Abstract
Background and Aims
Cultivated flax (Linum usitatissimum) is known to have undergone a whole-genome duplication around 5–9 million years ago. The aim of this study was to investigate whether other whole-genome duplication events have occurred in the evolutionary history of cultivated flax. Knowledge of such whole-genome duplications will be important in understanding the biology and genomics of cultivated flax.
Methods
Transcriptomes of 11 Linum species were sequenced using the Illumina platform. The short reads were assembled de novo and the DupPipe pipeline was used to look for signatures of polyploidy events from the age distribution of paralogues. In addition, phylogenies of all paralogues were assembled within an estimated age window of interest. These phylogenies were assessed for evidence of a paleopolyploidy event within the genus Linum.
Key Results
A previously unknown paleopolyploidy event that occurred 20–40 million years ago was discovered and shown to be specific to a clade within Linum containing cultivated flax (L. usitatissimum) and other mainly blue-flowered species. The finding was supported by two lines of evidence. First, a significant change of slope (peak) was shown in the age distribution of paralogues that was phylogenetically restricted to, and ubiquitous in, this clade. Second, a large number of paralogue phylogenies were retrieved that are consistent with a polyploidy event occurring within that clade.
Conclusions
The results show the utility of multi-species transcriptomics for detecting whole-genome duplication events and demonstrate that that multiple rounds of polyploidy have been important in shaping the evolutionary history of flax. Understanding and characterizing these whole-genome duplication events will be important for future Linum research.
Keywords: Linum usitatissimum, flax, paleopolyploidy, whole-genome duplication, transcriptomics, genomic phylogenetics, species phylogeny, gene trees, paralogue age distribution
INTRODUCTION
Polyploidy, the duplication of whole genomes, is an important evolutionary force that is especially prevalent in plants (Otto and Whitton, 2000). Recent study has revealed that all angiosperms have undergone at least two rounds of ancient whole-genome duplication (Jiao et al., 2011) in addition to several younger, lineage-specific events (Jiao et al., 2012). These events are thought to have been very important in the evolutionary diversification of flowering plants (Adams and Wendel, 2005; Soltis et al., 2009; Jiao et al., 2012). In addition to these ancient polyploidy events, recent whole-genome duplications are very common in most extant plant lineages (Otto and Whitton, 2000; Adams and Wendel 2005; Wood et al., 2009). This is especially the case for the majority of the world's most important crop species, in which polyploidy seems to be particularly prevalent (Adams and Wendel, 2005). However, the genome complexity caused by genome duplications can be troublesome for crop genomics, for instance in genome-wide association studies of polyploid crops (Harper et al., 2012). It is therefore of considerable importance to characterize fully the genome history of crop species in order to take this into account in crop research.
The flax genus (Linum) contains about 180 species that are spread across six continents. It is thought to have originated about 46 million years ago (MYA), making it a relatively old genus (McDill et al., 2009; McDill and Simpson, 2011). The genus is divided into numerous sections. A large, predominately blue-flowered clade contains the sections Dasylinum and Linum, while the group of yellow-flowered flaxes contains the sections Cathartolinum, Linopsis and Syllinum (McDill et al., 2009; McDill and Simpson, 2011). Cultivated flax (Linum usitatissimum) is an important source of high-quality fibres (Mohanty et al., 2000) and seed oil (Green, 1986). The oil has industrial uses as well as considerable perceived health benefits (Singh et al., 2011). Its genome was recently sequenced (Wang et al., 2012), resulting in the discovery that it had undergone a whole-genome duplication around 5–9 MYA. As an extension to this finding we wished to examine the possibility that another, older, Linum-specific polyploidy event might have occurred some time earlier in the evolutionary history of cultivated flax. Until now, this hypothesis could not be tested due to insufficient genomic data from related species. In this study we use transcriptome sequences of 11 Linum species to identify and characterize whole-genome duplication events in the evolutionary history of cultivated flax.
MATERIALS AND METHODS
Illumina sequencing and de novo assembly
Total RNA from several tissue types was extracted and used to make Illumina sequencing libraries using methods described by Johnson et al. (2012). The libraries were multiplexed and sequenced on an Illumina HiSeq platform using paired-end chemistry. Quality trimming of the Illumina reads is described in the Methods section of Johnson et al. (2012). All species used in this study had a single library constructed from a pooled RNA sample from various tissue types, except for Linum usitatissimum and L. perenne, which had certain tissue types separated into individual libraries. However, they were later combined into single-species libraries, since analyses of individual tissue libraries did not change any findings of the paper. A total of 12 species were used in this study: 11 Linum species and Bischofia javanica (Phyllanthaceae), which was used as an outgroup in the phylogenetic analyses due to its relatively close relationship with the Linaceae (Xi et al., 2012).
The short Illumina reads were assembled in a de novo fashion using Trinity v.r2013-02-25 (Grabherr et al., 2011) with the program's default settings. In order to reduce the number of almost identical sequences in the assembly, contigs with a sequence similarity higher than 98 % were clustered together using the CD-HIT-EST program in the CD-HIT package v.4·6 (cd-hit-est flags: -c 0·99 -l 299 -d 0) (Li et al., 2001; Li and Godzik, 2006). This step also removed all contigs shorter than 300 bp from the assembly.
Identification of orthologues and phylogenetic analyses of Linum
Transcripts were then translated into their corresponding amino acid sequences using prot4EST (Wasmuth and Blaxter, 2004). Prot4EST uses BlastX (Altschul et al., 1997) for certain parts of its amino acid translation pipeline and therefore requires a BLAST database in amino acid format. For this purpose we used the annotated protein sequences from the published flax (L. usitatissimum) genome (Wang et al., 2012), which can be downloaded from Phytozome (Goodstein et al., 2012). The best amino acid transcript from each cluster was determined by its length and number of internal stop codons, using a Python script, and was chosen as the cluster's representative in the assembly. OrthoMCL v.2·0·3 (Li et al., 2003) was used to identify orthologous sequences in the assemblies. An e-value of 1E − 5 was used in the all versus all BlastP step of the OrthoMCL pipeline, and percentMatchCutoff was set to 50 and evalueExponentCutoff to −5 in the orthomclPairs step. Other parts of OrthoMCL were executed with the pipeline's default settings.
A phylogeny of the Linum species used in this study was constructed using a selected subset of the orthologue groups generated by OrthoMCL. A Python script was used to extract orthologue groups that matched the following criteria: (1) the orthologue group had to contain contigs from all species but (2) could not contain more than one contig from each species (i.e. putative single copy genes). These singleton orthologue groups were used in the subsequent phylogenetic analyses. The amino acid and nucleotide sequences of transcripts in these orthologue groups were extracted using a Python script. MAFFT v.705b (Katoh and Standley, 2013) was used to align the amino acid sequences using the -auto flag. The alignments were then converted to their corresponding nucleotide sequence using RevTrans v.1·4 (Wernersson and Pedersen, 2003). Leading and trailing gaps were removed from the alignments using a Python script and then trimmed further with trimAl v.1·2 (Capella-Gutiérrez et al., 2009) with the -automated1 flag. Alignments smaller than 300 bp were discarded from further analyses. The appropriate model of nucleotide substitution for the trimmed nucleotide alignments was determined with jModelTest v.2·1·1 (Guindon and Gascuel, 2003; Darriba et al., 2012) using the Akaike information criterion. These alignments were used to generate species phylogenies using two methods.
All nucleotide alignments were concatenated into a single matrix that was analysed with MrBayes v.3·2·1 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). Each alignment was defined as a separate partition in the matrix and a Python script was used to incorporate the jModelTest outputs into the MrBayes block of the matrix, ensuring that the appropriate base substitution model was used for every alignment. Model parameters were unlinked across all partitions. Bischofia javanica was used as the outgroup to confirm the rooting of the analyses. Two runs were started with 2 000 000 generations and burn-in set at 25 %. Convergence of the analyses was checked by running two independent chains and manually inspecting traces using Tracer (http://tree.bio.ed.ac.uk/software/tracer/).
A second species phylogeny was constructed using the STAR method (species tree estimation using average ranks of coalescences) (Liu et al., 2009), which is based on the multispecies coalescent model (Rannala and Yang, 2003). The STAR method uses the average ranks of coalescent events in a collection of gene trees to construct a single species topology using a distance method. First we generated individual gene trees for each of the previously mentioned trimmed nucleotide alignment, using RAxML v.7·2·6 (Stamatakis, 2006) with 10 search replicates. Bischofia javanica was set as the outgroup to confirm the rooting of the phylogenetic analysis. The appropriate model of nucleotide substitution for each alignment was parsed from jModelTest outputs using a Python script. The star.sptree function in the phyBASE v.1·3 package (Liu and Yu, 2009), under R v.2·15·3 (Ihaka and Gentleman, 1996), was used to generate a single species topology from all gene trees. Branch lengths were estimated and added to the STAR tree using GARLI v.2·0 (Zwickl, 2006), by optimizing the model parameters of the concatenated matrix, which were initially estimated by jModelTest v.2·1·1 (Guindon and Gascuel, 2003; Darriba et al., 2012). Node support of the STAR tree was acquired by generating 1000 multilocus bootstrap replicates (Seo, 2008) with the bootstrap.mulgene function in phyBASE, analysing each bootstrap replicate with PhyML v.3·0 (Guindon et al., 2010) and constructing 1000 STAR trees in phyBASE. A consensus of the 1000 STAR trees was generated using the consense program in the PHYLIP package v.3·69 (Felsenstein, 2005). The transcriptome to phylogeny methods described above are implemented in a pipeline (T2Phy) under development by one of us (S.S.).
Whole-genome duplication inference from age distributions of paralogues
The DupPipe pipeline (Barker et al., 2008) was used to look for evidence of whole-genome duplication events in the 11 Linum species. The pipeline inferred pairs of paralogues within the transcriptome assemblies and estimated their divergence based on synonymous substitution rates (Ks), which are used as a proxy for the age of the duplicated gene pair. DupPipe uses a discontinuous MegaBlast (Zhang et al., 2000; Ma et al., 2002) within each assembly to cluster contigs into gene families based on 40 % sequence similarity over at least 300 bp. The appropriate reading frame was estimated by comparing each pair of sequences with a large set of protein sequences (Wheeler et al., 2007) using BlastX (Altschul et al., 1997). Each nucleotide sequence was then paired with its best protein hit and only genes with a minimum of 30 % similarity over at least 150 sites were retained for further analyses. Genwise 2·2·2 (Birney et al., 2004) was used to align each gene to its best protein hit in order to determine the correct reading frame. The nucleotide contigs were then converted to their corresponding amino acid sequence using the highest-scoring DNA–protein alignment from GeneWise. Alignments of duplicated gene pairs were generated using MUSCLE 3·6 (Edgar, 2004) and the aligned amino acids were converted back to DNA sequences using RevTrans 1·4 (Wernersson and Pedersen, 2003). Ks values (synonymous substitution rates) for each duplicate gene pair were estimated using the codeml program in the PAML package (Yang, 1997) under the F3 × 4 model (Goldman and Yang, 1994). Duplicated gene pairs that could be identified as transposable elements were removed from the dataset. Due to concerns that extremely low Ks values were noise caused by alternative splicing or assembly artefacts, all Ks values lower than 0·001 were removed from the dataset. Furthermore, we excluded all Ks values larger than 2 in order to minimize saturation effects (Vanneste et al., 2013).
The number of significant features, i.e. peaks, in the age distributions of gene duplicates was estimated using SiZer (Chaudhuri and Marron, 1999). SiZer looks for significant changes in kernel density by performing a Gaussian smoothing with a wide range of bandwidths and has been extensively used in the investigation of paleopolyploidy (Barker et al., 2008, 2009; Shi et al., 2010). SiZer identified two significant peaks in some of the transcriptome assemblies, which were then analysed further using mixture models. EMMIX (McLachlan et al., 1999) was used to fit a model with two normal distributions using maximum likelihood in order to estimate the position of the two peaks in the dataset. Mixture models are useful in characterizing peaks generated by paleopolyploidy events, since their distribution is expected to be roughly Gaussian (Blanc and Wolfe, 2004; Schlueter et al., 2004). We used the EMMIX function in the EMMIX R package v.1·0·18 [downloaded from http://www.maths.uq.edu.au/~gjm/mix_soft/EMMIX_R/index.html (19 December 2013)] on log-transformed Ks values, with two normally distributed components.
Inference of a whole-genome duplication event from phylogenetic analysis of paralogues
To establish phylogenetic evidence for the putative paleopolyploidy event inferred by DupPipe, phylogenies of all orthologue groups containing a paralogue pair with a Ks value between 0·4 and 1·0 were constructed. This range was established based on two lines of evidence. First, results from the mixture models produced by EMMIX (McLachlan et al., 1999) demonstrated that the median of the paleopolyploidy event was around 0·68. We incorporated this estimate in the modelling of the effects a polyploidy event at that time would have on the Ks distribution, using the R scripts provided in Cui et al. (2006). The modelling showed that the effects of a polyploidy event around Ks 0·68 would be most significantly noticed around Ks 0·4–0·8. Second, we examined the results from Gaussian smoothing with SiZer, which were largely in agreement with the modelling results. However, in order to be inclusive, we increased the upper Ks limit by 0·2 in order to investigate more paralogues phylogenetically. Amino acid and nucleotide sequences of orthologue groups containing the paralogues were extracted from all species using a Python script and amino acid alignments were generated using MAFFT v.705b (Katoh and Standley, 2013) with the -auto flag. The alignments were converted to their corresponding DNA sequences using RevTrans v1·4 (Wernersson and Pedersen, 2003) and trimmed using trimAl v.1·2 (Capella-Gutiérrez et al., 2009) with the -automated1 flag. Phylogenies from the trimmed nucleotide alignments were inferred using RAxML v.7·2·6 (Stamatakis, 2006), with ten search replicates and the GTR + gamma model of nucleotide substitution. Node support of each tree was estimated using the 50 % majority rule consensus of 100 bootstrap replicates from RAxML. Phylogenies were converted to NEXUS format and combined into a singe file using a Python script, which could be analysed using Dendroscope V.3·2·8 (Huson and Scornavacca, 2012).
The paralogue gene trees were inspected manually and split into two major groups based on the phylogenetic pattern observed. We first separated phylogenies that were uninformative regarding a polyploidy event in the Linum genus. Those phylogenies included orthologue groups that contained multiple gene families and paralogues that were likely generated by multiple tandem duplications or over-assembly. These also included orthologous groups that had a large number of missing taxa and showed strong phylogenetic conflict within the yellow- or blue-flower clades. The remaining phylogenies showed strong phylogenetic patterning of paralogues and are therefore potentially informative regarding a polyploidy event in the evolutionary history of the Linum species. The only consistent pattern observed in these kinds of trees was the occurrence of a single clade of yellow-flowered species and two blue-flowered clades. We excluded any of these phylogenies that had excessive numbers of missing taxa, i.e. fewer than two yellow-flowered species, or if either of the blue-flowered clades contained fewer than three species.
RESULTS
Illumina sequencing and de novo assembly
The Illumina sequencing yielded between 19 and 83 million high-quality paired-end reads per species (Table 1). The de novo assembly of these reads resulted in 22 416–48 269 contigs larger than 300 bp per library.
Table 1.
Number of reads acquired per species and tissue type information
| Species | Clade (Family) | Tissue sequenced | Number of reads (read length) |
|---|---|---|---|
| Bischofia javanica | Phyllanthaceae | Young leaves | 2·33 × 107 (90 bp) |
| Linum flavum | Yellow (Linaceae) | Stem apex, shoot, leaves | 2·42 × 107 (90 bp) |
| Linum macraei | Yellow (Linaceae) | Stem apex, shoot, leaves | 2·65 × 107 (90 bp) |
| Linum strictum | Yellow (Linaceae) | Stem apex, shoot, leaves, flowers | 2·76 × 107 (90 bp) |
| Linum tenuifolium | Yellow (Linaceae) | Stem apex, shoot, leaves | 2·79 × 107 (90 bp) |
| Linum hirsutum | Blue (Linaceae) | Stem apex, shoot, leaves | 2·97 × 107 (90 bp) |
| Linum perenne1 | Blue (Linaceae) | Stem apex, shoot, leaves, flower | 5·62 × 107 (90 bp) |
| Linum lewisii | Blue (Linaceae) | Stem apex, shoot, leaves | 2·92 × 107 (90 bp) |
| Linum leoni | Blue (Linaceae) | Stem apex, shoot, leaves | 2·78 ×107 (90 bp) |
| Linum grandiflorum | Blue (Linaceae) | Stem, flower buds, leaves, flowers | 1·89 × 107 (90 bp) |
| Linum bienne | Blue (Linaceae) | Stem apex, shoot, leaves, flowers, | 2·28 × 107 (90 bp) |
| Linum usitatissimum1 | Blue (Linaceae) | Stem apex, stem | 8·31 × 107 (90 bp) |
1Species with more than one library sequenced (two L. perenne and three L. usitatissimum).
Phylogenetic analysis of Linum
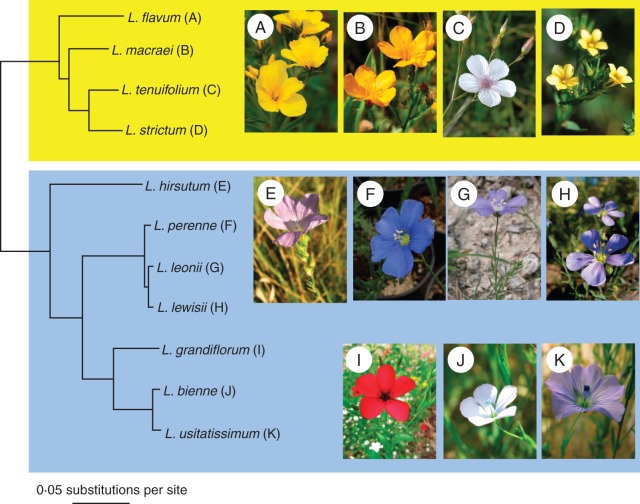
A total of 34 894 orthologous groups were identified by OrthoMCL, of which 413 were used to generate a phylogeny for the 11 Linum species. In these 413, all species had a single contig representing each group. The average alignment contained 546 characters (s.d. 212·97) and the combined length of all alignments was 225 891 characters. The two methods used (Bayesian analysis of a concatenated matrix and the coalescence-based STAR method) retrieved identical topologies, very similar branch lengths and a 100 % support value for every node in the phylogeny. As the two trees were very similar, only the STAR tree is presented here (Fig. 1). It is of interest to note that this phylogeny is almost identical to the most recently published Linum phylogeny (McDill et al., 2009).
Fig. 1.
STAR phylogeny of the 11 Linum species constructed from 413 gene trees. Branch lengths were estimated with GARLI and all nodes on the tree have 100 % bootstrap support. The tree shows the two major clades of Linum species studied here and their dominant flower colour. The tree was rooted with Bischofia javanica (Phyllanthaceae; taxon not shown here).
The 11 Linum species were split by a long branch into two major clades: (1) a mostly yellow-flowered clade containing L. flavum, L. macraei, L. strictum and L. tenuifolium; and (2) a predominantly blue-flowered clade that comprises all other species (Fig. 1). There were two exceptions to this flower colour rule, one in each clade, where L. tenuifolium flowers are white or pale pink and L. grandiflorum is red-flowered. However for simplicity's sake, all subsequent reference to these clades will be based on their dominant flower colour: yellow (y) and blue (b).
Paralogue age distributions
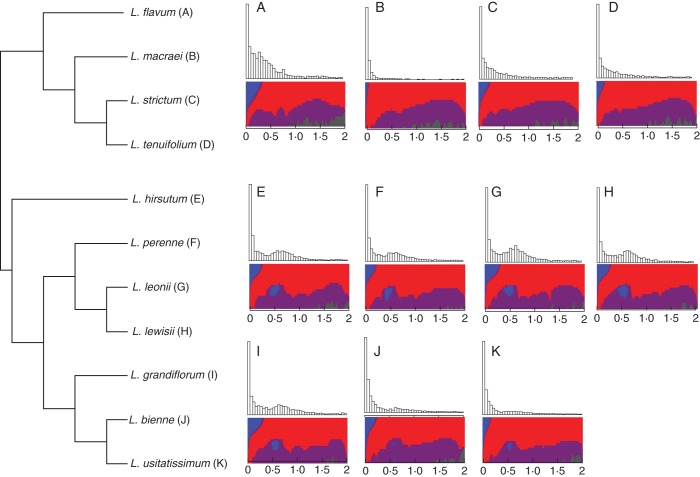
The possible presence of paleopolyploidy events in the evolutionary history of the 11 Linum species was first investigated by identifying paralogues within each transcriptome assembly and analysing their age distribution. An average of 3017 paralogue pairs (s.d. 1809) were identified in each of the transcriptome assemblies. Visualization of the duplicate age distribution revealed two types of patterns in genome evolution. Seven out of 11 Linum species showed a noticeable increase in Ks value frequency (i.e. a shallow peak) around 0·6 (Fig. 2E–K), compared with the L-shaped curve expected in species that have not undergone a whole-genome duplication recently in their evolutionary history (Blanc and Wolfe, 2004). However, four of our species showed a contrasting pattern, with no visible change of slope around Ks 0·6 (Fig. 2A–D). When this pattern is compared with the Linum phylogeny, it becomes clear that the peak at Ks 0·6 is restricted to species belonging to the blue clade (Fig. 2 and Supplementary Data Fig. S1), while being uniformly absent in the yellow clade. The presence of this peak in all blue clade species around the same Ks value strongly suggests a unique polyploidy event in an ancestor of this clade some time after the split from the yellow-flowered Linum species.
Fig. 2.
Cladogram, estimated with the STAR method, of the 11 Linum species (left), with their corresponding duplicate age distributions and SiZer plots (right). The SiZer plots are placed underneath each paralogue age distribution (the x-axis being Ks values). Different bandwidths used in the Gaussian smoothing of the Ks values are plotted on the y-axis of the SiZer plots and an optimal binning of the Ks values is plotted on the x-axes. These plots are composed of four colours: blue represents a significant increase in Ks value density, red represents a significant decrease in density, purple represents regions where there is no significant increase or decease in density, and grey areas represent insufficient data. Peaks in duplicate age distributions generated by paleopolyploidy events are characterized by the SiZer plots as blue areas flanked by red and purple areas, generally located in the middle of the y-axis. The position on the x-axis depends on the age of the duplication event. The blue areas around Ks 0·68 in all the SiZer plots of the blue-flowered Linum species represent statistical evidence supporting the occurrence of a polyploidy event.
The statistical significance of the changes in slopes in the duplicate age distributions was tested with SiZer (Chaudhuri and Marron, 1999). The results of these analyses are shown in the form of SiZer plots, positioned underneath their corresponding duplicate distribution in Fig. 2. A pattern consistent with a paleopolyploidy event was observed in all of the blue-flowered Linum species but in none of the species belonging to the yellow-flowered clade (Fig. 2) (see the legend of Fig. 2 for a detailed description of the SiZer plots). The evidence for a paleopolyploidy event is represented by blue areas in the SiZer plots in the bottom half of Fig. 2E–K. These blue areas correspond to a significant peak (P < 0·05) in the frequency of duplicate pairs in all species that is centred around Ks 0·6.
In order to determine the age of this putative paleopolyploidy event more precisely, mixture models with two normally distributed components were fitted to each of the paralogue age distributions of the blue-flowered Linum species. The mixture models fitted one component to the first peak in the age distribution, around Ks 0·01, which reflected the continuous birth and death model of gene evolution generated by frequently occurring single-gene duplications (Blanc and Wolfe, 2004). The second component was fitted to the older peak with an average median of Ks 0·68. Using this Ks value and two commonly used measures of the synonymous mutation rate (Koch et al., 2000; Lynch and Conery, 2000), the polyploidy event may be estimated to have occurred 23–42 MYA.
Phylogenetic analysis of paralogues
A total of 767 orthologous groups containing paralogues (henceforth referred to simply as paralogue groups) were extracted and their phylogenies manually inspected for evidence of whole-genome duplication. Most of the paralogue groups (587) were uninformative with regard to the presence or absence of a polyploidy event as the pattern of duplication showed no obvious phylogenetic pattern. About 260 paralogue groups clearly consisted of multiple gene families clustered together into a single group. The source of paralogy in the remaining 327 uninformative groups was likely to be from (1) multiple tandem duplications or assembly artefacts (239 groups), (2) incorrect orthology inference (62) or (3) numerous missing taxa (26) (see Table 2).
Table 2.
Summary of the patterns observed in paralogue phylogenies
| Group | Putative source of paralogy (comments) | n (frequency) |
|---|---|---|
| WGD uninformative | More than one gene family | 260 (33·9 %) |
| - | Small scale duplication | 239 (31·1 %) |
| - | Unknown (strong phylogenetic conflict, likely due to multiple tandem duplications or assembly artefact) | 62 (8·0 %) |
| - | Unknown (too many missing taxa to determine pattern of paralogy) | 26 (3·4 %) |
| WGD informative | WGD (strong phylogenetic pattern of paralogy consistent with a WGD event) | 180 (23·6 %) |
| Total | 767 |
WGD, whole-genome duplication.
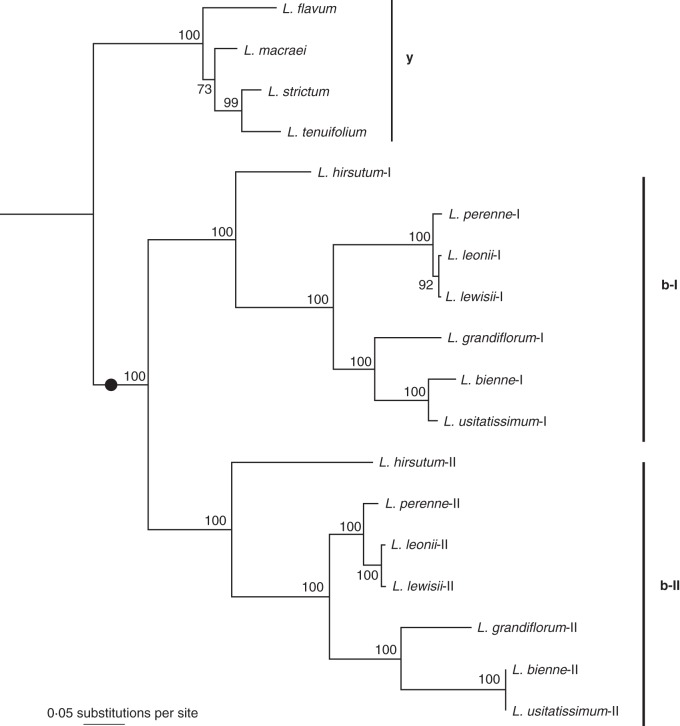
The remaining 180 (23·5 %) trees showed clear phylogenetic patterning of gene duplication and were therefore potentially informative of whole-genome duplication events. Remarkably, all 180 trees were consistent with the whole-genome duplication event proposed here in that they divided into three clades, the yellow-flowered species (clade y) and two duplicate clades of the blue-flowered species (clades b-I and b-II), each reflecting the organismal phylogeny of the component species and implying a gene duplication in the blue species but not in the yellow. Figure 3 shows an example of such a phylogeny (from a chaperone-like gene) and it shows three well-supported clades: one is composed of the yellow-flowered Linum species and two blue-flowered Linum clades are observed. Furthermore, the blue-flowered clades are sisters to each other, and when combined are sister to the yellow-flowered Linum species. Finally, individual species within each of the three clades followed the species phylogeny in Fig. 1. This phylogeny did not, however, show evidence for the later polyploidy event that is specific to L. bienne/L. usitatissimum (for expected reasons, see Discussion). It is also important to note that very few of the 180 paralogue phylogenies were as complete as shown in Fig. 3. Most of them had some missing genes or taxa, which is not surprising due to the nature of transcriptomic data. Nevertheless, all were consistent with a shared whole-genome duplication event in the evolutionary history of the blue-flowered Linum species, and this is the most likely cause for the large number of phylogenies supporting the pattern in Figure 3.
Fig. 3.
A phylogeny of orthologous groups that is consistent with a polyploidy event occurring on the branch leading to the blue-flowered Linum (black dot). The species relationship within each of the clades is consistent with the species phylogeny (Fig. 1). The tree was rooted on its midpoint for visualization purposes and bootstrap support is shown near the nodes of the phylogeny. Y indicates the yellow-flowered clade, b indicates the blue-flowered 1 clades. Based on a BlastX search on Phytozome (Goodstein et al., 2012) using the L. usitatissimum contigs, this gene appears to be a Co-chaperone-like protein in the GrpE family. L. usitatissimum-I corresponds to the gene Lus10029654 and L. usitatissimum-II matches Lus1002803 in the genome assembly of cultivated flax (L. usitatissimum). The best hit of both paralogues in the Arabidopsis thaliana genome assembly is AT5G17710.
DISCUSSION
Consistency of date estimation
Our results demonstrate that blue-flowered Linum species (sections Linum and Dasylinum) (McDill et al., 2009) underwent a whole-genome duplication after they split from the rest of Linum. This split occurred near the base of the genus and has previously been estimated to have occurred 41–46 MYA (McDill et al., 2009; McDill and Simpson, 2011). The former study also estimated the age of the most recent common ancestor of the blue-flowered clade to be 29–32 million years. We estimate the duplication event to have occurred 23–42 MYA, which is consistent with the independently derived phylogenetic estimates of divergence within the genus Linum. The difficulty of accurately inferring the age of ancient duplication events is well established (Doyle and Egan, 2010; Vanneste et al., 2013), so the close correspondence between our observations and the dates given by McDill et al. (2009) provides some confidence in the reliability of the dating. It also raises the possibility that species diversification in the blue-flowered clade may have been driven, at least in part, by the whole-genome duplication event, as has been suggested for other groups (Vamosi and Dickinson, 2006; Soltis et al., 2009).
Relationship between the two polyploidy events in the evolution of cultivated flax (Linum usitatissimum)
It is important to note that the paleopolyploidy event described here was not obvious in the single-species analysis of duplicated genes using the fully sequenced genome of cultivated flax (Wang et al., 2012). This highlights the importance of using a phylogenomic approach, as has been shown elsewhere (Jiao et al., 2011; Van de Peer, 2011). Cultivated flax belongs to the blue-flowered clade and therefore shares the whole-genome duplication event. The Ks and SiZer plots of L. usitatissimum and L. bienne, its sister species (Fig. 2J, K) do indeed show clear peaks around Ks 0·68, but they are not as large as in the other blue-flowered species (Fig. 2E–I). We do not know the reason for this, but it may be related to the fact that L. usitatissimum and L. bienne underwent an independent polyploidy event 5–9 MYA (Wang et al., 2012). It would be interesting to investigate whether this later ‘mesopolyploidy event’ (sensu Guerra, 2008; Schubert and Lysak, 2011) caused accelerated duplicate gene loss, thereby attenuating the signal from the palaeopolyploidy event. If this is true, it follows that it might be more difficult to pinpoint individual polyploidy events in lineages that have undergone multiple whole-genome duplication events.
There is little evidence in the present study for the later polyploidy event specific to Linum bienne/Linum usitatissimum. This is to be expected as the present study focused on the detection of ancient events in multiple species, and used entirely Illumina short-read data in contradistinction to the analyses included in Wang et al. (2012), which used the completely assembled flax genome and full-length cDNA. Focusing on ancient events allowed us to be conservative in excluding very closely related duplicates in case of assembly error or other artefacts (important because of our use of relatively low-depth short-read data), and this is likely to have had the effect of diminishing or eliminating the signal from the more recent event. This illustrates the importance of specifically targeted studies to discover genomic events at different periods of evolutionary history.
Polyploidy and chromosome number
This ancient polyploidy event described here is not evident from an examination of the published chromosome numbers of the species. However, as this is an ancient event (∼30 MYA) the absence of a chromosomal signature is not surprising. Whole-genome duplication events have been divided into three classes based on chromosomal repatterning (Guerra, 2008; Schubert and Lysak, 2011). As chromosome repatterning requires time, these classes are usually correlated with age. The classes are: (1) neopolyploidy, in which chromosomes are still in multiples of related diploids, with little if any, chromosome repatterning (usually very recent, Holocene, <11 000 years ago, to Pleistocene, <2·5 MYA); (2) mesopolyploidy, in which there may be some chromosome number reduction and chromosome repatterning, but polyploidy is still suggested by higher chromosome number (usually early Pleistocene to late Tertiary, <10 MYA); and (3) palaeopolyploidy, in which there is complete diploidization and considerable chromosome number reduction (usually Tertiary or older, >10 MYA). The whole-genome duplication event described here obviously falls into the last category and chromosome number reduction is to be expected as a normal pattern of palaeopolyploids.
Use of multiple species and multiple sources of inference in pinpointing polyploidy events
These findings demonstrate the limitations of relying solely on the results of age distribution plots from a single species in the inference of whole-genome duplication events in its evolutionary history. We furthermore argue that an analysis of Ks distributions from multiple species, when combined with phylogenetic reconstruction of paralogues and good taxon sampling, is a very powerful approach for discovering and characterizing paleopolyploidy events.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We would like to thank Charles Hefer (University of British Columbia) for his extensive assistance in getting several of the computer programs used in this study working. We also thank Armando Geraldes (University of British Columbia) for critically reading the manuscript and providing very useful comments. We also thank the following for use of flower photographs as part of the illustration in Fig. 1: Franck Le Driant (L. strictum and L. tenuifolium), Oliver Pichard (L. leonii) and Stefan Lefaner (L. flavum). We are grateful to D. E. Soltis (University of Florida) for kindly making available the transcriptome of Bischofia. We would like to thank Western Canada Research Grid (Westgrid) for access to their high-performance computing resources, which were very useful in parts of the data analysis of this paper. We gratefully acknowledge funding from The Natural Sciences and Engineering Research Council of Canada (NSERC) to Q.C. (Discovery Grant Program) and S.S., as well as the University of British Columbia for partial funding to S.S. We also thank Genome Alberta One Thousand Plants Project for funding.
LITERATURE CITED
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8:135–41. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Kane NC, Matvienko M, et al. Multiple paleopolyploidizations during the evolution of the Compositae reveal parallel patterns of duplicate gene retention after millions of years. Molecular Biology and Evolution. 2008;25:2445–2455. doi: 10.1093/molbev/msn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME. Paleopolyploidy in the Brassicales: analyses of the cleome transcriptome elucidate the history of genome duplications in arabidopsis and other Brassicales. Genome Biology and Evolution. 2009;1:391–399. doi: 10.1093/gbe/evp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Research. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri P, Marron JS. SiZer for exploration of structures in curves. Journal of the American Statistical Association. 1999;94:807–823. [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, et al. Widespread genome duplications throughout the history of flowering plants. Genome Research. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9 doi: 10.1038/nmeth.2109. 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Egan AN. Dating the origins of polyploidy events. New Phytologist. 2010;186:73–85. doi: 10.1111/j.1469-8137.2009.03118.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5 doi: 10.1186/1471-2105-5-113. 113. doi:10·1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Seattle: Department of Genome Sciences, University of Washington; 2005. PHYLIP (Phylogeny Inference Package) version 3·6. Distributed by the author. [Google Scholar]
- Goldman N, Yang ZH. Codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research. 2012;40 doi: 10.1093/nar/gkr944. D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AG. Genetic control of polyunsaturated fatty acid biosynthesis in flax (Linum usitatissimum) seed oil. Theoretical and Applied Genetics. 1986;72:654–661. doi: 10.1007/BF00289004. [DOI] [PubMed] [Google Scholar]
- Guerra M. Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenetic and Genome Research. 2008;120:339–350. doi: 10.1159/000121083. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3·0. Systematic Biology. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Harper AL, Trick M, Higgins J, et al. Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nature Biotechnology. 2012;30:798–802. doi: 10.1038/nbt.2302. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Systematic Biology. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Leebens-Mack J, Ayyampalayam S, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biology. 2012;13 doi: 10.1186/gb-2012-13-1-r3. R3. doi:10·1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MTJ, Carpenter EJ, Tian Z, et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PloS One. 2012;7 doi: 10.1371/journal.pone.0050226. e50226. doi:10·1371/journal.pone.0050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Molecular Biology and Evolution. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L. Phybase: an R package for species tree analysis. Bioinformatics. 2010;26:962–963. doi: 10.1093/bioinformatics/btq062. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Pearl DK, Edwards SV. Estimating species phylogenies using coalescence times among sequences. Systematic Biology. 2009;58:468–477. doi: 10.1093/sysbio/syp031. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Ma B, Tromp J, Li M. PatternHunter: faster and more sensitive homology search. Bioinformatics. 2002;18:440–445. doi: 10.1093/bioinformatics/18.3.440. [DOI] [PubMed] [Google Scholar]
- McDill J, Simpson BB. Molecular phylogenetics of Linaceae with complete generic sampling and data from two plastid genes. Botanical Journal of the Linnean Society. 2011;165:64–83. [Google Scholar]
- McDill J, Repplinger M, Simpson BB, Kadereit JW. The phylogeny of Linum and Linaceae subfamily Linoideae, with implications for their systematics, biogeography, and evolution of heterostyly. Systematic Botany. 2009;34:386–405. [Google Scholar]
- McLachlan GJ, Peel D, Basford KE, Adams P. The EMMIX software for the fitting of mixtures of normal and t-components. Journal of Statistical Software. 1999;4:1–14. [Google Scholar]
- Mohanty AK, Misra M, Hinrichsen G. Biofibres, biodegradable polymers and biocomposites: an overview. Macromolecular Materials and Engineering. 2000;276:1–24. [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics. 2003;164:1645–1656. doi: 10.1093/genetics/164.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schlueter JA, Dixon P, Granger C, et al. Mining EST databases to resolve evolutionary events in major crop species. Genome. 2004;47:868–876. doi: 10.1139/g04-047. [DOI] [PubMed] [Google Scholar]
- Schubert I, Lysak M. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends in Genetics. 2011;27:207–216. doi: 10.1016/j.tig.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Seo T-K. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Molecular Biology and Evolution. 2008;25:960–971. doi: 10.1093/molbev/msn043. [DOI] [PubMed] [Google Scholar]
- Shi T, Huang H, Barker MS. Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Annals of Botany. 2010;106:497–504. doi: 10.1093/aob/mcq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Mridula D, Rehal J, Barnwal P. Flaxseed: a potential source of food, feed and fiber. Critical Reviews in Food Science and Nutrition. 2011;51:210–222. doi: 10.1080/10408390903537241. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Vamosi JC, Dickinson TA. Polyploidy and diversification: a phylogenetic investigation in Rosaceae. International Journal of Plant Sciences. 2006;167:349–358. [Google Scholar]
- Van de Peer Y. A mystery unveiled. Genome Biology. 2011;12 doi: 10.1186/gb-2011-12-5-113. 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Van de Peer Y, Maere S. Inference of genome duplications from age distributions revisited. Molecular Biology and Evolution. 2013;30:177–190. doi: 10.1093/molbev/mss214. [DOI] [PubMed] [Google Scholar]
- Wang Z, Hobson N, Galindo L, et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant Journal. 2012;72:461–473. doi: 10.1111/j.1365-313X.2012.05093.x. [DOI] [PubMed] [Google Scholar]
- Wasmuth JD, Blaxter ML. prot4EST: translating expressed sequence tags from neglected genomes. BMC Bioinformatics. 2004;5:187. doi: 10.1186/1471-2105-5-187. doi:10·1186/1471-2105-5-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG. RevTrans-Constructing alignments of coding DNA from aligned amino acid sequences. Nucleic Acids Research. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2007;35 doi: 10.1093/nar/gkl1031. D5–D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences of the USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Ruhfel BR, Schaefer H, et al. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proceedings of the National Academy of Sciences of the USA. 2012;109:17519–17524. doi: 10.1073/pnas.1205818109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in the Biosciences. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. USA: University of Texas at Austin; 2006. PhD Thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.