Abstract
The genome of the budding yeast Saccharomyces cerevisiae was the first completely sequenced from a eukaryote. It was released in 1996 as the work of a worldwide effort of hundreds of researchers. In the time since, the yeast genome has been intensively studied by geneticists, molecular biologists, and computational scientists all over the world. Maintenance and annotation of the genome sequence have long been provided by the Saccharomyces Genome Database, one of the original model organism databases. To deepen our understanding of the eukaryotic genome, the S. cerevisiae strain S288C reference genome sequence was updated recently in its first major update since 1996. The new version, called “S288C 2010,” was determined from a single yeast colony using modern sequencing technologies and serves as the anchor for further innovations in yeast genomic science.
Keywords: Saccharomyces cerevisiae, model organism, reference sequence, genome release, S288C
Research regarding the genetics of yeast began in earnest during the 1930s and 1940s, with the pioneering work of Øjvind Winge (Szybalski 2001) and the work of Carl Lindegren (Lindegren 1949). The 16 chromosomes of Saccharomyces cerevisiae comprise the first completely finished eukaryotic genome and were sequenced in the early 1990s by an international consortium of researchers from 19 countries working in 94 laboratories using several different sequencing methods and technologies (Goffeau et al. 1996). The genome sequence is that of strain background S288C, and the strains used for the sequencing were predominantly AB972 (ATCC 76269) and FY1679 (ATCC 96604), two strains isogenic with S288C. Some sections of chromosome III were sequenced from XJ24-4a, A364A (ATCC 204626), and DC5 (ATCC 64665), and a small portion of chromosome XIV was taken from strain A364A (Table 1).
Table 1. Original Saccharomyces cerevisiae genome sequencing project.
| Chromosome | Length (bp) | Sequencing Coordinator | Original Straina | Sequencing Methodology | Initial Release | Updated Versions |
|---|---|---|---|---|---|---|
| I | 230,218 | H. Bussey, Canada | AB972 | Manual, automated | April 1995 | 11 |
| II | 813,184 | H. Feldmann, Germany | S288C | Diverse methods in collaborating laboratories | December 1994 | 13 |
| III | 316,620 | S. Oliver, England | XJ24-24a, AB972, A364A, DC5 | Diverse methods in collaborating laboratories | May 1992 | 5 |
| IV | 1,531,933 | C. Jacq, France | AB972, FY1679 | Automated | May 1997 | 10 |
| B. Barrell, England | ||||||
| M. Johnston, United States | ||||||
| R. Davis, United States | ||||||
| V | 576,874 | R. Davis, United States | AB972 | Automated | May 1997 | 1 |
| VI | 270,161 | Y. Murakami, Japan | AB972 | Automated shotgun, primer walking | July 1995 | 3 |
| VII | 1,090,940 | H. Tettelin, Belgium | FY1679 | Diverse methods in collaborating laboratories | May 1997 | 9 |
| VIII | 562,643 | M. Johnston, United States | AB972 | Diverse methods in collaborating laboratories | September 1994 | 4 |
| IX | 439,888 | B. Barrell, England | AB972 | Shotgun, primer walking | May 1997 | 0 |
| X | 745,751 | F. Galibert, France | FY1679 | Diverse methods in collaborating laboratories | September 1996 | 10 |
| XI | 666,816 | B. Dujon, France | FY1679 | Diverse methods in collaborating laboratories | June 1994 | 9 |
| XII | 1,078,177 | J. Hoheisel, Germany | AB972 | Diverse methods in collaborating laboratories | May 1997 | 4 |
| M. Johnston, United States | ||||||
| XIII | 924,431 | B. Barrell, England | AB972 | Automated shotgun, primer walking | May 1997 | 1 |
| XIV | 784,333 | P. Philippsen, Switzerland | FY1679, A364A | Diverse methods in collaborating laboratories | April 1996 | 8 |
| XV | 1,091,291 | B. Dujon, France | FY1679 | Manual, automated, shotgun, primer walking | May 1997 | 5 |
| XVI | 948,066 | A. Goffeau, Belgium | AB972 | Automated shotgun, primer walking | May 1997 | 2 |
| H. Bussey, Canada | ||||||
| R. Davis, United States | ||||||
| B. Barrell, England | ||||||
| M. Johnston, United States |
The original Saccharomyces cerevisiae genome sequencing project was a worldwide collaboration and chromosome sequences were subsequently updated independently numerous times before the recent major genome update.
All strains are derived from S288C.
Here, we recount the genealogical history of S288C and the key derivative strains AB972 and FY1679. We also discuss the early S. cerevisiae sequencing efforts of the 1990s. Finally, we describe the resequencing and update of the S. cerevisiae reference genome.
Materials and Methods
Provenance of S288C
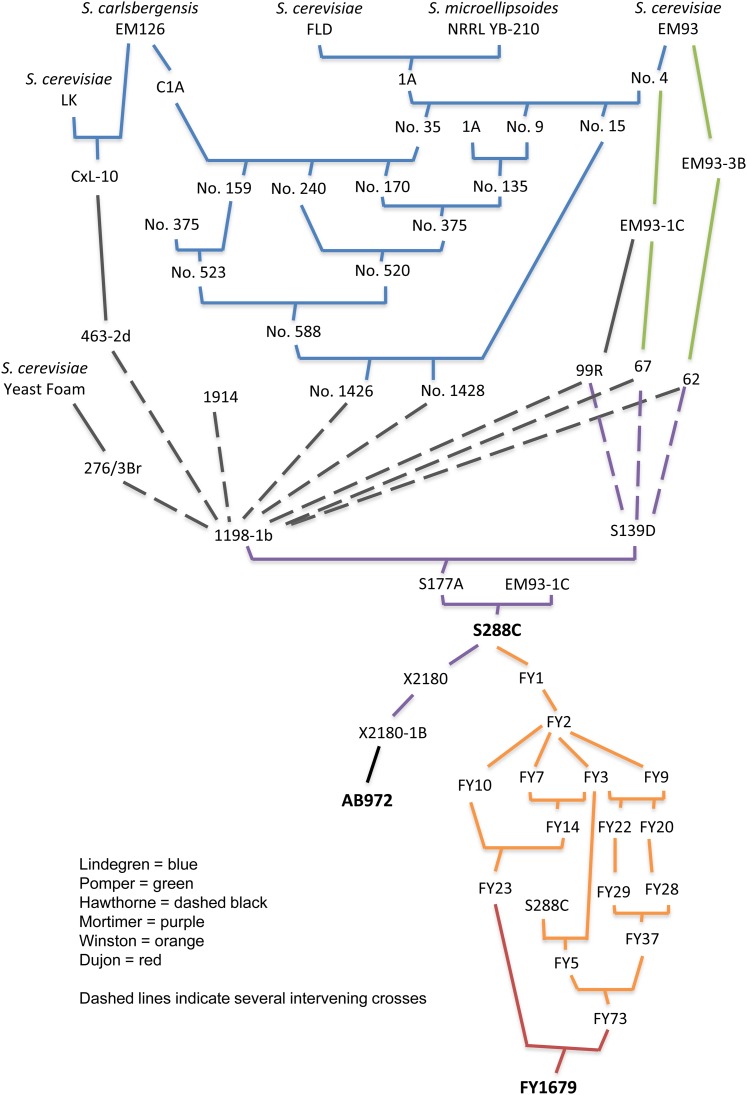
S288C is a common gal2 mutant haploid laboratory strain with a long history of use in genetic and molecular biology studies. S288C has a complex genealogy; it is a contrived strain produced through numerous deliberate crosses, first by Carl Lindegren, and in later years by Robert Mortimer (Figure 1). Almost 90% of the S288C gene pool is from strain EM93, isolated by Emil Mrak in 1938 from a rotting fig collected outside the town of Merced in California’s Central Valley (Mortimer and Johnston 1986). Lindegren obtained Mrak’s EM93 for use in a laborious project to develop fertile breeding stocks for his genetic studies concerning the fermentation of different carbohydrates (Lindegren 1949). Lindegren obtained from L. J. Wickerham a culture of Saccharomyces microellipsoides (NRRL YB-210), a diploid-weak galactose and melibiose fermenter, but a nonfermenter of maltose. Wickerham had isolated NRRL YB-210 from a rotting banana that had been collected in Costa Rica in 1942 (C. Kurtzman, personal communication). Lindegren made a hybrid between haploid derivatives of NRRL YB-210 and his own isolate of FLD, a strong galactose and maltose fermenter that Lindegren referred to as a “standard legitimate diploid strain of commercial baking yeast” (Lindegren 1949). The hybrid between the NRRL YB-210 and FLD haploid derivatives produced only one viable ascospore (“1A”), which was completely incapable of fermenting galactose (Figure 1). Lindegren shared his yeast strains widely with other researchers. Seymour Pomper obtained from Lindegren an alpha mating–type haploid segregant of EM93 (by then renamed EM93-1C) as well as an “a” mating–type segregant of EM93 (EM93-3B). Pomper called these strains by the numbers 67 and 62 (Pomper 1952; Pomper and Burkholder 1949; Mortimer and Johnston 1986). Reaume and Tatum also obtained through Lindegren an alpha mating–type derivative of Mrak’s EM93 (EM93-1C) and used it in studies of spontaneous and induced nutritional deficiencies (Reaume and Tatum 1949). Mortimer obtained from Reaume strain 99R, an “adenineless” derivative of EM93-1C, and used 99R in his own studies of yeast genetics and isolation of the strain S288C.
Figure 1.
Genealogy of Saccharomyces cerevisiae strains S288C, AB972, and FY1679.
Mortimer developed S288C to be used as a parental strain for the isolation of biochemical mutants. The immediate parents of S288C are EM93-1C (obtained by Cornelius Tobias from Lindegren and renamed SC7) and S177A, an EM93 derivative. One parent of S177A was S139D, which was derived by Mortimer entirely from Mrak’s EM93 via Reaume’s strain 99R and Pomper’s strains 67 and 62 (Mortimer and Johnston 1986). The other parent of S177A was 1198-1b, derived by Donald Hawthorne for genetic studies of the fermentation of sucrose, maltose, galactose, and alpha-methylglucoside (Hawthorne 1956). Hawthorne isolated 1198-1b through many crosses of a wide variety of strains, including Reaume’s strain 99R, Pomper’s strains 67 and 62, Lindegren’s 1426 and 1428, Ephrussi’s 276/3Br, which was a derivative of commercial baking strain Yeast Foam (Ephrussi et al. 1949), Lindegren’s 1914, and Roman and Douglas’s 463-2d, which was derived from Lindegren’s diploid CxL-10. The strain CxL-10 was derived by Lindegren from a cross between Mrak’s Saccharomyces carlsbergensis EM126 (a melibiose fermenter isolated from a rotting fig) and a commercial baking yeast called LK. The strain 1914 was deduced by Mortimer to have been isolated by Lindegren from crosses with the same set of strains that produced his 1426 and 1428 (Figure 1) (Lindegren 1949; Mortimer and Johnston 1986). It is no accident that S288C is widely used in laboratory settings. Mortimer bred it to be nonflocculent and to have only minimal nutrient requirements, including biotin, nitrogen, glucose, and various salts and trace elements (Mortimer and Johnston 1986).
The early maps
All the crosses performed in the previous decades by Lindegren, Hawthorne, and others to investigate inheritance of mating type, nutritional requirements, metal resistance, and the fermentation of various sugars had ultimately led to the isolation of Saccharomyces strain S288C, but they also enabled construction of the first genetic and physical yeast chromosome maps. Lindegren published the first genetic maps of four chromosomes (Lindegren 1949). Lindegren and Lindegren (1951) presented maps of five chromosomes and introduced the Roman numeral system of yeast chromosome designations still in use today. In 1959, Lindegren and colleagues published more extensive maps for Saccharomyces, including nine chromosome arms with centromeres and one syntenic group without a centromere, stating that the haploid number of chromosomes was “at least seven” (Lindegren et al. 1959). A year later, Hawthorne and Mortimer (1960) published their first map, which included 10 chromosomes and maintained the Roman numeral designations of the Lindegrens from the map published in 1951. Mortimer and Schild published the first comprehensive genetic map of S. cerevisiae in 1980, which included 17 chromosomes and discussed the possible existence of an eighteenth (Mortimer and Schild 1980). Klapholz and Esposito (1982) soon showed that “chromosome XVII” was actually the left arm of chromosome XIV. Soon thereafter, Kuroiwa et al. (1984) used fluorescent staining of meiotic nuclei to demonstrate the presence of 16 chromosomes. By the publication of the 11th edition of Mortimer’s map, the S. cerevisiae haploid chromosome number of 16 had become established and accepted (Mortimer et al. 1992).
The original genome project
Like Lindegren, and in keeping with the collegial atmosphere that characterizes the yeast research community, Mortimer shared his S288C strain with other researchers. Fred Winston and colleagues (Winston et al. 1995) used gene replacement to develop a set of yeast strains isogenic to S288C but repaired for GAL2, which also contained nonreverting mutations in several genes commonly used for selection in the laboratory environment (URA3, TRP1, LYS2, LEU2, HIS3). Winston shared derivatives of this set, FY23 (mating type “a”) and FY73 (mating type “alpha”), with Bernard Dujon, who mated the strains to make the diploid FY1679, which was used in the original genome sequencing project (Figure 1) (Thierry et al. 1990). A cosmid library made from FY1679 was used for sequencing chromosomes VII, X, XI, XIV, and XV (Table 1) (Tettelin et al. 1997; Galibert et al. 1996; Dujon et al. 1994; Philippsen et al. 1997; Dujon et al. 1997). FY1679 was also used for sequencing the mitochondrial DNA, which was not part of the nuclear genome project and was determined separately (Foury et al. 1998). AB972 (Link and Olson 1991) is an ethidium bromide–induced ρ° derivative of Mortimer’s X2180-1B (obtained via Elizabeth Jones), itself a haploid derivative of strain X2180, which was made by self-diploidization of S288C (Figure 1) (Olson et al. 1986; Mortimer and Johnston 1986; Riles et al. 1993). AB972 was used in the original sequencing project as source DNA for chromosomes I, III, IV, V, VI, VIII, IX, XII, XIII, and XVI (Table 1) (Bussey et al. 1995; Oliver et al. 1992; Jacq et al. 1997; Dietrich et al. 1997; Murakami et al. 1995; Johnston et al. 1994; Churcher et al. 1997; Johnston et al. 1997; Bowman et al. 1997; Bussey et al. 1997). Portions of chromosome III were also taken from strains XJ24-24a, A364A, and DC5 (Oliver et al. 1992). Chromosome II was sequenced directly from strain S288C (Feldmann et al. 1994).
The sequencing project was launched in 1989, and it initially focused on chromosome III, which was chosen because plasmid and phage DNA libraries, as well as a physical map, were already available (Goffeau and Vassarotti 1991; Dujon 1996). Chromosome III was divided into contiguous overlapping fragments ∼10 kbp in length and distributed to 35 laboratories in 10 European countries (Vassarotti and Goffeau 1992). Each laboratory was allowed to apply sequencing strategies and methods of its own choosing, as long as they adhered to standards agreed on by the consortium (Goffeau and Vassarotti 1991). In the early years of the project, final sequences for the first completed chromosomes were deposited into the data library at the Martinsried Institute for Protein Sequences (MIPS) under the direction of H. Werner Mewes. MIPS provided initial sequence data coordination, warehousing, and analysis (Dujon 1992). As the project progressed, sequences for other chromosomes were deposited at the DNA DataBank of Japan (DDBJ), the European Molecular Biology Laboratory (EMBL), and GenBank, the National Institutes of Health (NIH) sequence database. The original annotation and maintenance of the chromosomal sequences were provided by each chromosome sequencing group (Vassarotti et al. 1995), MIPS, and SGD. In the early to mid 2000s, the data warehousing, annotation, and maintenance duties were assumed completely by SGD, which has participated in the annotation and maintenance of the genome sequence for the past 20 years.
The genome project had identified approximately 6000 protein-coding genes, many of unknown function (Goffeau et al. 1996); during this process, the researchers involved recognized the necessity for a stable systematic nomenclature. Through a series of consortium meetings in the late 1980s and early 1990s, members devised and revised the system currently in use today. Open reading frames (ORFs) were initially labeled with “Y” for yeast, alphabetical letters for chromosome (such as C for chromosome III), labeled with “L” or “R” for the left or right chromosome arm, and labeled with sequential numbers indicating their order within the specific plasmid or cosmid clone used for sequencing (B. Dujon, personal communication). This system allowed the numbering for different portions of chromosomes sequenced in different laboratories to be determined independently. The first published uses of this systematic nomenclature were from Thierry et al. (1990), who presented the sequence of an 8.2-kb segment of chromosome III with names starting at YCR521, and from Jacquier et al. (1992), who reported the sequence of a 10.7-kb section of chromosome XI with names starting at YKL500. Once the sequences of full chromosomes were being completed, it became clear that continuous numbering for each entire chromosome would be preferable. As a result, ORFs were renumbered starting with 1 at the centromere, then incrementing by +1 on each arm moving toward the telomere. The issue of strandedness for each ORF was subsequently addressed through the addition of the “W” or “C” suffix (for Watson or Crick). This systematic nomenclature was used in publication of the first complete sequence of a eukaryotic chromosome, that of yeast chromosome III (Oliver et al. 1992). The brevity and utilitarian nature of this nomenclature system have ensured its continued use and success.
Surveys of the new genome sequence
Having the complete genome sequence available accelerated the progress of comprehensive surveys of different types of chromosomal elements that were already underway. Several different groups had been studying centromere structure (Hieter et al. 1985; Hegemann and Fleig 1993). Genomic mapping of replication origins (autonomously replicating sequences) had become an active area of investigation (Deshpande and Newlon 1992; Shirahige et al. 1993; Yang et al. 1999). Louis et al. (1994) were studying the mosaic structure of yeast telomeres. Moving forward, the complete sequence now enabled genomic surveys of different types of genes. Both Lowe and Eddy (1997) and Percudani et al. (1997) identified the entire set of transfer RNAs (tRNAs). Kim et al. (1998) identified hundreds of retrotransposon insertions. Planta and Mager (1998) determined the complete list of cytoplasmic ribosomal protein genes. Lowe and Eddy (1999) screened the genome for small nucleolar RNA (snoRNA) genes. In the 50 years after Lindgren first published manual drawings of four yeast chromosomes containing eight metabolic markers and one mating locus (Lindegren 1949), the yeast genome map had become almost fully populated.
What is a reference genome?
Since its inception two decades ago, yeast genomics has been built around the single reference genome of S288C. The original idea was the production of a single consensus representative S. cerevisiae genome against which all other yeast sequences could be measured. The reference genome serves as the scaffold on which to hang other genomic sequences, and the foundation on which to build different types of genomic datasets. Whereas the first genome took years to complete, through the efforts of the large international consortium described, the sequences of dozens of genomes have been determined in the past several years (Engel and Cherry 2013). As sequencing has become more widespread, less novel, and, above all, less expensive, decoding entire genomes has become less daunting. New genomes now take only days to sequence to full and deep coverage and are assembled quickly, by individuals or small groups, through comparison to the reference, which is an invaluable guide for the annotation of newly sequenced genomes.
It is becoming increasingly clear that the genome of a species can contain a great deal of complexity and diversity. A reference genome can vary significantly from that of any individual strain or isolate and therefore serves as the anchor from which to explore the diversity of allele and gene complements and to explore how these differences contribute to metabolic and phenotypic variation. In the pharmaceutical industry, knowledge of the yeast reference genome helps drive the development of strains tailored to specific purposes, such as the production of biofuels, chemicals, and therapeutic drugs (Runguphan and Keasling 2013). In the beverage industry, it aids in the fermentation of beers, wines, and sakes with specific attributes, such as desired flavor profiles or reduced alcohol (Engel and Cherry 2013). We have seen the advantage afforded the yeast and genetics communities because of the early availability of an S. cerevisiae reference genome. The great facilitation of scientific discoveries and breakthroughs is without question (Botstein and Fink 2011).
Maintenance of the genome annotation
The original genomic sequence and its annotation have been publicly available and tested by researchers around the world for the past 20 years. During that time, large numbers of corrections to the sequence and its annotation were proposed or published, and many of those were incorporated into the original reference genome sequence of 1996. New genes and other chromosomal features have been identified and added to the annotation, whereas others have been changed or deprecated (Fisk et al. 2006).
In the past several years, changes to the reference genome became less frequent as we moved toward a more stable and “correct” reference sequence. During the 5 yr spanning 2006–2010, 29 small sequence changes and 116 annotation updates were made. In addition, 576 new features were added to the genome annotation, including various ORFs, noncoding RNAs, mating cassette domains, autonomously replicating sequences (ARS), ARS consensus sequences (ACS), and 5′ untranslated region (UTR) introns. Clear descriptions of sequence and annotation changes for affected regions are available from the Locus History and Chromosome History pages of SGD (Table 4). SGD has always made new data available in a timely manner, such that before the recent major update of the entire genome, updates to individual chromosomes were made and released independently. As a result, between the original genome sequence and this new reference, SGD released 95 individual updated versions of the 16 nuclear chromosomes (Table 1). Whereas some chromosome sequences were never edited before now (e.g., chromosome IX), others changed several times over the 15-year period. Chromosome III, which had been sequenced before any other chromosomes as a pilot project from DNA libraries prepared from four different S288C-derivative strains (AB972, XJ24-24a, A364A, DC5), was completely resequenced in the late 1990s from strain FY1679 by the laboratories of G. Volckaert and G. Valle, who submitted the sequence to GenBank/EMBL but did not otherwise publish the revision.
Table 4. Some of the different data types that can be accessed at SGD.
| Type of Information | URL at SGD |
|---|---|
| Downloads site | http://downloads.yeastgenome.org/ |
| DNA and protein sequences | http://downloads.yeastgenome.org/sequence |
| Dates of genome releases | http://downloads.yeastgenome.org/sequence/S288C_reference/dates_of_genome_releases.tab |
| Protein sequences updated in R64.1.1 | http://www.yeastgenome.org/archive/ChangedProteins-2011.shtml |
| History of all sequence and annotation updates | http://www.yeastgenome.org/cgi-bin/chromosomeHistory.pl |
| History of sequence and annotation updates for specific loci | http://www.yeastgenome.org/cgi-bin/locusHistory.pl |
| All chromosome sequence changes | http://downloads.yeastgenome.org/sequence/S288C_reference/all_chromosome_sequence_changes.tab |
| LiftOver chain files | http://downloads.yeastgenome.org/sequence/S288C_reference/genome_releases/liftover/ |
| Yeast strain genomes | http://downloads.yeastgenome.org/sequence/strains |
The latest S. cerevisiae version R64 genomic reference sequence (also known as S288C 2010) was determined in a single laboratory from a single colony of S288C-derivative strain AB972. This clone was from a stored isolate from the original AB972 strain used by Linda Riles to create the DNA libraries for some chromosomes in the original genome project (Table 1). Recent advances in the development of DNA sequence technologies have allowed the genome to be decoded from a single individual, in this case a single yeast colony, so that the reference genome truly is a single genome. In this article, we describe the sequence and annotation changes made to the S. cerevisiae reference genome in the first major update to the yeast genomic sequence.
The “S288C 2010” S. cerevisiae reference sequence version was determined from an individual AB972 yeast colony. Strain AB972 was obtained from M. Olson (Olson et al. 1986; Link and Olson 1991). Genomic DNA was isolated using standard protocols (Amberg et al. 2005). DNA was sheared and library construction was achieved with the Illumina TruSeq DNA Sample Prep kit. Illumina HiSequation 36-base sequencing was used. Data were generated as FASTQ files. Alignment and mapping of sequence reads to the previous version of the reference genome sequence (release R63.1.1, 2010-01-05) were accomplished using the Burrows-Wheeler Aligner (BWA) (Li and Durbin 2009). The resequencing covered only unique areas of the genome. Regions of repetitive sequence, including some microsatellites, transposable elements, telomeric regions, tRNA genes, and other miscellaneous repeats and GC-rich regions, together accounting for approximately 10% of the genome, were excluded from the analysis because sequence coverage was low or reads were of suboptimal quality. Using standard sequence quality scores, low-quality mismatches with the reference genome sequence version R63.1.1 were ignored. Only high-quality discrepancies were individually investigated through careful manual assembly and editing. The genome coordinates of each feature were updated using the LiftOver software tool available from UCSC Genome Bioinformatics (Hinrichs et al. 2006). Polymorphisms in coding regions were inspected manually to exclude a number of dubious calls and further refined by expert analysis to ensure the proper placement of start and stop codons. Sequence and annotation differences were checked against the published literature for any previous reports.
Results
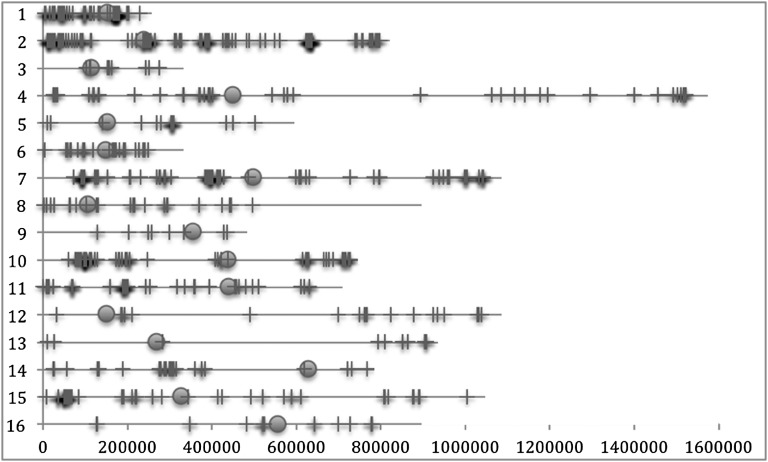
We compared the new genome sequence to our previous version and corrected the sequence according to these results. The sequences of all 16 nuclear chromosomes were updated, with changes occurring in a nonrandom distribution (Figure 2). A number of coding sequences were changed, resulting in amino acid sequence changes to 194 proteins and silent changes in 42 ORFs (Supporting Information, Table S1). This represents approximately 3% of protein coding genes. Other updated features included one 5′ UTR intron, two ncRNAs, two tRNAs, 16 ARSs, one retrotransposon, one long terminal repeat (LTR), three telomeres, and 232 intergenic regions (Table 2). The largest sequence change was a 352-nucleotide insertion on chromosome XI in the intergenic region between ORFs PMU1/YKL128C and MYO3/YKL129C. Chromosome XI was originally sequenced from strain FY1679. It is unclear whether this difference represents real variation between strains AB972 and FY1679 or if it is an artifact of the construction or distribution of the cosmid library used for sequencing by the various participating laboratories (Dujon et al. 1994). Numbers of changed regions in each of the different chromosomes did not correlate with chromosome length (r = 0.253) or sequencing technology used. It is important to note that the quality of the original 1996 genome sequence was very high regardless of which sequencing technology (manual using Maxam-Gilbert or Sanger methods or automated using ABI sequencers) or assembly method (computational assembly or manual piecemeal integration of each cosmid sequence) was used. This is a testament to the care taken by the dozens of individuals working on each chromosome during the original genome sequencing project.
Figure 2.
Chromosomal distribution of sequence changes. The sequences of all 16 nuclear chromosomes were updated, with changes between the previous genome version R63 and the current genome version R64 unevenly distributed throughout the genome. The X axis indicates chromosomal coordinates. Circles indicate centromeres.
Table 2. Numerous features on the 16 nuclear chromosomes were updated in the latest genome release.
| Chromosome | Intergenic | ORF | Silent | Intron | 5′ UTR Intron | ncRNA | tRNA | ARS | Retrotransposon | LTR | Telomere |
|---|---|---|---|---|---|---|---|---|---|---|---|
| I | 17 | 17 | 2 | 1 | |||||||
| II | 36 | 45 | 9 | 1 | 1 | 5 | |||||
| III | 6 | 4 | 1 | ||||||||
| IV | 23 | 15 | 1 | ||||||||
| V | 7 | 5 | 1 | ||||||||
| VI | 13 | 8 | 3 | 1 | 1 | 1 | 1 | ||||
| VII | 30 | 23 | 6 | 1 | 2 | ||||||
| VIII | 11 | 11 | 1 | 1 | 4 | 1 | |||||
| IX | 4 | 2 | 1 | ||||||||
| X | 24 | 26 | 5 | 3 | 1 | 1 | |||||
| XI | 11 | 17 | 1 | 2 | 1 | ||||||
| XII | 11 | 9 | 3 | ||||||||
| XIII | 5 | 4 | 1 | ||||||||
| XIV | 9 | 15 | 2 | 1 | |||||||
| XV | 19 | 15 | 6 | 1 | |||||||
| XVI | 6 | 5 | 1 | ||||||||
| Total | 232 | 221 | 42 | 7 | 1 | 2 | 2 | 16 | 1 | 1 | 3 |
The sequences of various features on the 16 nuclear chromosomes were updated in the latest genome release R64.1.1. In addition to 194 altered protein sequences, 42 ORFs underwent silent coding changes. Other updated features included one 5′ UTR intron, two ncRNAs, two tRNAs, 16 ARSs, one retrotransposon, one LTR, three telomeres, and 232 intergenic regions.
The original 1996 reference genome sequence was determined primarily from AB972 and FY1679, both derivatives of S288C (Figure 1). The “S288C 2010” sequence reported here, determined from AB972, has been compared to a subsequently produced genome sequence of FY1679. There are approximately four SNPs per 100,000 bp between the “S288C 2010” reference and FY1679 (G. Song and B. Dunn, personal communication) demonstrating that these two strains are virtually indistinguishable.
Genome versioning system
SGD has instituted a new versioning system, and this latest sequence update constitutes Genome Release 64.1.1 (released February 3, 2011, and still the latest version as of the writing of this article). In this nomenclature, the first number represents the reference sequence release, the second number represents the feature coordinate release within that sequence release, and the third number represents the annotation release. The reference sequence release number increments only when the reference sequence changes because of nucleotide insertion, deletion, or substitution. Moving forward, we anticipate such sequence change events will be exceedingly rare. The second number, the feature coordinate release number, will increment only when existing annotated features are altered (such as changing the translation start of an ORF) or when new genes or other chromosomal features are added. The last number, the annotation release, will increment when significant functional, Gene Ontology (GO), or phenotype information is added or updated. Although SGD biocurators typically add or update functional, GO, and phenotype information on a daily basis, once the versioning system is completely active, the annotation releases will be incremented once per week with the production of new files available from the SGD Downloads site (http://downloads.yeastgenome.org).
As stated, SGD released 95 total individual updated versions of the 16 nuclear chromosomes between the original release and this latest major update. Those chromosome sequence updates were released as independent events, although different chromosomes were sometimes updated on the same day. SGD has applied the new genome versioning system retroactively, such that chromosomal updates that were released simultaneously are now batched into a single genome version. The initial version is considered 1.1.1, and the 95 updated chromosome sequences correspond to Genome Releases 2.1.1 (released July 27, 1997) through 63.1.1 (released January 5, 2010) (Table 3). Of particular interest are SGD Genome Releases that correspond to genome versions used in the UCSC Genome Browser (http://genome.ucsc.edu/). UCSC release sacCer1 uses R27.1.1, sacCer2 uses R61.1.1, and sacCer3 matches the latest release, R64.1.1. SGD also provides LiftOver chain files between all previous releases and the current version of the S288C reference sequence, which allow researchers to convert data based on previous versions to current coordinates (for URL see Table 4).
Table 3. SGD genome versioning system.
| Genome Release | Date | Chromosome sequences updated |
|---|---|---|
| R1.1.1 | 1996-07-31 | Initial release of 16 nuclear chromosomes |
| R2.1.1 | 1997-07-27 | II, III, X, XIV |
| R3.1.1 | 1997-07-30 | XII |
| R4.1.1 | 1997-08-11 | XV |
| R5.1.1 | 1998-05-21 | III |
| R6.1.1 | 1998-09-13 | I, II |
| R7.1.1 | 1999-01-28 | XIV |
| R8.1.1 | 1999-02-06 | Mitochondriona |
| R9.1.1 | 1999-02-10 | IV |
| R10.1.1 | 1999-03-12 | XI |
| R11.1.1 | 1999-04-22 | II |
| R12.1.1 | 1999-04-26 | II |
| R13.1.1 | 2000-01-21 | VIII |
| R14.1.1 | 2000-03-16 | V |
| R15.1.1 | 2000-04-21 | IV |
| R16.1.1 | 2000-09-13 | III |
| R17.1.1 | 2001-05-29 | II, VI |
| R18.1.1 | 2001-05-31 | VII, XI, XV |
| R19.1.1 | 2001-06-12 | XII |
| R20.1.1 | 2001-06-29 | X |
| R21.1.1 | 2002-12-19 | I, IV |
| R22.1.1 | 2003-01-03 | II, VII, X |
| R23.1.1 | 2003-01-09 | VII, XI, XV |
| R24.1.1 | 2003-01-10 | XI |
| R25.1.1 | 2003-09-26 | VI |
| R26.1.1 | 2003-09-29 | I, II, X, XVI |
| R27.1.1 | 2003-10-01 | IV |
| R28.1.1 | 2003-12-15 | I |
| R29.1.1 | 2004-01-12 | II |
| R30.1.1 | 2004-01-14 | I |
| R31.1.1 | 2004-01-23 | II, VII |
| R32.1.1 | 2004-01-24 | I |
| R33.1.1 | 2004-01-27 | II |
| R34.1.1 | 2004-01-30 | I, II |
| R35.1.1 | 2004-02-01 | VIII |
| R36.1.1 | 2004-02-06 | IV, VI, XII |
| R37.1.1 | 2004-02-13 | IV, X, XI, XV |
| R38.1.1 | 2004-02-20 | III, X, XIV |
| R39.1.1 | 2004-02-27 | XIII |
| R40.1.1 | 2004-07-09 | II |
| R41.1.1 | 2004-07-16 | II, VII |
| R42.1.1 | 2004-07-23 | I, IV, VII, XI, XIV, XVI |
| R43.1.1 | 2004-07-26 | VIII |
| R44.1.1 | 2005-11-03 | XIV |
| R45.1.1 | 2005-11-07 | XIV |
| R46.1.1 | 2005-11-08 | VIII |
| R47.1.1 | 2005-11-23 | VII |
| R48.1.1 | 2005-12-02 | VII, XI |
| R49.1.1 | 2005-12-16 | XI |
| R50.1.1 | 2006-01-06 | XV |
| R51.1.1 | 2006-01-13 | III, XII |
| R52.1.1 | 2006-01-20 | I, IV, X |
| R53.1.1 | 2006-04-14 | IV |
| R54.1.1 | 2006-10-06 | X |
| R55.1.1 | 2006-11-10 | XIV |
| R56.1.1 | 2007-04-06 | I |
| R57.1.1 | 2007-12-12 | VII |
| R58.1.1 | 2008-03-05 | I |
| R59.1.1 | 2008-06-03 | XI |
| R60.1.1 | 2008-06-04 | X |
| R61.1.1 | 2008-06-05 | IV |
| R62.1.1 | 2009-02-18 | X |
| R63.1.1 | 2010-01-05 | XIV |
| R64.1.1 | 2011-02-03 | All nuclear chromosomes |
SGD has instituted a genome versioning system. There are a total of 95 individual updated versions of the 16 nuclear chromosomes between the original release and this latest major update (R64.1.1).
The mitochondrial chromosome was not part of the original genome project and was determined separately (Foury et al. 1998).
Discussion
With this comprehensive update in place, we anticipate very few sequence changes in the future. The increased quality of the new reference is such that we will handle all future reported differences as variations from this reference, rather than as sequencing “errors.” However, because a reference genome must take into consideration the best representation for that organism rather than simply the sequence of one individual, SGD will consider the possibility of accommodating small changes, if those changes increase the utility of the reference sequence. For example, we have allowed sequence updates that result in bringing two neighboring, nonfunctional ORFs into a single corrected reading frame to form a complete, functional coding region (such as FLO8/YER109C) (Liti et al. 1996). As new genomic sequences of other direct derivatives of S288C become available, differences between these additional S288C-derived genomic sequences from the reference will be detailed as alleles or as observed variation.
New technologies and approaches are now pushing S. cerevisiae annotation past the limits of a system based exclusively on a single reference sequence. Current sequencing methods have made possible the determination of the genomic sequences of hundreds of S. cerevisiae wild and laboratory strains. Comparative genomics of different sequences provides an expanded understanding of the full genetic constituent parts of a species and helps in the definition of conserved regions and in the identification of cryptic sequence features such as binding sites and noncoding RNAs. Different S. cerevisiae genomes vary not only in specific nucleotide sequence but also in their complements of genes; many genes are lost or gained as isolated populations adapt to their environment (Sliwa and Korona 2005; Lopez-Maury et al. 2008; Gordon et al. 2009; Lin and Li 2011). For example, the S288C reference sequence is missing several genes that are well-characterized in other strains, such as XDH1 (xylitol dehydrogenase) (Wenger et al. 2010), KHS1 (killer toxin) (Goto et al. 1991), TAT3 (tyrosine transporter) (Omura et al. 2007), and BIO1 (pimeloyl-CoA synthetase) (Hall and Dietrich 2007). The composition of several gene families also differs between strains; for instance, S288C includes only one (SUC2) of an eight-member invertase gene family (Carlson and Botstein 1983). SGD currently includes these genes in the database designated as “not in the systematic sequence of S288C” (Hirschman et al. 2006).
To address this variety in genome content, we are working toward the development of a virtual S. cerevisiae pan-genome that will contain all the genes found within all sequenced S. cerevisiae strains and wild isolates. The S. cerevisiae pan-genome contains hundreds of genes that are found in some strains but not in others. A pan-genome more accurately describes the full genetic complement of a species and will, in the future, provide a valuable resource for the annotation of newly determined budding yeast genomes and for the functional analysis and comparison of observed variation within S. cerevisiae.
We have also begun integrating into the database the complete genomic sequences of other S. cerevisiae strains (Engel and Cherry 2013). We currently provide precomputed protein and coding DNA alignments (ClustalW) for each ORF, as well as ORF-specific dendrograms, which depict the degree of similarity of that ORF sequence among the set of strains in which it was identified. Furthermore, we continue to associate information regarding sequence variation with functional effects and phenotypic variations (Engel et al. 2010). The genomes of the various strains have already been incorporated into the Basic Local Alignment Search Tool (BLAST) datasets, available for searching against genomic and coding DNA, as well as protein sequences. All the strain DNA and protein sequences are available for download so that researchers can perform their own analyses (http://downloads.yeastgenome.org/sequence). New tools are being developed that will provide access to this compendium of allelic and variation information and will allow any determined genomic sequence to be compared with the reference strain, as well as with the sequences of other widely used and commonly studied S. cerevisiae strains.
In their review of the publication of the complete genomic sequence of S. cerevisiae two decades ago, Clayton et al. (1997) lauded it as an enormous achievement and turned our gaze toward the future. Now, with a modern stable S. cerevisiae reference genome in place and a fresh appreciation of the inherent differences between strains, the next trend in yeast genomic science will focus on the elucidation and documentation of sequence variation and the biological and evolutionary consequences thereof. Yeast genomics has entered a new era and, once again, the greatest significance lies in the work that is yet to come.
DNA accession numbers
The accession numbers for the 16 S. cerevisiae nuclear chromosomes and mitochondrial genome within the Reference Sequence (RefSeq) collection at the National Center for Biotechnology Information (NCBI) are as follows: NC_001133; NC_001134; NC_001135; NC_001136; NC_001137; NC_001138; NC_001139; NC_001140; NC_001141; NC_001142; NC_001143; NC_001144; NC_001145; NC_001146; NC_001147; NC_001148; and NC_001224.
Supplementary Material
Acknowledgments
Bernard Dujon, Andre Goffeau, Peter Philippsen, Fred Winston, Frederick (Fritz) Roth, and the yeast research community and genome sequencing coordinators have all been helpful in the preparation of this manuscript and/or in discussions of the reference genome changes. The Saccharomyces Genome Database project is funded by a U41 grant from the National Human Genome Research Institute at the U.S. National Institutes of Health (HG001315). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Human Genome Research Institute or the National Institutes of Health.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Amberg D. C., Burke D. J., Strathern J. N., 2005. Methods in yeast genetics: A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Botstein D., Fink G. R., 2011. Yeast: An experimental organism for 21st century biology. Genetics 189: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S., Churcher C., Badcock K., Brown D., Chillingworth T., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XIII. Nature 387: 90–93 [PubMed] [Google Scholar]
- Bussey H., Kaback D. B., Zhong W., Vo D. T., Clark M. W., et al. , 1995. The nucleotide sequence of chromosome I from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92: 3809–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey H., Storms R. K., Ahmed A., Albermann K., Allen E., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XVI. Nature 387: 103–105 [PubMed] [Google Scholar]
- Carlson M., Botstein D., 1983. Organization of the SUC gene family in Saccharomyces. Mol. Cell. Biol. 3: 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher C., Bowman S., Badcock K., Bankier A., Brown D., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome IX. Nature 387: 84–87 [PubMed] [Google Scholar]
- Clayton R. A., White O., Ketchum K. A., Venter J. C., 1997. The first genome from the third domain of life. Nature 387: 459–462 [DOI] [PubMed] [Google Scholar]
- Deshpande A. M., Newlon C. S., 1992. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 4305–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich F. S., Mulligan J., Hennessy K., Yelton M. A., Allen E., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome V. Nature 387: 78–81 [PMC free article] [PubMed] [Google Scholar]
- Dujon B., 1992. Altogether now—sequencing the yeast genome. Curr. Biol. 2: 279–281 [DOI] [PubMed] [Google Scholar]
- Dujon B., 1996. The yeast genome project: what did we learn? Trends Genet. 12: 263–270 [DOI] [PubMed] [Google Scholar]
- Dujon B., Alexandraki D., Andre B., Ansorge W., Baladron V., et al. , 1994. Complete DNA sequence of yeast chromosome XI. Nature 369: 371–378 [DOI] [PubMed] [Google Scholar]
- Dujon B., Albermann K., Aldea M., Alexandraki D., Ansorge W., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XV. Nature 387: 98–102 [PubMed] [Google Scholar]
- Engel, S. R., and J. M. Cherry, 2013 The new modern era of yeast genomics: community sequencing and the resulting annotation of multiple Saccharomyces cerevisiae strains at the Saccharomyces Genome Database, 2013. Article ID bat012; 10.1093/ database/bat012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S. R., Balakrishnan R., Binkley G., Christie K. R., Costanzo M. C., et al. , 2010. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 38: D433–D436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi B., Hottinguer H., Tavlitski J., 1949. Action de l’acriflavine sur les levures. II. Etude genetique du mutant “petite colonie.” Ann. Inst. Pasteur (Paris) 76: 419–450 [Google Scholar]
- Feldmann H., Aigle M., Aljinocvic G., Andre B., Baclet M. C., et al. , 1994. Complete DNA sequence of yeast chromosome II. EMBO J. 13: 5795–5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk D. G., Ball C. A., Dolinski K., Engel S. R., Hong E. L., et al. , 2006. Saccharomyces cerevisiae S288C genome annotation: a working hypothesis. Yeast 23: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Roganti T., Lecrenier N., Purnelle B., 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440: 325–331 [DOI] [PubMed] [Google Scholar]
- Galibert F., Alexandraki D., Baur A., Boles E., Chalwatzis N., et al. , 1996. Complete nucleotide sequence of Saccharomyces cerevisiae chromosome X. EMBO J. 15: 2031–2049 [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Vassarotti A., 1991. The European project for sequencing the yeast genome. Res. Microbiol. 142: 901–903 [DOI] [PubMed] [Google Scholar]
- Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., et al. , 1996. Life with 6000 genes. Science 274: 546–567 [DOI] [PubMed] [Google Scholar]
- Gordon J. L., Byrne K. P., Wolfe K. H., 2009. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 5: e1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K., Fukuda H., Kichise K., Kitano K., Hara S., 1991. Cloning and nucleotide sequence of the KHS killer gene of Saccharomyces cerevisiae. Agric. Biol. Chem. 55: 1953–1958 [PubMed] [Google Scholar]
- Hall C., Dietrich F. S., 2007. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics 177: 2293–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D. C., 1956. The genetics of galactose fermentation in Saccharomyces hybrids. C. R. Trav. Lab. Carlsberg., Ser. Physiol. 26: 149–160 [Google Scholar]
- Hawthorne D. C., Mortimer R. K., 1960. Chromosome mapping in Saccharomyces: centromere-linked genes. Genetics 45: 1085–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann J. H., Fleig U. N., 1993. The centromere of budding yeast. Bioessays 15: 451–460 [DOI] [PubMed] [Google Scholar]
- Hieter P., Pridmore D., Hegemann J. H., Thomas M., Davis R. W., et al. , 1985. Functional selection and analysis of yeast centromeric DNA. Cell 42: 913–921 [DOI] [PubMed] [Google Scholar]
- Hinrichs A. S., Karolchik D., Baertsch R., Barber G. P., Bejerano G., et al. , 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34: D590–D598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J. E., Balakrishnan R., Christie K. R., Costanzo M. C., Dwight S. S., et al. , 2006. Genome Snapshot: a new resource at the Saccharomyces Genome Database (SGD) presenting an overview of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 34: D442–D445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq C., Alt-Morbe J., Andre B., Arnold W., Bahr A., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome IV. Nature 387: 75–78 [PubMed] [Google Scholar]
- Jacquier A., Legrain P., Dujon B., 1992. Sequence of a 10.7 kb segment of yeast chromosome XI identifies the APN1 and the BAF1 loci and reveals one tRNA gene and several new open reading frames including homologs to RAD2 and kinases. Yeast 8: 121–132 [DOI] [PubMed] [Google Scholar]
- Johnston M., Andrews S., Brinkman R., Cooper J., Ding H., et al. , 1994. Complete nucleotide sequence of Saccharomyces cerevisiae chromosome VIII. Science 265: 2077–2082 [DOI] [PubMed] [Google Scholar]
- Johnston M., Hillier L., Riles L., Albermann K., Andre B., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XII. Nature 387: 87–90 [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Vanguri S., Boeke J. D., Gabriel A., Voytas D. F., 1998. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464–478 [DOI] [PubMed] [Google Scholar]
- Klapholz S., Esposito R. E., 1982. Chromosomes XIV and XVII of Saccharomyces cerevisiae constitute a single linkage group. Mol. Cell. Biol. 2: 1399–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T., Kojima H., Miyakawa I., Sando N., 1984. Meiotic karyotype of the yeast Saccharomyces cerevisiae. Exp. Cell Res. 153: 259–265 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Li W.-H., 2011. Expansion of hexose transporter genes was associated with evolution of aerobic fermentation in yeasts. Mol. Biol. Evol. 28: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindegren C. C., 1949. The Yeast Cell: Its Genetics and Cytology. Education Publishers Inc., St. Louis, Missouri [Google Scholar]
- Lindegren C. C., Lindegren G., 1951. Linkage relationships in Saccharomyces of genes controlling the fermentation of carbohydrates and the synthesis of vitamins, amino acids and nucleic acid components. Indian Phytopathol. 4: 11–20 [Google Scholar]
- Lindegren C. C., Lindegren G., Shult E. E., Desborough S., 1959. Chromosome maps of Saccharomyces. Nature 183: 800–802 [DOI] [PubMed] [Google Scholar]
- Link A. J., Olson M. V., 1991. Physical map of the Saccharomyces cerevisiae genome at 110-kilobase resolution. Genetics 127: 681–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti H., Styles C. A., Fink G. R., 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144: 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Maury L., Marguerat S., Bahler J., 2008. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 9: 583–593 [DOI] [PubMed] [Google Scholar]
- Louis E. J., Naumova E. S., Lee A., Naumov G., Haber J. E., 1994. The chromosome end in yeast: Its mosaic nature and influence on recombinational dynamics. Genetics 136: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R., 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R., 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171 [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D., 1980. Genetic map of Saccharomyces cerevisiae. Microbiol. Rev. 44: 519–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Johnston J. R., 1986. Genealogy of principal strains of the Yeast Genetic Stock Center. Genetics 113: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer, R. K., C. R. Contopoulou, and J. S. King, 1992 Genetic and physical maps of Saccharomyces cerevisiae, edition 11. Yeast 8: 817–902. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Naitou M., Hagiwara H., Shibata T., Ozawa M., et al. , 1995. Analysis of the nucleotide sequence of chromosome VI from Saccharomyces cerevisiae. Nat. Genet. 10: 261–268 [DOI] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., et al. , 1992. The complete DNA sequence of yeast chromosome III. Nature 357: 38–46 [DOI] [PubMed] [Google Scholar]
- Olson M. V., Dutchik J. E., Graham M. Y., Bordeur G. M., Helms C., et al. , 1986. Random-clone strategy for genomic restriction mapping in yeast. Proc. Natl. Acad. Sci. USA 83: 7826–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura F., Hatanaka H., Nakao Y., 2007. Characterization of a novel tyrosine permease of lager brewing yeast shared by Saccharomyces cerevisiae strain RM11–1a. FEMS Yeast Res. 7: 1350–1361 [DOI] [PubMed] [Google Scholar]
- Percudani R., Pavesi A., Ottonello S., 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 268: 322–330 [DOI] [PubMed] [Google Scholar]
- Philippsen P., Kleine K., Pohlmann R., Dusterhoft A., Hamberg K., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature 387: 93–98 [PubMed] [Google Scholar]
- Planta R. J., Mager W. H., 1998. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14: 471–477 [DOI] [PubMed] [Google Scholar]
- Pomper S., 1952. Purine-requiring and pyrimidine-requiring mutants of Saccharomyces cerevisiae. J. Bacteriol. 63: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomper S., Burkholder P. R., 1949. Studies on the biochemical genetics of yeast. Proc. Natl. Acad. Sci. USA 35: 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume S. E., Tatum E. L., 1949. Spontaneous and nitrogen mustard-induced nutritional deficiencies in Saccharomyces cerevisiae. Arch. Biochem. 22: 331–338 [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., et al. , 1993. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics 134: 81–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runguphan W., Keasling J. D., 2013. Metabolic engineering of saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab Eng. pii S1096–S7176(13)00067–0. 10.1016/j.ymben.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Shirahige K., Iwasaki T., Rashid M. B., Ogasawara N., Yoshikawa H., 1993. Location and characterization of autonomously replicating sequences from Chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 5043–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwa P., Korona R., 2005. Loss of dispensable genes is not adaptive in yeast. Proc. Natl. Acad. Sci. USA 102: 17670–17674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szybalski W., 2001. My road to Øjvind Winge, the father of yeast genetics. Genetics 158: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Agostoni Carbone M. L., Albermann K., Albers M., Arroyo J., et al. , 1997. The nucleotide sequence of Saccharomyces cerevisiae chromosome VII. Nature 387: 81–84 [PubMed] [Google Scholar]
- Thierry A., Fairhead C., Dujon B., 1990. The complete sequence of the 8.2 kb segment left of MAT on Chromosome III reveals five ORFs, including a gene for a yeast ribokinase. Yeast 6: 521–534 [DOI] [PubMed] [Google Scholar]
- Vassarotti A., Goffeau A., 1992. Sequencing the yeast genome: the European effort. Trends Biotechnol. 10: 15–18 [DOI] [PubMed] [Google Scholar]
- Vassarotti A., Dujon B., Mordant P., Feldmann H., Mewes W., et al. , 1995. Structure and organization of the European Yeast Genome Sequencing Network. J. Biotechnol. 41: 131–137 [DOI] [PubMed] [Google Scholar]
- Wenger J. W., Schwartz K., Sherlock G., 2010. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 6: e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., Ricupero-Hovasse S. L., 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Yang C., Theis J. F., Newlon C. S., 1999. Conservation of ARS elements and chromosomal DNA replication origins on Chromosomes III of Saccharomyces cerevisiae and S. carlsbergensis. Genetics 152: 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.