Abstract
Oropharyngeal squamous cell carcinoma (OPSCC) is associated with human papillomavirus (HPV). HPV-positive OPSCC is considered a distinct molecular entity with a better prognosis than HPV-negative cases of OPSCC. However, the exact pathogenic mechanisms underlying the differences in clinical and molecular behavior between HPV-positive and HPV-negative OPSCC remain poorly understood. Epigenetic events play an important role in the development of cancer. Hypermethylation of DNA in promoter regions and global hypomethylation are 2 epigenetic changes that have been frequently observed in human cancers. It is suggested that heterogeneous epigenetic changes play a role in the clinical and biological differences between HPV-positive and HPV-negative tumors. Unraveling the differences in methylation profiles of HPV-associated OPSCC may provide for promising clinical applications and may pave the road for personalized cancer treatment. This systematic review aims to assess the current state of knowledge regarding differences in promoter hypermethylation and global methylation between HPV-positive and HPV-negative OPSCC.
Keywords: head and neck cancer, oropharynx, epigenetic, HPV, methylation, systematic review
Background
Epigenetic alterations refer to heritable changes in genes expression that occur without changes in the underlying DNA sequence.1,2 Methylation of DNA is an epigenetic modification and can occur at sites where cytosine is followed by guanine (CpG dinucleotide). Epigenetic regulation plays a central role in embryogenesis and cell type differentiation.3-6 Hypermethylation of CpG dinucleotides, particularly in promoter regions, and global hypomethylation are both frequently observed in human cancer. Promoter hypermethylation (further denoted “promoter methylation”) of tumor suppressor genes is the best characterized epigenetic change and leads to transcriptional silencing of the gene.7-9 In cancer, gene silencing by promoter methylation may occur even more frequently than structural inactivation of genes by mutations.4,8 In contrast to genetic events, DNA methylation is reversible and, therefore, an attractive target for new therapeutic strategies.10 The effect of genomic hypomethylation is less clear. It is thought to contribute to chromosomal instability and to activate the expression of proto-oncogenes.4,5,7,11
Head and neck squamous cell carcinoma (HNSCC) is the sixth leading cancer in the world with a reported incidence of over 600 000 new cases a year. Traditional risk factors for HNSCC are excessive alcohol and tobacco consumption. In the last decades, preventive strategies targeting these risk factors have resulted in a significant decline in the incidence of HNSCC. The greatest decrease was observed in the incidence of carcinomas of the larynx and hypopharynx. In contrast to these promising figures, the incidence of oropharynx squamous cell carcinoma (OPSCC) is currently increasing, particularly in young patients without a history of smoking or excessive alcohol consumption. This absence of traditional risk factors suggests the involvement of an additional risk factor.12-14 During the past years, human papillomavirus (HPV) has been identified as an additional independent risk factor for the development of HNSCC, particularly OPSCC. HNSCC testing positive for HPV, in particular HPV type 16, is considered a distinct molecular and clinical entity with a better response to radiotherapy and overall survival when compared with HPV-negative cases of HNSCC.15-17 One of the most important pathogenic mechanisms underlying these differences in clinical behavior pertains to the inactivation of tumor suppressor gene p53. In HPV-negative tumors inactivation of p53 is mainly due to mutations in the gene, whereas in HPV-positive tumors, inactivation of p53 occurs through oncoprotein E6, leaving the gene functionally intact.18-20
Therefore, wild type p53 can potentially be re-activated after irradiation, causing apoptosis of tumor cells. Cell line studies have recently proven that this pathway results in the improved outcome of HPV-positive OPSCC.21 In addition, studies investigating gene expression in HPV-positive and HPV-negative HNSCC show distinct transcriptomic patterns, which might be partly due to viral E6- and E7-mediated alterations in the genome and epigenome.22-25 One group of different expressed genes is that of DNA methyltransferases, involved in regulation of DNA methylation.26,27
Differences in methylation profiles between HPV-positive and HPV-negative OPSCC have been explored in many studies, though most promoter methylation studies have evaluated only a limited number of genes and did not focus on solely the oropharynx. This is important as HPV has a distinct association with the oropharynx. A few studies reported epigenome-wide analysis of DNA methylation in HPV-positive and HPV-negative HNSCC. In order to summarize current knowledge regarding differences in methylation profiles between HPV-positive and HPV-negative OPSCC, we conducted a systematic review aimed at delineating future research directions. Evidence provided by this review could enhance characterization of methylation profiles of HPV-positive and HPV-negative OPSCC and may thus lead to the introduction of new biomarkers in the light of personalized cancer care.
Results
Search results
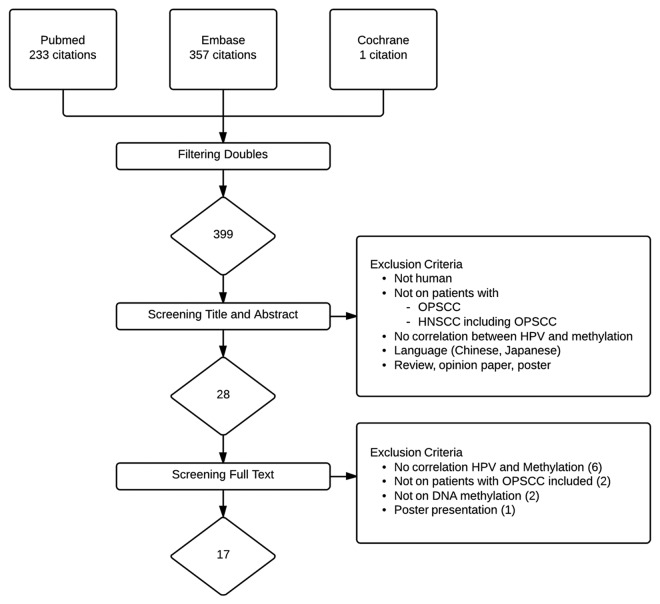
Combined PubMed, Embase, and Cochrane searches retrieved 399 unique articles (Fig. 1). After screening titles and abstracts, 28 articles remained. The remaining 28 articles were retrieved in full text for formal review. Seventeen articles met the inclusion criteria and were eligible for further analysis.28-44 The main reasons for exclusion were duplicates and studies not investigating correlation between HPV status and methylation status in OPSCC.
Figure 1.
Flowchart of included and excluded studies.
Study characteristics
The number of patients included for methylation analysis across all studies varied from 5 to 350. Four studies dealt with OPSCC only28,33,34,44. In other publications studying overall HNSCC, OPSCC subgroup size ranged from 9% to 80% of the total study population.29-32,35-43 There was only one study that reported the distribution of average age, nicotine and alcohol consumption between HPV-positive and HPV-negative tumors of which promoter methylation was evaluated.34 The mean age in HPV-positive tumors was 56.9 compared with 58.4 in HPV-negative tumors. In the group of HPV-positive tumors, 38% of the patients had ever smoked in comparison with 62% of HPV-negative tumors. Excessive alcohol consumption was more frequent in the patients with HPV-negative tumors (62% vs. 10% in HPV-positive tumors). Eight studies detected methylation in HPV-positive and HPV-negative primary tumors using formalin-fixed paraffin-embedded (FFPE) specimen.28,29,32,34-37,40 Six used fresh frozen (FF) tissue or both.31,38,41-44 Three studies did not report used material.30,33,39 The majority of studies used polymerase chain reaction (PCR) to detect HPV 16 DNA. Three studies 34,41,43 used PCR and immunohistochemistry (IHC) and Poage et al. used multiplex serology.38 Sartor et al. did not describe their method to detect HPV DNA.40 There was a wide variation in percentage of HPV positivity between studies ranging from 7% to 56%. The studies used varying techniques to determine gene methylation; most studies used methylation-specific PCR (MSP), while other studies used combined bisulfite restriction analysis (COBRA),28,32 pyrosequencing analysis (PMA),38,39 bead array method (BAM),29,34,40 or quantitative MSP (qMSP).33,42,44 Thirteen studies evaluated promoter methylation in selected genes in association with HPV status.28-31,33-37,41-44 Three studies investigated global methylation in association with HPV status.32,38,39 One study examined both promoter methylation and global methylation, in association with HPV status.40 Promoter methylation was interpreted differently among studies. Several studies used a methylation cut-off point to determine hypermethylation,42,43 while others compared methylation percentages or mean methylation differences percentage.29,44 Some studies compared the tumor methylation with methylation status of adjacent normal tissue.28 Most studies did not mention how methylation was interpreted.30-41 The main features of the 17 studies eligible for systematic review are summarized in Table 1.
Table 1. Characteristics of studies included in research analysis.
| Year | Country | First author | N | Tumor location (% OPSCC) | Material | HPV detecting method | HPV -positive (%) | Method | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 2010 | USA | Bennett28 | 16 | Oropharynx | FFPE | PCR (E6) | 56 | COBRA | PH |
| 2013 | USA | Colacino29 | 68 | HNSCC (47) | FFPE | PCR (E6) | 35*** | BAM | PH |
| 2003 | USA | Dong30 | 46 | HNSCC (57) | NA | PCR (E6,E7) | 35 | MSP | PH |
| 2012 | Brasil | Freitas Cordeira- Silva31 | 45 | Oral cavity and oropharynx (9) | FFPE, FF | Nested PCR | 7 | MSP | PH |
| 2008 | USA | Furniss32 | 193 | HNSCC (NA) | FFPE | PCR (L1) | 24 | COBRA | GM |
| 2013 | Sweden | Gubanova33 | 5 | Oropharynx | NA | PCR (L1, E6, E7) | 40 | qMSP | PM |
| 2013 | Germany | Kostareli44 | 170* | Oropharynx | FF | PCR | 25 | qMSP | PM |
| 2013 | UK | Lechner34 | 32 | Oropharynx | FFPE | IHC, qPCR (E6) | 56 | BAM | PM |
| 2006 | USA | Marsit35 | 350 | HNSCC (25) | FFPE | PCR (L1) | 19 | MSP | PM |
| 2008 | USA | Marsit36 | 340 | HNSCC (25)*** | FFPE | PCR (L1) | 26 | MSP | PM |
| 2008 | USA | O’Regan37 | 24 | Oral cavity and oropharynx (46) | FFPE | PCR (L1) | 42 | MSP | PM |
| 2011 | Germany | Poage38 | 138 | HNSCC (20)* * | FF | Multiplex serology E6 | 12 | PMA | GM |
| 2009 | USA | Richards39 | 26 | HNSCC (31) | NA | PCR (E6) | 31 | PMA | GM |
| 2011 | USA | Sartor40 | 49 46 |
HNSCC (65) Oral cavity and oropharynx (NA) |
FFPE NA |
NA NA |
43 40 |
BAM NA |
PM GM |
| 2009 | USA | Taioli41 | 88 | Oral cavity and oropharynx (43) | FF | IHC, Nested PCR (GP5+/6+) | 24 | MSP | PM |
| 2011 | Germany | Weiss42 | 55 | HNSCC (78) | FF.FFPE | PCR (E6, E7) | 46 | qMSP | PM |
| 2013 | Germany | Weiss43 | 34 | HNSCC (80) | FF, FFPE | IHC, PCR (GP5+/6+) | 44 | MSP | PM |
N, number of patients studied; NA, not available; FFPE, formalin-fixed paraffin embedded; FF, fresh frozen; IHC, immunohistochemistry; COBRA, combined bisulfite restriction analysis; BAM, Bead array method; MSP, methylation specific polymerase chain reaction; qMSP, quantitative methylation specific polymerase chain reaction; PMA, pyrosequencing analysis; PH, promoter hypermethylation; GM, global methylation. *Initial screened in 15 samples and validated in two larger cohorts. **Tumor site pharynx not further subdivided, ***HPV positive includes also HPV 18, 35, and 59.
Gene promoter methylation and HPV-status
A summary of genes with promoter methylation values showing significant differences between HPV-positive and HPV-negative tumors among included studies is shown in Table 2.
Table 2. Genes with DNA promoter methylation values significantly associated with HPV status of tumor in included studies.
| Hallmark45 | Gene | locus | Gene function | Hypermethylated in | P value | Reference |
|---|---|---|---|---|---|---|
| 1. Sustaining proliferative signaling | CCNA1 | 13q13.3 | Regulator Cell cycle | HPV-positive |
P < 0.01 p < 0.05 |
Weiss ‘11 Colacino |
| GRB7 | 17q12 | Regulator cell growth and migration | HPV-positive | P < 0.001 | Colacino | |
| RASSF1 | 3p21 | RAS pathway regulation | HPV-negative |
P < 0.05 P < 0.05 NS NS |
Colacino Dong Weiss ‘11 Taioli |
|
| STAT5a | 17q11.2 | Stimulating proliferation and preventing apoptosis | HPV-negative | P < 0.05 | Colacino | |
| ALDH1A2 | 15q22.1 | Regulates synthesis of retinoic acid; major role in regeneration and differentiation | HPV-negative | P < 0.001 | Kostareli | |
| OSR2 | 8q22.2 | Mediates expression of downstream genes involved in cell proliferation and the cell cycle | HPV-negative | P < 0.001 | Kostareli | |
| (2) Activating invasion and metastasis | SYBL1 | Xq28 | MT1-MMP-dependent matrix degradation | HPV-positive | P < 0.05 | Colacino |
| SPDEF | 6p21.3 | Enhance cell motility, invasion, and growth | HPV-negative | P < 0.05 | Colacino | |
| TIMP3 | 2q12.1-q13.2|22q12.3 | Inhibition of tumor growth and angiogenesis | HPV-positve | P < 0.05 | Weiss ‘11 | |
| SFRP4 | 7p14.1 | WnT pathway antagonist | HPV-positive | P < 0.05 | Marsit | |
| CDH11 | 16q22 | Cell-cell adhesion | HPV-positive | P > 0.05 | Colacino | |
| (3) Genome instability and mutation | MGMT | 10q26 | Guanine alkylation repair/ DNA repair | HPV-negative |
P < 0.05 NS NS |
Colacino Weis’11 Taioli |
| (4) Resisting cell death | ESR2 | 14q23.2 | Positive regulation of apoptotic process | HPV-negative | P < 0.05 | Colacino |
| (5) Tumor promoting inflammation | JAK3 | 19p13 | Cytokine receptor-mediated intracellular signal transduction | HPV-positive | P < 0.05 | Colacino |
| (6) Deregulating cellular energetics | HSD17B12 | 11p11.2 | E1/E2 conversion, fatty acid elongation | HPV-negative | P < 0.05 | Colacino |
| TUSC3 | 8p22 | Regulating glycosylation efficiency | HPV-positive | P < 0.05 | Colacino | |
| (7) Evading growth suppressors | RUNX1T1 | 8q22 | Interact with DNA-bound transcription factors | HPV-positive | P < 0.05 | Colacino |
| TCF21 | 6q23-q24 | Cell fate Differentiation | HPV-positive |
P < 0.05* NS |
Weiss ‘13 Weiss ‘11 | |
| IRX4 | 5p15.3 | Interaction with vitamin D receptor | HPV-positive | P < 0.001 | Kostareli | |
| GATA4 | 10q26.11 | Promote cell differentiation | HPV-positive | P < 0.001 | Kostareli | |
| GFRA1 | 10q26.11 | Activation of the RET tyrosine kinase receptor | HPV-positive | P < 0.001 | Kostareli |
NS, not significant; *Significant in univariate analysis.
Four studies investigated promoter methylation status in correlation with HPV in OPSCC.28,33,34,44 Bennet et al. explored promoter methylation in 5 selected genes (IRX1, EBF3, SLC5A8, SEPT9, and FUSSEL18) in a group of 16 OPSCC.28 No statistical difference was found in promoter methylation for these 5 genes between the HPV-positive and HPV-negative tumors. Gubanavo et al. hypothesized that better response to chemotherapy and radiotherapy in HPV-positive OPSCC may be due to the ability of cancer cells to respond to DNA damage.33 An important protein in DNA damage response is SMG-1. Methylation status of SMG-1 was investigated in 3 HPV-negative and 2 HPV-positive OPSCC. The mean prevalence of promoter methylation in HPV negative lesions was 5% and in HPV-positive lesions was 43%. The study of Kostareli et al. investigated genome wide promoter methylation in a cohort of 15 OPSCC.44 OPSCC positive for DNA and RNA of HPV were considered as HPV-positive. Promoter methylation levels of 15 genes (ALDH1A2, FKBP4, GDNF, OSR2, PROX1, WIF1, BDNF, EOMES, GATA4, GFRA1, GRIA4, HOCA13, IRX4, SOX1, TBX5) were significantly different between HPV-positive and HPV-negative OPSCC. Methylation status of 5 genes (ALDH1A2, OSR2, GATA4, GFRA4, and IRX4) was further validated in 2 independent cohorts of respectively 85 patients and 70 patients. Both cohorts consisted of patients with OPSCC with known HPV status. In both cohorts, promoter methylation of the 5 selected genes were significantly different between HPV-positive and HPV-negative tumors. Lechner et al. evaluated promoter methylation status of individual CpGs in 32 OPSCC, which were validated in cell lines and samples using 2 different analysis methods.34 HPV-positive tumors showed significantly more hypermethylation, most prominently at promoter regions. Promoter methylation was found in 43 genes according to 3 data sets including cadherin genes.
Nine articles studied promoter methylation status in correlation with HPV status in HNSCC. All 9 included the oropharynx as a subsite.29-31,35-37,41-43 Colacino et al. included 68 HNSCC tumor samples of which 47% was obtained from the oropharynx.29 DNA methylation was measured at 1505 CpG sites across 807 genes and correlated with HPV status. Promoter methylation in 13 CpG sites was significantly associated with HPV status: CCNA1, GRB7, CDH11, RUNX1T1, SYBL1, and TUSC3 were found to be more frequently methylated in HPV-positive tumors. SPDEF, RASSF1, STAT5a, MGMT, ESR2, JAK3, and HSD17D12 were hypomethylated in HPV-positive tumors. Dong et al. investigated promoter methylation of RASSF1A in 46 HNSCC samples and found a significant inverse correlation between RASSF1A promoter methylation and HPV positivity comparable to the results of Colacino et al.29,30 The study of De Freitas Coreidro-Silva et al. included 45 oral cavity and oropharyngeal SCC of which 9% derived from oropharynx with low HPV prevalence.31 Four genes (CDKN2A, EDNRB, RUNX3, and SFN) were selected for determining their promoter methylation status. No association was observed between promoter methylation and HPV status. O’Regan et al. evaluated promoter methylation of CDKN2A in association with HPV status in a cohort of 24 oral cavity and oropharyngeal SCC.37 No correlation was found between HPV status and promoter methylation, concurrent to the results of previous studies.30,31 A third study by Taioli et al. also investigated promoter methylation of CDKN2A in a study population of 88 oral cavity and oropharyngeal SCC. No correlation was observed between HPV status and promoter methylation status.41 However, it was remarkable that all of the CDKN2A promoters of HPV-positive tumors were not methylated. Methylation of 2 other genes, MGMT and RASSF, were simultaneously analyzed in the same study. For both genes no association was found between HPV status and promoter methylation, in contrast to the results of Dong et al. and Colacino et al.29,30 The study of Marsit et al. (2006) was the largest study with 350 patients with HNSCC of which 25% derived from oropharynx.35 Methylation of 4 soluble frizzled receptor protein (SFRP) genes was determined. Methylation of SFRP4 was found to be significantly associated with HPV status. In another study, Marsit et al. (2008) found no association between promoter methylation of cadherin 1 type 1 (CDH1) and HPV status in a cohort of 340 HNSCC patients.36 Weiss et al. (2011) studied promoter methylation of 12 genes (TIMP3, CDH1, CDKN2A, DAPK1, TCF21, CD44, MLH1, MGMT, RASSF1, CCNA1, LARS2, CEPBA) in a cohort of 55 HNSCC, mostly derived from oropharynx.42 HPV-positive tumors showed significant promoter methylation of CCNA1 and TIMP3. Promoter methylation patterns of MGTMT and RASSF1 showed no correlation with HPV status, comparable to the results of Tailoli et al. but in contrast to Dong et al. and Colacino et al.29,30,41 The second included study by Weiss et al. (2013) evaluated TCF21 promoter methylation in 34 HNSCC of which 80% derived from oropharynx.43 Promoter methylation frequency of TCF21 was found to be higher in HPV positive tumors, although results from multivariate analysis were not statistically significant.
Four studies investigated correlation of gene promoter methylation and clinical outcome.29,36,41,44 Kosterali et al. found HPV-related promoter methylation state of 5 genes (ALDH1A2, OSR2, GATA4, GFRA4, and IRX4), which correlated with progression free survival (PSF) and overall survival (OS) in OPSCC. A scoring system was developed, ranging from 0 to 5 where the favorable prognostic pattern scored 1 per gene and the unfavorable pattern scored 0 per gene. A score ranging from 3 to 5 correlated with a good clinical outcome and a low score of 0 to 2 correlated with worse outcome. The first line of therapy was 73% surgery, 22% a combination of radiotherapy and chemotherapy and, in 5%, unknown. This methylation signature score was validated and confirmed in 2 independent cohorts, independent of treatment and HPV status.44 Taioli et al. studied the methylation of 3 tumor suppressor genes in correlation with OS and tumor recurrence in oral and pharyngeal cancer. Most patients had surgery (50%) or surgery in combination with radiotherapy (44.3%) as initial therapy. MGMT promoter methylation was found to be significantly associated with poorer outcome, indicating that MGMT may serve as a prognostic biomarker.41 In the multivariate analyses adjustments were done for treatment. Colacino et al. investigated methylation at individual CpG sites in correlation with 3 y OS. From included patients, 45% received a combination of radiotherapy and chemotherapy and 21% underwent additional surgery. Notch 1 was found to be the strongest predictor for improved survival in this study.29 Marsit et al. (2008) reported that hypermethylation of CDH1 was an independent predictor for improved OS in HNSCC. Treatment of the patients in this cohort was not reported.36
Global DNA methylation and HPV-status
Four studies investigated global DNA methylation in correlation with HPV status in HNSCC including the oropharynx.32,38-40 Furniss et al. studied global methylation status in 193 HNSCC using the relative methylation status of long interspersed repeat sequences (LINE-1) retrotransposon (LRE1).32 Unfortunately, Furniss et al. did not report tumor location in studied HNSCC; therefore, the percentage of oropharynx carcinomas is unclear. HPV presence showed no association with methylation of LRE1. The study of Richards et al. evaluated global methylation represented by levels of LINE-1 methylation in 26 HNSCC, of which 31% derived from the oropharynx.39 HPV-positivity showed a positive correlation with normal level of LINE methylation. Sartor et al. studied differences in methylation patterns between HPV-positive and HPV-negative tumors in 49 oral cavity and oropharynx SCC in comparison with 4 cell lines (2 HPV-negative OPSCC, 1 HPV-positive OPSCC, and 1 cervix carcinoma).40 Overall, HPV-positive tumors showed significantly higher levels of CpG island methylation in tested samples compared with HPV-negative tumors. Unfortunately, differences in methylation patterns of promoter regions in investigated genes between HPV-positive and HPV-negative SCC were not reported for primary tumors. Another cohort of 34 oral cavity and oropharynx SCC was evaluated for global methylation using LINE-1 elements. In line with results from Richards et al., there was a significant association between HPV-positive tumors and higher levels of LINE-1 methylation elements.39 In the study of Poage et al., global methylation of 138 HNSCC was assessed by pyrosequencing of LINE-1 and Aluyb8 elements, and by luminometric methylation assay (LUMA).38 The HPV prevalence was low in this study and 20% of HNSCC was derived from the pharynx. The exact amount of oropharyngeal tumors in this study was not reported. HPV status showed a significant correlation with an increase in methylation of LINE-1 elements as reported in previous studies and with LUMA methylation. Results of the 4 studies are summarized in Table 3. Only Furniss et al. investigated the correlation between global methylation and clinical outcome. A poorer survival was found (P < 0.07, not significant) among HPV-negative individuals with lower LRE1 methylation. There was no association with survival and LRE1 methylation in patients with HPV-positive tumors. The distribution of various therapies was not mentioned.32
Table 3. Global measures of methylation in association with HPV status.
| Study | Method | Hypomethylated in | P value |
|---|---|---|---|
| Furniss et al.32 | LRE1 | HPV-negative | NS |
| Richards et al.39 | LINE-1 | HPV-negative | P < 0.05 |
| Sartor et al.40 | LINE-1 | HPV-negative | P < 0.01 |
| Poage et al.38 | LINE-1 LUMA |
HPV-negative HPV-negative |
P < 0.05 P < 0.05 |
LINE-1, long interspersed nucleotide elements; LUMA, luminometric methylation assay; NS, not significant.
Discussion
Epigenetic events are important in carcinogenesis of head and neck squamous cell carcinoma.8 Unraveling the role of epigenetics in HPV-positive and HPV-negative OPSCC is vital for a better understanding of differences in clinical behavior and carcinogenesis between both tumors as well as identification of potential biomarkers for treatment and prognosis.46,47 Additionally, it will result in better classification of patients with OPSCC and different treatment of HPV-positive and HPV-negative tumors. We systematically reviewed the literature for studies assessing the association between promoter methylation or global methylation and HPV status of OPSCC. To our knowledge, this is the first systematic review to determine differences in methylation profiles between HPV-positive and HPV-negative OPSCC.
Gene promoter methylation and HPV-status
Hypermethylation of promoter region is an important mechanism for gene silencing and thought to be an early event in carcinogenesis.9 Several tumor suppressor genes in HNSCC are identified to be frequently hypermethylated. These methylation patterns persist and usually increase during disease progression which makes them a suitable tool to obtain predictive or prognostic information.48 The first striking point of our search was the sparseness of literature studying promoter methylation of OPSCC in correlation with HPV status. Only 4 studies investigated promoter methylation of individual genes in a study population of only OPSCC,28,33,34,44 which is known to be associated with HPV.14-16 Because of the lack of studies investigating promoter methylation in association with HPV status in solely OPSCC, we decided to include studies investigating this association in a mixture of subsites in head and neck regions, including the oropharynx. Thirteen extra articles were included29-32,35-43 of which 9 studied promoter methylation in individual genes.29-31,35-37,41-43 Although numerous genes have been investigated, only the genes CCNA1, RASSF1, TCF21, CDH1, p16, and MGMT were analyzed in two or more studies. Promoter methylation of CDH1 and p16 showed very similar results and was not associated with HPV 16 in all studies investigating those genes.31,36,37,41,42 Noteworthy, 2 studies investigating promoter methylation of p16 showed no methylation of p16 in any of the HPV-positive samples,37,41 which is in line with overexpression of p16 in HPV-positive tumors.49 Both studies investigating CCNA1 showed similar results, despite differences in analysis method and small sample size, suggesting promoter methylation of CCNA1 might be induced by HPV 16.29,42 Studies examining promoter methylation of RASSF1, TCF21, and MGMT in association with HPV status, showed varying results.29,31,41,42 These variations most probably result from the differences in sensitivity of the methods used for methylation analyses and composition of the patient cohorts. Overall, HPV-positive tumors seem to have a greater association with promoter methylation compared with HPV-negative tumors. This could be explained by overexpression and increased activation of DNA methyltransferases DNMT1 and DNMT3b, which are both key proteins in regulation of DNA methylation. These events may be due to suppression of tumor suppressor genes p53 and retinoblastoma protein (Rb) by HPV oncoproteins E6 and E7.26,27,50,51 However, there was a wide variation in reported HPV prevalence between studies. In the selected studies, the prevalence of HPV ranged from 7% to 56%. These varying results may be due to geographical and ethnical differences among studied populations, variation in origin of studied material, as well as different detection methods. Epidemiological studies have demonstrated differences in HPV prevalence among countries, reaching over 90% of HPV-positive OPSCC in Scandinavian countries52-54 Additionally, HPV has a greater association with the oropharynx than with other subsites of the head and neck region, such as the oral cavity or larynx.15 Thirteen studies included for review studied a mixture of subsites in the head and neck region, including the oropharynx. This variety in localization resulted in a lower prevalence of HPV-positive tumors. Another important explanation for heterogeneity of results of selected studies is the variation in used detection methods and examined tissue (Table 1). HPV-positive DNA does not imply a biologically active virus directly contributing to carcinogenesis. HPV only plays a role in carcinogenesis when the viral oncogenes E6 and E7 are expressed.55,56 In addition, Kosterali et al. showed, with respect to aberrant methylation changes, that tumors positive for solely HPV DNA were more similar to HPV negative tumors than tumors positive for both HPV DNA and RNA. This confirms the importance of E6/E7 RNA for the carcinogenic process.44 A validated algorithm based on a combination of p16 IHC and GP5+/6+ PCR has the highest sensitivity and specificity for detection of biological active HPV in FFPE and is currently considered the golden standard.57 Some, though not all, of the selected studies used this method to determine HPV status.41,43 Most studies used PCR methods for HPV-amplification, which are highly sensitive and may lead to over identification of the virus. Moreover, the variety in molecular assays used (each having its own sensitivity and specificity) most probably contributed to variations in HPV prevalence. In addition PCR techniques are more efficient on FF than on FFPE, because fixation leads to shorter DNA fragments.58
In these combined analyses, genes involved in evading growth suppression and activating invasion and metastasis were found to be more frequently methylated in HPV-positive OPSCC compared with HPV-negative OPSCC. HPV-negative OPSCCs were more frequently methylated in genes involved in pathways of genome instability and in genes resisting cell death. Limited differences in methylation between HPV-positive and HPV-negative OPSCC could be observed in gene expression and proteome, because most selected studies did not test the possible effects of this promoter methylation on gene and protein expression. Better understanding of these effects of methylation could provide for a structured framework of the molecular processes of HPV-driven OPSCC carcinogenesis. Weiss et al. (2011) reported that HPV positivity showed more methylation of the promoter region of TCF21 gene with lower TCF21 protein expression levels, indicating that HPV induces silencing of this gene by promoter methylation.42 TCF21 is known to regulate cell fate differentiation through epithelial-mesenchymal transition (EMT), a function that has been shown to be deficient in carcinomas.59 In contrast, the relationship between CCNA1 promoter hypermethylation and protein expression is not fully understood. Several studies showed no effect of promoter methylation of CCNA1 on protein and gene expression in HPV-positive HNSCC23,60 Surprisingly a strong correlation was found between HPV-positivity and overexpression of CCNA1 protein. This suggests that HPV induces both promoter hypermethylation and overexpression of CCNA1. However, Sartor et al. observed hypermethylation and lower protein expression of CCNA1 in HPV-positive cell lines in comparison with HPV-negative cell lines.40 GATA4, GRIA4, and IRX4 showed inversed correlation with transcription levels, suggesting that HPV induces promoter methylation and, consecutively, decreases transcription levels.44 GATA4 and IRX4 are related to signaling and metabolism of retinoid, which plays a role in cell differentiation and cell growth.61 The same inversed relation was found for ALDH1A2 and OSR2, which are also involved in retinoid metabolism and associated with hypermethylation in HPV-negative OPSCC.23,44 MGMT, a DNA repair protein which protects the cellular genome against mutagenic actions, is associated with hypermethylation in HPV-negative HNSCC. According to Srivenugopal et al., MGMT is downregulated in HPV-positive HNSCC, indicating that this effect in HPV-positive HNSCC is induced by another mechanism than promoter methylation.62 Overall, little is known about the correlation between HPV-related gene promoter methylation and its effect on gene and protein expression. This exact relationship is difficult to investigate as epigenetic silencing by methylation on protein and gene expression is complicated by other alterations and crosstalk.
Survival
Clinical biomarkers predicting overall survival, disease free survival or tumor recurrence are vital for the improvement of individualized cancer care. The presence of active HPV 16 in OPSCC is an important predictor for therapy response and survival.63 However, the exact mechanism underlying this difference in clinical behavior between HPV-positive and HPV-negative OPSCC remains poorly understood. Epigenetic changes as promoter methylation and global hypomethylation could be key elements in this mechanism, in addition to inactivation of p53 by oncoproteins E6 and E7. As discussed previously, associations have been found between HPV status and epigenetic changes as promoter methylation and global methylation. Additionally, Kosteraly et al. found HPV-related promoter status of 5 genes, which correlated with overall survival and progression free survival. The prognostic signature was validated in two external cohorts and appeared to be a powerful independent predictor for overall survival and progression free survival in OPSCC. This indicates that promoter methylation of HPV-related genes may be responsible for different clinical behavior of HPV-positive and HPV-negative OPSCC. Four of these genes, ALDH1A2, OSR2, GATA4, and IRX4, are involved in retinoid metabolism, suggesting that retinoid acid may play an important role in carcinogenesis of OPSCC.44 Three other studies investigated promoter methylation and clinical outcome. However, in these publications the reported associations did not contain genes with DNA methylation values significantly associated with HPV status. Also, study populations consisted of OPSCC and other HNSCC.29,36,41
Recent studies showed the involvement of global hypomethylation in the pathogenesis of HNSCC.7,11,12 In cancer, abnormal demethylation in repetitive elements leads to loss of silencing. No statistically significant correlation between global methylation and survival was found in the only study investigating this correlation.32
Global DNA and HPV-status
Four studies investigated the association between global DNA methylation and HPV-status.32,38-40 Frequent hypomethylation of repetitive elements as LINE-1 is representative for global hypomethylation. This is frequently seen in various types of cancer and is correlated with chromosomal instability.12,64 Three studies showed a significant increase in hypomethylation of LINE-1 elements in HPV-negative tumors,38-40 while Furniss et al. found no association between LINE-1 methylation and HPV status.32 This could be explained by differences in use of methylation assay and study population. Overall, these studies indicate that there is more maintenance of global methylation and less genomic instability in HPV-positive tumors. Although the exact mechanism of global hypomethylation remains poorly understood, there are several theories explaining how this phenomenon occurs during carcinogenesis. One of them states that altered regulation of methylation by DNMT1 is due to decreased expression or mutation, resulting in global DNA hypomethylation. Another theory explains global hypomethylation through overexpression of the splice variant DNMT3b4, which lacks DNA methyltransferase activity, resulting in global DNA hypomethylation.65-67 Any of these hypotheses could explain differences in global methylation between HPV-positive and HPV-negative tumors.
Limitations
The results of this systematic review should be viewed within the constraints of several limitations. First, evidence is sparse and most studies evaluated few genes without validation and limited overlap between the genes in the different studies. Second, in only 4 studies the cohorts consisted explicitly of OPSCC. Third, the investigated genes, used methods for methylation assay and HPV detection varied between included studies. Fourth, large variations in interpretation of promoter methylation are reported among the various studies, which makes it difficult to compare results. Fifth, all included studies were performed retrospectively with non-consecutive enrolment of patients. This could potentially lead to biased estimations of prevalence of studied molecular characteristics. Because of this heterogeneity in methods and analyses, statistical pooling was not permitted. Hence, no overall promoter and global methylation levels could be calculated. Only results derived from the various studies could be used, which resulted in low statistical power due to the small sample size of most studies. As mentioned before, quality assessment for molecular studies are currently lacking and were therefore not performed in our systematic review. In the future, there is need for sufficient quality scoring systems for these studies, especially due to the growing demand for personalized cancer care. As such, specific molecular characteristics of tumors and biomarkers are used for clinical decision-making in the treatment of oncologic patients. In conclusion, according to the current available evidence, HPV status in OPSCC was found to be associated with hypermethylation of promoter regions of 21 genes. Promoter methylation of 13 genes was correlated with HPV-positivity, whereas 8 genes were hypermethylated in HPV-negative tumors. LINE-1 hypomethylation was more common in HPV-negative tumors compared with HPV-positive tumors. These results show that there are differences in methylation profiles between HPV-positive and HPV-negative tumors. This suggests that HPV-positive tumors are to a greater extent driven by methylation alterations in the promoter region than HPV-negative tumors, in which global hypomethylation is more frequently observed. Moreover, HPV-positive tumors showed increased methylation in genes involved in evading growth suppression and in genes involved in activating invasion and metastasis. HPV-negative tumors showed this effect in pathways of genome instability and in genes resisting cell death. We advise caution in interpreting these results because of the small number of candidate genes with overlap and the methodological differences between studies. Comparison and validation of these results was thus impaired. For future research we recommend adequately designed studies with study populations of explicitly OPSCC, uniform protocols for interpretation of promoter methylation and validation in independent cohorts to evaluate these promising results on a larger scale.
Materials and Methods
Search strategy
A systematic search was conducted in the PubMed, Embase and Cochrane databases for original articles published until 12th of June 2013. Search terms used were “oropharynx cancer,” “head and neck cancer,” “epigenetic” and their synonyms in title and abstract fields (Supplemental Materials). Citations and references of selected articles and reviews were checked to identify potentially missed relevant studies. Two authors (P.M.W.K. and R.N.) independently screened all titles and abstracts of the retrieved search for selection using predefined inclusion and exclusion criteria. Subsequently, full text of relevant studies was screened for a more detailed selection. If disagreement was encountered a 3rd doctor would decide on the disagreement.
Study selection
Inclusion criteria
Studies were selected on the basis of (1) full text publication with detailed description of used methods, (2) original reports containing data on assessment of DNA methylation status in oropharynx SCC or HNSCC with subsite oropharynx included, (3) containing a correlation between DNA methylation and HPV status of tumors (4) Using primary tumors to identify promoter methylation, because cell lines have a different methylation pattern than primary tumors.68
Exclusion criteria
Used exclusion criteria were (1) duplicate articles that contained all or some of the original publication data, (2) reviews, book chapters, cases reports, editorials, oral presentations, technical notes and poster presentations, (3) articles which included subsites other than head and neck without subgroup analysis.
Data extraction and analysis
Using a standardized data extraction form we extracted first author, year of publication, sample size, country, detecting methods, tumor location, percentage HPV positive tumors, distribution of average age, current nicotine and alcohol consumption between HPV-positive and HPV-negative tumors, testing material and outcome on methylation in association with HPV status and survival from each study. In addition data on assessment of methylation status in correlation with HPV status were extracted. Data extraction was performed independently by 2 authors (P.M.W.K and R.N.), disagreement was resolved by discussion. No quality assessment with reference to clinical study guidelines (random inclusion, etc.) was performed due to the nature of the included studies.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
SMW is funded by the Dutch Cancer Society (clinical fellowship: 2011–4964). The authors like to thank W.T.M. Blokland for meaningful discussions regarding methylation methods used in the summarized studies and S. Kalkman for critical reading of the manuscript.
Glossary
Abbreviations:
- HPV
human papillomavirus
- OPSCC
oropharyngeal squamous cell carcinoma
- HNSCC
head and neck squamous cell carcinomas
- CpG island
cytosine is followed by phosphorylated guanine
- FFPE
using formalin-fixed paraffin-embedded
- FF
fresh frozen
- PCR
polymerase chain reaction
- IHC
immunohistochemistry
- MSP
methylation-specific PCR
- COBRA
combined bisulfite restriction analysis
- PMA
pyrosequencing analysis
- BAM
bead array method
- qMSP
quantitative methylation-specific PCR
- SFRP
soluble frizzled receptor protein
- CDH1
Cadherin 1 type 1
- PSF
progression free survival
- LRE1
LINE1 retrotransposable element 1
- LINE-1
long interspersed nucleotide elements
- LUMA
luminometric methylation assay
- Rb
retinoblastoma protein
- EMT
epithelial-mesenchymal transition
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/26881
References
- 1.Tsai HC, Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 2011;21:502–17. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–63. doi: 10.1016/S0959-437X(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 5.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 6.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994;4:255–9. doi: 10.1016/S0959-437X(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 7.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stebbing J, Bower M, Syed N, Smith P, Yu V, Crook T. Epigenetics: an emerging technology in the diagnosis and treatment of cancer. Pharmacogenomics. 2006;7:747–57. doi: 10.2217/14622416.7.5.747. [DOI] [PubMed] [Google Scholar]
- 9.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–74. doi: 10.1016/S0168-9525(99)01971-X. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 11.Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J Nutr. 2002;132(Suppl):2424S–9S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- 12.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 13.Smith EM, Rubenstein LM, Haugen TH, Hamsikova E, Turek LP. Tobacco and alcohol use increases the risk of both HPV-associated and HPV-independent head and neck cancers. Cancer Causes Control. 2010;21:1369–78. doi: 10.1007/s10552-010-9564-z. [DOI] [PubMed] [Google Scholar]
- 14.Curado MP, Hashibe M. Recent changes in the epidemiology of head and neck cancer. Curr Opin Oncol. 2009;21:194–200. doi: 10.1097/CCO.0b013e32832a68ca. [DOI] [PubMed] [Google Scholar]
- 15.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–9. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillison ML. Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol. 2004;31:744–54. doi: 10.1053/j.seminoncol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–9. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 18.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin Cancer Res. 2009;15:6758–62. doi: 10.1158/1078-0432.CCR-09-0784. [DOI] [PubMed] [Google Scholar]
- 20.Lechner M, Frampton GM, Fenton T, Feber A, Palmer G, Jay A, Pillay N, Forster M, Cronin MT, Lipson D, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimple RJ, Smith MA, Blitzer GC, Torres AD, Martin JA, Yang RZ, Peet CR, Lorenz LD, Nickel KP, Klingelhutz AJ, et al. Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 2013;73:4791–800. doi: 10.1158/0008-5472.CAN-13-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohavanichbutr P, Houck J, Fan W, Yueh B, Mendez E, Futran N, Doody DR, Upton MP, Farwell DG, Schwartz SM, et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135:180–8. doi: 10.1001/archoto.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA. Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur J Cancer. 2007;43:415–32. doi: 10.1016/j.ejca.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlecht NF, Burk RD, Adrien L, Dunne A, Kawachi N, Sarta C, Chen Q, Brandwein-Gensler M, Prystowsky MB, Childs G, et al. Gene expression profiles in HPV-infected head and neck cancer. J Pathol. 2007;213:283–93. doi: 10.1002/path.2227. [DOI] [PubMed] [Google Scholar]
- 25.Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:701–9. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 26.Leonard SM, Wei W, Collins SI, Pereira M, Diyaf A, Constandinou-Williams C, Young LS, Roberts S, Woodman CB. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis. 2012;33:1286–93. doi: 10.1093/carcin/bgs157. [DOI] [PubMed] [Google Scholar]
- 27.Burgers WA, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–5. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett KL, Lee W, Lamarre E, Zhang X, Seth R, Scharpf J, Hunt J, Eng C. HPV status-independent association of alcohol and tobacco exposure or prior radiation therapy with promoter methylation of FUSSEL18, EBF3, IRX1, and SEPT9, but not SLC5A8, in head and neck squamous cell carcinomas. Genes Chromosomes Cancer. 2010;49:319–26. doi: 10.1002/gcc.20742. [DOI] [PubMed] [Google Scholar]
- 29.Colacino JA, Dolinoy DC, Duffy SA, Sartor MA, Chepeha DB, Bradford CR, McHugh JB, Patel DA, Virani S, Walline HM, et al. Comprehensive analysis of DNA methylation in head and neck squamous cell carcinoma indicates differences by survival and clinicopathologic characteristics. PLoS One. 2013;8:e54742. doi: 10.1371/journal.pone.0054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong SM, Sun DI, Benoit NE, Kuzmin I, Lerman MI, Sidransky D. Epigenetic inactivation of RASSF1A in head and neck cancer. Clin Cancer Res. 2003;9:3635–40. [PubMed] [Google Scholar]
- 31.de Freitas Cordeiro-Silva M, Stur E, Agostini LP, de Podestá JR, de Oliveira JC, Soares MS, Mendonça EF, Gouvea SA, Von Zeidler SV, Louro ID. Promoter hypermethylation in primary squamous cell carcinoma of the oral cavity and oropharynx: a study of a Brazilian cohort. Mol Biol Rep. 2012;39:10111–9. doi: 10.1007/s11033-012-1885-4. [DOI] [PubMed] [Google Scholar]
- 32.Furniss CS, Marsit CJ, Houseman EA, Eddy K, Kelsey KT. Line region hypomethylation is associated with lifestyle and differs by human papillomavirus status in head and neck squamous cell carcinomas. Cancer Epidemiol Biomarkers Prev. 2008;17:966–71. doi: 10.1158/1055-9965.EPI-07-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gubanova E, Brown B, Ivanov SV, Helleday T, Mills GB, Yarbrough WG, Issaeva N. Downregulation of SMG-1 in HPV-positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin Cancer Res. 2012;18:1257–67. doi: 10.1158/1078-0432.CCR-11-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lechner M, Fenton T, West J, Wilson G, Feber A, Henderson S, Thirlwell C, Dibra HK, Jay A, Butcher L, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med. 2013;5:15. doi: 10.1186/gm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsit CJ, McClean MD, Furniss CS, Kelsey KT. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int J Cancer. 2006;119:1761–6. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 36.Marsit CJ, Posner MR, McClean MD, Kelsey KT. Hypermethylation of E-cadherin is an independent predictor of improved survival in head and neck squamous cell carcinoma. Cancer. 2008;113:1566–71. doi: 10.1002/cncr.23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Regan EM, Toner ME, Finn SP, Fan CY, Ring M, Hagmar B, Timon C, Smyth P, Cahill S, Flavin R, et al. p16(INK4A) genetic and epigenetic profiles differ in relation to age and site in head and neck squamous cell carcinomas. Hum Pathol. 2008;39:452–8. doi: 10.1016/j.humpath.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Poage GM, Houseman EA, Christensen BC, Butler RA, Avissar-Whiting M, McClean MD, Waterboer T, Pawlita M, Marsit CJ, Kelsey KT. Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer. Clin Cancer Res. 2011;17:3579–89. doi: 10.1158/1078-0432.CCR-11-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards KL, Zhang B, Baggerly KA, Colella S, Lang JC, Schuller DE, Krahe R. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS One. 2009;4:e4941. doi: 10.1371/journal.pone.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sartor MA, Dolinoy DC, Jones TR, Colacino JA, Prince ME, Carey TE, Rozek LS. Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6:777–87. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taioli E, Ragin C, Wang XH, Chen J, Langevin SM, Brown AR, Gollin SM, Garte S, Sobol RW. Recurrence in oral and pharyngeal cancer is associated with quantitative MGMT promoter methylation. BMC Cancer. 2009;9:354. doi: 10.1186/1471-2407-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss D, Basel T, Sachse F, Braeuninger A, Rudack C. Promoter methylation of cyclin A1 is associated with human papillomavirus 16 induced head and neck squamous cell carcinoma independently of p53 mutation. Mol Carcinog. 2011;50:680–8. doi: 10.1002/mc.20798. [DOI] [PubMed] [Google Scholar]
- 43.Weiss D, Stockmann C, Schrödter K, Rudack C. Protein expression and promoter methylation of the candidate biomarker TCF21 in head and neck squamous cell carcinoma. Cell Oncol (Dordr) 2013;36:213–24. doi: 10.1007/s13402-013-0129-5. [DOI] [PubMed] [Google Scholar]
- 44.Kostareli E, Holzinger D, Bogatyrova O, Hielscher T, Wichmann G, Keck M, Lahrmann B, Grabe N, Flechtenmacher C, Schmidt CR, et al. HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J Clin Invest. 2013;123:2488–501. doi: 10.1172/JCI67010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–66. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 47.Verma M, Srivastava S. Epigenetics in cancer: implications for early detection and prevention. Lancet Oncol. 2002;3:755–63. doi: 10.1016/S1470-2045(02)00932-4. [DOI] [PubMed] [Google Scholar]
- 48.Demokan S, Dalay N. Role of DNA methylation in head and neck cancer. Clin Epigenetics. 2011;2:123–50. doi: 10.1007/s13148-011-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.König F, Krekeler G, Hönig JF, Cordon-Cardo C, Fischer G, Korabiowska M. Relation between human papillomavirus positivity and p16 expression in head and neck carcinomas--a tissue microarray study. Anticancer Res. 2007;27(1A):283–8. [PubMed] [Google Scholar]
- 50.Au Yeung CL, Tsang WP, Tsang TY, Co NN, Yau PL, Kwok TT. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep. 2010;24:1599–604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- 51.Dyson N, Howley PM, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–7. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 52.Näsman A, Attner P, Hammarstedt L, Du J, Eriksson M, Giraud G, Ahrlund-Richter S, Marklund L, Romanitan M, Lindquist D, et al. Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden: an epidemic of viral-induced carcinoma? Int J Cancer. 2009;125:362–6. doi: 10.1002/ijc.24339. [DOI] [PubMed] [Google Scholar]
- 53.Deng Z, Hasegawa M, Matayoshi S, Kiyuna A, Yamashita Y, Maeda H, Suzuki M. Prevalence and clinical features of human papillomavirus in head and neck squamous cell carcinoma in Okinawa, southern Japan. Eur Arch Otorhinolaryngol. 2011;268:1625–31. doi: 10.1007/s00405-011-1515-0. [DOI] [PubMed] [Google Scholar]
- 54.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, Brakenhoff RH. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96:998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 56.Smeets SJ, Braakhuis BJ, Abbas S, Snijders PJ, Ylstra B, van de Wiel MA, Meijer GA, Leemans CR, Brakenhoff RH. Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene. 2006;25:2558–64. doi: 10.1200/JCO.2011.36.4596. [DOI] [PubMed] [Google Scholar]
- 57.Smeets SJ, Hesselink AT, Speel EJ, Haesevoets A, Snijders PJ, Pawlita M, Meijer CJ, Braakhuis BJ, Leemans CR, Brakenhoff RH. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121:2465–72. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 58.Specht K, Richter T, Müller U, Walch A, Werner M, Höfler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–29. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu J, Richardson JA, Olson EN. Capsulin: a novel bHLH transcription factor expressed in epicardial progenitors and mesenchyme of visceral organs. Mech Dev. 1998;73:23–32. doi: 10.1016/S0925-4773(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 60.Weiss D, Koopmann M, Basel T, Rudack C. Cyclin A1 shows age-related expression in benign tonsils, HPV16-dependent overexpression in HNSCC and predicts lower recurrence rate in HNSCC independently of HPV16. BMC Cancer. 2012;12:259. doi: 10.1186/1471-2407-12-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gudas LJ. Emerging roles for retinoids in regeneration and differentiation in normal and disease states. Biochim Biophys Acta. 2012;1821:213–21. doi: 10.1016/j.bbalip.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivenugopal KS, Ali-Osman F. The DNA repair protein, O(6)-methylguanine-DNA methyltransferase is a proteolytic target for the E6 human papillomavirus oncoprotein. Oncogene. 2002;21:5940–5. doi: 10.1038/sj.onc.1205762. [DOI] [PubMed] [Google Scholar]
- 63.Salazar CR, Smith RV, Garg MK, Haigentz M, Jr., Schiff BA, Kawachi N, Anayannis N, Belbin TJ, Prystowsky MB, Burk RD, et al. Human Papillomavirus-Associated Head and Neck Squamous Cell Carcinoma Survival: A Comparison by Tumor Site and Initial Treatment. Head Neck Pathol. 2013 doi: 10.1007/s12105-013-0486-4. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capellà G, Ribas M, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 65.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–62. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Wild L, Flanagan JM. Genome-wide hypomethylation in cancer may be a passive consequence of transformation. Biochim Biophys Acta. 2010;1806:50–7. doi: 10.1016/j.bbcan.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc Natl Acad Sci U S A. 2002;99:10060–5. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hennessey PT, Ochs MF, Mydlarz WW, Hsueh W, Cope L, Yu W, Califano JA. Promoter methylation in head and neck squamous cell carcinoma cell lines is significantly different than methylation in primary tumors and xenografts. PLoS One. 2011;6:e20584. doi: 10.1371/journal.pone.0020584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.