Abstract
Background and Aims
Double B-box zinc finger (DBB) proteins are recently identified plant transcription regulators that participate in the response to sodium chloride-induced stress in arabidopsis plants. Little is known regarding their subcellular localization and expression patterns, particularly in relation to other osmotic constraints and the day/night cycle. This study investigated natural variations in the amount of a Solanum DBB protein, SsBBX24, during plant development, and also under various environmental constraints leading to cell dehydration in relation to the circadian clock and the time of day.
Methods SsBBX24
transcript and protein abundance in various organs of phytotron-grown Solanum tuberosum and S. sogarandinum plants were investigated at different time points of the day and under various osmotic constraints. The intracellular location of SsBBX24 was determined by western blot analysis of subcellular fractions.
Key Results
Western blot analysis of SsBBX24 protein revealed that it was located in the nucleus at the beginning of the light period and in the cytosol at the end, suggesting movement (‘trafficking’) during the light phase. SsBBX24 gene expression exhibited circadian cycling under control conditions, with the highest and lowest abundances of both transcript and protein occurring 8 and 18 h after dawn, respectively. Exposing Solanum plants to low temperature, salinity and polyethylene glycol (PEG), but not to drought, disturbed the circadian regulation of SsBBX24 gene expression at the protein level. SsBBX24 transcript and protein accumulated in Solanum plants in response to salt and PEG treatments, but not in response to low temperature or water deficit. Most interestingly, the time of the day modulated the magnitude of SsBBX24 expression in response to high salt concentration.
Conclusions
The interplay between circadian rhythm and osmotic constraints in the regulation of the expression of a Solanum DBB transcriptional regulator is demonstrated. It is proposed that stress-dependent, post-transcriptional mechanisms alter the regulation by the circadian clock of the amount of SsBBX24.
Keywords: Circadian clock, Double B-box protein, gating, osmotic stress, salinity, Solanum tuberosum, Solanum sogarandinum
INTRODUCTION
During their life cycle, plants are frequently exposed to a number of environmentally induced abiotic stress factors that constrain growth and productivity. Plants respond to these stress factors by altering a wide range of cellular processes, thus enabling some adaptation to a varying environment. This complex process of reprogramming involves a number of physiological and biochemical alterations, including changes in plant cell wall composition, in membrane structure and function, and also in primary and secondary metabolism (Murelli et al., 1995; Rajashekar and Lafta, 1996; Knight, 2000; Örvar et al., 2000). The control of gene expression plays a key role in this reprogramming. Consistently, hundreds of genes encoding various types of transcription factor are involved in the regulation of plant responses to environmental stresses via the transcriptional regulation of target genes (Riechmann et al., 2000; Montiel et al., 2004). The transcription factors DREB1 and DREB2 are the main regulators of plant responses to cold and water deficit, including some that may be regulatory (Yamaguchi-Shinozaki and Shinozaki, 2006; Chinnusamy et al., 2010). Zinc finger proteins form a relatively large family of transcription regulators in plants (Takatsuji et al., 1999). They are important components in the regulation of plant growth and development, and also participate in responses to biotic and abiotic stresses. On the basis of the structural and functional characteristics of individual members of this family, they are arranged into several distinct subfamilies. Among them, the Double B-box (DBB) subfamily represents a small group of zinc finger proteins containing two tandem B-box domains, the best characterized members of which are called STO (salt tolerance) and STH2 (salt tolerance homologue 2). The typical features of DBB proteins are the presence of two tandem zinc-binding B-box domains located at the N-terminal end and a VPDLG motif, located at the C-terminal region, which plays a role in protein–protein interaction as well as in nuclear localization (Kurup et al., 2000; Holm et al., 2001; Robson et al., 2001). The arabidopsis STO was originally identified as complementing the growth defect of a yeast calcineurin-deficient mutant, which shows a salinity-sensitive phenotype (Lippuner et al., 1996). Unexpectedly, STO gene expression is not induced by salt treatment in arabidopsis plants (Lippuner et al., 1996; Nagaoka and Takano, 2003). However, STO overexpression enhances root growth of arabidopsis in high-salt conditions (Nagaoka and Takano, 2003). In addition, STO is able to interact with the CEO1/RCD1 protein (Belles-Boix et al., 2000) and, as a consequence, negatively regulates the expression of a wide range of stress-related genes. CEO1/RCD1 has recently been characterized as a new component in the response of plants to severe salinity (Katiyar-Agarwal et al., 2006). Interestingly, another arabidopsis DBB protein homologous to STO, called STH2, is able to interact with the well-characterized photomorphogenic transcription factor HY5 (LONG HYPOCOTYL 5) in both yeast and plants. HY5 is a bZIP transcription factor that promotes early photomorphogenesis (seedling development) in the light (Datta et al., 2007).
Transcriptional analysis of several DBB genes has revealed circadian-dependent regulation of their expression (Kumagai et al., 2008). The circadian rhythm generated by an internal clock persists in continuous light or dark conditions with an approximately 24-h period. This endogenous timekeeper in living organisms acts as a mechanism measuring the duration of day and night periods (Harmer, 2009). In plants, the circadian rhythm regulates numerous processes, from leaf movements to cyclic expression of the mRNAs of many genes. With regard to cold acclimation, CBF transcription factors and several cold-regulated (COR) genes are circadian-regulated (Harmer et al., 2000; Cook et al., 2004). One result of circadian regulation is that the accumulation of transcript induced by the stimulus can be strongly influenced by the time of day, referring to the time point (hour) in a diel (24-h period), at which the stimulus is applied. This occurrence is known as gating (Fowler et al., 2005; Bieniawska et al., 2008; Wilkins et al., 2010). Thus, the time point throughout the day (the time of day) appears as an emerging key element modifying plant responses to environmental constraints at the molecular level. However, the understanding of this process remains poor, particularly regarding DBB transcription factors.
In this study we investigated the relationship between the circadian clock and ionic and osmotic constraints in the control of the expression of a Solanum gene designated SsBBX24 and encoding a protein belonging to the Double B-box family. We show that, under normal growth conditions, the expression of SsBBX24 undergoes circadian regulation at both the mRNA and the protein level. We provide evidence that circadian-regulated cycling of the SsBBX24 protein is maintained in drought conditions but substantially reduced in plants exposed to cold, high salt conditions and PEG treatment. In addition, we reveal circadian clock gating of the salt-induced expression of SsBBX24.
MATERIALS AND METHODS
Plant material, growth conditions and environmental constraints
Solanum tuberosum ‘Ursus’ and S. sogarandinum ‘PI 230510’ plants, line 2 were both propagated for two weeks in vitro on solid MS medium at 20/15 °C (day/night) under a photon flux density (PFD) of150 μmol photons m−2 s−1 and a 14-h photoperiod, as previously described (Rorat and Irzykowski, 1996), and in a phytotron under a PFD of 250 μmol photons m−2 s−1 at 21/18 °C (day/night) and a 14-h photoperiod (Rorat et al., 2004). Under these conditions the term Zeitgeber time (ZT) refers to experimental time (Johnson et al., 2003; Beneragama and Goto, 2010). The ZT0 point corresponds to light-on (initiation of experimental dawn) and the ZT14 point to light-off (initiation of experimental dusk 14 h after light-on in control day/night conditions). For cold treatment, 2-week-old phytotron-grown plants were exposed for 20 min, 4 and 8 h and 1, 3 and 7 days at 4/3 °C (day/night) under a PFD of 250 μmol photons m−2 s−1. For salt treatment, 2-week-old phytotron-grown plants were watered with a nutrient solution containing 0·2 m NaCl (osmotic potential −0·99 MPa) at 25 °C for 8 h and 1 and 3 days under the same PFD. For osmotic stress, 2-week-old phytotron-grown plants were watered with a solution containing 20 % PEG-6000 (w/v) (osmotic potential −0·49 MPa) at 25 °C for 3 days to complete saturation of the soil. A gradually increasing water deficit was applied to phytotron-grown plants by withholding watering for ∼11 days, during which the soil water content (SWC) decreased from 80 % to 10 % and sometimes 1 day longer (10 % +1), and then plants were rewatered for 6 h or 3 days up to 80 % SWC. SWC was calculated relative to 100 % soil water capacity by weighing the pots at regular intervals. To determine soil water capacity, two pots were filled with soil, wetted until saturation, then allowed to dry completely at 80 °C for at least 24 h and weighed. SWC was calculated as the percentage of maximal soil water capacity. Samples of drought-stressed plants were collected at dawn (ZT2). Leaf relative water content (RWC) was determined as described by Pruvot et al. (1996). Additionally, one set of plants grown under a 14-h photoperiod were transferred to continuous light for 24 h at 20/15 °C (day/night) and the second set to darkness for 24 h. In these conditions the term ‘subjective day’ refers to experimental time from ZT0 to ZT14 and ‘subjective night’ to experimental time from ZT14 to ZT0. Two independent experiments were performed for each environmental constraint and two independent plant samples per experiment were used for each experiment.
Isolation of cold-induced transcripts
Total cellular RNA was prepared from control and cold-hardened plantlets using the guanidine isothiocyanate method as described (Rorat et al., 1997). For RNA isolation, the two samples collected at each time point were combined. A cDNA library was constructed from poly(A+) mRNA prepared using a Uni-ZAP-cDNA synthesis kit (Stratagene) and cold-induced cDNA was screened through two steps of differential screening as described previously (Kiełbowicz-Matuk et al., 2007).
DNA sequencing was performed using the dideoxynucleotide chain-termination method, BigDye™ and dRhodamine Terminator Cycle DNA Sequencing Kits (ABI Prism) and an ABI Prism 310 Genetic Analyzer. Partial nucleotide sequences of the 5′ and 3′ terminals of cDNA clones were identified with the NCBI BLAST program (http:/www.ncbi.nlm.nih.gov/BLAST/; Altschul et al., 1997). The theoretical isoelectric point (pI) and the predicted molecular mass of SsBBX24 were calculated using the DNAStart program. Amino acid sequences were aligned using the ClustalW program, version 1·81 (Thompson et al., 1994).
Semi-quantitative RT–PCR
Total RNA was prepared from organs of Solanum plants grown in the phytotron using TRIzol Reagent (Invitrogen). Equal amounts of total RNA were treated with RNase-free DNase I. First-strand cDNA was synthesized from 1 μg of total RNA using universal oligo (dT)23 primer and 200 units of Superscript II Reverse Transcriptase (Invitrogen) at 42 °C for 1 h. The resulting single-strand cDNA was amplified using specific primers for the SsBBX24 gene (forward, 5′-CCAGGCCTATGAAGATCCAGTGTGATGTGTGTGA; reverse, 5′-TTCTGCAGTCAACCAAGATCTGGGACAGTAAA) based on the SsBBX24 sequence. The S. tuberosum elongation factor (EF-1-α) gene was amplified with the specific primers 5′-TGCCCCCGGACACAGAGACTT (forward) and 5′-CCGGGGAGTGCCTCCTGGAG (reverse) and used as a control housekeeping gene (Nicot et al., 2005). PCR products were sampled after 23, 25, 28, 31, 34, 37, 40 and 43 cycles to determine the linearity requirement for semi-quantitative RT–PCR analysis. The optimal number of cycles was in the same range for SsBBX24 and EF-1-α RNA, thus allowing quantification on the same gel. PCR was performed using a thermal cycler (DNA Engine, BioRad) through 28 cycles of 94 °C for 35 s, 60 °C for 35 s and 72 °C for 1 min followed by 72 °C for 10 min. The expected RT–PCR products (702 and 606 bp for SsBBX24 and EF-1-α, respectively) were separated by electrophoresis in 1 % agarose gels. The bands were quantified with Gel-Pro Analyzer 3·1 software (Media Cybernetics). Mean values ± s.d. from three replicates are presented. Semi-quantitative RT–PCR was performed twice from independent biological samples, and similar results were obtained. The ratio between SsBBX24 and EF-1-α RNAs was calculated to normalize variation in terms of sample concentration and reaction efficiency.
Preparation of protein extracts
Soluble proteins were extracted from young leaves (including very young developing leaves and terminal shoots), well-expanded leaves, old leaves, open flowers (including stamens, pistils, petals and sepals), stems, rhizomes, roots and tubers in a buffer containing 50 mm Tris–HCl, pH 8·0, 2 % β-mercaptoethanol and 1 mm PMSF, as described by Rey et al. (2005).
Cytosolic proteins were prepared from young leaves and stems of S. sogarandinum and S. tuberosum plants grown in the phytotron in a buffer containing 0·3 m sucrose, 50 mm Tris–HCl, pH 8·0, 10 mm MgCl2, 1 mm EDTA and 1 mm PMSF. The homogenate was filtered through a two-layer Miracloth (Calbiochem) and centrifuged at 3000 g for 10 min at 4 °C. The cleared supernatant was then centrifuged at 20 000 g for 20 min at 4 °C and collected. Cytosolic proteins were precipitated with four volumes of 100 % acetone.
Proteins from nucleus-enriched fractions were isolated at 2 °C from Solanum plants according to a modified version of the method of Liu et al. (2001). Nuclei were isolated as follows. Fifteen grams of young leaves or 30 g of stems was homogenized using a Virtis TEMPEST VirTishear homogenizer in 200 ml buffer containing 50 mm Tris–HCl, pH 8·0, 10 mm MgCl2, 1 mm PMSF, 0·8 m sucrose, 5 % glycerol, 2 mm spermine and 1 % Triton X-100. The crude homogenate was filtered through two layers of Miracloth and two layers of nylon paper (mesh 80 μm, Bionovo) and pelleted by centrifugation (3800 g) at 4 °C for 15 min. The pellet was suspended in washing buffer (50 mm Tris–HCl, pH 8·0, 10 mm MgCl2, 1 mm PMSF, 0·8 m sucrose, 5 % glycerol, 1 % Triton X-100) and centrifuged (10 000 g) at 0 °C for 15 min on a 2·3 m sucrose cushion solution. The purified nucleus pellet was then washed four times in 20 ml of washing buffer at 4 °C and pelleted at 3000 g for 10 min. The quality of isolated nuclei was verified using light microscopy following staining with DAPI at 1 μg ml−1. To extract nuclear proteins, isolated nuclei were resuspended in a buffer containing 50 mm Tris–HCl, pH 8·0, 150 mm NaCl, 1 mm PMSF, 1 % sodium deoxycholate, 1 % SDS and 0·5 % Triton X-100, and incubated at 4 °C for 1 h, and then centrifuged at 14 000 g for 10 min. Nuclear proteins were precipitated with four volumes of 100 % acetone.
Chloroplasts were prepared from leaves, and proteins were isolated as described by Pruvot et al. (1996) with some modifications. About 30 leaves (15 g) were homogenized using a Waring Blender (two 15-s bursts at maximum speed) in 200 ml of ice-cold isolation buffer containing 50 mm Tris–HCl, pH 8·0, 0·33 m sorbitol, 2 mm EDTA, 1 mm MgCl2, 2 mm ascorbic acid and 5 mm cysteine. After filtration of the homogenate through three layers of Miracloth and centrifugation (2000 g) for 10 min at 4 °C, the supernatant was discarded and the green pellet carefully suspended in washing buffer (50 mm Tris–HCl, pH 8·0, 0·33 m sorbitol and 2 mm EDTA) and layered onto a Percoll step gradient composed of two layers of washing buffer containing 90 and 25 % Percoll. After centrifugation (4000 g, 4 °C, 10 min), intact chloroplasts were collected at the interphase, washed three times in 20 ml washing buffer and pelleted at 2000 g for 10 min. Finally, the chloroplast pellet was suspended in 50 mm Tris–HCl, pH 8·0, 150 mm NaCl, 1 % sodium deoxycholate, 1 % SDS and 1 % NP-40, incubated for 30 min at 4 °C and centrifuged at 3000 g for 10 min. Chloroplast proteins were precipitated with four volumes of 100 % acetone.
Mitochondria were prepared from tubers, and proteins were isolated using a modified version of the procedure of Eubel et al. (2007). All steps were performed at 2 °C. Tubers (300 g) were homogenized using a Waring Blender in ice-cold isolation buffer (500 ml) containing 0·4 m sucrose, 50 mm Tris–HCl, pH 8·0, 2 mm EDTA, 5 mm cysteine, 5 mm ascorbic acid, 0·1 % BSA, 1 % PVP- 40 and 2·5 mm PMSF. The homogenate was filtered through three layers of Miracloth and centrifuged at 3000 g for 10 min, centrifuged twice at 2200 g for 2 min (4 °C) and again for 10 min at 15 000 g. The pellet was gently suspended in washing buffer (0·4 m sucrose, 50 mm Tris–HCl, pH 8·0, 2 mm EDTA) and centrifuged twice at 3000 g for 10 min and 15 000 g for 20 min. After centrifugation, the pellet, which represented a crude mitochondrial preparation, was suspended in washing buffer and layered onto a Percoll step gradient composed of three layers of washing buffer containing 50, 26 and 18 % Percoll. After centrifugation (27 000 g, 50 min), mitochondria were collected at the 26/50 % interphase. To remove Percoll, purified mitochondria were centrifuged twice in 20 ml washing buffer at 15 000 g for 10 min. The mitochondrial pellet was suspended in a buffer containing 50 mm Tris–HCl, pH 8·0, 150 mm NaCl, 1 mm PMSF, 1 % sodium deoxycholate, 1 % SDS and 0·5 % Triton X-100, incubated for 30 min and centrifuged at 14 000 g for 10 min. Mitochondrial proteins were precipitated with four volumes of 100 % acetone.
Generation of a serum against SsBBX24 and western blot analysis
A 702-bp fragment of the coding region of SsBBX24 cDNA from nucleotides 320–1021 was cloned in the pQE-30Xa expression vector (Qiagen) to produce a recombinant protein fused to a 6× His tag in the M15[pRep4] E. coli strain. The tagged recombinant protein was purified on Ni-NTA resin (Qiagen) and used to raise rabbit polyclonal antibodies at Eurogentec (Belgium). The antibody concentration was determined using an indirect ELISA technique.
The protein content was determined using a modified Lowry method (Pierce). Proteins (10–40 μg) were separated using SDS–PAGE gels containing 13 % acrylamide and then electroblotted onto a 0·22 μm nitrocellulose membrane (Schleicher and Schuell, Germany) using a semi-dry blotting apparatus (BioRad). Membranes were stained with Ponceau red to ensure that equal protein amounts were transferred in each lane. Western blot analysis was performed using the serum raised against SsBBX24 diluted 1:500. Sera raised against histone H3, UDP-glucose pyrophosphorylase (UGPase), isocitrate dehydrogenase (IDH) and cytochrome c6 (Cyt c6) were purchased from Agrisera (Vännäs, Sweden) and used at dilutions of 1:5000, 1:1000, 1:5000 and 1:500, respectively. Bound antibodies were detected by subsequent incubation using biotinylated goat anti-rabbit secondary antibodies conjugated with Qdot 625–streptavidin (Invitrogen). Visualization of the proteins was performed by exposing membranes to standard UV light. Band intensity (relative) was analysed using densitometry, ImageMaster (Amersham Pharmacia) and Gel-Pro Analyzer 3·1 (Media Cybernetics, MD, USA) software. Mean values ± s.d. from three replicates are presented. Western analyses were performed using protein extracts originating from two independent sets of plants per condition, and at least two replicates were conducted for each condition.
RESULTS
SsBBX24 distribution in organs
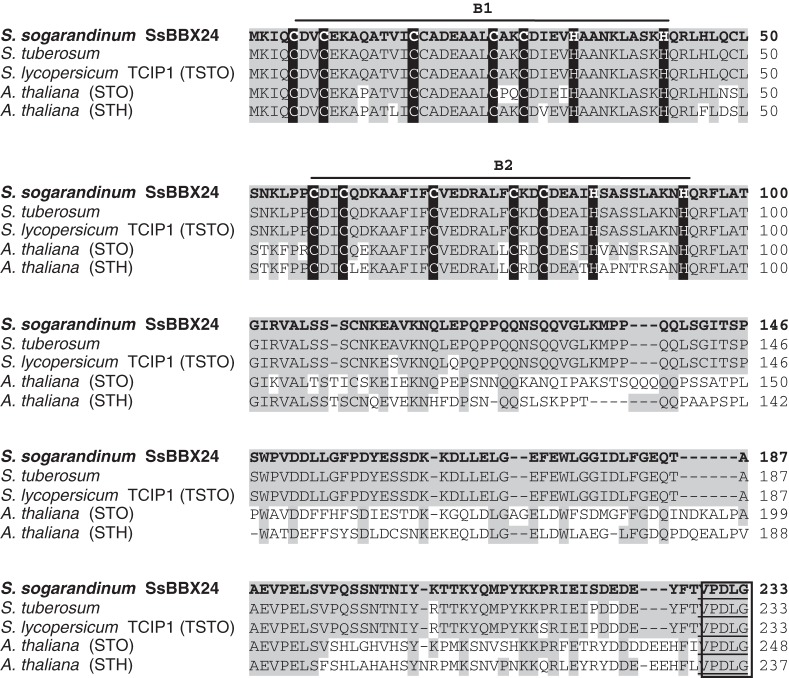
One clone, corresponding to a cold-induced mRNA and designated SsBBX24, was isolated by two-step differential screening from a cDNA library prepared from cold-treated S. sogarandinum plantlets (Kiełbowicz-Matuk et al., 2007). This clone encodes a protein displaying a calculated molecular mass of 25·8 kDa and sharing the highest identity with DBB proteins from S. tuberosum and S. lycopersicum (99 and 97 % identity, respectively) and with the STO (salt tolerance) and STH (salt tolerance homologue) proteins from Arabidopsis thaliana (59 and 58 % identity, respectively) (Fig. 1).
Fig. 1.
Alignment of Double B-box (DBB) zinc finger protein sequences from different plant species: SsBBX24 (in bold) from Solanum sogarandinum (ABC25454), S. tuberosum (ABA40448), S. lycopersicum (AAS67368; Ben-Naim et al., 2006), Arabidopsis thaliana STO (NP_172094) and A. thaliana STH (NP_565722). Alignment of DBB proteins was performed using ClustalW. Dashes indicate where the sequence was expanded to allow maximal alignment. Identical amino acids are shaded. The conserved cysteine and histidine residues are marked in black and the two putative B-Box domains, B1 and B2, are shown by lines. The conserved VPDLG motif is shown in a box.
To gain insight into the location of SsBBX24 in organs, the protein amount was investigated in various organs of 2-month-old S. tuberosum plants at the flowering stage, cultivated in the phytotron. Proteins at ∼27 and 26 kDa were specifically recognized by the serum, but not by the pre-immune serum, in bacterial cells and extracts from Solanum plants, respectively (Fig. 2A, and data not shown). The higher mass of the E. coli protein very likely originates from the presence of the His tag encoded by the pQE-30Xa expression vector. As shown in Fig. 2B, a substantial amount of SsBBX24 protein was observed in extracts from well-expanded leaves, young leaves and stems (lanes 1, 2 and 4). The protein amount was much smaller in old leaves, tubers and rhizomes (Fig. 2B, lanes 3, 5 and 6). In extracts from open flowers, the highest protein amount was noticed in stamens and pistils, and the protein was barely detected in petals and sepals (Fig. 2B, lanes 8–11). A similar SsBBX24 distribution was observed in the vegetative organs of 2-week-old S. sogarandinum plants (data not shown). These data reveal that, in Solanum species, SsBBX24 gene expression is differentially regulated at the protein level as a function of organ type during vegetative and reproductive development.
Fig. 2.
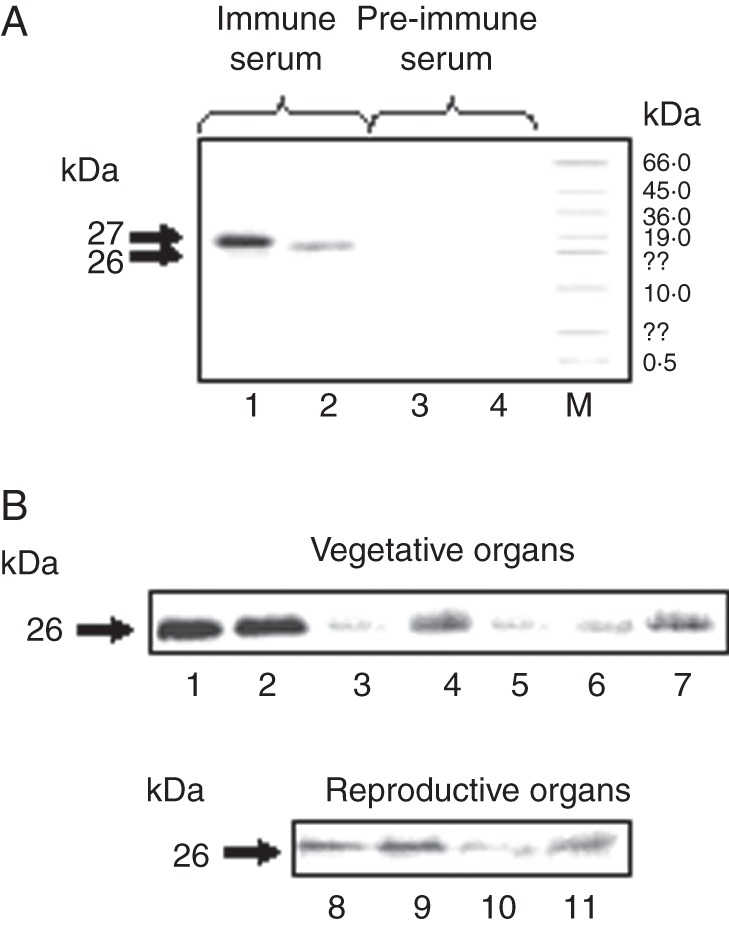
Distribution of SsBBX24 in organs of Solanum plants. (A) Immunodetection of the SsBBX24 protein using either serum raised against His tag-SsBBX24 or pre-immune serum. Lanes 1 and 3, purified recombinant His-tagged SsBBX24 protein produced in E. coli strain M15REP4; lanes 2 and 4, extracts from S. sogarandinum plants. M, molecular weight marker (Sigma). (B) Immunoblot analysis of SsBBX24 amount in vegetative and reproductive organs of 2-month-old S. tuberosum phytotron-grown plants. Lane 1, young leaves (including very young developing leaves and terminal shoots); lane 2, well-expanded leaves; lane 3, old leaves; lane 4, stems; lane 5, tubers; lane 6, rhizomes; lane 7, roots; lane 8, stamens; lane 9, pistils; lane 10, petals; lane 11, sepals.
Expression of the SsBBX24 gene is controlled by the circadian clock
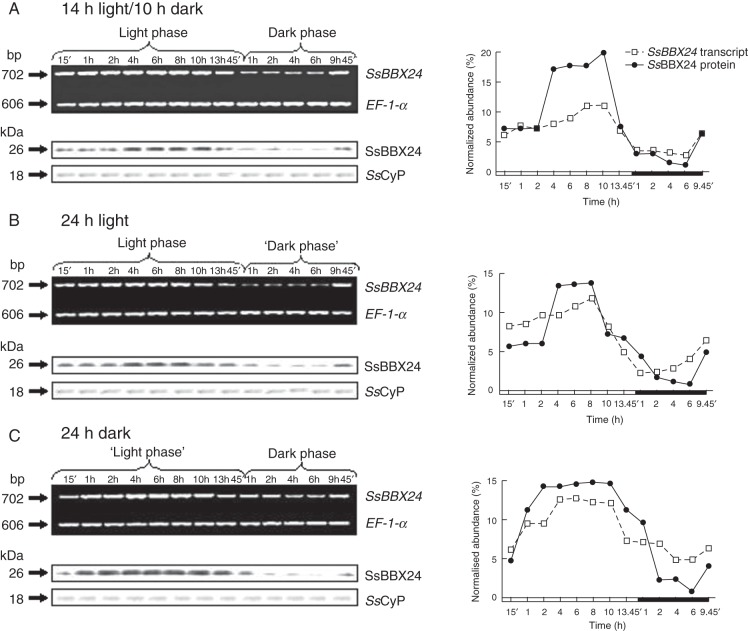
In subsequent experiments, we monitored SsBBX24 transcript and protein abundances in whole 2-week-old S. tuberosum plants during the light and dark phases under a 14-h photoperiod. Significant increases in both transcript and protein abundances were noticed during the light phase, with maxima after 8 h of light (Fig. 3A). In addition, a noticeable decrease in both mRNA and protein was observed at the beginning of the dark phase, with minima after 4 h in the darkness (Fig. 3A). To further characterize the regulation of SsBBX24 expression in relation to the circadian rhythm, we analysed transcript and protein abundances in plants subjected to continuous light or dark. Two-week-old phytotron plants, grown under a 14-h photoperiod, were separated into two sets and either exposed to light for 24 h or placed in the dark for the same period. Samples were collected at different times during the subjective light and dark cycles. RT–PCR and western analysis data clearly showed that the expression patterns of SsBBX24 under continuous light or dark were very similar to those observed under a normal day/night cycle (Fig. 3A–C). These data indicate that the expression of SsBBX24 is regulated by the circadian clock at both transcript and protein levels.
Fig. 3.
Circadian regulation of SsBBX24 expression. (A) Analysis of SsBBX24 transcript and protein abundances in 2-week-old S. tuberosum plants grown in a phytotron under a 14-h photoperiod at different time points during the light and dark phases. (B) Analysis of SsBBX24 transcript and protein abundances in 2-week-old S. tuberosum plants grown in a phytotron under a 14-h photoperiod and then transferred to continuous light for 24 h. Samples were collected at the indicated time points. (C) Analysis of SsBBX24 transcript and protein abundances in 2-week-old S. tuberosum plants grown in a phytotron under a 14-h photoperiod and then transferred to continuous dark for 24 h. Samples were collected at the indicated times. Semi-quantitative RT–PCR, western analysis and quantification of band intensity were performed as described in Materials and methods.
Subcellular location of SsBBX24
The subcellular location of SsBBX24 was investigated from the protein amount in cytosolic, nucleus-enriched, chloroplastic and mitochondrial fractions prepared from S. tuberosum leaves and stems. To ensure the purity of fractions, we used sera raised against (1) histone H3 (a nuclear marker), (2) UGPase, a cytosolic marker (Martz et al., 2002), (3) IDH, a mitochondrial marker, and (4) Cyt c6, a chloroplastic marker (Merchant and Quinn, 1998). The serum raised against histone H3 revealed one 17 kDa band in the nucleus-enriched fraction, but not in the three others (Fig. 4A). Similarly, single protein bands of 51, 45 and 15 kDa, corresponding to UGPase, IDH and Cyt c6, respectively, were specifically revealed in the cytosolic, mitochondrial and chloroplastic fractions (Fig. 4A). Western blot analysis clearly revealed the presence of SsBBX24 protein in cytosolic and nuclear fractions (Fig. 4A). Similar results were obtained when performing experiments using S. sogarandinum extracts (data not shown).
Fig. 4.
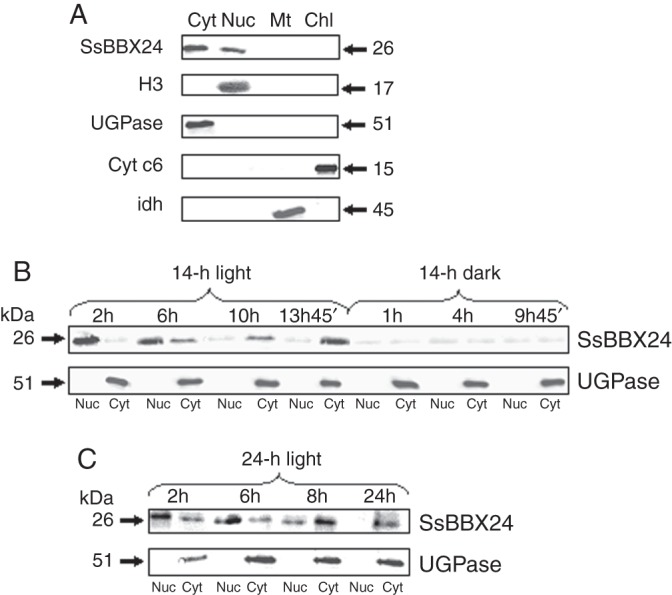
Subcellular location and light-dependent distribution of SsBBX24. (A) SsBBX24 subcellular localization. Western blot analysis of proteins from cytosolic (Cyt), nuclear (Nuc) and chloroplastic (Chl) fractions prepared from leaves and mitochondrial (Mt) fraction prepared from tubers of S. tuberosum plants was performed using sera raised against SsBBX24, histone H3, UDP-glucose pyrophosphorylase (UGPase), isocitrate dehydrogenase (IDH) and cytochrome c6 (Cyt c6). Proteins from chloroplastic and mitochondrial fractions (35 μg) and nucleus-enriched and cytosolic fractions (10 μg) were set for each lane and western blot analysis was carried out using SsBBX24, H3, UGPase, IDH and Cyt c6 antibodies. (B, C) Light-dependent distribution of SsBBX24 in 2-week-old phytotron-grown S. tuberosum plants under a normal light cycle (14 h light/10 h dark) (B) and under a 24-h light period (C). Samples were collected at the indicated time points of the subjective light/dark cycle. Protein samples from cytosol and nuclei were isolated and separated using SDS–PAGE. Thirty-five micrograms of protein was set per lane. UGPase and histone H3 antibodies were used as positive markers of cytosolic and nuclear fractions, respectively.
Distribution of SsBBX24 in cytosol and nucleus as a function of light
The distribution of the SsBBX24 protein in cytosol and nucleus was analysed in 2-week-old S. tuberosum plants grown under a normal cycle (14 h light/10 h dark), and then transferred to continuous light for 24 h. As shown in Fig. 4B, under a normal light cycle the highest SsBBX24 amounts were observed in nuclei at the beginning of the light phase and after 6 h in the light. Then, this amount gradually decreased and a concomitant increase in SsBBX24 abundance occurred in the cytosol. During the dark period of the normal cycle, the protein amount was much lower and equal in both nucleus and cytosol (Fig. 4B). Similarly, when plants were subjected to continuous light for 24 h, the highest SsBBX24 abundance in nuclei was observed at the same times, with a maximum reached after 6 h of light (Fig. 4C). The amount of SsBBX24 substantially decreased in nuclei and was barely detected at the end of the 24-h period of light exposure, whereas a strong increase in protein abundance was observed in the cytosol, the maximum being observed after 8 h of treatment (Fig. 4C). Using the cytosolic UGPase protein marker, no contamination was revealed in the cytosolic fraction (Fig. 4C).
Expression of the SsBBX24 gene in response to cold, drought, high salt and osmotic stress
As SsBBX24 was primarily isolated using differential screening of cDNA libraries prepared from in vitro control and cold-treated S. sogarandinum plantlets, we first investigated transcript abundance using semi-quantitative RT–PCR in 2-week-old S. tuberosum plants exposed to low temperature. When phytotron-grown plants were exposed to cold for 7 days, no change in transcript abundance was observed in apical parts, well-expanded leaves and stems (Fig. 5A and data not shown). In accordance with the RT–PCR data, no change in SsBBX24 protein amount was noticed for any organs of phytotron-grown plants subjected to cold for 7 days (Fig. 5B and data not shown). As a positive control, the amount of a lipid transfer protein, SsLTP1, reported to be induced at low temperature (Kiełbowicz-Matuk et al., 2008), was analysed in the same tissues. As shown in Fig. 5B, the amount of SsLTP1 protein increased substantially during the cold-hardening period. Similar results were obtained when performing experiments using S. sogarandinum extracts (data not shown).
Fig. 5.
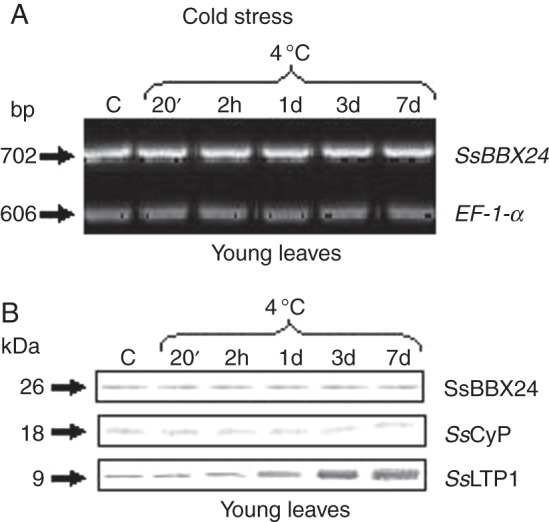
Expression of SsBBX24, SsLTP1 and SsCyP genes in S. tuberosum plants subjected to cold treatment. Semi-quantitative RT–PCR of SsBBX24 and EF-1-α transcript abundances (A) and western blot analysis of SsBBX24, SsCyP and SsLTP protein amounts (B) in young leaves of 2-week-old phytotron-grown S. tuberosum plants subjected to cold treatment (4/3 °C, day/night) for 20 min, 2 h and 1, 3 and 7 days. Lane C, control plants grown in vivo in a phytotron at 21/18 °C (day/night). RT–PCR and western blot analyses were performed as described in Materials and methods. The amounts of proteins set for detection of SsBBX24, SsCyP and SsLTP1 were 30, 10 and 40 μg per lane, respectively.
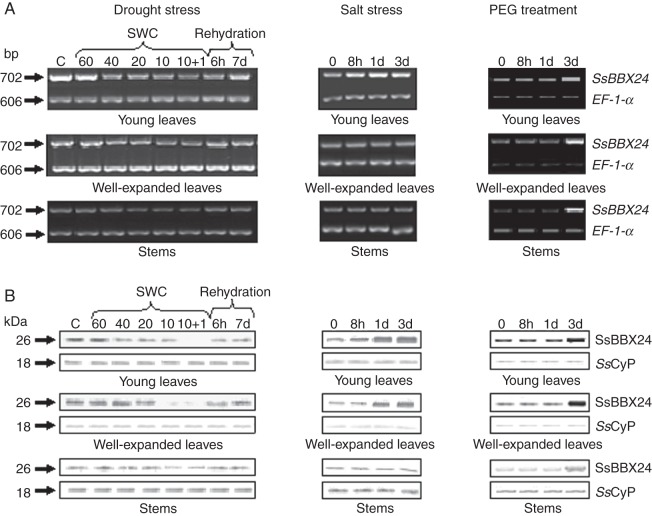
The lack of induction of SsBBX24 gene expression in response to cold and the induction of SsBBX24 homologue during salt stress in arabidopsis prompted us to expose plants to other osmotic stress conditions. Phytotron-grown S. tuberosum plants were therefore subjected to water deficit by withholding watering. During the first 5 days, the plants remained turgid. However, after 10 days without watering, when the soil water content had decreased to 10 %, old leaves exhibited severe wilting and turned yellow, whereas young leaves were moderately wilted (data not shown). In water-deprived plants, the amounts of SsBBX24 transcript and protein decreased in all organs tested when the soil water content dropped from 40 to 10 % (Fig. 6A, B). It is noteworthy that, following 6 h of rehydration, the abundances of mRNA and protein increased substantially in leaves but not in stems. Similar results were obtained when performing experiments using S. sogarandinum plants (data not shown).
Fig. 6.
Expression of the SsBBX24 gene in various organs (young leaves, well-expanded leaves and stems) of 2-week-old phytotron-grown S. tuberosum plants subjected to water deficit, salt treatment (0·2 m NaCl) and PEG treatment (20 % PEG-6000, w/v). Lane C, control plants grown in a phytotron at 80 % soil water content (SWC). Semi-quantitative RT–PCR (A) and western blot analysis (B) were performed as described in Materials and methods.
SsBBX24 expression was then investigated in various organs of S. tuberosum plants watered with a nutrient solution containing 0·2 m NaCl. When plants were exposed for 3 days to salt, no chlorosis was apparent in young and well-expanded leaves, but some old leaves displayed pale green areas (data not shown). The abundance of SsBBX24 transcript increased only in young leaves after 8 h of treatment (Fig. 6A). An increase in protein amount occurred in young leaves and well-expanded leaves, with maxima after 3 days of exposure to salt (Fig. 6B). At that time, SsBBX24 protein amount in young leaves and well-expanded leaves of treated plants was 4-fold higher than in control conditions (data not shown). In stems, no substantial change in SsBBX24 protein amount was observed (Fig. 6B). Similar results were obtained using S. sogarandinum plants (data not shown).
We then monitored SsBBX24 expression in various organs of Solanum plants watered for 3 days with a solution containing 20 % PEG-6000. Substantial accumulations of SsBBX24 transcript and protein were noticed in all tested organs. It is noteworthy that the variations in protein amount appeared later than those recorded in plants treated with NaCl (Fig. 6A, B). After 3 days, SsBBX24 protein amount in well-expanded leaves was 6-fold higher in PEG-treated plants compared with controls.
Cold, salinity and PEG modify regulation of SsBBX24 gene expression by the circadian clock
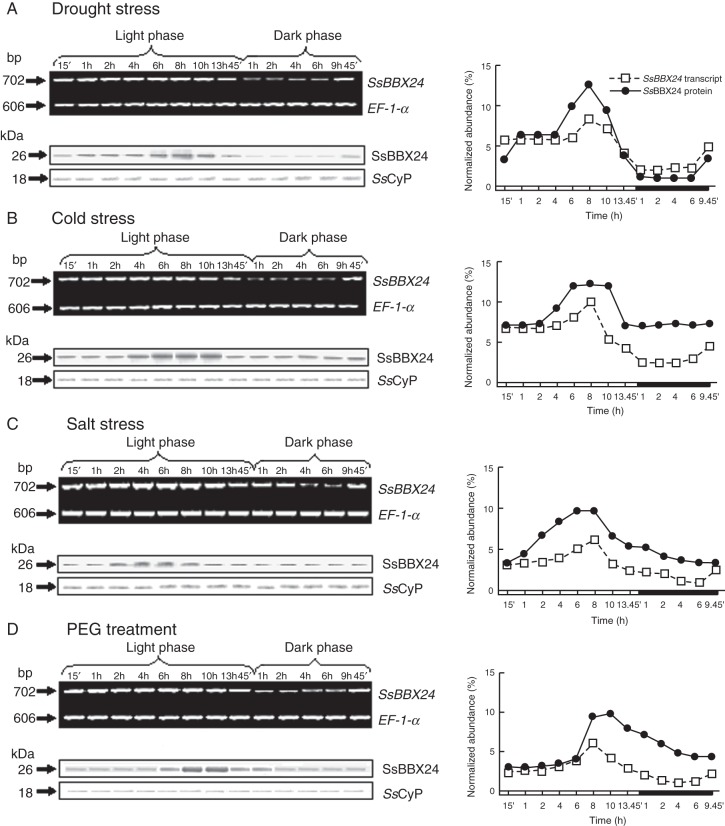
In order to uncover whether environmental constraints could influence the circadian regulation of SsBBX24 expression, we investigated transcript and protein abundances in whole 2-week-old Solanum plants entrained in a 14-h photoperiod and subjected to various environmental conditions: (1) cold (4/3 °C, day/night) for 24 h; (2) water deficit for 6 days (20 % soil water content and RWC of expanded leaves of ∼80 %); (3) exposure to salt (0·2 m NaCl) or (4) PEG-6000 (20 %) for 3 days. Under all stress conditions, a progressive increase in SsBBX24 transcript abundance was noticed during the light phase, similar to that seen in control conditions, the maxima being observed after 8 h of light. A noticeable decrease in transcript abundance was also recorded at the beginning of the dark phase in all treatments (Fig. 7A–D).
Fig. 7.
Circadian regulation of SsBBX24 expression under osmotic stress conditions. Analysis of SsBBX24 transcript and protein abundances in 2-week-old S. tuberosum plants grown under a 14-h photoperiod and subjected to water deficit (soil water content 20 %) (A), low temperature (4/3 °C, day/night) for 7 days (B), salt treatment (0·2 m NaCl) for 3 days (C) or 20 % PEG-6000 treatment for 3 days (D). Samples were collected at the indicated times during the light and dark phases. Semi-quantitative RT–PCR and western blot analysis were performed as described in Materials and methods.
The amount of SsBBX24 protein was analysed during the day/night cycle in plants subjected to these various environmental conditions. When plants were subjected to water deficit, no change in comparison with control conditions occurred in the circadian regulation of SsBBX24 protein amount (compare Fig. 7A with Fig. 3A). In contrast, a marked change in the regulation of protein amount by the circadian cycle occurred in plants exposed to cold, salinity or PEG treatments: during the dark period protein abundance was substantially higher than under control and water stress conditions. Consequently, the ratio between maximal and minimal SsBBX24 protein amounts (after 8 h of light and after 4 h of darkness, respectively) was much higher in control and drought stress conditions (∼15-fold) (Figs 3A and 7A) than for cold, salinity and PEG treatments (∼2-fold) (Fig. 7B–D). Taken together, these data indicate that the amounts of SsBBX24 transcript and protein are differentially regulated as a function of the circadian clock and environmental conditions.
Salt-induced expression of SsBBX24 gene is gated by the circadian clock
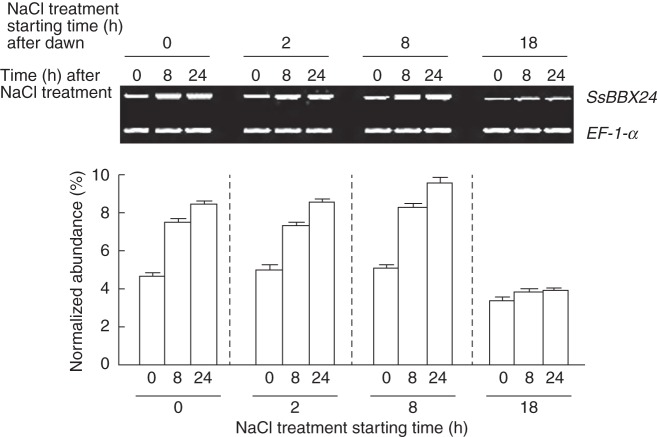
Because SsBBX24 transcript accumulates in young leaves of Solanum plants in response to high salt levels (Fig. 6A), we investigated whether the time of day (i.e. the time point in a 24-h period) at which plants are subjected to salt treatment could change the SsBBX24 expression pattern. Solanum tuberosum plants were grown for 2 weeks under a 14-h photoperiod (14 h light/10 h dark). The plants were then exposed to continuous light from ZT0 and treated at ZT0, ZT2, ZT8 or ZT18 with 0·2 m NaCl until saturation of the soil for 0, 8 and 24 h. These time points for treatments were chosen based on previous results showing maximal SsBBX24 transcript accumulation 8 h after subjective dawn (ZT8) and a minimal level 4 h after subjective dusk (i.e. ZT18, 18 h after subjective dawn), under normal growth conditions (Fig. 3A). Our results clearly reveal that the induction of SsBBX24 gene expression following 24 h of treatment was strongly dependent on the time of salt application. Indeed, induction of SsBBX24 expression was pronounced when salt was applied 0 and 2 h after dawn and maximal when the treatment was performed 8 h after dawn. In sharp contrast, there was no significant change in transcript abundance when plants maintained in continuous light were treated 4 h after subjective dusk, i.e. at ZT18 (Fig. 8).
Fig. 8.
Gating by circadian clock of salt-induced SsBBX24 gene expression. Solanum tuberosum plants were grown for 2 weeks under a 14-h photoperiod (14 h light/10 h dark) and then exposed to continuous light for 42 h from 0 h ZT time and treated at ZT0, ZT2, ZT8 or ZT18 with 0·2 m NaCl for 0, 8 and 24 h until saturation of soil. Semi-quantitative RT–PCR was performed as described in Materials and methods. The timing of dawn was determined subjectively.
DISCUSSION
SsBBX24 amount is differentially regulated by environmental conditions leading to cell dehydration
Transcription factors belonging to the zinc finger family are involved in plant responses to stress (Rizhsky et al., 2004; Davletova et al., 2005; Vogel et al., 2005). The transcript abundance of many zinc finger proteins is elevated under various stress conditions, including cold, drought, salt, wounding and oxidative treatments, as shown by transcription profiling (Kiełbowicz-Matuk, 2012). Among these zinc finger proteins, DBB proteins are very poorly characterized and knowledge of their participation in responses stress is still in its infancy. Our analysis reveals no change in SsBBX24 transcript and protein abundances in phytotron-grown Solanum plants exposed to cold conditions (Fig. 5A, B). In the case of water deficit, SsBBX24 transcript and protein abundances decreased in all organs tested and rapidly increased following rehydration (Fig. 6A). Conversely, high salinity and osmotic stress induced by PEG treatment resulted in significant increases in SsBBX24 protein amount in most aerial organs of the two Solanum species studied (Fig. 6B and data not shown). Lippuner et al. (1996) concluded that in arabidopsis the abundance of STO transcript does not increase in response to salt treatment, a conclusion that is not consistent with our data. In the experiment reported by Lippuner et al. (1996), arabidopsis plants were grown in vitro for 12 days under continuous fluorescent light, whereas we used 2-week-old phytotron-grown plants exposed to a 14-h photoperiod. Thus, the very different culture conditions very likely explain this discrepancy. Furthermore, it is worth mentioning that the conditions used in our study were much more physiologically relevant, since Solanum plants were grown on soil. Taken together, these results clearly indicate that low temperature, in contrast to salt, PEG and drought, does not modify SsBBX24 expression at the protein level. The data reveal for the first time that stress-regulated pathways control the expression of DBB genes in plants. Since previous observations have revealed the participation of DBB subfamily members in the transduction of light signals in plants (Chang et al., 2008; Kumagai et al., 2008), we presume that these transcriptional regulators act downstream in pathways integrating various types of environmental signal.
Distribution of SsBBX24 between cytosol and nucleus is light-dependent
In the present study we have shown that light plays an important role in the subcellular distribution of SsBBX24. At the beginning of the light phase the protein is present in the nucleus, where it accumulates for approximately the first 6 h. Then, the protein is very likely relocated from the nucleus to the cytosol, where a significant increase in the amount of SsBBX24 occurs (Fig. 4B), with a maximum occurring at the end of the light period. The data are not in full agreement with those reported for the arabidopsis STO:eGFP fusion protein expressed under the control of the CaMV35S promoter (Indorf et al., 2007). Using one transgenic line overexpressing STO:eGFP, Indorf et al. (2007) analysed the location of the STO fusion protein both before transfer to light and during light exposure. Microscopic analysis revealed a nuclear location of STO:eGFP dependent on light and its duration, since no or a weak green fluorescent protein (GFP) signal was recorded in the dark or following a 24-h exposure to light, respectively, whereas a signal was observed at the beginning of the light phase. Furthermore, western blot analysis using polyclonal antibodies raised against GFP confirmed the presence of the STO:eGFP protein only at the beginning of light treatment and demonstrated its absence at other time points of the day. The discrepancy between our results and those obtained by Indorf et al. (2007) may result from the use by these authors of a GFP–STO fusion protein, which might lead to modified subcellular compartmentalization and/or sensitivity to degradation. Furthermore, as these authors also used a very particular material (very young dark-adapted seedlings), we conclude that our results, which were derived from adult plants grown in physiological conditions, are more reliable regarding the subcellular distribution of SsBBX24 and its relocation to the cytosol at the end of the light period. On the other hand, we observed reduced SsBBX24 protein amounts in the cytosol during the dark phase (Fig. 4B). This could indicate degradation to a basal level with similar low amounts in both cytosol and nucleus. It is also noteworthy that the decreased SsBBX24 amount in the dark is consistent with the circadian profile, showing downregulation of SsBBX24 expression at the beginning of the dark phase under a daily light/dark cycle (Fig. 3A). The existence of light-dependent shuttles between nucleus and cytosol is well documented for proteins such as cryptochrome CRY1 (Yang et al., 2000). In the dark, most of the fusion β-glucuronidase (GUS)–CRY1 C-terminal domain (CCT1) protein is located in the nucleus. In contrast, under light conditions the fusion protein disappears from the nucleus and simultaneously appears in the cytosol. In addition, there is evidence that CRY1 interacts in vitro in the nucleus with various proteins, including COP1 (CONSTITUTIVE PHOTOMORPHOGENIC PROTEIN 1) (Yang et al., 2001) and SPA1 (SUPPRESSOR OF PHYTOCHROME A) (Liu et al., 2011). Hence, it is tempting to infer that the light-dependent nuclear location of SsBBX24 might similarly lead both to interaction with nuclear partners acting downstream in light- or stress-related signalling pathways and to participation in the transcriptional regulation of gene expression during the first hours of the light phase.
Interplay between circadian regulation of SsBBX24 gene expression and osmotic stress type
Although the expression of nearly one-third of genes in arabidopsis is likely regulated by the circadian clock (Covington et al., 2008), few plant transcriptome studies have directly examined the interdependence between circadian clock components and the complex network underlying plant responses to abiotic constraints like cold (Fowler et al., 2005; Bieniawska et al., 2008; Nakamichi et al., 2009), drought (Wilkins et al., 2009, 2010) and salinity (Kant et al., 2008). Our research has focused on the impact of constraints leading to cell dehydration on the cyclic expression of SsBBX24 in Solanum plants and, on the other hand, on the effect of the time of day of salt application on SsBBX24 transcript abundance. Our data clearly reveal that cold, salinity and PEG affect the circadian-regulated amount of SsBBX24 protein. In such stress conditions, only a slight decrease in SsBBX24 protein amount is observed in the dark period compared both with normal growth conditions (Fig. 3A) and with water deficit (Fig. 7A). Modulation of circadian oscillations in SsBBX24 by stress-related signals, including low temperature, salinity and PEG, may indicate the existence of a non-transcriptional mechanism depending on environmental constraints and controlling, for instance, the stability of the protein. The importance of non-transcriptional mechanisms in the maintenance of circadian rhythms has been reported recently by O'Neill et al. (2011).
We have demonstrated that circadian clock gates the salt induction of SsBBX24 expression. Our results reveal a tight and complex interplay between high salt and the circadian clock in the regulation of SsBBX24 expression, since salt modulates the clock phase at the protein level and the clock gates the response to salt at the transcript level in the dark. There is increasing evidence that the biological clock regulates the stress response pathways by modifying the perception of environmental signals. Indeed, the time of day is considered a major factor modifying responses to cold, drought and oxidative stresses, and also to ABA (Bieniawska et al., 2008; Wilkins et al., 2010; Lai et al., 2012). Cold gating affects the level of expression changes of more than 50 transcription factors, including CBF1, CBF2, CBF3 and ZAT12 (Cook et al., 2004; Fowler et al., 2005; Guy et al., 2008). Our report provides the first evidence of such a mechanism in saline conditions for a DBB translational regulator. A better understanding of the molecular bases of the regulation of SsBBX24 gene expression is needed to understand how salt-related factors interfere with circadian clock components.
Conclusions
The results presented here emphasize not only the circadian clock-dependent regulation of SsBBX24 expression, but also the interplay between circadian the clock and osmotic constraints in the regulation of SsBBX24 protein amount. Based on the data showing altered circadian regulation of SsBBX24 protein amount during the day/night cycle in salt- or cold-treated plants, we conclude that stress-dependent post-transcriptional mechanisms play a key role in the circadian control of SsBBX24 expression. We suggest that, under specific stress conditions, degradation of SsBBX24 is slowed in the dark. This would maintain a pool of protein immediately available at the beginning of the light period, thus favouring relocation to the nucleus, where it could interact with other protein partners to create a complex modulating the expression of stress-related genes (Holm et al., 2001; Datta et al., 2008) and thus to improve the plant's responses to environmental constraints.
ACKNOWLEDGEMENTS
The authors wish to thank Tomasz Skrzypczak for helpful assistance in producing a recombinant SsBBX24 protein and Dr Bartosz Szabała for valuable advice and assistance with the isolation of cell organelles. We thank Translmed (Cedar Hill, TX, USA), a proofreading and copyediting company, for help in copyediting and linguistic consultation regarding the manuscript. This study was supported in part by Polish Ministry of Science Higher Education grant no. PBZ-MNiSW-2/3/2006.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles-Boix E, Babiychuk E, Montażu MV, Inzé D, Kushnir S. CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Letters. 2000;482:19–24. doi: 10.1016/s0014-5793(00)02016-0. [DOI] [PubMed] [Google Scholar]
- Beneragama CK, Goto K. When does subjective day come under 24-h light/dark cycles? The case of circadian rhythms of UV-C resistance and timing of cell division in Euglena gracilis. International Journal of Botany. 2010;6:28–34. [Google Scholar]
- Ben-Naim O, Eshed R, Parnis A, et al. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant Journal. 2006;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Spinoza C, Schlereth A, Suplice R, Hincha DK, Hannah MA. Disruption of the arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiology. 2008;147:263–279. doi: 10.1104/pp.108.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Li YH, Chen LT, et al. LZF1, a HY5-regulated transcriptional factor, functions in arabidopsis de-etiolation. Plant Journal. 2008;54:205–219. doi: 10.1111/j.1365-313X.2008.03401.x. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK, Sunkar R. Gene regulation during cold stress acclimation in plants. Methods in Molecular Biology. 2010;639:39–55. doi: 10.1007/978-1-60761-702-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2004;101:15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biology. 2008;9 doi: 10.1186/gb-2008-9-8-r130. pR130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-Box protein in arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007;19:3242–3255. doi: 10.1105/tpc.107.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, et al. LZF1/SALT TOLERANCE HOMOLOG3, an arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell. 2008;20:2324–2338. doi: 10.1105/tpc.108.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in arabidopsis. Plant Physiology. 2005;139:847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Heazlewood JL, Millar AH. Isolation and subfractionation of plant mitochondria for proteomic analysis. Methods in Molecular Biology. 2007;355:49–62. doi: 10.1385/1-59745-227-0:49. [DOI] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow MF. Low temperature induction of arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiology. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy C, Kaplan F, Kopka J, Selbig J, Hincha DK. Metabolomics of temperature stress. Physiologia Plantarum. 2008;32:220–235. doi: 10.1111/j.1399-3054.2007.00999.x. [DOI] [PubMed] [Google Scholar]
- Harmer SL. The circadian system in higher plants. Annual Review of Plant Biology. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, et al. Orchestrated transcription of key pathways in arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of arabidopsis COP1. EMBO Journal. 2001;20:118–127. doi: 10.1093/emboj/20.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodríguez-Franco M. Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant Journal. 2007;51:563–574. doi: 10.1111/j.1365-313X.2007.03162.x. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Elliott JA, Foster R. Entrainment of circadian programs. Chronobiology International. 2003;20:741–774. doi: 10.1081/cbi-120024211. [DOI] [PubMed] [Google Scholar]
- Kant P, Gordon M, Kant S, et al. Functional-genomics-based identification of genes that regulate arabidopsis responses to multiple abiotic stresses. Plant Cell & Environment. 2008;31:697–714. doi: 10.1111/j.1365-3040.2008.01779.x. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Zhu J, Kim K, et al. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2006;103:18816–18821. doi: 10.1073/pnas.0604711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbowicz-Matuk A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Science. 2012;185–186:78–85. doi: 10.1016/j.plantsci.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Kiełbowicz-Matuk A, Rey P, Rorat T. The abundance of a single domain cyclophilin in Solanaceae is regulated as a function of organ type and high temperature and not by other environmental constraints. Physiologia Plantarum. 2007;131:387–398. doi: 10.1111/j.1399-3054.2007.00968.x. [DOI] [PubMed] [Google Scholar]
- Kiełbowicz-Matuk A, Rey P, Rorat T. The organ-dependent abundance of a Solanum lipid transfer protein is up-regulated upon osmotic constraints and associated with cold acclimation ability. Journal of Experimental Botany. 2008;59:2191–2203. doi: 10.1093/jxb/ern088. [DOI] [PubMed] [Google Scholar]
- Knight H. Calcium signaling during abiotic stress in plants. International Review of Cytology. 2000;195:269–324. doi: 10.1016/s0074-7696(08)62707-2. [DOI] [PubMed] [Google Scholar]
- Kumagai T, Ito S, Nakamichi N, et al. The common function of a novel subfamily of B-Box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Bioscience, Biotechnology, and Biochemistry. 2008;72:1539–1549. doi: 10.1271/bbb.80041. [DOI] [PubMed] [Google Scholar]
- Kurup S, Jones HD, Holdsworth MJ. Interactions of the developmental regulator ABI3 with proteins identified from developing arabidopsis seeds. Plant Journal. 2000;21:143–155. doi: 10.1046/j.1365-313x.2000.00663.x. [DOI] [PubMed] [Google Scholar]
- Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JH, Dijkwel PP. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proceedings of the National Academy of Sciences of the USA. 2012;109:17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V, Cyert MS, Gasser CS. Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. Journal of Biological Chemistry. 1996;271:12859–12866. doi: 10.1074/jbc.271.22.12859. [DOI] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes & Development. 2011;25:1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. EFL3 encodes a circadian clock-regulated nuclear protein that functions in an arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martz F, Wilczynska M, Kleczkowski LA. Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochemical Journal. 2002;367:295–300. doi: 10.1042/BJ20020772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S, Quinn JM. Copper-responsive gene expression in photosynthetic microorganisms. Methods in Enzymology. 1998;297:263–279. doi: 10.1016/s0076-6879(98)97020-3. [DOI] [PubMed] [Google Scholar]
- Montiel G, Gantet P, Jay-Allemand C, Breton C. Transcription factor networks, pathways to the knowledge of root development. Plant Physiology. 2004;136:3478–3485. doi: 10.1104/pp.104.051029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murelli C, Rizza F, Albini FM, Dulio A, Terzi V, Cattivelli L. Metabolic changes associated with cold-acclimation in contrasting cultivars of barley. Physiologia Plantarum. 1995;94:87–93. [Google Scholar]
- Nagaoka S, Takano T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in arabidopsis. Journal of Experimental Botany. 2003;54:2231–2237. doi: 10.1093/jxb/erg241. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, et al. Transcript profiling of an arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant and Cell Physiology. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT–PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- O'Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örvar BL, Sangwan V, Omann F, Dhindsa RS. Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant Journal. 2000;23:785–794. doi: 10.1046/j.1365-313x.2000.00845.x. [DOI] [PubMed] [Google Scholar]
- Pruvot G, Cuiné S, Peltier G, Rey P. Characterization of a novel drought-induced 34-kDa protein located in the thylakoids of Solanum tuberosum L. plants. Planta. 1996;198:471–479. doi: 10.1007/BF00620065. [DOI] [PubMed] [Google Scholar]
- Rajashekar CB, Lafta A. Cell-wall changes and cell tension in response to cold acclimation and exogenous abscisic acid in leaves and cell cultures. Plant Physiology. 1996;111:605–612. doi: 10.1104/pp.111.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P, Cuiné S, Eymery F, et al. Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant Journal. 2005;41:31–42. doi: 10.1111/j.1365-313X.2004.02271.x. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in arabidopsis. Journal of Biological Chemistry. 2004;279:11736–11743. doi: 10.1074/jbc.M313350200. [DOI] [PubMed] [Google Scholar]
- Robson F, Costa MM, Hepworth SR, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant Journal. 2001;28:619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- Rorat T, Irzykowski W. Changes in mRNA population during cold acclimation in two potato lines of Solanum sogarandinum differing by their cold hardiness. Acta Physiologiae Plantarum. 1996;18:25–32. [Google Scholar]
- Rorat T, Irzykowski W, Grygorowicz WJ. Identification and expression of novel cold induced genes in potato (Solanum sogarandinum) Plant Science. 1997;124:69–78. [Google Scholar]
- Rorat T, Grygorowicz WJ, Irzykowski W, Rey P. Expression of KS-type dehydrins is primarily regulated by factors related to organ type and leaf developmental stage during vegetative growth. Planta. 2004;218:878–885. doi: 10.1007/s00425-003-1171-8. [DOI] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Molecular Biology. 1999;9:1073–1078. doi: 10.1023/a:1006184519697. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of arabidopsis. Plant Journal. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM. Genotype and time of day shape the Populus drought response. Plant Journal. 2009;60:703–715. doi: 10.1111/j.1365-313X.2009.03993.x. [DOI] [PubMed] [Google Scholar]
- Wilkins O, Brautigam K, Campbell MM. Time of day shapes arabidopsis drought transcriptomes. Plant Journal. 2010;63:715–727. doi: 10.1111/j.1365-313X.2010.04274.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR. The C termini of arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR. The signalling mechanism of arabidopsis CRY1 involves direct interaction with COP1. Plant Cell. 2001;13:2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]