCs) from tectal neurons in animals with or without VC. Visually-evoked retinotectal responses and recurrent tectal circuit activity are significantly greater when recorded 1–3 hours after VC (Figure 7A,B). We electroporated tadpoles with CPEB-MO 2 hours before VC and recorded visually-evoked responses within 3 hours afterwards. Acute CPEB-MO blocked VC-induced potentiation of visually-evoked responses (Figure 7A,B).
Figure 7.
Conditioning-dependent synaptic and structural plasticity requires acute CPEB synthesis
A. Visual stimulus-induced excitatory compound synaptic currents (eCSCs) from control, VC and VC + CPEB-MO animals at light intensities of 10, 20, 250 cd/m2. B. VC-induced increases in early (<100ms) and late (>100ms) components of CSCc are significantly inhibited by CPEB-MO. Control and Ctrl-MO data were combined because they are not significantly different (Early eCSCs (nA*s): 10 cd/m2: Control = 0.88±0.24nA; Ctrl-MO = 0.87±0.21nA, P=0.97; 20 cd/m2: Control = 1.91±0.68nA; Ctrl-MO = 1.02±0.19nA, P=0.26; 300 cd/m2: Control = 2.47±0.73nA; Ctrl-MO = 1.15±0.22nA, P=0.14. Late eCSC’s: 10 cd/m2: Control = 4.74±1.63nA; Ctrl-MO = 5.57±1.04nA, P=0.66; 20 cd/m2: Control = 20.34±8.9nA; Ctrl-MO = 7.18±1.32nA, P=0.12; 300 cd/m2: Control = 29.45±12.33nA; Ctrl-MO = 10.52±3.39nA, P=0.12.). MO’s were electroporated 2 h before recording. N=7–8 neurons/group) C. Drawings of representative neurons imaged in vivo over 4 h in animals exposed to ambient light, VC, VC+Ctrl-MO and VC+CPEB-MO. MO’s were electroporated 2 h before imaging. D. VC significantly increases total dendritic branch length (TDBL) in control and Ctrl-MO neurons, but not in CPEB-MO neurons. *P<0.05; **P<0.01.
In vivo time lapse imaging of optic tectal neuronal dendrites shows VC-induced increase in dendritic arbor elaboration (Figure 7C), as previously described (Sin et al., 2002). Electroporating CPEB-MO 2 h before time-lapse imaging specifically blocked the VC-induced structural plasticity of tectal cell dendrites (Figure 7C,D). Together, these data suggest that acutely decreasing CPEB synthesis simultaneously blocks VC-induced tectal cell structural plasticity, synaptic plasticity in retinotectal circuit and behavioral plasticity.
Discussion
This study demonstrates that visual avoidance behavior in Xenopus is plastic and increases after visual conditioning. We demonstrate that a signaling pathway including NMDAR, GluAR, CaMKII and CPEB is required for VC-induced plasticity of visual avoidance behavior. Furthermore, the behavioral plasticity requires proteins whose synthesis is induced by VC. By combining AHA incorporation, BONCAT and MudPIT, we identified the newly synthesized brain proteome labeled over 24 h in the developing brain in vivo. We show that CPEB, CaMKII, MEK1, HDAC3 and GAD65 are among the AHA-labeled proteins whose de novo synthesis is increased in response to VC. Finally, we show that delivery of translation-blocking morpholinos for CPEB immediately prior to conditioning blocks behavioral, synaptic and structural plasticity in the tectal circuit. We expect that improved strategies we present here to label and identify newly synthesized proteins from the intact brain of behaving animals by MudPIT and Western blot will be broadly applicable to a variety of experimental systems and conditions.
In vivo labeling and screening of newly synthesized proteins
Since the original observations that protein synthesis inhibitors block learning in goldfish and mice (Agranoff and Klinger, 1964; Flexner et al., 1963), considerable effort has been devoted to identify proteins synthesized during plasticity, starting with labeling newly synthesized proteins with radioactive amino acids (Duffy et al., 1981; Shashoua, 1977a, b). More recent work has used stable isotope labeling with amino acids in cell culture (SILAC) combined with mass spectrometry to identify and quantify differences in brain protein expression (Konzer et al., 2013; Liao et al., 2008), however, isotope labeling for global proteomic analysis does not distinguish newly synthesized proteins from pre-existing proteins, because they are chemically identical. BONCAT labeling in neurons followed by detection of biotinylated proteins by Western blot has permitted gross quantification of global changes in levels of protein synthesis and identification of specific candidate proteins (Dziembowska et al., 2012; Hinz et al., 2012; Hodas et al., 2012).
A previous study in which Zebrafish embryos were exposed animals to 4 mM AHA in embryo medium demonstrated a gradual accumulation of AHA-biotin labeled proteins over 72 h (Hinz et al., 2012). To increase the temporal resolution of the AHA labeling and to compete with endogenous methionine, which interferes with AHA incorporation into nascent proteins, we injected 150–500 mM AHA into the brain ventricle. A single ventricular injection of AHA is sufficient to label new proteins in vivo within 1–2 hour after injection. The increased temporal resolution of AHA-labeling was key to our ability to identify proteins whose synthesis is rapidly regulated by VC.
AHA biotinylation by click chemistry permits enrichment of newly synthesized proteins for subsequent analysis by mass spectrometry (Choi et al., 2012; Hodas et al., 2012). In previous studies, biotinylated proteins were enriched by incubating complex protein mixtures with Neutravidin resin. The bound proteins, including biotinylated proteins and proteins that are non-specifically or indirectly retained on the resin, were digested with protease and resultant peptides were identified by MudPIT. This protocol makes it difficult to identify peptides that are directly modified by the incorporation of the AHA, because they are a minority of peptides in the complex mixture applied to MS/MS. To address this problem of signal to noise, we performed the biotinylation click reaction with biotin-alkyne, which cannot be cleaved by proteases, and digested the protein mixture with trypsin prior to incubation with Neutravidin resin. This optimized the enrichment of AHA-biotinylated peptides from the total lysate, resulting in samples in which the majority of peptides identified by MS/MS were directly modified by AHA-biotin. Consequently, our improved sample preparation, together with search algorithms targeted for the mass shift conferred by the AHA-biotin tag, increased our confidence in the identification of newly synthesized proteins by enriching the sample in peptides that included AHA-biotin, by directly identifying the modified peptides, and by excluding false positives from the candidate list that arise by calling proteins based on unmodified peptides.
We identified over 990 proteins labeled by AHA incorporation in the brain. We note that proteins concerned with protein synthesis and degradation are represented in the AHA-labeled protein population, including translational machinery, elongation factors, mRNA binding proteins, ribosomal proteins, proteosomal proteins and ubitiquitin ligases. This indicates that protein homeostasis itself is under dynamic control by factors controlling protein synthesis and degradation. Many AHA-labeled proteins synthesized within the 24 h window are cytoskeletal proteins, which are known to play an active role in cell proliferation, the development of neuronal structures, and in structural modifications pertaining to synaptogenesis and synaptic plasticity. Similarly, proteins that regulate membrane dynamics, such as GTPases, were also highly represented in the AHA-labeled sample, consistent with the major structural modifications of neurons, glia and neural progenitor cells during neuronal development and circuit assembly in the tadpole stages used in this study. Proteins related to axon outgrowth, cell adhesion and axon guidance are AHA-labeled, consistent with a predominance of these circuit-assembly events in tadpole brain. These data indicate that even within a relatively short period of 24 hours, cells in the developing central nervous system support considerable turnover of structural proteins and proteins related to membrane structural dynamics, as well as proteins specifically required for neuronal development and building neural circuits. Of particular interest for this study, we note that synaptic proteins, including neurotransmitter receptors, calcium channels, vesicular transport proteins, vesicular proteins, such as SNAP25, are AHA-labeled, suggesting active assembly and plasticity of synaptic components. Another functional category of AHA-labeled proteins relevant to the current study is signaling proteins, including protein kinases, CaMKII, MAP kinases and protein phosphatases. These data demonstrate that identification of AHA-biotin labeled proteins provides an overview of cellular processes subject to control by protein homeostasis and also allow targeted evaluation of the role of protein synthesis in specific events such as behavioral plasticity.
Experience-dependent synaptic and behavioral plasticity
Here we show that a tectally-mediated behavior, the visual avoidance response, is plastic for at least 24 h after VC. The 30-minute VC protocol is sufficient to induce plasticity, and reveals the gradual time-course of expression of plasticity. Brief VC also resulted in delayed expression of BDNF-dependent changes in synaptic plasticity and a behavioral readout of increased visual acuity (Schwartz et al., 2011), consistent with the gradual or delayed expression of plasticity that we observe. VC appears to induce plasticity through several activity-regulated transcriptional pathways (Chen et al., 2012b; Schwartz et al., 2009) and CPEB-mediated translational regulation reported here and previously (Bestman and Cline, 2008), which may operate in parallel. Maintenance of behavioral plasticity was comparable across the different induction protocols, suggesting that comparable plasticity was induced with different VC protocols. Furthermore, VC increases the strength of retinotectal synaptic transmission and recurrent tectal activity (Figure 7), supporting with the idea that potentiation of central synapses that process information prior to output to motor systems is a fundamental mechanism of behavioral plasticity.
Numerous studies support a step-wise model in which synaptic activity-mediated membrane depolarization causes a rise in calcium, which in turn activates divergent calcium-dependent signaling pathways that result in both rapid and local changes in synaptic efficacy (Lisman et al., 2012; Malenka, 2003), as well as changes in gene expression and protein synthesis, which in turn affect neural circuit plasticity (West and Greenberg, 2011). In particular, visual avoidance plasticity is induced by a highly conserved signaling pathway for synaptic and behavioral plasticity (Griffith et al., 1993; Lisman et al., 2002; Liu and Zukin, 2007; Man, 2011). Our data showing that application of NMDAR antagonists only during conditioning blocked behavioral plasticity are consistent with the prevailing model of distinct sequential mechanisms for induction and expression of plasticity. Interestingly, blockade of calcium-permeable GluAR with JST during or within the first hour after VC inhibited plasticity, suggesting that induction of visual avoidance plasticity includes a sensitive period for calcium influx through GluAR that extends after VC. Calcium entry thought GluNR and GluAR engages different downstream signaling pathways, likely reflecting distinct spatiotemporal features of calcium signaling (Man, 2011). In addition, plasticity of transmission through GluNR and GluAR differs (Liu and Zukin, 2007). For instance, VC regulates transmission through calcium-permeable GluAR by increasing intracellular polyamines (Aizenman et al., 2002), which effectively enhances responses to convergent co-activate synaptic inputs by a postsynaptic mechanism of use-dependent relief of polyamine block of calcium-permeable AMPA receptors (Bowie and Mayer, 1995; Rozov et al., 1998). Therefore, exposure to JST after VC may block behavioral plasticity by interfering with the enhanced detection of coactive convergent input and resultant signaling from calcium-permeable GluAR. VC-induced plasticity is mediated by increased retinotectal synaptic transmission, likely due to increased synaptic GluAR trafficking (Haas et al., 2006). Visual experience can also induce persistent neural circuit refinement in a spike-timing dependent manner (Engert et al., 2002; Mu and Poo, 2006; Pratt et al., 2008). Repeated visual stimuli improve visual acuity by a mechanism depending on BDNF-mediated facilitation of retinotectal synapses (Schwartz et al., 2011), consistent with the demonstrated role of BDNF in retinotectal connectivity (Cohen-Cory et al., 2010) and our mass spectrometry results that BDNF is labeled by AHA incorporation. Together these studies suggest that induction and expression of synaptic and behavioral plasticity in response to VC may operate through highly overlapping and conserved molecular and cellular mechanisms.
Rapid CPEB signaling
Our data indicate that VC increased translation of transcripts including CaMKII, MEK1, GAD65. Some of the proteins whose synthesis was increased with VC have CPE sites and are likely CPEB targets. Others are likely effector proteins, such as GAD65, a known activity-regulated protein that increases GABA synthesis. Finally other proteins whose translation is regulated by VC are kinases such as MAP kinase, which themselves are pleiotropic in their influence of synaptic and behavioral plasticity. Importantly, blocking CPEB translation acutely during VC blocks behavioral plasticity, as well as synaptic and structural plasticity. This suggests that protein translation dependent regulation of plasticity occurs more rapidly than previously thought. Furthermore, based on the conservation of CPEB signaling, our study suggests that rapid temporal control of CPEB function may regulate plasticity in other experimental systems.
Experimental Procedures
Animals and transfection
All protocols were approved by the Animal Care and Use Committee at the Scripps Research Institute. Xenopus laevis tadpoles were purchased and reared as previously described (Bestman and Cline, 2008). Stage 47–48 animals were anesthetized in 0.02% MS-222 (Tricane methanesulfonate, Sigma, St. Louis, MO) and the tectum was electroporated with plasmids (1–2 μg/μl) or morpholinos (1 μM) (Haas et al., 2002).
Visual avoidance assay and visual conditioning
The visual avoidance assay was conducted on individual tadpoles as described (Shen et al., 2011) (Supplemental Methods).
Electrophysiology
In vivo whole cell recordings of visually-evoked responses were collected and analyzed as described (Shen et al., 2011) (Supplemental Methods).
Imaging
In vivo time-lapse 2 photon imaging and analysis were conducted as described (Bestman and Cline, 2008) (Supplemental Methods).
Click chemistry and analysis of AHA labeling by Western blots
AHA (L-azidohomoalaine, 150–500 mM, pH 7.4, Anaspec) or PBS was injected into the tectal ventricle of anesthetized stage 47/48 tadpoles. Animals recovered from anesthesia for 30 minutes before they were used for experiments. For analysis of AHA-biotin tagged proteins by Western blots, tecta were dissected from 60 to 100 anesthetized animals for each group and processed for click chemistry labeling (Dieterich et al., 2007; Speers and Cravatt, 2009; Weerapana et al., 2007), as described in Supplemental Methods.
Click chemistry and analysis of AHA labeling by tandem mass spectrometry
For tandem mass spectrometry (MS/MS), AHA injections were done once a day for two days and brains of 1200–1500 animals were dissected 1–2 hours after the second injection. Brains were homogenized in PBS containing 0.5% SDS in PBS/protease inhibitor (PI; Roche, complete ULTRA Tablets, Mini, EDTA-free Protease Inhibitor cocktail tablets) followed by sonication with a probe sonicator. Samples were boiled for 10 min at 96–100°C and adjusted to 0.2 % TritonX-100 and then chilled. For each 400 μl reaction, 1.5 mg of total protein was used with 1.7 mM Triazole ligand (Invitrogen) in 4:1 tBuOH/DMSO (Sigma), 50 mM CuSO4 (Sigma), 5mM Biotin Alkyne (Invitrogen) and 50 mM TCEP (Sigma) added in sequence. The reaction proceeded for 1–2 hours at room temperature. Excess reagents were removed with methanol/chloroform precipitation. Precipitated protein (15mg) was air dried and resuspended in 100 μl of 0.2% ProteaseMAX (Promega, Madison, WI) and then 100ul of 8M urea was added. The solution was reduced with 5 mM TCEP for 20 min at 37°C, and then reduced with 10 mM IAA for 20 min in the dark at room temperature. Next, 150 μl of 50 mM ammonium bicarbonate and 2.5 μl of 1% ProteaseMAX were added prior to the addition of 200 μg trypsin. The sample was digested for three hours at 37°C in a shaking incubator. The peptides were desalted as previously described (Villen and Gygi, 2008) and dried with a speed-vac prior to AHA-peptide enrichment. The peptides were resuspended in 1 ml PBS and incubated with 200 μl Neutravidin beads (Pierce) at room temperature for 2 hours. Beads were washed with PBS and the peptides were eluted with elution buffer (0.1% TFA/0.1% formic acid/70% acetonitrile in H2O) for the MS/MS analysis.
Multidimensional Protein Identification Technology (MudPIT)
The eluted AHA-biotin peptides were pressure-loaded onto a 250 μm i.d. capillary with a kasil frit containing 2 cm of 10 μm Jupiter C18 material (Phenomenex, Ventura, CA) followed by 2 cm 5 μm Partisphere strong cation exchanger (Whatman, Clifton, NJ). This loading column was washed with buffer containing 95% water/5% acetonitrile/0.1% formic acid. After washing, a 100 μm i.d. capillary with a 5 μm pulled tip packed with 15 cm of 4 μm Jupiter C18 material (Phenomenex, Ventura, CA) was attached to the loading column with a union, and the entire split-column (loading column union analytical column) was placed inline with an Agilent 1100 quaternary HPLC (Palo Alto, CA) and analyzed using a modified 12-step separation described previously (Washburn et al., 2001). The buffer solutions used were 5% acetonitrile/0.1% formic acid (buffer A), 80% acetonitrile/0.1% formic acid (buffer B), and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (buffer C). Step 1 consisted of a 60 min gradient from 0–100% buffer B. Steps 2–11 had the following profile: 3 min of 100% buffer A, 5 min of X% buffer C, a 10 min gradient from 0–10% buffer B, and a 105 min gradient from 15–100% buffer B. The buffer C percentages (X) were 10, 15, 20, 25, 30, 35, 40, 45, 50, 60% respectively for the 12-step analysis. For the final two steps, the gradient contained: 5 min of 100% buffer A, 5 min of 100% buffer C, a 10 min gradient from 0–15% buffer B, and a 105 min gradient from 15–100% buffer B.
As peptides eluted from the microcapillary column, they were electrosprayed directly into an LTQ-OrbitrapXL hybrid mass spectrometer (ThermoFinnigan, Palo Alto, CA) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan FT mass spectrum (300–1600 m/z) at 60,000 resolution followed by 10 data-dependent IT MS/MS spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. Application of mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcaliber datasystem.
Analysis of Tandem Mass Spectra
MS/MS spectra were analyzed using the following software analysis protocol. MS/MS spectra remaining after filtering were searched with the Prolucid (Xu et al., 2006) against the UniProt Xenopus laevis 01-01-2011 concatenated to a decoy database in which the sequence for each entry in the original database was reversed (Peng et al., 2003). All searches were parallelized and performed on a Beowulf computer cluster consisting of 100 1.2 GHz Athlon CPUs (Sadygov et al., 2002). A static modification of 57.02146 on cysteine and a differential modification of 523.2749 on methionine were searched. Prolucid results were assembled and filtered using the DTASelect (version 2.0) program (Cociorva et al., 2007; Tabb et al., 2002). DTASelect 2.0 uses a linear discriminant analysis to dynamically set XCorr and DeltaCN thresholds for the entire dataset to achieve a user-specified false discovery rate (FDR). In addition, the modified peptides were required to be fully tryptic, less than 5ppm deviation from peptide match, and a FDR at the spectra level of 0.01. The FDRs are estimated by the program from the number and quality of spectral matches to the decoy database. For this analysis, the AHA-modified protein and peptide FDRs were 0.81 and 0.49 respectively. Raw data are available at http://uther.scripps.edu/published/AHA_invivo_Xenopus/raws/, and can be accessed with Xcalibur 1.0 software for use with Thermo mass spectrometers. Cellular localization as annotated by Gene Ontology was determined by STRAP (Software Tool for Researching Annotations of Proteins) (Bhatia et al., 2009). Figure 4 represents the percentage of genes with annotated localizations and some genes were annotated to multiple compartments.
Statistical tests
All data are presented as mean ± SEM. Data are considered significantly different when p values are less than 0.05. Where noted, two-tailed Student’s t-test was used for comparisons of two groups and ANOVA with Newman-Keuls test was used for comparisons of multiple groups. Experiments and analysis were performed blind to the experimental conditions.
Supplementary Material
Acknowledgments
We thank Dr. Ben Cravatt and Jonathan Hulce for help optimizing the click chemistry. The work was supported by NIH EY011261, MH 91676 and MH099799, Dart Neuroscience, LLC., the Nancy Lurie Marks Family Foundation and an endowment from the Hahn Family Foundation to HTC and MH067880 and P41 GM103533 to JRY and NSFC KG 31271176 to WS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agranoff BW, Klinger PD. Puromycin Effect on Memory Fixation in the Goldfish. Science. 1964;146:952–953. doi: 10.1126/science.146.3646.952. [DOI] [PubMed] [Google Scholar]
- Aizenman CD, Munoz-Elias G, Cline HT. Visually driven modulation of glutamatergic synaptic transmission is mediated by the regulation of intracellular polyamines. Neuron. 2002;34:623–634. doi: 10.1016/s0896-6273(02)00674-8. [DOI] [PubMed] [Google Scholar]
- Atkins CM, Nozaki N, Shigeri Y, Soderling TR. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J Neurosci. 2004;24:5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty KE, Tirrell DA. Two-color labeling of temporally defined protein populations in mammalian cells. Bioorganic & medicinal chemistry letters. 2008;18:5995–5999. doi: 10.1016/j.bmcl.2008.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learning & memory. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- Best MD. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48:6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. Application of Antisense Morpholino Oligonucleotides to Study Xenopus Brain Development and Plasticity. In: Schecter S, editor. Brain Development. New York: Springer; 2013. [Google Scholar]
- Bhatia VN, Perlman DH, Costello CE, McComb ME. Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal Chem. 2009;81:9819–9823. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D, Mayer ML. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15:453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wu JK, Lin HW, Pai TP, Fu TF, Wu CL, Tully T, Chiang AS. Visualizing long-term memory formation in two neurons of the Drosophila brain. Science. 2012a;335:678–685. doi: 10.1126/science.1212735. [DOI] [PubMed] [Google Scholar]
- Chen SX, Cherry A, Tari PK, Podgorski K, Kwong YK, Haas K. The transcription factor MEF2 directs developmental visually driven functional and structural metaplasticity. Cell. 2012b;151:41–55. doi: 10.1016/j.cell.2012.08.028. [DOI] [PubMed] [Google Scholar]
- Choi KY, Lippert DN, Ezzatti P, Mookherjee N. Defining TNF-alpha and IL-1beta induced nascent proteins: combining bio-orthogonal non-canonical amino acid tagging and proteomics. Journal of immunological methods. 2012;382:189–195. doi: 10.1016/j.jim.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Cociorva D, DLT, Yates JR. Validation of tandem mass spectrometry database search results using DTASelect. In: Baxevanis Andreas D, et al., editors. Current protocols in bioinformatics. Unit 13. Chapter 13. 2007. p. 14. [DOI] [PubMed] [Google Scholar]
- Cohen-Cory S, Kidane AH, Shirkey NJ, Marshak S. Brain-derived neurotrophic factor and the development of structural neuronal connectivity. Developmental neurobiology. 2010;70:271–288. doi: 10.1002/dneu.20774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nature neuroscience. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nature protocols. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Lee RH, Xu H, Yang S, Pratt KG, Cao V, Song YK, Nurmikko A, Aizenman CD. Visual avoidance in Xenopus tadpoles is correlated with the maturation of visual responses in the optic tectum. J Neurophysiol. 2009;101:803–815. doi: 10.1152/jn.90848.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy C, Teyler TJ, Shashoua VE. Long-term potentiation in the hippocampal slice: evidence for stimulated secretion of newly synthesized proteins. Science. 1981;212:1148–1151. doi: 10.1126/science.7233208. [DOI] [PubMed] [Google Scholar]
- Dziembowska M, Milek J, Janusz A, Rejmak E, Romanowska E, Gorkiewicz T, Tiron A, Bramham CR, Kaczmarek L. Activity-dependent local translation of matrix metalloproteinase-9. J Neurosci. 2012;32:14538–14547. doi: 10.1523/JNEUROSCI.6028-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert F, Tao HW, Zhang LI, Poo MM. Moving visual stimuli rapidly induce direction sensitivity of developing tectal neurons. Nature. 2002;419:470–475. doi: 10.1038/nature00988. [DOI] [PubMed] [Google Scholar]
- Flexner JB, Flexner LB, Stellar E. Memory in mice as affected by intracerebral puromycin. Science. 1963;141:57–59. doi: 10.1126/science.141.3575.57. [DOI] [PubMed] [Google Scholar]
- Griffith LC, Verselis LM, Aitken KM, Kyriacou CP, Danho W, Greenspan RJ. Inhibition of calcium/calmodulin-dependent protein kinase in Drosophila disrupts behavioral plasticity. Neuron. 1993;10:501–509. doi: 10.1016/0896-6273(93)90337-q. [DOI] [PubMed] [Google Scholar]
- Haas K, Jensen K, Sin WC, Foa L, Cline HT. Targeted electroporation in Xenopus tadpoles in vivo--from single cells to the entire brain. Differentiation. 2002;70:148–154. doi: 10.1046/j.1432-0436.2002.700404.x. [DOI] [PubMed] [Google Scholar]
- Haas K, Li J, Cline HT. AMPA receptors regulate experience-dependent dendritic arbor growth in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12127–12131. doi: 10.1073/pnas.0602670103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewapathirane DS, Dunfield D, Yen W, Chen S, Haas K. In vivo imaging of seizure activity in a novel developmental seizure model. Experimental neurology. 2008;211:480–488. doi: 10.1016/j.expneurol.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Hinz FI, Aizenberg M, Tushev G, Schuman EM. Protein synthesis-dependent associative long-term memory in larval zebrafish. J Neurosci. 2013;33:15382–15387. doi: 10.1523/JNEUROSCI.0560-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz FI, Dieterich DC, Tirrell DA, Schuman EM. Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS chemical neuroscience. 2012;3:40–49. doi: 10.1021/cn2000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodas JJ, Nehring A, Hoche N, Sweredoski MJ, Pielot R, Hess S, Tirrell DA, Dieterich DC, Schuman EM. Dopaminergic modulation of the hippocampal neuropil proteome identified by bioorthogonal noncanonical amino acid tagging (BONCAT) Proteomics. 2012;12:2464–2476. doi: 10.1002/pmic.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Molecular cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nature neuroscience. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- Konzer A, Ruhs A, Braun H, Jungblut B, Braun T, Kruger M. Stable isotope labeling in zebrafish allows in vivo monitoring of cardiac morphogenesis. Molecular & cellular proteomics : MCP. 2013;12:1502–1512. doi: 10.1074/mcp.M111.015594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Park SK, Xu T, Vanderklish P, Yates JR., 3rd Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15281–15286. doi: 10.1073/pnas.0804678105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nature reviews Neuroscience. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nature reviews Neuroscience. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Zukin RS. Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends in neurosciences. 2007;30:126–134. doi: 10.1016/j.tins.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Malenka RC. Synaptic plasticity and AMPA receptor trafficking. Annals of the New York Academy of Sciences. 2003;1003:1–11. doi: 10.1196/annals.1300.001. [DOI] [PubMed] [Google Scholar]
- Man HY. GluA2-lacking, calcium-permeable AMPA receptors--inducers of plasticity? Current opinion in neurobiology. 2011;21:291–298. doi: 10.1016/j.conb.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian OK, Asiedu MN, Tillu DV, Peebles KA, Yan J, Ertz N, Dussor GO, Price TJ. IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. Journal of Neuroscience. 2010;30:15113–15123. doi: 10.1523/JNEUROSCI.3947-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y, Poo MM. Spike timing-dependent LTP/LTD mediates visual experience-dependent plasticity in a developing retinotectal system. Neuron. 2006;50:115–125. doi: 10.1016/j.neuron.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Nedivi E. Molecular analysis of developmental plasticity in neocortex. Journal of neurobiology. 1999;41:135–147. [PMC free article] [PubMed] [Google Scholar]
- Ngo JT, Tirrell DA. Noncanonical amino acids in the interrogation of cellular protein synthesis. Accounts of chemical research. 2011;44:677–685. doi: 10.1021/ar200144y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oruganty-Das A, Ng T, Udagawa T, Goh EL, Richter JD. Translational control of mitochondrial energy production mediates neuron morphogenesis. Cell metabolism. 2012;16:789–800. doi: 10.1016/j.cmet.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Pratt KG, Dong W, Aizenman CD. Development and spike timing-dependent plasticity of recurrent excitation in the Xenopus optic tectum. Nature neuroscience. 2008;11:467–475. doi: 10.1038/nn2076. [DOI] [PubMed] [Google Scholar]
- Rajan I, Witte S, Cline HT. NMDA receptor activity stabilizes presynaptic retinotectal axons and postsynaptic optic tectal cell dendrites in vivo. Journal of neurobiology. 1999;38:357–368. doi: 10.1002/(sici)1097-4695(19990215)38:3<357::aid-neu5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Richter JD. Translational control of synaptic plasticity. Biochemical Society transactions. 2010;38:1527–1530. doi: 10.1042/BST0381527. [DOI] [PubMed] [Google Scholar]
- Rozov A, Zilberter Y, Wollmuth LP, Burnashev N. Facilitation of currents through rat Ca2+-permeable AMPA receptor channels by activity-dependent relief from polyamine block. The Journal of physiology. 1998;511(Pt 2):361–377. doi: 10.1111/j.1469-7793.1998.361bh.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, Yates JR., 3rd Code developments to improve the efficiency of automated MS/MS spectra interpretation. J Proteome Res. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- Schwartz N, Schohl A, Ruthazer ES. Neural activity regulates synaptic properties and dendritic structure in vivo through calcineurin/NFAT signaling. Neuron. 2009;62:655–669. doi: 10.1016/j.neuron.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Schwartz N, Schohl A, Ruthazer ES. Activity-Dependent Transcription of BDNF Enhances Visual Acuity during Development. Neuron. 2011;70:455–467. doi: 10.1016/j.neuron.2011.02.055. [DOI] [PubMed] [Google Scholar]
- Shashoua VE. Brain protein metabolism and the acquisition of new behaviors. II. Immunological studies of the alpha, beta and gamma proteins of goldfish brain. Brain research. 1977a;122:113–124. doi: 10.1016/0006-8993(77)90666-7. [DOI] [PubMed] [Google Scholar]
- Shashoua VE. Brain protein metabolism and the acquisition of new patterns of behavior. Proceedings of the National Academy of Sciences of the United States of America. 1977b;74:1743–1747. doi: 10.1073/pnas.74.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, McKeown CR, Demas JA, Cline HT. Inhibition to excitation ratio regulates visual system responses and behavior in vivo. J Neurophysiol. 2011;106:2285–2302. doi: 10.1152/jn.00641.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419:475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF. Activity-Based Protein Profiling (ABPP) and Click Chemistry (CC)-ABPP by MudPIT Mass Spectrometry. Current protocols in chemical biology. 2009;1:29–41. doi: 10.1002/9780470559277.ch090138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nature protocols. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature biotechnology. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nature protocols. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harbor perspectives in biology. 2011:3. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley PF, Cole AJ, Murphy TH, Christy BA, Nakabeppu Y, Baraban JM. Synaptic regulation of immediate-early genes in brain. Cold Spring Harbor symposia on quantitative biology. 1990;55:213–223. doi: 10.1101/sqb.1990.055.01.023. [DOI] [PubMed] [Google Scholar]
- Wu G, Malinow R, Cline HT. Maturation of a central glutamatergic synapse. Science. 1996;274:972–976. doi: 10.1126/science.274.5289.972. [DOI] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Stabilization of dendritic arbor structure in vivo by CaMKII. Science. 1998;279:222–226. doi: 10.1126/science.279.5348.222. [DOI] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Xu T, Venable JD, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JRI. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Paper presented at: Molecular & cellular proteomics : MCP.2006. [Google Scholar]
- Yang YY, Grammel M, Raghavan AS, Charron G, Hang HC. Comparative analysis of cleavable azobenzene-based affinity tags for bioorthogonal chemical proteomics. Chemistry and Biology. 2010;17:1212–1222. doi: 10.1016/j.chembiol.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.