Abstract
Iddm14 (formerly Iddm4) is a non-MHC-linked genetic locus associated with autoimmune diabetes. Its effects have been well-documented in BB-derived rats in which diabetes is either induced by immunologic perturbation or occurs spontaneously. The role of Iddm14 in non-BB rat strains is unknown. Our goal was to extend the analysis of Iddm14 in new diabetes-susceptible strains and to identify candidate genes in the rat Iddm14 diabetes susceptibility locus that are common to these multiple diabetic strains. To determine if Iddm14 is important in strains other than BB, we first genotyped a (LEW.1WR1 × WF)F2 cohort in which diabetes was induced by perturbation with polyinosinic:polycytidylic acid. We found that Iddm14 is a major determinant of diabetes susceptibility in LEW.1WR1 rats. We then used nucleotide sequencing to establish a strain distribution pattern of polymorphisms (insertions, deletions, and single nucleotide polymorphisms [SNPs]) that predicts susceptibility to diabetes in a panel of inbred and congenic rats. Using the positional information from the congenic strains and the new linkage data, we identified a susceptibility haplotype in the T-cell receptor Vβ chain (Tcrb-V) locus. This haplotype includes Tcrb-V13, which is identical in five susceptible strains but different in resistant WF and F344 rats. We conclude that Iddm14 is a powerful determinant of both spontaneous and induced autoimmune diabetes in multiple rat strains, and that Tcrb-V13 SNPs constitute a haplotype of gene elements that may be critical for autoimmune diabetes in rats.
Introduction
Type 1-like autoimmune diabetes (T1D) is relatively common in different strains of rats (Ellerman and Like 2000; Mordes et al. 2007). As with mice and humans, the strongest T1D susceptibility locus in rats maps to the major histocompatibility complex (MHC). With only one exception (Penhale et al. 1990), T1D-susceptible rats express the RT1B/Du MHC class II haplotype. Viral antibody-free (VAF) BBDP rats develop spontaneous T1D with a penetrance greater than 90%. They are also congenitally lymphopenic due to a mutation in the Gimap5 gene on chromosome 4 (RNO4) (Hornum et al. 2002; MacMurray et al. 2002). Genetic crosses have shown that deficiency in peripheral T cells is necessary, but not sufficient, for the expression of spontaneous T1D in BB rats (Mordes et al. 2007). Coisogenic nonlymphopenic BBDR rats develop autoimmune diabetes only after immunologic perturbation (Mordes et al. 2007).
Both BBDP and BBDR rats have been used to identify a susceptibility locus, Iddm14 (previously designated Iddm4), that is highly significantly associated with the development of T1D in backcross animals treated with polyinosinic:polycytidylic acid (poly I:C) and an antibody that depletes ART2+ regulatory T cells (Tregs) (Martin et al. 1999a, b). Poly I:C is a ligand of toll-like receptor 3 and MDA5 (melanoma-differentiation-associated gene 5) (Kato et al. 2006). Iddm14, on RNO4, is a dominant, but incompletely penetrant, non-MHC determinant of T1D in BB rats. Introgression of F344 rat genomic DNA into the Iddm14 region in congenic BB rats generates diabetes resistance (Fuller et al. 2006), as does the substitution of DNA from WF rats in WF.Iddm14 congenic rats (Mordes et al. 2002). Iddm14 is also important for virus-induced T1D in BBDR rats (Blankenhorn et al. 2005). Using subinterval congenic WF.Iddm14 strains that narrowed the Iddm14 interval, we identified the T-cell receptor β-chain variable (Tcrb-V) genes as attractive candidates for Iddm14 (Blankenhorn et al. 2007).
Spontaneous diabetes also occurs in certain MHC congenic LEW rats that express RT1 class II u alleles. Both the LEW.1AR1-iddm rat (RT1.AaB/DuCu) (Lenzen et al. 2001) and the LEW.1WR1 rat (RT1.AuB/DuCa) (Mordes et al. 2005) develop spontaneous diabetes. Their peripheral lymphoid phenotypes are normal, including the fraction of ART2+ Tregs. Treatment with poly I:C alone at doses comparable to those used in BBDR rat studies induces diabetes in 80–100% of rats from the LEW.1WR1 strain (Mordes et al. 2005; Wedekind et al. 2005).
The goals of the present study were to determine the relationship of Iddm14 to diabetes susceptibility and resistance in rats other than the BB, specifically LEW.1WR1 rats (Mordes et al. 2005), and to identify candidate genes or haplotype blocks (Cuppen 2005) that could account for the role of Iddm14 in determining diabetes susceptibility. We show formally by gene mapping that the Iddm14 locus is a determinant of T1D susceptibility in LEW.1WR1 rats, making the LEW.1WR1 haplotype informative for Iddm14 candidate gene identification. Using both the LEW.1WR1 and subinterval congenic WF.Iddm14 strains, we have identified a susceptibility haplotype in the Tcrb-V locus. The susceptibility haplotype includes Tcrb-V13, which is identical in five susceptible strains but different in resistant WF and F344 rats. We propose that Iddm14-mediated diabetes resistance could be due to recessive protective mutations in Tcrb-V genes.
Materials and methods
Animals and DNA
F344 rats were obtained from Charles River, Inc. (Wilmington, MA). BBDP/Wor, LEW.1WR1, and PVG.R8 rats were obtained from BRM, Inc. (Worcester, MA). DR.F344/lyp rats were bred at the University of Washington, Seattle (Fuller et al. 2006). BBDR/Wor, WF.ART2, and WF.Idd-m14.ART2a rats (all RT1u/u, ART2a) were obtained from colonies maintained at the University of Massachusetts. WF.ART2 congenic rats were developed by us and differ from ordinary WF animals in that they express at least one copy of the “a” rather than the “b” allotype of the ART2 T-cell alloantigen on chromosome 1 (Blankenhorn et al. 2005; Mordes et al. 2002), making it possible to deplete them of ART2+ regulatory cells with available reagents. Both WF.ART2a/a homozygotes and WF.ART2a/b heterozygotes were bred, and their use in individual experiments is specified where necessary. The original WF breeding stock (ART2b/b) was obtained from Charles River Laboratories. WF.Iddm14.ART2a/b double congenic rats bear a small interval of BBDR chromosome 4 on the WF background and were developed as described (Blankenhorn et al. 2005, 2007; Martin et al. 1999a, b). Cohorts of (WF.ART2 × WF.Iddm14) congenic rats with progressively smaller Iddm14 intervals were bred by repetitive rounds of mating using a marker-assisted selection protocol as described (Blankenhorn et al. 2007; Mordes et al. 2002). (LEW.1WR1 × WF.ART2a/a)F2 rats were bred in our colony. Animals were housed in viral antibody-free conditions, confirmed monthly to be serologically free of rat pathogens (Mordes et al. 2002), and maintained in accordance with institutional guidelines and recommendations in the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, Commission on Life Sciences and National Research Council 1996).
Most DNA samples were prepared from tail snips and screened using microsatellite markers that define the Iddm14 and ART2 intervals plus a selected panel of additional microsatellite loci as described (Blankenhorn et al. 2005, 2007; Mordes et al. 2002). DNA from spontaneously diabetic KDP rats (Yokoi et al. 2002) was the gift of Dr. Fraser Scott (University of Ottawa, Ottawa, Ontario, Canada).
Induction of diabetes
Diabetes in WF.Iddm14.ART2a rats was induced using the protocol employed in previous mapping studies of this locus (Mordes et al. 2002). Briefly, 28–32-day-old rats were treated with cytotoxic DS4.23 anti-ART2.1 mAb (25 μg of purified mAb 5 times per week) and poly I:C (2.5 μg/g 3 times per week). Treatment was continued for 40 days or until diagnosis of diabetes. Animals were screened three times weekly for glycosuria, and diabetes was diagnosed on the basis of a plasma glucose concentration greater than 250 mg/dl (One-Touch Handheld Glucose Analyzer, Johnson & Johnson, Milpitas, CA). Diabetes in (LEW.1WR1 × WF)F2 rats was induced with poly I:C alone at a dose of 1 μg/g 3 times per week.
Sequencing
Primers were designed to flank-selected members of the Tcrb-V gene family from the leader sequences to the heptamer-nonamer joining signals (Williams and Gutman 1989). Primers were added to genomic DNA templates and then subjected to PCR using Platinum Taq DNA Polymerase High Fidelity Supermix (Invitrogen, Carlsbad, CA) for 40 cycles at 0.2 μM final primer concentration. PCR samples were purified and sequenced by GeneWiz (www.genewiz.com) from a dye-terminator reaction using an internal variable region primer. Sequences were aligned and compared with the Multiple Sequence Alignment program by CLUSTAL (http://align.genome.jp).
Results
Diabetes in the LEW.1WR1 rat segregates with Iddm14
To determine if Iddm14 is a determinant of diabetes susceptibility in LEW.1WR1 rats, (LEW.1WR1 × WF.ART2a)F2 rats were treated with poly I:C alone as described in Methods. This treatment protocol induces autoimmune diabetes in 100% of LEW.1WR1 rats (Mordes et al. 2005) but few WF rats (Ellerman and Like 2000). Of the 75 rats in the F2 cohort, 29 (39%) became diabetic within 40 days of induction. All rats were typed for Iddm14 using the D4Got43 microsatellite marker that is located near the peak of the Iddm14 QTL interval (Blankenhorn et al. 2005). As shown in Table 1, the distribution of Iddm14 and diabetes was absolute: all 29 diabetic rats bore at least one allele of the LEW-derived Iddm14 marker (p <0.01).
Table 1.
Association of Iddm14 genotype and diabetes susceptibility in (LEW.1WR1 × WF.ART2a/a)F2 rats
| Iddm14 genotype | Number diabetic (%) | Number nondiabetic (%) | Totals |
|---|---|---|---|
| L/L | 10 (50) | 10 (50) | 20 |
| L/W | 19 (43) | 25 (57) | 44 |
| W/W | 0 (0) | 11 (100) | 11 |
| Totals | 29 (39) | 46 (61) | 75 |
L = LEW.1WR1-origin allele of Iddm14; W = WF.ART2a/a-origin allele
(LEW.1WR1 × WF)F2 rats were bred, treated with poly I:C to induce diabetes, and genotyped to determine the origin of Iddm14 alleles as described in Methods. All diabetic animals were found to have inherited at least one LEW.1WR1-origin allele of Iddm14 (p <0.01)
Analysis of Tcrb-V genes and haplotypes in diabetes-susceptible and -resistant rat strains
Our previous linkage analyses have demonstrated that the supported interval for Iddm14 is between 67,962,305 and 69,392,848 bases on rat chromosome 4 (Blankenhorn et al. 2007). The T-cell receptor β-chain variable region gene family is of particular interest as a potential source of candidate genes. It is located in the middle of the supported interval for Iddm14. In preliminary studies we observed a difference in the ability of four T1D-susceptible rats (BBDR, KDP, PVG.R8, and LEW.1WR1) and one resistant rat (WF) to express Tcrb-V4 (Blankenhorn et al. 2007 and data not shown). Sequencing of Tcrb-V4 revealed an amber stop codon at amino acid position 107 in the 3′ end of the WF allele. This stop codon is also present in the rat genome project-type strain BN, which is known to be a diabetes-resistant animal (Klöting et al. 2003). We showed by quantitative PCR that Tcrb-V4 is, in fact, not expressed in T cells from WF rats (data not shown). As Iddm14 encompasses the entire TCRBV region, however, we elected to study the remaining TCR variable genes for nucleotide polymorphisms.
As shown in Table 2, we constructed a strain distribution pattern (SDP) for diabetes itself and for the multiple single nucleotide polymorphisms (SNPs) in the Tcrb-V region of the Iddm14 interval for a panel of rat strains. These now included three rat strains associated with resistance to T1D and five with documented susceptibility. The expanded panel of resistant strains included not only WF (Martin et al. 1999a), but also BN (Klöting et al. 2003) and F344 (Fuller et al. 2006; Klaff et al. 1999). The susceptible strains included BBDR (Mordes et al. 2007), BBDP (Blankenhorn et al. 2005), LEW.1WR1 (Mordes et al. 2005), PVG.R8 (Ellerman and Like 2000), and KDP (Yokoi et al. 2002). KDP and PVG.R8 rats have not been subjected to linkage analyses that would have the ability to identify their requirement for Iddm14.
Table 2.
Single nucleotide polymorphism strain distribution pattern in the Tcrb-V gene family
| Nucleotide | LEW | DR | DP | PVG | KDP | WF | F344 | BN | Vβ | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| Diabetes | S* | S* | S* | S | S | R* | R* | R | ||
| 67,744,276 | C | C | T | C | T | gnl|ti|896738376 | ||||
| 67,776,972 | C | C | T | C | T | J492097 | ||||
| 67,958,211 | T | T | C | T | C | rat101_007_p22.p1cb_228 | ||||
| 67,971,934 | G | G | A | G | A | rat104_023_e09.q1ca_288 | ||||
| 68,242,582 | T | C | T | C | WKYc89h05_s1_460 | |||||
| 68,306,418 | C | C | C | T | C | SHRSPa62d12_r1_653 | ||||
| 68,607,310 | C | C | A | C | A | gko-72l19_fp2_b1_249 | ||||
| 68,658,071 | T | T | C | C | C | J663774 | ||||
| 68,771,571 | T | T | C | C | C | WKYc99a04_s1_542 | ||||
| 68,693,725 | C | T | N | N | N | T | T | T | 4 | |
| 68,694,230 | C | C | C | C | C | G | C | G | 4 | SNP creates stop codon |
| 68,695,608 | N | G | G | G | G | A | G | A | 16 | |
| 68,695,656 | N | G | C | C | C | G | C | G | 16 | |
| 68,703,875 | T | T | T | T | T | T | C | T | 10 | |
| 68,704,035 | T | T | T | T | T | C | T | C | 10 | |
| 68,704,044 | A | A | A | A | A | G | A | G | 10 | |
| 68,704,057 | A | A | A | A | A | G | A | G | 10 | |
| 68,704,095 | A | A | A | A | A | G | A | G | 10 | |
| 68,707,986 | C | G | C | C | C | G | C | G | 1 | |
| 68,708,164 | AAG | AAG | AAG | 1 | In-frame insertion | |||||
| 68,708,234 | T | C | T | T | T | C | T | C | 1 | |
| 68,708,242 | 35** | 35** | 35** | 35** | 35** | del | 35** | del | 1 | Deletion → frameshift |
| 68,708,245 | TGA | TGA | 1 | Frameshift → stop codon | ||||||
| 68,773,516 | A | G | G | J875134 | ||||||
| 68,799,483 | G | T | G | G | G | T | G | T | 8.4 | |
| C | T | C | C | C | T | T | T | 8.4 | Indel | |
| 68,805,643 | A | A | A | A | A | A | C | A | 8.2 | |
| 68,805,656 | T | C | T | T | T | C | G | C | 8.2 | |
| 68,805,677 | T | T | T | T | T | T | C | T | 8.2 | |
| 68,805,713 | A | T | A | A | A | T | A | T | 8.2 | |
| 68,805,751 | A | G | A | A | A | G | G | G | 8.2 | |
| 68,805,800 | A | A | A | A | A | A | G | A | 8.2 | |
| 68,805,850 | A | A | A | A | A | A | G | A | 8.2 | |
| 68,805,856 | G | G | G | G | G | G | A | G | 8.2 | |
| 68,819,273 | G | A | G | G | G | A | A | A | 8.1 | |
| 68,819,329 | G | A | G | G | G | G | G | G | 8.1 | |
| 68,819,360 | C | T | C | C | C | T | C | T | 8.1 | |
| 68,819,456 | A | A | A | T | A | A | A | A | 8.1 | |
| 68,819,486 | A | A | A | G | A | A | A | A | 8.1 | |
| 68,819,531 | A | G | A | G | A | G | G | G | 8.1 | |
| 68,825,401 | T | T | T | T | T | T | C | T | 13 | |
| 68,825,406 | C | C | C | C | C | T | T | T | 13 | |
| 68,825,428 | C | C | C | C | C | T | T | T | 13 | |
| 68,825,594 | T | T | T | T | T | C | C | C | 13 | |
| 68,825,600 | C | C | C | C | C | G | G | G | 13 | |
| 68,825,621 | C | C | C | C | C | G | G | G | 13 | |
| 68,825,628 | C | C | C | C | C | A | A | A | 13 | |
| 68,825,654 | G | G | G | G | G | C | C | C | 13 | |
| 68,825,687 | A | A | A | A | A | G | G | G | 13 | |
| 68,825,731 | T | 13 | Insertion → stop at 68,825,743 | |||||||
| 68,830,734 | G | T | G | G | G | T | G | T | 12 | |
| 68,850,939 | G | A | N | G | G | G | G | G | 9 | |
| T | T | N | T | T | C | T | C | 9 | ||
| 68,864,236 | G | G | G | G | G | G | A | G | 6 | |
| 68,884,395 | G | G | G | G | G | G | T | G | 19 | |
| 68,884,422 | A | A | A | A | A | A | C | A | 19 | |
| 68,903,863 | T | T | T | T | T | T | G | T | 20 | |
| 68,906,016 | T | T | T | A | T | T | T | T | 17 | |
| 68,925,654 | G | G | G | T | G | T | gnl|ti|896772869 | |||
| 68,970,132 | A | G | N | A | A | A | A | A | 7 | |
| 68,970,173 | T | C | N | T | T | T | T | T | 7 | |
| 69,099,999 | G | T | J596338 | |||||||
| 69,128,798 | C | C | C | T | T | T | rdahl-76h11_rp2_b1_35 | |||
| 69,183,306 | D4Rat26 | |||||||||
| 69,316,394 | G | G | G | A | G | A | J1293540 | |||
| 69,316,636 | T | T | T | C | T | C | WKY-G-j-54f11_r1_299 | |||
| 69,571,744 | T | G | G | G | T | G | J1268582 | |||
| 69,626,785 | T | A | A | A | T | A | DS-g-c-09h07_r1_143 | |||
| 70,127,658 | C | T | T | C | C | T | rat103_031_m09.p1ca_683 |
DNA from five rat strains with known susceptibility to autoimmune diabetes (T1DM-S) and three strains with documented resistance (T1DM-R) was prepared and genes in the Tcrb-V region were sequenced as described in Methods. Shown are informative single nucleotide polymorphisms (SNPs) that distinguish susceptible and resistant strains. In the case of genes encoding Tcrb-V1, Tcrb-V4, and Tcrb-V13, polymorphisms leading to the generation of premature stop codons are indicated. The Tcrb-V13 haplotype block that uniformly distinguishes susceptible and resistant strains is indicated by shading. Nucleotide locations in the rat genome are indicated in the left column.
Denotes strains with documented Iddm14-dependent susceptibility or resistance to diabetes (see text); other alleles have not been tested for Iddm14 status.
Denotes normal sequence here is CTCATGTTTCTCTACAATTTTCAAAAACTGGCTAG
Our SNP analysis encompassed Tcrb-V1, 2, 3, 4, 5.1 and 5.2, 6, 7, 8.1, 8.2, 8.3, and 8.4, 9, 10, 11, 12, 13, 15, 16, 17, 18, 19, and 20. These comprise the majority (22) of the 24 Tcrb-V family members identified either by previous studies (Smith et al. 1991) or by our own manual bioinformatics analysis using a sequencing contig from the BN genome strain known to contain all the TCRB elements (NW_047690). There were multiple SNP and insertion/deletions (“indels”) among these sequences (Table 2).
This SDP analysis not only confirmed our previous observations concerning the allelic variance of Tcrb-V4 but it also revealed numerous additional SNP alleles, among them a 35-bp deletion that disrupts the Tcrb-V1 reading frame in both WF and BN rats. Surprisingly, however, the resistant F344 rat was found to share the same Tcrb-V1 and
Tcrb-V4 alleles that are present in our panel of five T1D susceptible strains
Additional analysis revealed that among all the changes, the Tcrb-V element with a SDP that most closely resembles the diabetes SDP is the complex set of polymorphisms in Tcrb-V13. Eight of the SNPs form an allele that is dichotomous and distinguishes those rats susceptible to T1D (including all the strains for which Iddm14 is critical) from those that are resistant to T1D. Furthermore, the T1D-resistant F344 rat has a resistant WF-like allele that contains an additional SNP (indel). This SNP creates a frameshift that leads to a stop codon, making Tcrb-V13 a pseudogene in this strain. The lack of expression of Tcrb-V13 was noted previously (Stienekemeier et al. 2000), and our sequence confirms the basis for its absence. The Tcrb-V13 alleles in BN and WF rats have been designated BV13S1A2, and LEW rats carry the BV13S1A1 allele; the allele in F344 rats has been designated BV13S1A3P (Stienekemeier et al. 2000). In addition, DNA from “DR.F344/lyp” diabetes-resistant congenic rats (Fuller et al. 2006) showed Tcrb-V1, Tcrb-V4, and Tcrb-V13 SNPs identical to those in commercial F344 rats (data not shown).
Finally, a phylogenetic analysis of the BV13S1 genes from the eight rat strains considered here associates the Iddm14 resistance haplotype withT1D resistance. SNPs in the three resistant strains are clustered in an evolutionary group that is clearly distinguished from the group to which the five susceptible strains belong (Fig. 1).
Fig. 1.
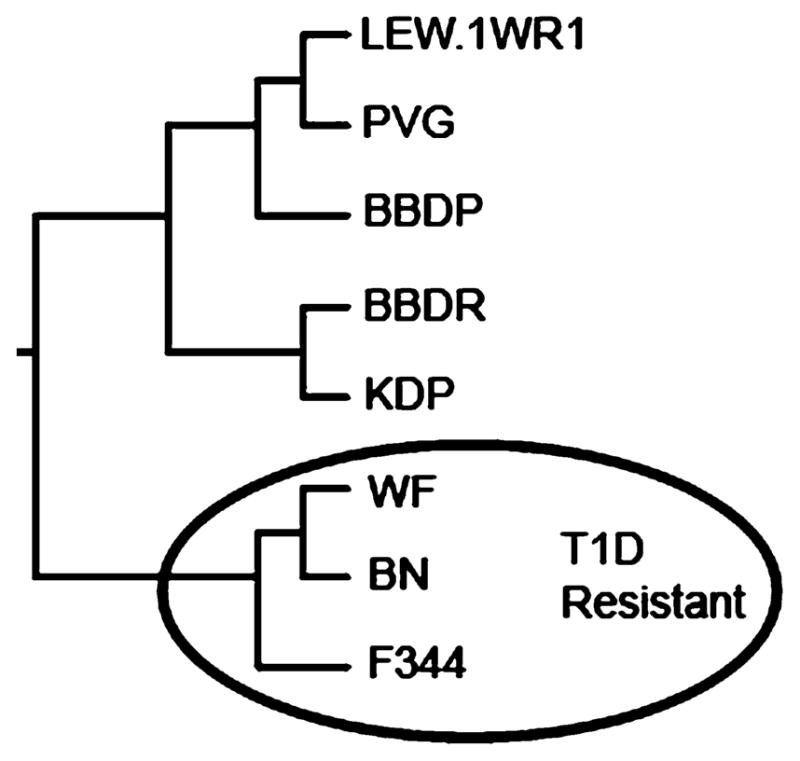
Neighbor-joining tree showing phylogenetic relationship of the TCR Vβ13 sequences for the eight rat strains we studied; the circled strains are diabetes resistant
Discussion
The data presented here confirm and extend our hypothesis that the Iddm14 autoimmune diabetes locus in the rat is a powerful determinant of disease penetrance and that Iddm14-mediated diabetes resistance may be due to recessive protective mutations in Tcrb-V genes. Previous studies of (WF × BBDP) × WF (Martin et al. 1999a) and (WF × BBDR) × WF (Martin et al. 1999b) backcrosses established Iddm14 as a major determinant of diabetes susceptibility common to both BBDP and BBDR rats (Martin et al. 1999a, b). Subsequent studies have confirmed the stability of our phenotype through an N11 generation congenic in which Iddm14 is mapped to an interval no greater than approximately 2.5 Mb on chromosome 4 (Blankenhorn et al. 2007).
The present work extends our previous findings to five additional strains, including LEW.1WR1, that vary in their susceptibility to T1D. The data are consistent with a role for the Tcrb-V13 TCR element in T1D that is induced in susceptible animals by immune perturbation. They are also consistent with the resistance to spontaneous T1D of DR.F344/lyp rats bearing an F344-derived Iddm14 interval (Fuller et al. 2006), indicating that Iddm14 is important for both induced and spontaneous T1D.
Iddm14 is incompletely penetrant and expression of diabetes is certain to be sensitive to genetic background. Consistent with this view of sensitive dependence on genetic background are several observations. First, in previous studies, not all F2 or backcross rats bearing permissive alleles of Iddm14 became diabetic. Second, an unexpected reduction in spontaneous type 1 diabetes incidence in a BBDP.WF congenic line was found to be the result of WF alleles at a novel locus (Iddm24) on chromosome 8 (Wallis et al. 2007). Third, we (J. M. Fuller et al., unpublished) and others (Hornum et al. 2007) have noted that WF.Iddm14.Iddm2 congenic rats do not develop diabetes despite the presence of three high-risk diabetes susceptibility loci, Iddm1 (the MHC), Iddm2 (Gimap5, the cause of lymphopenia), and Iddm14. Considering the last two points together, it is interesting to speculate that WF-origin alleles at another locus such as Iddm24 might also contribute to the absence of spontaneous diabetes in WF.Iddm14.Iddm2 congenics.
Our discovery that Tcrb-V1, 4, and 13 are associated with resistance to diabetes in three strains (WF, F344, and BN) that bear a related haplotype at Iddm14 suggests that when present in the context of a single high-risk MHC class II haplotype, the Tcrb-V gene family is a major determinant of disease probability. In the rat, with a single unusual exception (Penhale et al. 1990), all strains with documented susceptibility to autoimmune diabetes express the RT1B/Du class II MHC haplotype (Awata et al. 1995). Extrapolating from human and mouse data, it has always been assumed that redundancy among cognate rat TCRs would preclude reliance on any one family of TCR α or β chains for disease susceptibility. As a corollary, it was further assumed that the resistance of many RT1u rat strains to diabetes was dependent on other, unspecified “background” genes. Nonetheless, in our analysis, Tcrb-V13 alone could account for the resistance of WF, F344, and BN rats, as all three share the BV13S1A2 (Stienekemeier et al. 2000) sequence of Tcrb-V13. This holds true for systems in which autoimmune diabetes occurs as the result of immunologic perturbation (Mordes et al. 2007; the present data) as well as spontaneously (Fuller et al. 2006). The F344 rat in addition harbors a stop codon that makes the BV13A3 gene product nonfunctional (Stienekemeier et al. 2000). Whether the Tcrb-V13 allele alone predisposes to T1D in the rat or whether other mutations such as those we have discovered in Tcrb-V1 and Tcrb-V4 can also contribute to susceptibility will require functional studies. With respect to functionality, it is noteworthy that the BV13S1A2 allele has been shown to skew the TCR repertoire and substantially alter the CD4+ to CD8+ ratio of Vβ13+ T cells (Stienekemeier et al. 2000).
The elements of the TCR form the basis of antigen recognition by T cells, which is critical for diabetes pathogenesis, but there has been limited support for the importance of the TCR in existing genetic studies of NOD mouse and human T1D. Our discovery that TCR β chains are strong candidate determinants of diabetes susceptibility in the rat is therefore of potential importance for understanding the mechanisms of T1D etiology and pathogenesis. This finding should spur further detailed study of human and mouse T1D genetics.
In the rat, our Iddm14 candidates function as recessive protective alleles. Control rats that rarely or never become diabetic are homozygous for an Iddm14 haplotype that does not make either Tcrb-V1 or Tcrb-V4 (WF, BN), or does not make Tcrb-V13 (F344). This situation is less likely to occur and be detected in outbred human populations. In addition, the human data sets that fail to implicate the TCR are based on either classical genome mapping or SNP haplotyping. Neither method is ideal for detecting recessive protective alleles because “resistant” individuals would have to be homozygous for the loss of several key Tcrb-V elements, as we propose is important in the rat. Furthermore, stratification by HLA may be required for detection of TCR effects in human T1D.
In the NOD mouse studies, no TCR chain family has been linked to T1D, but a role for a limited TCRβ repertoire could nonetheless be important. Several analyses of T-cell repertoire, especially in early prediabetic stages, implicate an oligoclonal T-cell response by NOD mice (Haskins 2005; Quinn et al. 2006). Interestingly, Tcrb-V1 and Tcrb-V4, which are homologous in mouse and rat, are frequently found in this limited primary T-cell repertoire (Haskins 2005; Quinn et al. 2006), and Tcrb-V4+ diabetogenic T-cell clones, including BDC2.5, are abundant (Haskins 2005). NOD and C57Bl/6 mice share alleles at most Tcrb loci (phenome.jax.org) and, thus, Idd-congenic strains based on differences between NOD and C57Bl/6 would be unlikely to show linkage to the Tcrb region. The one major direct linkage test of a role for Tcrb-V used a parental strain (SJL) that has a major deletion of Tcrb-V and proved only that murine Tcrb-V5, 8, 9, 11, 12, and 13 are not required for development of insulitis or spontaneous diabetes (Shizuru et al. 1991). However, NOD8.3 TCR transgenic mice, in which the majority of T cells are skewed toward expression of an IGRP-specific Vβ8+ TCR, develop disease with a much more rapid onset (Dudek et al. 2006; Verdaguer et al. 1996).
We suggest that inbred rat strains have retained more TCR polymorphism and diversity than have inbred mice. If so, they may serve as better models of human Tcrb-V usage than mice. Autoimmunity-prone rats may offer the opportunity to test in depth the hypothesis that Tcrb-V chain variation contributes to T1D susceptibility or severity across a range of genetically susceptible strains.
We recognize that a Tcrb-V haplotype, although a plausible cluster of candidate genes, remains only a candidate haplotype. Among the candidate genes in the Iddm14 interval are many trypsinogen genes, olfactory genes, caspase 2, and taste receptors. In addition, even if Tcrb-V4, 1, and 13 elements contribute susceptibility, the data do not exclude the possibility that other Tcrb-V family members can be used in the autoimmune recognition of islet antigens, as has been demonstrated in the NOD mouse, where little if any Tcrb-V oligoclonality is observed after diabetes onset (Galley and Danska 1995). Nonetheless, it its intriguing to note both that many published diabetogenic murine T-cell clones express Tcrb-V4 and that the sequence of several of these mouse β chains is remarkably similar to rat Tcrb-V4 (Haskins 2005; Quinn et al. 2006; Sherman et al. 1987).
Studies to determine if Tcrb-V1, 4, and/or 13 are all constituents of an Iddm14 haplotype will require new resources: antibodies to rat Vβ family members, an analysis of Tcrb-V gene usage in the pancreas of rats that are becoming diabetic, TCR spectratyping, or the application of RNA interference technology to knock down rat Tcrb-V4 expression. Efforts to develop the needed resources are underway in our laboratories.
In conclusion, our data suggest that the TCR β-chain usage may contribute to the penetrance of both spontaneous and induced autoimmune diabetes in genetically susceptible rats of multiple different strains. Dissection of the TCR gene region will provide mechanistic analyses to uncover the possible role of the MCH/TCR interaction in the development of β-cell-specific autoimmunity.
Acknowledgments
We thank Vikranth Mulkanoor for assistance with the nucleic acid sequencing. This work was supported in part by grants DK36024 (DLG), DK49106 (DLG, JPM, EPB), and DK25306 (JPM), ADA grants 7-08-RA-106 (JPM) and 7-06-RA-14 (EPB), AI42386 (JMF, AL), and Center Grant DK32520 from the National Institutes of Health. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Contributor Information
John P. Mordes, Department of Medicine, University of Massachusetts Medical School, Worcester, MA 01655, USA
Laura Cort, Email: libby@drexelmed.edu, Department of Microbiology and Immunology, Center for Immunogenetics and Inflammatory Diseases, Drexel University College of Medicine, Philadelphia, PA 19129, USA.
Elaine Norowski, Department of Medicine, University of Massachusetts Medical School, Worcester, MA 01655, USA.
Jean Leif, Department of Medicine, University of Massachusetts Medical School, Worcester, MA 01655, USA.
Jessica M. Fuller, Department of Medicine, University of Washington, Seattle, WA 98195, USA. Department of Clinical Sciences (Malmö), Clinical Research Center, Lund University, Malmö, Sweden
Åke Lernmark, Department of Medicine, University of Washington, Seattle, WA 98195, USA. Department of Clinical Sciences (Malmö), Clinical Research Center, Lund University, Malmö, Sweden.
Dale L. Greiner, Department of Medicine, University of Massachusetts Medical School, Worcester, MA 01655, USA
Elizabeth P. Blankenhorn, Email: eblanken@drexelmed.edu, Department of Microbiology and Immunology, Center for Immunogenetics and Inflammatory Diseases, Drexel University College of Medicine, Philadelphia, PA 19129, USA
References
- Awata T, Guberski DL, Like AA. Genetics of the BB rat: association of autoimmune disorders (diabetes, insulitis, and thyroiditis) with lymphopenia and major histocompatibility complex class II. Endocrinology. 1995;136:5731–5735. doi: 10.1210/endo.136.12.7588330. [DOI] [PubMed] [Google Scholar]
- Blankenhorn EP, Rodemich L, Martin-Fernandez C, Leif J, Greiner DL, et al. The rat diabetes susceptibility locus Iddm4 and at least one additional gene are required for autoimmune diabetes induced by viral infection. Diabetes. 2005;54:1233–1237. doi: 10.2337/diabetes.54.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhorn EP, DeScipio C, Rodemich L, Cort L, Leif JH, et al. Refinement of the Iddm4 diabetes susceptibility locus reveals TCRVbeta4 as a candidate gene. Ann NY Acad Sci. 2007;1103:128–131. doi: 10.1196/annals.1394.020. [DOI] [PubMed] [Google Scholar]
- Cuppen E. Haplotype-based genetics in mice and rats. Trends Genet. 2005;21:318–322. doi: 10.1016/j.tig.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Dudek NL, Thomas HE, Mariana L, Sutherland RM, Allison J, et al. Cytotoxic T-cells from T-cell receptor transgenic NOD8.3 mice destroy beta-cells via the perforin and Fas pathways. Diabetes. 2006;55:2412–2418. doi: 10.2337/db06-0109. [DOI] [PubMed] [Google Scholar]
- Ellerman KE, Like AA. Susceptibility to diabetes is widely distributed in normal class IIu haplotype rats. Diabetologia. 2000;43:890–898. doi: 10.1007/s001250051466. [DOI] [PubMed] [Google Scholar]
- Fuller JM, Kwitek AE, Hawkins TJ, Moralejo DH, Lu W, et al. Introgression of F344 rat genomic DNA on BB rat chromosome 4 generates diabetes-resistant lymphopenic BB rats. Diabetes. 2006;55:3351–3357. doi: 10.2337/db06-0715. [DOI] [PubMed] [Google Scholar]
- Galley KA, Danska JS. Peri-islet infiltrates of young non-obese diabetic mice display restricted TCR β-chain diversity. J Immunol. 1995;154:2969–2982. [PubMed] [Google Scholar]
- Haskins K. Pathogenic T-cell clones in autoimmune diabetes: more lessons from the NOD mouse. Adv Immunol. 2005;87:123–162. doi: 10.1016/S0065-2776(05)87004-X. [DOI] [PubMed] [Google Scholar]
- Hornum L, Rømer J, Markholst H. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of iddm1. Diabetes. 2002;51:1972–1979. doi: 10.2337/diabetes.51.6.1972. [DOI] [PubMed] [Google Scholar]
- Hornum L, Lundsgaard D, Markholst H. PolyI:C induction of diabetes is controlled by Iddm4 in rats with a full regulatory T cell pool. Ann NY Acad Sci. 2007;1110:65–72. doi: 10.1196/annals.1423.008. [DOI] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources, Commission on Life Sciences and National Research Council. Guide for the Care and Use of Laboratory Animals. National Academy Press; Washington, DC: 1996. p. 119. [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Klaff LS, Koike G, Jiang JJ, Wang YL, Bieg S, et al. BB rat diabetes susceptibility and body weight regulation genes colocalize on Chromosome 2. Mamm Genome. 1999;10:883–887. doi: 10.1007/s003359901108. [DOI] [PubMed] [Google Scholar]
- Klöting I, van den Brandt J, Klöting N, Radovic B. Alleles of diabetes-resistant BN rats contribute to insulin-dependent type 1 diabetes mellitus. J Autoimmun. 2003;20:119–123. doi: 10.1016/s0896-8411(02)00111-7. [DOI] [PubMed] [Google Scholar]
- Lenzen S, Tiedge M, Elsner M, Lortz S, Weiss H, et al. The LEW.1AR1/Ztm-iddm rat: a new model of spontaneous insulin-dependent diabetes mellitus. Diabetologia. 2001;44:1189–1196. doi: 10.1007/s001250100625. [DOI] [PubMed] [Google Scholar]
- MacMurray AJ, Moralejo DH, Kwitek AE, Rutledge EA, Van Yserloo B, et al. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (lan)-related gene. Genome Res. 2002;12:1029–1039. doi: 10.1101/gr.412702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AM, Blankenhorn EP, Maxson MN, Zhao M, Leif J, et al. Non-major histocompatibility complex-linked diabetes susceptibility loci on chromosomes 4 and 13 in a backcross of the DP BB/Wor rat to the WF rat. Diabetes. 1999a;48:50–58. doi: 10.2337/diabetes.48.1.50. [DOI] [PubMed] [Google Scholar]
- Martin AM, Maxson MN, Leif J, Mordes JP, Greiner DL, et al. Diabetes-prone and diabetes-resistant BB rats share a common major diabetes susceptibility locus, iddm4: Additional evidence for a “universal autoimmunity locus” on rat chromosome 4. Diabetes. 1999b;48:2138–2144. doi: 10.2337/diabetes.48.11.2138. [DOI] [PubMed] [Google Scholar]
- Mordes JP, Leif J, Novak S, DeScipio C, Greiner DL, et al. The iddm4 locus segregates with diabetes susceptibility in congenic WF.iddm4 rats. Diabetes. 2002;51:3254–3262. doi: 10.2337/diabetes.51.11.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes JP, Guberski DL, Leif JH, Woda BA, Flanagan JF, et al. LEW.1WR1 rats develop autoimmune diabetes spontaneously and in response to environmental perturbation. Diabetes. 2005;54:2727–2733. doi: 10.2337/diabetes.54.9.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes JP, Poussier P, Rossini AA, Blankenhorn EP, Greiner DL. Rat models of type 1 diabetes: Genetics, environment, and autoimmunity. In: Shafrir E, editor. Animal models of diabetes: frontiers in research. CRC Press; Boca Raton, FL: 2007. pp. 1–39. [Google Scholar]
- Penhale WJ, Stumbles PA, Huxtable CR, Sutherland RJ, Pethick DW. Induction of diabetes in PVG/c strain rats by manipulation of the immune system. Autoimmunity. 1990;7:169–179. doi: 10.3109/08916939008993389. [DOI] [PubMed] [Google Scholar]
- Quinn A, McInerney M, Huffman D, McInerney B, Mayo S, et al. T cells to a dominant epitope of GAD65 express a public CDR3 motif. Int Immunol. 2006;18:967–979. doi: 10.1093/intimm/dxl033. [DOI] [PubMed] [Google Scholar]
- Sherman DH, Hochman PS, Dick R, Tizard R, Ramachandran KL, et al. Molecular analysis of antigen recognition by insulin-specific T-cell hybridomas from B6 wild-type and bm12 mutant mice. Mol Cell Biol. 1987;7:1865–1872. doi: 10.1128/mcb.7.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuru JA, Taylor-Edwards C, Livingstone A, Fathman CG. Genetic dissection of T cell receptor Vβ gene requirements for spontaneous murine diabetes. J Exp Med. 1991;174:633–638. doi: 10.1084/jem.174.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LR, Kono DH, Theofilopoulos AN. Complexity and sequence identification of 24 rat V beta genes. J Immunol. 1991;147:375–379. [PubMed] [Google Scholar]
- Stienekemeier M, Hofmann K, Gold R, Herrmann T. A polymorphism of the rat T-cell receptor beta-chain variable gene 13 (BV13S1) correlates with the frequency of BV13S1-positive CD4 cells. Immunogenetics. 2000;51:296–305. doi: 10.1007/s002510050623. [DOI] [PubMed] [Google Scholar]
- Verdaguer J, Yoon JW, Anderson B, Averill N, Utsugi T, et al. Acceleration of spontaneous diabetes in TCR-β-transgenic nonobese diabetic mice by β-cell cytotoxic CD8+ T cells expressing identical endogenous TCR-α chains. J Immunol. 1996;157:4726–4735. [PubMed] [Google Scholar]
- Wallis RH, Wang K, Dabrowski D, Marandi L, Ning T, et al. A novel susceptibility locus on rat chromosome 8 affects spontaneous but not experimentally induced type 1 diabetes. Diabetes. 2007;56:1731–1736. doi: 10.2337/db06-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind D, Weiss H, Jorns A, Lenzen S, Tiedge M, et al. Effects of polyinosinic-polycytidylic acid and adoptive transfer of immune cells in the Lew.1AR1-iddm rat and in its coisogenic LEW.1AR1 background strain. Autoimmunity. 2005;38:265–275. doi: 10.1080/08916930500114321. [DOI] [PubMed] [Google Scholar]
- Williams CB, Gutman GA. T cell receptor beta-chain genes in the rat. Availability and pattern of utilization of V gene segments differs from that in the mouse. J Immunol. 1989;142:1027–1035. [PubMed] [Google Scholar]
- Yokoi N, Komeda K, Wang HY, Yano H, Kitada K, et al. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat Genet. 2002;31:391–394. doi: 10.1038/ng927. [DOI] [PubMed] [Google Scholar]


