Abstract
Autophagy is a catabolic process that normally utilizes the lysosome. The far-reaching implications of this system in disease are being increasingly understood. Studying autophagy is complicated by its role in cell survival and programmed cell death and the involvement of the canonical marker of autophagy, Atg8/LC3, in numerous non-autophagic roles. The malaria parasite, Plasmodium, has conserved certain aspects of the autophagic machinery but for what purpose has long remained a mystery. Major advances have recently been gained and suggest a role for Atg8 in apicoplast maintenance, degradation of heme inside the food vacuole, and possibly trafficking of proteins or organelles outside the parasite membrane. Autophagy may also participate in programmed cell death under drug treatment or as a selective tool to limit parasite load. We review the current findings and discuss discrepancies in the field of autophagy in the Plasmodium parasite.
Keywords: Plasmodium, apicoplast, heme, Atg, digestive vacuole
Introduction
Classically, autophagy has been defined as a eukaryotic system for degradation of cellular material inside the lysosome. The best-studied form, macroautophagy, encapsulates the cytoplasmic material inside a double-membrane vesicle, the autophagosome that fuses with the lysosome. Once considered a nonselective bulk process, autophagy is now appreciated as a way to quickly respond to shifting cellular conditions and to selectively degrade damaged organelles, proteins, nucleic acids, and pathogens. Interest in autophagy has surged in the past five years and is now seen as a therapeutic target in many diseases including cancer, neurodegeneration, and infectious disease [1–3], where not just host autophagy but that of the eukaryotic pathogen plays a critical role in infection [4]. While much was known about the role of autophagy in the protozoans Leishmania, Trypanosoma, and Toxoplasma [5–7], the story was less conclusive for the malaria parasite Plasmodium, which kills nearly one million people each year [8], as well as the related species, Theileria and Eimeria. However, great advances have been made in the last year.
The first mention of plasmodial autophagy in the literature was in the 1970s by D.C. Warhurst and colleagues. They conducted extensive studies showing increased pigment clumping in “autophagic vacuoles” following chloroquine (CQ) treatment in blood stage parasites [9, 10]. In the 2000s, drug treatment was again reported to induce autophagic vacuoles and an autophagic death [11, 12]. However, Klionsky, a leading researcher in autophagy, has warned against the visualization of autophagosomes through electron microscopy as the sole evidence for an autophagic process [13]. The difficulty in culturing certain stages of the parasite, genetically manipulating the organism to make conditional knockouts, and to clone and express genes recombinantly all have contributed to a void of knowledge in this area. The last year witnessed a vast expansion in our understanding of autophagic pathways in the Plasmodium parasite.
Plasmodium biology
The Plasmodium parasite has evolved distinctive biological mechanisms to invade and replicate within different cell types in both the mosquito vector and mammalian host as part of its life cycle. It is necessary to understand Plasmodium biology if one is to study autophagy in the context of this unique unicellular eukaryote. During a mosquito blood meal, elongated sporozoites are injected into the host and invade liver cells. Within a hepatocyte the sporozoite transforms into a round trophozoite and organelles used for the invasion process are expelled [14, 15]. The parasite uses a form of asexual replication, schizogony, to produce hundreds to thousands of teardrop-shaped merozoites. Extensive remodeling of the host cell creates a merosome, which contains the merozoites that are released into the bloodstream to continue the next stage of infection, the symptomatic erythrocytic stage [16, 17]. A merozoite invades a red blood cell (RBC), develops into the ring stage form, converts into a trophozoite and then undergoes schizogony to produce more merozoites for sequential rounds of invasion and replication. During the trophic stage, hemoglobin is taken up from the host RBC cytoplasm and transported to the food vacuole (FV), a lysosome-like compartment where it is degraded to provide amino acids for parasite growth. Some merozoites are shunted from this cycle to produce gametocytes, which are subsequently taken up by a mosquito during a blood meal, continuing the Plasmodium life cycle.
Overview of Macroautophagy
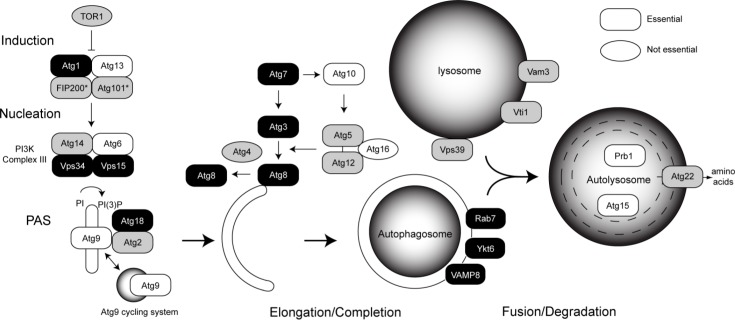
Although over 30 AuTophaGy (Atg) proteins have been identified in yeast, not all are essential to macroautophagy (Figure 1). Autophagy induction in yeast (and mammals) begins with the Atg1 (ULK1 in mammals) protein kinase complex, which is inhibited by the target of rapamycin, TORC1 (mTOR in mammals) complex under homeostasis. Nutrient deprivation or rapamycin treatment alleviates the negative regulation on the Atg1 (ULK1) complex [18]. Atg1 and Atg13 form a complex with Atg31/Atg17/Atg29/Atg11 in yeast and FIP200/Atg101 in mammals at the pre-autophagosomal structure (PAS) [19, 20]. Atg17 has been implicated as the most upstream scaffold for protein recruitment to the PAS [21]. Phosphorylation of substrates by Atg1 (ULK1) activates the Phosphatidyl-Inositol 3-phosphate Kinase (PI3K) class III complex, composed of Vps34, Atg14, Vps15/p150, and Atg6 (Beclin1 in mammals) which converts phosphatidylinositol (PI) into phosphatidylinositol triphosphate (PI(3)P) at the site of the PAS [22]. This creates a binding site for Atg18 (WIPIs in mammals) and Atg2 at the isolation membrane, also requiring association with the transmembrane protein Atg9 [23]. Elongation of the membrane and closure of the autophagosome requires covalent attachment of the C-terminal glycine of Atg8 (LC3 in mammals) to phosphatidylethanolamine (PE) in the phagophore membrane [24]. Atg8 is a ubiquitin-like protein (Ubl) that requires the protease Atg4, an E1-activating enzyme, Atg7, and an E2-conjugating enzyme, Atg3 for lipidation [25, 26]. Another autophagy Ubl protein, Atg12, is covalently attached to Atg5, requiring Atg7 as well, and its own E2, Atg10 [27]. The Atg12-Atg5 conjugate forms a complex with Atg16 to drastically increase conjugation of Atg8, presumably by forming a scaffold with Atg3 at the membrane [28]. The autophagosome seals and fuses with endocytic compartments and the lysosome, where the material is broken down and recycled back to the cytoplasm.\
Figure 1.
Conservation of autophagy proteins in Plasmodium. Proteins essential for macroautophagy are depicted as rectangles. Ovals represent proteins whose essentiality is not known. Black indicates strong evidence for homology (e-value less than 1e-10 in initial BLAST within PlasmoDB and reciprocal identification using PSI-BLAST with e-value less than 1e-20). Gray indicates homology less clear (initial e-value cutoff of 0.012 and identification of initial query in either PSI- or DELTA-BLAST with e-value that may not be significant). White indicates no orthologue was identified. *Human proteins FIP200 and Atg101 thought to be functional homologues of yeast Atg31/Atg17Atg29.
To obtain an overview of the system, we performed bioinformatics analysis to identify putative orthologues of autophagy proteins in Plasmodium falciparum. While such lists have been published, our analysis focused on the minimal machinery needed for macroautophagy in yeast [4, 6, 29–31]. The coordination of events in macroautophagy is diagrammed in Figure 1, with putative homologues identified in Plasmodium colored in black and less reliable identifications in gray. Although several hits were identified for the nutrient sensor TOR, these hits have clearer homology to Phosphatidyl-Inositol 4-Kinases. Of the proteins involved in autophagy induction, Atg1 and its mammalian binding partners, FIP200 and Atg101 appear to have orthologues in Plasmodium, while Atg13 appears absent. The C-terminal Atg13-binding domain is also missing from PfAtg1. It contains several potential Atg8-interacting motifs, which mediate Atg1 binding to Atg8 in an Atg13-independent manner in yeast and humans [32]. PfVps34 exists in P. falciparum and is one of the better-characterized proteins. Vps15 and Atg14, also members of the PI3K complex were identified, but Atg6 was not.
Atg9 is an essential transmembrane protein for autophagy thought to supply Golgi-derived membranes to the PAS during initiation [33, 34]. None of the significant BLAST hits for Atg9 are predicted to contain any of its six transmembrane domains. Atg18 likely has a homologue in Plasmodium and contains the FRRG motif necessary for binding to PI(3)P and Atg18-Atg2 recruitment to the PAS as well as WD40 repeats for beta propeller formation. Atg18 is thought to aid recruitment of Atg8 to the PAS and protect it from premature Atg4 cleavage [35]. A putative homologue for Atg18's binding partner, Atg2, was also identified but is annotated as a putative rhoptry protein.
Atg8, Atg7, and Atg3 orthologues are present. Atg8 contains a C-terminal glycine in Plasmodium [36]. Similar Atg8 homologs have been identified in plants and other protists including Toxoplama [37, 38]. Therefore, the putative Atg4 orthologue is likely important for recycling improperly conjugated Atg8 [39]. Putative homologues for the Ubl, Atg12 and its target, Atg5, were identified. However, the C-terminal glycine of PfAtg12 and its E2 enzyme needed for covalent attachment are absent. It is unclear if Atg16, which forms a 350 kDa complex with Atg12-Atg5 is present. It is essential in vivo but not in vitro for Atg8 lipidation [28, 40].
Fusion of both autophagosomes and endosomes with the lysosome requires Rab7 and various SNAREs [41, 42]. Apicomplexans contain a basic set of Rabs, including Rab7 [43]. Putative homologues for essential SNARES including Vti1b and VAMP8 also exist in Plasmodium. The lipase for breaking down the autophagosome membrane, Atg15, and the vacuolar proteinase Prb1 are absent [44, 45]. However, the parasite contains numerous falcipains in the food vacuole for degrading hemoglobin. The lysosomal amino acid efflux pump Atg22 is present [46]. More information, including the identity of hits is in supplementary table 1 (ST1).
Methods
The genome sequence of P. falciparum was published in 2002 and is stored along with various data associated with each gene in the Plasmodium Genome database (PlasmoDB) [47, 48]. The amino acid coding sequence of Saccharomyces cerevisiae and Homo sapiens ATG genes were queried for putative homologues using the WU-BLAST program within PlasmoDB. Hits were identified based on the Expect (e)-value of the BLAST search, as well as annotation in PlasmoDB, any functional knowledge, conservation among all the plasmodial species sequenced, and the length of the hit in comparison to the query. Of note, the most likely homologue did not necessarily have the highest e-value in the initial PlasmoDB BLAST search. Additionally the species used as the query had a major impact on the returned BLAST hits. Often the same hit was identified with both the human and yeast homologue query but sometimes a strong hit was only identified with one species. In a few cases other species were used as a query (ST1). An inverse BLAST was performed with the potential hits using NCBI Position-Specific Iterated (PSI) BLAST against the S. cerevisiae or H. sapiens non-redundant protein sequence database, and if that failed, with the Domain Enhanced Lookup Time Accelerated (DELTA) BLAST. Hits were additionally screened in Pfam to see if they contained the domains or motifs important to that homologue's function, such as coiled-coil domains and lipid binding motifs. For transmembrane proteins, the TMHMM server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to compare predicted membrane topology.
Localization of the autophagy marker, Atg8
Atg8/LC3 is the classical autophagy marker as it is covalently attached to the pre-autophagosome membrane at an early stage and stays conjugated to the autophagosome during fusion with the lysosome. In yeast and mammals, Atg8/LC3 has a diffuse cytoplasmic distribution under nutrient rich conditions that becomes punctate upon autophagy induction [25]. The lipidated fraction of Atg8 can also be visualized as a faster migrating band with high percentage or urea-contaning gels in SDS-PAGE [26].
Several groups have examined the localization of Atg8 in different Plasmodium species and stages. Under normal conditions, P. berghei liver and P. falciparum blood stage Atg8 are in punctae that are nearby and partially colocalize with the apicoplast marker, acyl-carrier protein (ACP) [30, 31, 49]. All members of the phylum Apicomplexa, with the exception of Cryptospiridium, contain apicoplasts, plastid organelles analogous to chloroplasts in plants, acquired through endosymbiosis. The organelle is essential to Plasmodium survival in both asexual stages and is the site for synthesis of iron-sulfur clusters, isoprenoids, and fatty acids [50]. Localization to other organelles, such as mitochondria, rhoptries, micronemes, and dense granules was negative in the schizont, while transgenic Atg8 lacking the C-terminal glycine for lipidation showed diffuse localization, validating the results of these studies [30]. In hepatocytes, PbAtg8 localization along the apicoplast was observed 14 hours post-invasion and increased with organelle size throughout the liver stage [49].
Roepe and Sinai also observe punctate PfAtg8 in blood stage trophozoites, but co-localization with ACP, or other organelle markers, was not assessed [5]. Interestingly, when cells were transferred to media lacking amino acids and glucose for 6 hours, PfAtg8 redistributes to a wider punctate staining that seems to be inside the host red blood cell cytoplasm. Similar re-localization was seen in treatment with high levels of CQ [5]. Langsley and colleagues also starved blood stage parasites in media lacking amino acids. At 4 hours, they observed localization of PfAtg8 to double-membrane structures also containing PfRab7 that were near or inside the food vacuole, visualized with electron micrographs, implicating the food vacuole as the final digestive compartment in a Plasmodium autophagic-like process [51].
PfAtg8 shows elongated tubular localization in oocytes obtained from mosquito midgut and sporozoites from the mosquito salivary glands, reminiscent of apicoplast morphology under normal cell conditions [49].
Expression data for autophagy genes
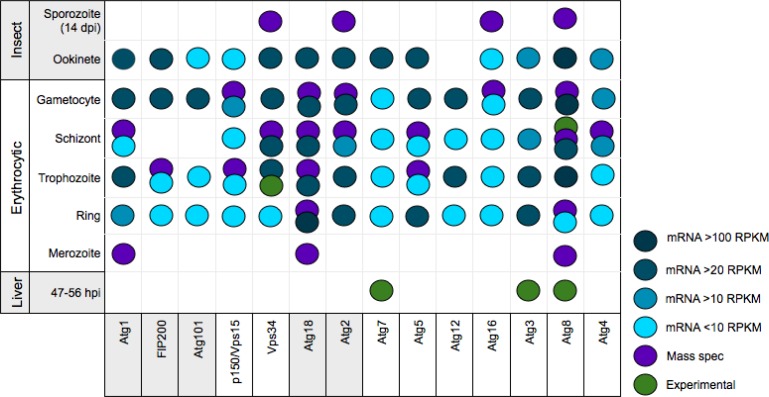
PlasmoDB houses RNA-Seq data during the blood stage for each gene [52]. Additionally, several widespread mass spectrometry studies have provided data on protein expression in the liver and blood stages [53–58]. There is also specific transcript, immunoblotting and/or immunofluorescence data for a small subset of autophagy genes [30, 31, 49, 59]. Evidence for expression from these various sources is compiled in Figure 2 and supports an important role for autophagy proteins during the late blood stage, where both hemoglobin digestion and apicoplast growth is maximal. Less data is available for non-blood stages. Atg8, Atg3, and Atg7 are transcribed during all life cycles of P. berghei [49].
Figure 2.
Compiled expression data of autophagy proteins during Plasmodium life cycle. RNA-Seq and mass spectrometry data from PlasmoDB and data from published studies is compiled. All data is for P. falciparum homologues (ST1), except liver stage data from P. berghei.
Potential Functions
Strictly speaking, confirmation of macroautophagy in Plasmodium would require 1. the presence of Atg8-labelled double-membrane vesicles, 2. fusion of these vesicles with a lysosomal-like compartment, and 3. breakdown and recycling of the autophagosome cargo. Recently, Atg8 has been observed on double-membrane vesicles near and inside the food vacuole, an acidic compartment that most likely serves as a lysosome in the parasite, providing intriguing evidence for macroautophagy [51]. Additionally, there is evidence of autophagic-like and distinct functions utilizing the autophagic machinery. The unique biology of the parasite as well as the adaptability of the machinery in other organisms for non-autophagic purposes makes alternative roles quite feasible and even likely [60]. Atg8's essentiality in Plasmodium is intriguing, as Atg8 knockouts are viable in yeast, which suggests an alternative or additional vital function(s) in the parasite [24]. In the highly related Apicomplexa, Toxoplasma, the E2 conjugating enzyme for TgAtg8, TgAtg3, is also essential [38]. Here we will present current thinking for the role of Atg8 and the autophagy machinery in Plasmodium, which is summarized in Figure 3.
Figure 3.
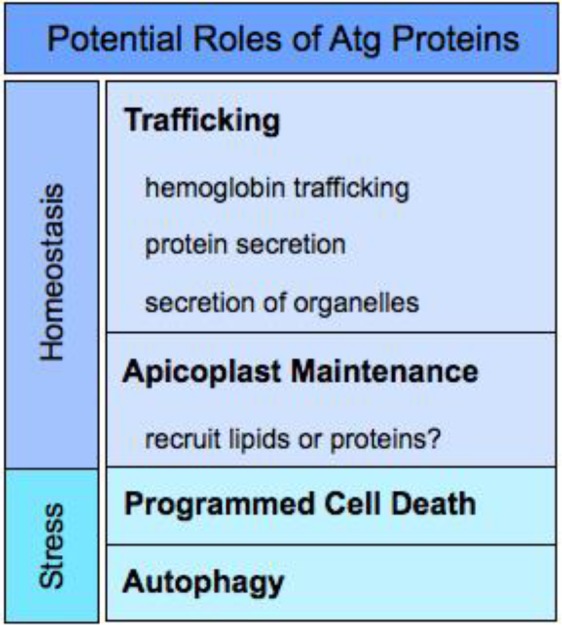
Potential Roles for Atg proteins in Plasmodium.
Vesicular Trafficking
Protein Secretion
The Plasmodium parasite initiates extensive remodeling of the host cell upon infection by exporting its own proteins into the host cell. Plasmodium is encapsulated by a parasitopherous vacuole within host cells. Proteins targeted for export to the host cytosol or plasma membrane must be trafficked out of the parasite plasma membrane and the PV, necessitating novel pathways for protein secretion. It is possible that exophagy, where proteins inside autophagosomes are re-routed to the plasma membrane is involved [61]. Exophagy of Acb1 in yeast utilizes the autophagosomal machinery as well as GRASP65 and the t-SNARE, Sso1, which have putative homologues in Plasmodium [62, 63]. In the current model, autophagosomes fuse with multivesicular bodies to form amphisomes. Exophagy utilizes the COPII machinery but bypasses Golgi processing and is packaged into autophagosomes that fuse with plasma membrane [64].
A recent review suggests that vesicles originating at the ER via the formation of PI(3)P are destined for export to the host cytoplasm [65]. Vps34, also known as type III PI3K, is a lipid kinase at the ER that produces PI(3)P on the outer leaflet, important for recruitment of proteins to the phagophore and proper curvature of the autophagosome. PfVps34 purified from parasite lysate has kinase activity that can be inhibited by the well-characterized class III PI3K complex inhibitor, wortmannin. Endogenous PfVps34 localizes to the food vacuole and vesicular structures near the host plasma membrane in the blood stage, partially co-localizing with the VARC protein, a virulence factor in P. falciparum that is exported to the erythrocyte surface [59]. One of the more characterized routes of protein export beyond the PV utilizes a PEXEL motif for recognition. Recently, PI(3)P was shown to bind the PEXEL motif at PI(3)P-enriched regions in the ER [66].
We previously identified the small GTPase sar1 in a pull-down with tagged PfAtg8 and blood stage P. falciparum lysate [36]. Sar1 is a component of the COPII vesicle machinery for trafficking between the ER and Golgi. Sar1 was shown to have similar localization to the ER and small vesicle compartments in P. falciparum blood stages as in yeast, at the ER site for secretion of proteins [67, 68]. The role of other Atg proteins in protein secretion is not known. As will be discussed below, PfAtg8 localization in the RBC cytoplasm is reported, but only under non-homeostatic conditions and co-localization with VARC or the Maurer's cleft, an intermediate compartment for protein export in the RBC cytoplasm, was not examined [5].
Organelle expulsion
The Plasmodium parasite undergoes major morphological changes in the early liver stage, converting from a motile, slender sporozoite into a round trophozoite. During this transformation, organelles used for liver cell invasion, including the inner membrane complex, rhoptries, and micronemes, are expelled from the parasites [14]. Treatment of Plasmodium sporozoites with 3-methyladenine (3-MA), which targets Vps34, needed for PI(3)P production and phagophore formation, delays conversion into trophozoites [69]. Electron micrographs of liver stage parasites revealed double-membraned structures morphologically similar to autophagosomes containing micronemes leading to the hypothesis that exophagy of organelles could be important for differentiation in the liver stage in the absence of an extensive intracellular degradative system [6].
Trafficking to food vacuole
Chloroquine (CQ) is a lysosomatropic agent that prevents autophagosome-lysosome fusion [70, 71]. In other studied Eukarya, CQ causes an accumulation of Atg8 lipidated to autophagosomes in starvation and non-starvation conditions [71]. A longstanding antimalarial, CQ accumulates in the food vacuole and prevents hemozoin dimerization and hemoglobin digestion [72], starving the parasite of amino acids. Varying responses are observed in Plasmodium upon CQ treatment and this appears to be due in part to the concentration that is used and the duration studied. In CQ sensitive strains, such as 3D7, ten to 100 nM is cytostatic whereas micromolar amounts are cytocidal [73].
In early to mid-trophozoite stages, host cytoplasm is endocytosed, delivering hemoglobin to the food vacuole as a source for amino acids. It has long been reported that CQ treatment led to production of vesicles inside the Plasmodium parasite [10, 11], which were assumed to be autophagic vesicles. Extended incubation (12 hours) with 120 nM CQ leads to a build-up of hemoglobin-containing vesicles, likely by preventing fusion of these vesicles with the food vacuole, preventing hemoglobin digestion and starving the parasite of amino acids [74]. Sinai and Roepe report drastic re-localization of PfAtg8 from the parasite to the RBC host cytoplasm with either prolonged starvation or high cytocidal concentrations of CQ [5]. The authors hypothesize that PfAtg8 may be involved in vesicular trafficking of hemoglobin or other nutrients under starvation conditions.
In contrast, Kitamura et al. do not see a change in PfAtg8 localization or protein levels in response to CQ treatment as would be expected if Atg8-containing vesicles could not fuse with the food vacuole (FV) or if Atg8 was exported into the RBC cytoplasm. They used 100 and 300 nM concentrations of CQ, which are cytostatic, not cytocidal, and may not be sufficient to induce a lysosomatrophic effect, as 100 µM is typically used in mammalian cell lines to block autophagy [73]. The treatment period in the paper was also shorter than with Sinai and Roepe.
During the preparation of this review it was shown with immuno-EM that endogenous PfAtg8 was located at Rab7-containing vesicles near and inside the food vacuole in parasites starved of amino acids for 4 hours. The authors never saw PfAtg8 in the host cytoplasm [51]. Immunofluorescence, co-staining for anti-hemoglobin antiserum and anti-Atg8 antibodies, would elucidate whether PfAtg8 is involved in trafficking of hemoglobin. Due to the differences in P. falciparum strain sensitivity to CQ and different effects observed with varying concentrations and exposure times, amino acid deprivation, the canonical inducer of autophagy, is a more suitable tool to study this pathway in Plasmodium.
Additional support for autophagic functions in hemoglobin degradation is garnered by localization of PfVps34 to the food vacuole as well as vesicles in the host cytoplasm [59]. Strikingly, the PI3K inhibitor, wortmannin (10 µM, 5 hour treatment), caused accumulation of hemoglobin-containing endocytic vesicles within the parasite and subsequently less in the FV, indicating a defect in digestion concomitant with decreased parasite growth. They suggest the class III PI3K complex is important for hemoglobin endocytosis and trafficking to the FV in Plasmodium. While the effect of wortmannin on Plasmodium Atg8 was examined under normal conditions, it was not tested under starvation conditions to see if treatment prevented PfAtg8 localization to vesicles and the FV. However another PI3K inhibitor, 3-MA, inhibited re-localization of PfAtg8 to the host cytoplasm upon starvation or CQ treatment, implicating involvement of Atg8 in trafficking of theses hemoglobin-containing vesicles [75].
The malaria parasite does not obtain isoleucine from hemoglobin and is dependent on the host for this amino acid. Plasmodium can survive up to 72 hours without exogenous isoleucine, suggesting a survival mechanism is present. Since digestive vacuole proteases are essential to survival, an autophagic mechanism was suggested [76]. As mentioned previously, amino acid starvation induces a re-localization of Atg8 to either the periphery of the parasite or to vesicles near or within the food vacuole depending on the experimental conditions [5, 51]. It would be of great interest to determine if isoleucine deprivation alone induced co-localization between PfAtg8 and PfRab7 at the food vacuole.
The product of PI3K, PI(3)P, is enriched at both the apicoplast and the food vacuole and one of the few PI(3)P-binding proteins identified in Plasmodium, FCP protein, is trafficked to the FV using an unknown mechanism, which some suggest may be similar to the cytoplasm-to-vacuole targeting pathway in yeast [77]. However PfAtg8 re-localization away from the apicoplast appears to require a signal such as drug treatment or starvation and physiological relevance of these conditions is unclear. One could speculate a role for detoxification of drugs or increased uptake of nutrients through exophagy of importer proteins.
Programmed Cell Death
Programmed cell death (PCD) pathways are increasingly appreciated as an important way to modulate host-pathogen responses and limit parasite load [78]. Autophagy is a recognized mechanism of cell death in other eukarya where the balance between pro-survival and pro-death is tightly regulated [79]. Autophagic cell death has previously been reported for the protozoan parasites Toxoplasma and trypanosomes and less conclusively in Leishmania, but there is controversy as to the legitimacy of a regulated rather than incidental mechanism of cell death in protozoa [7, 80–83].
Treatment of P. falciparum erythrocytes with chloroquine (180 nM), SNAP, a nitric oxide donor, and staurosporine, an apoptosis inducing agent, led to cell death that was described as autophagic-like because of distinct vacuolarization of the parasite cytoplasm and presence of double-membrane structures, whereas the nuclear morphology remained in tact and other apoptotic markers were absent [11]. The morphology of this cell death is similar to that observed in retinal pigment epithelial cells upon treatment with much higher concentrations (120 µM) of CQ and is thought to be the mechanism for retinotoxicity associated with CQ. In this case, they also did not see hallmarks of apoptosis but did see increased lipidation of LC3 to autophagic vacuoles (AVs) and a block in degradation of the autophagy-specific substrate, p62 [84]. Autophagic vacuoles were also observed in P. berghei blood stage parasites dying after treatment with dihydroartemisinin [12]. In this study, the authors concluded, “Since the food vacuole membranes were affected, the nutrition of parasites was blocked and the autophagic vacuoles formed quickly as a result of amino-acid hunger of parasites.” However distribution of Atg8 was not investigated.
Recently, it has been reported that after invasion of hepatocytes, a significant fraction of P. berghei parasites die through an autophagic mechanism [30]. This was observed in cell culture and in mice, indicating it is a physiological mechanism. The process is restricted to the later stages of development. Eickel et al. observed double membrane structures that appeared similar to those reported by Meis and Verhave in 1985, then described as “membranous whorls” [15]. Yet, fluorescence microscopy revealed that PbAtg8 never localized to these vesicular structures and remained associated with the apicoplast, determined by colocalization with ACP, and its organellar fragments throughout the cell death event [30]. Treatment with rapamycin (500 nM), which potently induces autophagy in yeast through inhibition of TOR, decreased parasite size and changed apicoplast morphology, but did not affect PbAtg8 localization. However, there is no known TOR orthologue in Plasmodium, and the observed effect on parasite growth could be mediated through off-target effects or effects on host autophagy [85]. The authors speculate that autophagy occurs in the parasite but is independent of Atg8. Although Atg8 is the canonical autophagic marker, there are reports of macroautophagy that is independent of LC3 conjugation [86]. However this alternative pathway requires ULK1, Rab9, and Beclin 1 and Rab9 and Beclin 1 orthologues have not been identified in Plasmodium.
Apicoplast Maintenance
The apicoplast is vital to survival in both mammalian stages. A gradual increase in apicoplast size begins in the trophozoite stage. Dramatic elongation and branching of the organelle occurs in the late trophozoite/early schizont stage. Division of the apicoplast occurs late in the schizont and ultimately results in each merozoite containing one apicoplast [87]. If the autophagic machinery delivers lipids or proteins to the growing apicoplast membrane, expression of its component proteins is necessary by the trophozoite stage. As shown in Figure 2, this appears to be the case.
Pf/PbAtg8 localizes to the apicoplast in liver and blood stages under normal conditions. Electron micrographs and biochemical data indicate that Atg8 is conjugated to the outermost membrane of the apicoplast [31]. Plasmodium treated with chloramphenicol leads to loss of the apicoplast, which is normally lethal, but supplementation with isopentenylpyrophosphate allows the parasite to survive during the RBC stage [88]. Tomlins et al. observed PfAtg8 localization under these conditions. Strikingly, PfAtg8 was located at vesicular structures that also contained the apicoplast-targeted protein LipDH1, suggesting a role in trafficking [51].
Treatment of parasites with the Vps34 inhibitor, wortmannin (10 µM), did not change the localization of PfAtg8 at the apicoplast in the blood or liver stage suggesting Atg8 localization to the apicoplast is constitutive and not induced by the class III PI3K complex [31, 51]. However PfVps34 is also located at the apicoplast and its product, PI(3)P, is enriched at the apicoplast and nearby single-membrane vesicles, in addition to the food vacuole [89]. PI(3)P production is maximal in later blood stages and Tawk et al. suggest that PI(3)P vesicles transport lipids or proteins to the apicoplast, as is the case in T. gondii [90]. In agreement with this vital role, PI3K is essential in P. berghei [89]. As of now, there are no studies describing co-localization between PI3K and Atg8.
Interestingly, although PbAtg8 localized to the apicoplast in the liver stage, its E1 activating and E2 conjugating enzyme, Atg7 and Atg3, were located at the mitochondria, leading the authors to question whether they interact with PbAtg8 [30]. However, localization was only described at 52 h.p.i., at which point PbAtg8 is already lipidated to the outer membrane of the apicoplast [49]. It is difficult to imagine how Atg8 is lipidated without an activating and conjugating enzyme, other than the possibility that an E1 and E2 enzyme function has divergently evolved. Further localization studies in other stages and time points, as well as knockout of Atg3 and Atg7 would address these questions.
Current Model
During normal cellular conditions, the autophagy machinery, including Atg8 and a pool of Vps34 is localized to the apicoplast where it traffics proteins and/or lipids to the apicoplast and later recruits proteins/lipids for increased growth in preparation for cellular division. This does not require induction and occurs both in the liver and blood stage and most likely in the oocyst and sporozoite mosquito stages as well. This role most clearly explains PfAtg8 and Vps34's essential phenotype as autophagy would only be required under starvation conditions and protein trafficking to the membrane is most likely not essential, especially in in vitro conditions. During cases of extreme duress, such as starvation or drug treatment, the parasite induces an autophagic-like response that increases endocytosis of host cytoplasm and digestion in the food vacuole. If the stress conditions persist and the parasite cannot cope, the balance may be tipped to autophagic death. This may also be a mechanism to limit the number of parasites in the liver stage such that only the most robust parasites continue to the blood stage of infection. What little evidence exists suggests this cell death is independent of Atg8, but this remains to be further clarified [30].
Targeting Autophagy
It appears that Atg8, due to its unique ability to be covalently attached to membranes and to bind diverse proteins through adaptors has been conserved throughout Eukarya, even when other autophagy proteins are not. The list of non-autophagic functions for Atg8 homologues continues to grow and includes functions independent of membrane conjugation [91, 92]. Since Plasmodium Atg8 knockouts are nonviable and conditional knockouts are difficult in Plasmodium, a selective inhibitor preventing lipidation of Atg8 would be an invaluable tool that could also serve as a potential antimalarial therapeutic. Plasmodial Atg8's essentiality and our recently elucidated crystal structure make PfAtg8 a prime target for structure-guided drug design [36]. It would be of great interest to determine whether apicoplast morphology changes upon Atg8 inhibition, particularly during schizogony when it grows in size. Additionally, would parasites exhibit similar cell death by CQ when Atg8 is inhibited? Or conversely, would they be more susceptible?
While two independent studies concluded that PbAtg8 could not complement an Atg8 knockout in yeast [30, 49] a recent study showed PfAtg8 to be a functional homologue for ScAtg8 [30, 49, 51]. This discrepancy between the species is unexpected as P. berghei and P. falciparum Atg8 are 88% identical to each other and 40% identical to yeast Atg8. Transgenic PfAtg8 was not lipidated in mammalian cells, indicating PfAtg8 cannot complement the human homologue and PfAtg3 could not interact with human LC3 [36, 49]. We demonstrated that although PfAtg3 utilizes an Atg8-interacting motif similar to the human system, it also interacts through a short loop in PfAtg8 not present in humans [36]. Likely, other autophagy protein-protein interactions have similar divergences in their interaction surface and represent potential drug targets. The PfVps34 C-terminal region has 40% similarity and 23% identity to the human homologue but contains a substantial 1000 amino acid N-terminal extension with no known domain structure in Pfam. Perhaps its function at the apicoplast is mediated through this region? Unique insertions like these would serve as a platform for novel drug design against autophagic and trafficking pathways that increasingly seem important for multiple stages of the Plasmodium life cycle. However, their function needs to be elucidated.
Concluding Remarks
Remarkable progress has been made in the last year, dissecting the role of the autophagic machinery in Plasmodium, but many questions remain. The field is in agreement that Atg8 is located at the apicoplast, but it is not known if Atg8 is essential for the integrity of the organelle. Does Atg8 transport proteins, phospholipids, or both to the apicoplast and how is it targeted?
Starvation or CQ treatment leads to re-localization of Atg8 away from the apicoplast and is suggestive of a pro-survival mechanism under stress. But does this represent a typical autophagy pathway whereby parasitic proteins or organelles are recycled? The presence of Atg8 in the host cytoplasm suggests Atg8 is involved in trafficking that could include parasite protein export, host protein or heme import or even an attempt at detoxification of drugs such as CQ.
What, if anything, acts upstream of Atg1 to induce autophagy under starvation conditions? In yeast, cAMP-dependent protein kinase (PKA) signaling regulates Atg1 independent from TOR and may be a regulator in Plasmodium [93]. PfVps34 is phosphorylated in the parasite at a predicted PKA site [94]. PfeIK1, an orthologue for eukaryotic initiation factor 2α kinase, has been implicated in the amino acid starvation response in Plasmodium and has been suggested as a possible mediator of autophagy induction [5, 95]. At this time, there are more questions than answers, but it is encouraging to see that the field embraces autophagy in apicomplexa as a highly interesting research area. It can be expected that progress towards intervening molecules will be made and these will further facilitate biological research questions.
Supplementary Material
Acknowledgements
This work was partially funded through The Bloomberg Family Foundation and a Predoctoral fellowship from the Johns Hopkins Malaria Institute for A.U.P.H.
Competing Interests
The authors have declared that no competing interests exist.
References
- 1.Giuliani CM, Dass CR (2013) Autophagy and cancer: taking the ‘toxic’ out of cytotoxics. J Pharm Pharmacol 65: 777–789 [DOI] [PubMed] [Google Scholar]
- 2.Metcalf DJ, García-Arencibia M, Hochfeld WE, Rubinsztein DC (2012) Autophagy and misfolded proteins in neurodegeneration. Exp Neurol 238: 22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi AM, Ryter SW, Levine B (2013) Autophagy in human health and disease. N Engl J Med 368: 651–662 [DOI] [PubMed] [Google Scholar]
- 4.Brennand A, Gualdrón-López M, Coppens I, Rigden DJ, Ginger ML (2011) Autophagy in parasitic protists: unique features and drug targets. Molecular and Biochemical Parasitology 177: 83–99 [DOI] [PubMed] [Google Scholar]
- 5.Sinai AP, Roepe PD (2012) Autophagy in Apicomplexa: a life sustaining death mechanism?. Trends Parasitol 28: 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duszenko M, Ginger ML, Brennand A, Gualdron-Lopez M, Colombo MI (2011) Autophagy in protists. Autophagy 7: 127–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavine MD, Arrizabalaga G (2012) Analysis of monensin sensitivity in Toxoplasma gondii reveals autophagy as a mechanism for drug induced death. PLoS ONE 7: e42107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO (2011) World Malaria Report.
- 9.Homewood CA, Warhurst DC, Peters W, Baggaley VC (1972) Lysosomes, pH and the Anti-malarial Action of Chloroquine. Nature 235: 50–52 [DOI] [PubMed] [Google Scholar]
- 10.Warhurst DC, Thomas SC (1975) Pharmacology of the malaria parasite--a study of dose-response relationships in chloroquine-induced autophagic vacuole formation in Plasmodium berghei. Biochem Pharmacol 24: 2047–2056 [DOI] [PubMed] [Google Scholar]
- 11.Totino PR, Daniel-Ribeiro CT, Corte-Real S, de Fátima Ferreira-da-Cruz M (2008) Plasmodium falciparum: erythrocytic stages die by autophagic-like cell death under drug pressure. Exp Parasitol 118: 478–486 [DOI] [PubMed] [Google Scholar]
- 12.Chen PQ, Yuan J, Du QY, Chen L, Li GQ (2000) Effects of dihydroartemisinin on fine structure of erythrocytic stages of Plasmodium berghei ANKA strain. Acta Pharmacol Sin 21: 234–238 [PubMed] [Google Scholar]
- 13.Klionsky DJ (2011) The autophagosome is overrated!. autophagy 7: 353–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayabalasingham B, Bano N, Coppens I (2010) Metamorphosis of the malaria parasite in the liver is associated with organelle clearance. Cell Res 20: 1043–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meis JF, Verhave JP, Jap PH, Meuwissen JH (1985) Transformation of sporozoites of Plasmodium berghei into exoerythrocytic forms in the liver of its mammalian host. Cell Tissue Res 241: 353–360 [DOI] [PubMed] [Google Scholar]
- 16.Sturm A, Amino R, van de Sand C, Regen T, Retzlaff S (2006) Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science 313: 1287–1290 [DOI] [PubMed] [Google Scholar]
- 17.Graewe S, Rankin KE, Lehmann C, Deschermeier C, Hecht L (2011) Hostile takeover by Plasmodium: reorganization of parasite and host cell membranes during liver stage egress. PLoS Pathog 7: e1002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N (2010) Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 30: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y (2008) Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 19: 2039–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hara T, Takamura A, Kishi C, Iemura S, Natsume T (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki K, Kubota Y, Sekito T, Ohsumi Y (2007) Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12: 209–218 [DOI] [PubMed] [Google Scholar]
- 22.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP (2012) Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A 109: 2003–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y (2013) Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. [DOI] [PubMed] [Google Scholar]
- 24.Xie Z, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19: 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y (1998) A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem 273: 33889–33892 [DOI] [PubMed] [Google Scholar]
- 28.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282: 37298–37302 [DOI] [PubMed] [Google Scholar]
- 29.Rigden DJ, Michels PA, Ginger ML (2009) Autophagy in protists: Examples of secondary loss, lineage-specific innovations, and the conundrum of remodeling a single mitochondrion. Autophagy 5 [DOI] [PubMed] [Google Scholar]
- 30.Eickel N, Kaiser G, Prado M, Burda PC, Roelli M (2013) Features of autophagic cell death in Plasmodium liver-stage parasites. Autophagy 9: 568–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitamura K, Kishi-Itakura C, Tsuboi T, Sato S, Kita K (2012) Autophagy-Related Atg8 Localizes to the Apicoplast of the Human Malaria Parasite Plasmodium falciparum. PLoS ONE 7: e42977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS (2012) Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J 31: 3691–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T (2012) Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 198: 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ (2010) An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 190: 1005–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair U, Cao Y, Xie Z, Klionsky DJ (2010) Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem 285: 11476–11488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hain AU, Weltzer RR, Hammond H, Jayabalasingham B, Dinglasan RR (2012) Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J Struct Biol 180: 551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Besteiro S, Brooks CF, Striepen B, Dubremetz JF (2011) Autophagy Protein Atg3 is Essential for Maintaining Mitochondrial Integrity and for Normal Intracellular Development of Toxoplasma gondii Tachyzoites. Plos Pathogens 7: e1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu ZQ, Ni T, Hong B, Wang HY, Jiang FJ (2012) Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 8: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuma A, Mizushima N, Ishihara N, Ohsumi Y (2002) Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 277: 18619–18625 [DOI] [PubMed] [Google Scholar]
- 41.Gutierrez MG, Munafó DB, Berón W, Colombo MI (2004) Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 117: 2687–2697 [DOI] [PubMed] [Google Scholar]
- 42.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E (2004) Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 117: 4837–4848 [DOI] [PubMed] [Google Scholar]
- 43.Langsley G, van Noort V, Carret C, Meissner M, de Villiers EP (2008) Comparative genomics of the Rab protein family in Apicomplexan parasites. Microbes Infect 10: 462–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM (2001) Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem 276: 2083–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ (2006) Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell 17: 5094–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardner MJ, Hall N, Fung E, White O, Berriman M (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kissinger JC, Brunk BP, Crabtree J, Fraunholz MJ, Gajria B (2002) The Plasmodium genome database. Nature 419: 490–492 [DOI] [PubMed] [Google Scholar]
- 49.Jayabalasingham B, Voss C, Ehrenman K, Romano JD, Smith MEet al. (2013) Characterization of the Atg8-conjugation system in two Plasmodium species with special focus on the liver stage: Possible linkage between the apicoplastic and autophagic systems?Autophagy: In press; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM (2009) Type II fatty acid synthesis is essential only for malaria parasite late liver stage development. Cell Microbiol 11: 506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomlins AM, Ben-Rached F, Willams RAM, Proto WR, Coppens Iet al. (2013) Plasmodium falciparum Atg8 implicated in both autophagy and apicoplast formation Autophagy: In press; [DOI] [PubMed] [Google Scholar]
- 52.López-Barragán MJ, Lemieux J, Quiñones M, Williamson KC, Molina-Cruz A (2011) Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419: 520–526 [DOI] [PubMed] [Google Scholar]
- 54.Florens L, Liu X, Wang Y, Yang S, Schwartz O (2004) Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol Biochem Parasitol 135: 1–11 [DOI] [PubMed] [Google Scholar]
- 55.Silvestrini F, Lasonder E, Olivieri A, Camarda G, van Schaijk B (2010) Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics 9: 1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowyer PW, Simon GM, Cravatt BF, Bogyo M (2011) Global profiling of proteolysis during rupture of Plasmodium falciparum from the host erythrocyte. Molecular & cellular proteomics: MCP 10: M110.001636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O (2012) Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol 13: R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treeck M, Sanders JL, Elias JE, Boothroyd JC (2011) The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 10: 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaid A, Ranjan R, Smythe WA, Hoppe HC, Sharma P (2010) PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood 115: 2500–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramani S, Malhotra V (2013) Non-autophagic roles of autophagy-related proteins. EMBO Rep 14: 143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manjithaya R, Subramani S (2010) Role of autophagy in unconventional protein secretion. autophagy 6: 650–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF (2007) The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell 130: 524–534 [DOI] [PubMed] [Google Scholar]
- 63.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V (2010) Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abrahamsen H, Stenmark H (2010) Protein secretion: unconventional exit by exophagy. Curr Biol 20: R415–418 [DOI] [PubMed] [Google Scholar]
- 65.Römisch K (2012) Diversion at the ER: How Plasmodium falciparum exports proteins into host erythrocytes. F1000Res: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhattacharjee S, Stahelin RV, Speicher KD, Speicher DW, Haldar K (2012) Endoplasmic reticulum PI(3)P lipid binding targets malaria proteins to the host cell. Cell 148: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee MC, Moura PA, Miller EA, Fidock DA (2008) Plasmodium falciparum Sec24 marks transitional ER that exports a model cargo via a diacidic motif. Molecular microbiology 68: 1535–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adisa A, Frankland S, Rug M, Jackson K, Maier AG (2007) Re-assessing the locations of components of the classical vesicle-mediated trafficking machinery in transfected Plasmodium falciparum. Int J Parasitol 37: 1127–1141 [DOI] [PubMed] [Google Scholar]
- 69.Coppens I (2011) Metamorphoses of malaria: the role of autophagy in parasite differentiation. Essays Biochem 51: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rønning OW, Pettersen EO, Seglen PO (1979) Protein synthesis and protein degradation through the cell cycle of human NHIK 3025 cells in vitro. Exp Cell Res 123: 63–72 [DOI] [PubMed] [Google Scholar]
- 71.Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fitch CD (2004) Ferriprotoporphyrin IX, phospholipids, and the antimalarial actions of quinoline drugs. Life Sci 74: 1957–1972 [DOI] [PubMed] [Google Scholar]
- 73.Paguio MF, Bogle KL, Roepe P (2011) Plasmodium falciparum resistance to cytocidal versus cytostatic effects of chloroquine. Molecular and Biochemical Parasitology 178: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoppe HC, van Schalkwyk DA, Wiehart UI, Meredith SA, Egan J (2004) Antimalarial quinolines and artemisinin inhibit endocytosis in Plasmodium falciparum. Antimicrobial Agents and Chemotherapy 48: 2370–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gaviria D, Paguio MF, Turnbull LB, Tan A, Siriwardana Aet al. (2013) An Autophagy – Related Process is Associated with Cytocidal Chloroquine Resistance in Plasmodium falciparum Molecular microbiology: In revision [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Babbitt SE, Altenhofen L, Cobbold SA, Istvan ES, Fennell C (2012) Plasmodium falciparum responds to amino acid starvation by entering into a hibernatory state. Proc Natl Acad Sci U S A 109: E3278–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McIntosh MT, Vaid A, Hosgood HD, Vijay J, Bhattacharya A (2007) Traffic to the malaria parasite food vacuole: a novel pathway involving a phosphatidylinositol 3-phosphate-binding protein. J Biol Chem 282: 11499–11508 [DOI] [PubMed] [Google Scholar]
- 78.Lüder CG, Campos-Salinas J, Gonzalez-Rey E, van Zandbergen G (2010) Impact of protozoan cell death on parasite-host interactions and pathogenesis. Parasit Vectors 3: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsujimoto Y, Shimizu S (2005) Another way to die: autophagic programmed cell death. Cell Death Differ 12(Suppl 2): 1528–1534 [DOI] [PubMed] [Google Scholar]
- 80.Li FJ, Shen Q, Wang C, Sun Y, Yuan AY (2012) A role of autophagy in Trypanosoma brucei cell death. Cell Microbiol 14: 1242–1256 [DOI] [PubMed] [Google Scholar]
- 81.Ghosh D, Walton JL, Roepe P, Sinai A (2012) Autophagy is a cell death mechanism in Toxoplasma gondii. Cell Microbiol 14: 589–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Proto WR, Coombs GH, Mottram JC (2013) Cell death in parasitic protozoa: regulated or incidental?. Nat Rev Micro 11: 58–66 [DOI] [PubMed] [Google Scholar]
- 83.Bera A, Singh S, Nagaraj R, Vaidya T (2003) Induction of autophagic cell death in Leishmania donovani by antimicrobial peptides. Molecular and Biochemical Parasitology 127: 23–35 [DOI] [PubMed] [Google Scholar]
- 84.Yoon YH, Cho KS, Hwang JJ, Lee SJ, Choi JA (2010) Induction of lysosomal dilatation, arrested autophagy, and cell death by chloroquine in cultured ARPE-19 cells. Invest Ophthalmol Vis Sci 51: 6030–6037 [DOI] [PubMed] [Google Scholar]
- 85.van Dam TJ, Zwartkruis FJ, Bos JL, Snel B (2011) Evolution of the TOR pathway. J Mol Evol 73: 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T (2009) Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461: 654–658 [DOI] [PubMed] [Google Scholar]
- 87.Waller RF, McFadden GI (2005) The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol 7: 57–79 [PubMed] [Google Scholar]
- 88.Yeh E, DeRisi JL (2011) Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol 9: e1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tawk L, Chicanne G, Dubremetz JF, Richard V, Payrastre B (2010) Phosphatidylinositol 3-phosphate, an essential lipid in Plasmodium, localizes to the food vacuole membrane and the apicoplast. Eukaryotic Cell 9: 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tawk L, Dubremetz J, Montcourrier P, Chicanne G, Merezegue F (2011) Phosphatidylinositol 3-monophosphate is involved in toxoplasma apicoplast biogenesis. PLoS Pathog 7: e1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamura N, Oku M, Sakai Y (2010) Atg8 regulates vacuolar membrane dynamics in a lipidation-independent manner in Pichia pastoris. J Cell Sci 123: 4107–4116 [DOI] [PubMed] [Google Scholar]
- 92.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M (2010) Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7: 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK (2009) The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA 106: 17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lasonder E, Green JL, Camarda G, Talabani H, Holder AA (2012) The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase A signaling. J Proteome Res 11: 5323–5337 [DOI] [PubMed] [Google Scholar]
- 95.Fennell C, Babbitt S, Russo I, Wilkes J, Ranford-Cartwright L (2009) PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress-response to amino-acid starvation. Malar J 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.