Abstract
Cadmium (Cd) is prevalent in the environment yet understudied as a developmental toxicant. Cd partially crosses the placental barrier from mother to fetus and is linked to detrimental effects in newborns. Here we examine the relationship between levels of Cd during pregnancy and 5-methylcytosine (5mC) levels in leukocyte DNA collected from 17 mother-newborn pairs. The methylation of cytosines is an epigenetic mechanism known to impact transcriptional signaling and influence health endpoints. A methylated cytosine-guanine (CpG) island recovery assay was used to assess over 4.6 million sites spanning 16,421 CpG islands. Exposure to Cd was classified for each mother-newborn pair according to maternal blood levels and compared with levels of cotinine. Subsets of genes were identified that showed altered DNA methylation levels in their promoter regions in fetal DNA associated with levels of Cd (n = 61), cotinine (n = 366), or both (n = 30). Likewise, in maternal DNA, differentially methylated genes were identified that were associated with Cd (n = 92) or cotinine (n = 134) levels. While the gene sets were largely distinct between maternal and fetal DNA, functional similarities at the biological pathway level were identified including an enrichment of genes that encode for proteins that control transcriptional regulation and apoptosis. Furthermore, conserved DNA motifs with sequence similarity to specific transcription factor binding sites were identified within the CpG islands of the gene sets. This study provides evidence for distinct patterns of DNA methylation or “footprints” in fetal and maternal DNA associated with exposure to Cd.
Keywords: cadmium, cigarette smoke, cord blood, epigenetics, heavy metal, maternal blood, methylation, pregnancy
Introduction
Cadmium (Cd) is a heavy metal that ranks among the top ten chemicals in the Agency for Toxic Substances and Disease Registry priority list of hazardous substances.1 It is widespread in the environment, found in byproducts of industrial processes, contaminated water or soil, certain foods, and tobacco products.2 Cd is a known lung carcinogen and a putative carcinogen in other tissues including the liver, prostate, kidney, bladder, stomach, and pancreas.3-5 In addition to its role as a carcinogen, Cd has also been associated with other health endpoints, including developmental effects early in life. For example, prenatal Cd exposure has been inversely associated with fetal growth parameters such as newborn length, weight, height, and head circumference,6-13 as well as adverse cognitive developmental effects later in life.12,14
Health effects associated with moderate to low levels of Cd exposure are of growing concern, particularly among susceptible populations such as pregnant women and children.15 Highlighting this issue, among pregnant women studied in the Fourth National Report on Human Exposure to Environmental Chemicals (NHANES IV), 66% had detectable blood Cd levels with an average level of 0.22 μg/L.16 The presence of Cd in the blood is particularly concerning as it has a half-life ranging up to 10 y.17 Cd is a component of cigarette smoke and concurrent exposure occurs. In utero exposure to both Cd and cigarette smoke is associated with lower newborn birth weight6,9,18 where Cd has been suggested to be the component of cigarette smoke that affects fetal skeletal growth.19 Cotinine, the primary metabolite of nicotine and biomarker of cigarette smoke exposure, is a reliable measure of actual dose received with a half-life of less than 1 d.20,21 Thus, cotinine is a general measure of recent exposure to tobacco products, whereas Cd can represent long-term and/or historic tobacco exposure. For non-smokers, diet is the major source of Cd exposure.17
The mechanism(s) of action of the detrimental health effects related to prenatal Cd exposure are not well established. An epigenetic mechanism has been hypothesized,22 but is understudied. The addition or removal of methyl groups from cytosine at the 5′ position (5mC) is an epigenetic mechanism that plays a key role in mediating gene expression and subsequent biological processes,23 possibly contributing to subsequent health effects resulting from environmental toxicant exposure. Indeed there is evidence from animal and cell culture studies that Cd alters DNA methyltransferase activity and subsequently DNA methylation patterns.3,22,24-29 In addition, prenatal tobacco smoke exposure is a known modifier of DNA methylation patterns,30-32 and is therefore an important consideration in this study.
In an effort to understand potential impacts of prenatal Cd exposure, in the present study we assess DNA methylation in blood leukocytes and distinguish between Cd- and cotinine-associated changes in a cohort of mother-baby pairs from Durham County, North Carolina. Maternal cotinine and Cd levels were used to compare and contrast the DNA methylation levels associated with either contaminant, enabling the differentiation between Cd-specific patterns in DNA methylation from those associated with cotinine. DNA methylation changes of more than 16 000 promoter-based cytosine-phosphate-guanine (CpG) islands were assessed in fetal and maternal DNA associated with maternal and thus in utero Cd and cotinine levels.
Results
This study consisted of 34 subjects, 17 mother–newborn pairs, selected as a nested cohort from the CEHI Healthy Pregnancy, Healthy Baby study in Durham, North Carolina. The pairs were selected from the larger cohort based on stratified maternal blood Cd levels. Maternal and infant characteristics are presented in Table 1. The average maternal age was 28 y. Most of the women had more than one child (n = 13; 76.5%), inclusive of the infants described in this study. There were similar proportions of male (n = 9; 52.9%) and female children (n = 8; 47.1%). All newborns, with one exception, had a birth weight greater than 2500 g (range: 2495–3740 g). Levels of maternal Cd ranged from below the detection limit to 1.05 μg/L with an average maternal blood concentration of 0.44 μg/L. Ten women in the study had Cd levels above the NHANES median level in pregnant women and were classified as the “higher cadmium-exposed” group (See Materials and Methods). Maternal cotinine levels ranged from below the detection limit to 166.96 μg/L with an average of 14.5 μg/L. While 11 women had detectable blood levels of cotinine, only two had levels above 10 μg/L, a level associated with active or passive smoking activity.20 The two individuals with elevated cotinine reported active smoking during pregnancy. As we were also interested in the effect of exposure due to second-hand or inactive smoking, cotinine exposure was classified as any detectable level vs. none. Eleven women had a detectable level of cotinine and were classified as “cotinine-exposed.” The exposure data for each of the 17 mother-baby pairs is provided (Tables S1 and S2).
Table 1. Characteristics of mother-newborn pairs (n = 34 subjects).
| Mean ± SD (range) /N (%) | |
|---|---|
| Maternal cadmium (μg/L) | 0.44 ± 0.31 (0 - 1.05) |
| Maternal cotinine (μg/L) | 14.5 ± 42.23 (0 - 166.96) |
| Maternal age (years) | 28 ± 7 (19–42) |
| Maternal race* | |
| NHB | / 12 (70.6) |
| NHW | / 4 (23.5) |
| Other | / 1 (5.8) |
| Parity | |
| First | / 4 (23.5) |
| Second | / 5 (29.4) |
| Third or higher | / 8 (47.1) |
| Child’s Sex | |
| M | / 9 (52.9) |
| F | / 8 (47.1) |
| Birth weight (g) | 3210 ± 377 (2495–3740) |
This study intentionally over-sampled NHB women.
Regression analysis revealed that maternal Cd or cotinine levels did not vary significantly with respect to maternal age, race, parity, child’s sex, or birth weight. Exclusion of the low birth weight infant from the data set did not significantly affect the results (data not shown). Generally, younger women had higher levels of Cd, but this finding was not statistically significant (P > 0.05). Spearman rank correlation revealed a positive relationship between maternal serum cotinine and Cd (r = 0.45, P = 0.07).
Cd-associated gene-specific DNA methylation
Methylated maternal and newborn DNA was isolated using a methyl-CpG-binding domain protein complex (MBD2b/MBD3L1) and hybridized onto Affymetrix Human Promoter 1.0R arrays. These arrays contain more than 4.6 million probes that cover more than 25, 500 human gene promoter regions. Computational methods were used to summarize the DNA methylation levels at a gene-specific CpG island level annotated to the reference Human Genome 18 (HG18) as in our recent publications33,34 (see Materials and Methods). More than 16, 000 CpG islands were included in this study.
Prior to analysis for the exposures of interest, differences in average DNA methylation levels associated with maternal age, race, and infant sex were identified. Analysis of the maternal DNA revealed there was a significant difference in gene-specific methylation levels associated with maternal age (n = 596 genes) and race (n = 83 genes). In fetal DNA, there was a significant difference in gene-specific DNA methylation levels associated with maternal age (n = 39 genes), race (n = 949 genes), and infant sex (n = 176 genes) (Table S3).
In relation to newborn environmental exposures of Cd and cotinine, two gene sets were identified with significantly different average DNA methylation abundances in fetal DNA including a Cd-associated gene set (n = 61) and a cotinine-associated gene set (n = 366) (Fig. 1A and B; Table S4). Of these, one gene showed hypomethylation with increasing Cd, and five genes showed hypomethylation with respect to increasing cotinine (Fig. 1C). A total of 30 genes overlapped between the Cd and cotinine gene sets and all were hypermethylated (Fig. 1C; Table S4).
Figure 1. Heat map of genes with differential DNA methylation levels in fetal and maternal DNA associated with Cd (A), or cotinine (B). Venn diagram representing the total number of Cd-or cotinine-associated genes in fetal DNA (C) or maternal DNA (D). Heat maps represent average DNA methylation levels of exposure-associated gene sets. Data are z-score normalized for each gene. Individuals are ordered from left to right based on increasing level of exposure. Red indicates a relative increase in average DNA methylation and blue represents a relative decrease in average DNA methylation. In the Venn diagrams, the number in parentheses indicates the number of hypomethylated genes contained in each set. LE, lower exposed; HE, higher exposed; UE, unexposed; E, exposed.
In maternal DNA, distinct sets of Cd-associated (n = 92) or cotinine-associated genes (n = 134) were identified (Fig. 1A and B; Table S5). Of these, 11 of the 92 Cd-associated differentially methylated genes were hypomethylated, whereas four of 134 cotinine-associated genes were hypomethylated. There were no overlapping genes between the Cd- or cotinine-associated genes in maternal DNA (Fig. 1D).
A comparison of the Cd-associated genes between maternal and fetal DNA showed no overlap. Very few (n = 12) of the cotinine-associated genes were differentially methylated in both fetal and maternal DNA (data not shown). The majority of differentially methylated genes showed increasing promoter methylation with increasing Cd or cotinine level for both fetal and maternal DNA (Fig. 1A and B).
In addition to identifying genes that are differentially methylated and associated with Cd or cotinine levels, a comparison was also performed between the DNA methylomes of the women and their newborns. Regardless of environmental exposure, this comparison of the 34 individuals’ methylomes showed that there were 12, 820 genes significantly differentially methylated between fetal and maternal DNA that were not due to differences in maternal age, race, or infant sex (Fig. S1). All of the 12, 820 genes showed lower average methylation abundance in maternal DNA when compared with fetal DNA.
Differential methylation does not reflect shifts in leukocyte cell types
The Cd- and cotinine-associated gene sets were compared with known differentially methylated regions (DMRs) that correspond to shifts in the abundance of white blood cell types.35 Within a list of 500 genes with DMRs known to predict cell type in an adult population, 227 of these genes were contained on the MIRA array. No blood cell type-associated genes were contained in the maternal Cd- or cotinine-associated genes lists in this study. A single gene, UNC84 domain containing 1 (UNC84A), was present in the Cd-associated fetal DNA gene set and 4 genes: neutrophil cytosolic factor 4 (NCF4), CBP80/20-dependent translation initiation factor (KIAA0427), sorting nexin 8 (SNX8), and scavenger receptor class F (SCARF) were present in the cotinine-associated gene set in fetal DNA.
Biological functions are enriched among differentially methylated genes
The identified Cd- and cotinine-associated gene sets in mother–baby pairs were analyzed to determine whether they encode proteins with similar functionality in the cell. Each of the gene sets was analyzed for enriched biological functions using two independent methods (see Materials and Methods). The most significantly enriched biological functional categories were gene expression, cell cycle, cell death, and nervous system development (Table 2). Interestingly, while there was no overlap between the individual gene sets, genes that encode proteins that play a role in regulation of transcription were enriched among Cd-associated genes in both fetal and maternal DNA (P < 0.001). Apoptosis was an enriched biological process in the Cd and cotinine-associated gene sets identified as differentially methylated within both maternal and fetal DNA (P < 0.05). Specifically, there were 14 apoptosis-associated genes including proline rich 13 (PRR13) in the fetal gene set and 20 additional genes among the maternal gene set (Table S6).
Table 2. Enriched biological functions within the differentially methylated gene sets.
| Fetal DNA (P values) | Maternal DNA (P values) | ||||
|---|---|---|---|---|---|
| Category | Function Annotation | Cd a | Cotinine a | Cd a | Cotinine a |
| Gene expression | Regulation of transcription | 0.001* [0.01] | – | 0.001* [0.001] |
– |
| Tissue morphology | Adipose tissue quantity | 0.004 | – | – | – |
| Cancer | Hyperplasia | 0.004 | – | – | – |
| Lipid metabolismb | Lipid accumulation | 0.005 | – | – | – |
| Cell death | Apoptosis | 0.02 [0.05] |
0.001* | 1.2 × 10−4* [0.04] | 0.05 |
| Cell cycle | Delay in G1, interphase | – | 3.0 × 10−5* [0.003] | – | – |
| Nervous systemc | Neuronal quantity | – | 0.002 [0.03] |
5.2 × 10−5* [0.03] | – |
| Cell proliferation | Tumor proliferation | – | 0.002 | – | 0.03 |
| Protein degradation | Proteolysis | – | 0.003 [0.003] |
– | 0.01 |
| Infectious disease | Replication of virus | – | – | – | 0.01 |
| Neurological disease | Movement disorder | – | – | – | 0.02 |
P < 0.001; [ ] = P value of gene ontology terms in DAVID.aAssociated contaminant exposure in fetal or maternal DNA. bLipid metabolism, molecular transport, small molecule biochemistry. cNervous system development and function.
Common DNA motifs identified in gene sets
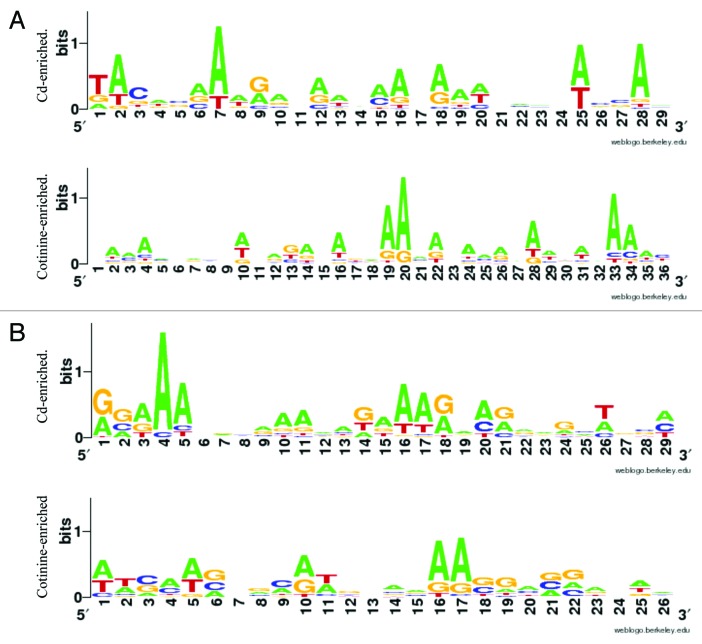
We hypothesized that the identified differentially methylated genes may contain common underlying sequences or motifs. To examine this, the promoter regions of the CpG islands for each of the differentially methylated genes were analyzed for statistically enriched common sequence patterns (e.g., motifs). The motifs were then compared with known transcription factor binding sites. Among the Cd- and cotinine-associated gene sets, significantly enriched motifs representing conserved DNA sequences were identified (Fig. 2). The conserved motifs showed sequence similarity to binding sites for several transcription factors including: transcription factor 7-like 1 (TCF7L1 also known as TCF3), metal-responsive transcription factor-1 (MTF-1), transcription factor AP-2-eplison (TCFAP2E), and serum response factor (SRF) (Table 3). Notably, for the Cd-associated gene sets identified both in maternal and fetal DNA, the enriched motifs had significant sequence similarity to the binding sites of MTF-1 and TCF7L1.
Figure 2. Significantly enriched DNA motifs identified within the Cd and cotinine-associated gene lists in fetal DNA (A) and maternal DNA (B) as identified by MEME. The motifs shown have standard orientation 5′ to 3′. Estimates of sequence conservation at each nucleotide position within the motif are shown where a higher bit score is represented by increased font size.
Table 3. Transcription factors with response element sequence similarity to the identified motifs within the Cd- or cotinine-associated gene sets.
| Fetal DNA (P values) | Maternal DNA (P values) | |||
|---|---|---|---|---|
| Transcription Factor | Cd a | Cotinine a | Cd a | Cotinine a |
| TCFAP2E | 2.3 × 10−5* | 9.2 × 10−4* | – | 4.9 × 10−3 |
| TCF7L1 | 6.7 × 10−5* | 2.0 × 10−5* | 5.1 × 10−4* | 1.5 × 10−4* |
| SRF | 2.2 × 10−4* | 3.2 × 10−4* | 1.5 × 10−3 | 5.0 × 10−3 |
| MTF1 | 3.1 × 10−4* | 1.6 × 10−4* | 3.5 × 10−4* | 2.0 × 10−3 |
P < 0.001. aAssociated contaminant exposure in fetal or maternal DNA.
Validation of MIRA DNA methylation results
Validation of the MIRA-based methylation results was performed using two methodologies for both fetal and maternal DNA. First using a gene-specific analysis, the promoter region of the apoptosis-associated PRR13 gene was targeted for methylation-specific quantitative polymerase chain reaction (qPCR)-based analysis. Primers were designed to amplify the promoter region comparable to the MIRA-assessed site (see Materials and Methods). DNA collected from 15 newborns representing a range of Cd levels was bisulfite converted and assessed. The data show a significant Spearman’s rank correlation (r = 0.53, P < 0.05) between the MIRA-based methylation assessment and the qPCR-based methylation for PRR13 (Fig. S2). Second, the results from the MIRA platform were compared with data obtained using the Illumina 450K platform using 2 maternal DNA samples. While the Spearman’s rank correlation for the genome-wide (n = 11, 347) assessment was not statistically significant (r = 0.01, P = 0.1), the methylation levels for 83 of the 92 Cd-specific genes that could be compared between platforms were significantly correlated (r = 0.35, P = 0.0009). A majority of the Cd-associated CpG islands (82%) had concordant relative hyper- or hypo-methylation on the 450K platform as compared with the MIRA assay (Table S5). Of the 20 Cd-associated genes enriched for apoptosis, 17 could be compared on both platforms and these had significant Spearman’s rank correlation (r = 0.41, P = 0.07).
Discussion
There is growing evidence that the prenatal environment may influence the burden of disease in adult life, and that this relationship is associated with epigenetic modifications altered during the prenatal period.23,36 The suggested impacts of Cd exposure include enzyme inhibition, generation of reactive oxygen species, and perturbation of apoptosis or cell cycle37; however, the ability to induce genomic instability without genotoxic action has implicated a possible epigenetic mechanism of Cd toxicity.22 Cd-associated genome-wide DNA methylation has not been previously assessed in human samples. Given the relationship of Cd with birth outcomes in humans6,9 and its known role as a mediator of DNA methyltransferase activity,22 we set out to identify whether patterns of differential promoter DNA methylation in fetal and maternal leukocyte DNA associated with exposure to Cd. The data were compared with cotinine-associated patterns of differential DNA methylation.
Here differences in gene-specific levels of DNA methylation were observed and linked to Cd exposure in utero. These genes were largely independent of those associated with cotinine. Overall, the majority of the differentially methylated genes showed increased or hypermethylation associated with Cd exposure. The Cd-associated genes were classified into their known ontologies, and transcription regulatory processes emerged as significantly enriched in both fetal and maternal DNA. Cell death, specifically apoptosis, was a significantly enriched function for each gene list. The altered Cd-associated DNA methylation in maternal DNA was validated using an alternate genome-wide approach, as well as for a subset of apoptosis-enriched genes. In addition, a gene-specific approach was used for PRR13 in fetal DNA, known to play a role in apoptosis.38 Cd has been shown in vitro to perturb pathways involved in inflammatory response, cell survival, apoptosis, tumorigenesis, and oxidative stress.22,39-42 In animal models, in utero exposure to Cd is associated with a wide range of cell cycle and proliferative genomic responses.43 The data presented here may support an epigenetic mechanism, namely DNA methylation, by which genes involved in transcriptional regulation and apoptosis could be influenced by Cd exposure. As RNA is not available for the specific study subjects analyzed here, gene expression analysis cannot be performed. It is important to note that it is not anticipated that all of the changes in DNA methylation will have functional consequence and impact gene expression.
Relative to Cd, cotinine was associated with a greater number of genes with differential DNA methylation in both maternal DNA and fetal DNA. Three previous studies of prenatal cigarette smoke exposure examined gene-specific DNA methylation.30-32 One study reported global hypomethylation with gene-specific hypermethylation of eight genes investigated30 while the other found largely gene-specific hypomethylation among 38 total genes.31 The third study reported 26 significant CpGs mapped to 10 genes with both hyper- and hypomethylation associated with plasma cotinine levels.32 Interestingly, none of the genes identified were common between the three studies and none are similarly reported in the present study. There are several factors that could potentially influence differences in observed DNA methylation patterns that include exposure type, exposure duration, tissue type (i.e., buccal vs. placental vs. cord blood), subject’s age at sampling, type of assay used, or other unaccounted for co-exposures.
It is important to mention that a woman’s current Cd levels may reflect prior exposure to cigarette smoke. Although we have accounted for cotinine exposure (a measure of recent and active smoking) in the analysis, it cannot be ruled out that some of the Cd-associated changes may be due to historic cigarette smoke exposure. Our data are supported by previous in vitro and ex vivo studies showing that Cd exposure led to hypermethylation of DNA after prolonged chronic exposures, whereas hypomethylation was present after acute Cd exposure.22,25,26
Our data highlight that regardless of exposure, there were significant differences in DNA methylation profiles between maternal and fetal samples that were not due to differences in maternal age, race, or infant sex. This is supported by a recent study that demonstrated that relative to newborn DNA, there is less methylation is observed among CpG island promoters in older individuals.15 The increased methylation levels in fetal DNA may have relevance for developmental biology. Further evaluation of these basic processes may increase our understanding of how early life exposures resulting in epigenetic shifts can have long-term health effects.
Of interest, many of the genes contained in the Cd and cotinine gene sets showed common sequences in their CpG islands within promoters. Among these conserved motif regions, binding sites for a common set of transcription factors were identified. The specific binding of transcription factors to target sites is a mechanism that protects CpG islands from methylation.44,45 Very recently studies have demonstrated that transcription factor binding results in local regions of low methylation and in contrast, absence of DNA-binding factors triggers the remethylation of local promoter regions,46,47 however this phenomenon has not been described related to environmental exposures. Here we identify conserved motifs with sequence similarity to binding sites for transcription factors including MTF-1. Notably, MTF-1 is known to respond to changes in cellular concentrations of multiple metals and coordinate expression of genes protective against metal toxicity.48,49 All of the identified transcription factors (e.g., TCF7L1, TCFAP2E, MTF-1, SRF) are known to regulate developmental processes within cells and represent targets for future investigation.50-53 Taken together, we hypothesize that these results suggest that patterns of DNA methylation that are associated with Cd may represent “footprints” indicating transcription factor presence or absence that occur during periods of DNA methylation.
As a limitation, the cord blood sampling done here is representative of newborn leukocyte DNA rather than potential target organ systems such as the kidney, liver, or bone. There are obvious ethical and technical reasons to use leukocyte DNA as a proxy for target tissue analysis. Moreover, the use of circulating white blood cells as proxies for disease has been shown.54,55 While patterns of DNA methylation can differ between white blood cell types,32,35,56 our data support that the Cd and cotinine gene sets are not simply due to shifts in blood cell types. There is increasing evidence that there are contaminant-specific changes to leukocyte DNA methylation associated with various environmental contaminant exposures.33,57,58 Future studies should aim to compare the Cd-associated changes here to tissue-specific changes.
In summary, the data from the present study provide evidence of Cd- and cotinine-associated patterns in DNA methylation present in the leukocyte DNA of newborns and their mothers. We identify gene-specific changes in DNA methylation levels associated with in utero Cd exposure, and distinguish these from methylation changes attributable to cotinine exposure, a general proxy measure for exposure to tobacco products. These distinct patterns of environmentally-associated DNA methylation alterations or “footprints” in fetal and maternal DNA may have functional consequences in the cell and warrant further research. Metal exposure continues to be an important area of public health concern for both maternal and child health. Increased education about the potential risks of environmental contaminants including Cd and cigarette smoke will be key to reducing and preventing harm to infants.
Materials and Methods
Study participants
The Children’s Environmental Health Initiative (CEHI) conducted a prospective cohort study of pregnant women living in Durham County, North Carolina from 2005–2011. This study is a key component of the Southern Center on Environmentally-Driven Disparities in Birth Outcomes (SCEDDBO), an interdisciplinary center aimed at understanding how environmental, social, and host factors jointly contribute to health disparities (http://cehi.snre.umich.edu/projects/sceddbo/). The CEHI Healthy Pregnancy, Healthy Baby study was reviewed and approved by the Institutional Review Boards at Duke University (Pro00007633) and the University of North Carolina (#09–0866). All women participating in this study consented for maternal venous and newborn cord blood collection for chemical and genetic analysis.
Women receiving prenatal care at either the Duke Obstetrics Clinic or the Durham County Health Department Prenatal Clinic were eligible to participate if they planned to deliver at Duke University Medical Center, were at least 18 y of age, were English-literate, lived in Durham County, and did not have a multi-fetal gestation or any known fetal genetic or congenital anomalies. Additional methods on subject recruitment, enrollment, and data collection have been described previously.59 Women were enrolled between 18 and 28 weeks of pregnancy, and demographic data were collected including maternal age, race, and parity, as well as child’s sex and birth weight. At delivery, biological samples including maternal venous blood and newborn cord blood were collected. From the CEHI study, we selected a nested subcohort of 17 mother-infant pairs stratified as above or below a maternal Cd level of 0.2 μg/L (see Supplemental Material).
Methylated CpG island recovery assay (MIRA)
Venous maternal blood and newborn cord blood samples were obtained at delivery. DNA was extracted using Qiagen’s PAXgene Blood DNA kit (Qiagen) according to manufacturer’s protocol. DNA was re-suspended in nuclease-free water and stored at –80 °C prior to DNA methylation assessment. CpG methylated DNA was collected using the MethylCollector Ultra Kit (Active Motif) and enriched DNA was amplified using the WGA3 kit (Sigma) according to manufacturer instructions with the following modification: 10 mM dATP, 10 mM dCTP, 10 mM dGTP, 8 mM dTTP, and 2 mM dUTP. Amplified DNA was then hybridized to the Affymetrix Human Promoter 1.0R arrays (Affymetrix) which assess over 4.6 million sites.
Statistical Analyses
Linear regression analyses were performed using the statistical package SAS 9.3 (SAS Institute Inc.) to examine relationships between maternal blood Cd levels, serum cotinine levels and demographic characteristics for women and children. Maternal demographics included age, race, and parity, and children’s demographics included sex and birth weight. The relationship between Cd and cotinine was assessed with linear regression as well as Spearman rank correlation. Cd and cotinine levels below detect were treated as zero.
For each of the study subjects, the DNA methylation abundance data obtained through the MIRA assay assessing 4.6 million probes were normalized using robust multi-chip average.60 The DNA methylation levels were then summarized at a gene-specific CpG island level annotated to the reference HG18 where islands were defined as in Davies et al.61-63 The resulting average methylation abundances for 16 421 CpG islands were compared using ANOVA (Partek Genomic Suite 6.4) where differential DNA methylation levels were assessed for each island and statistically defined as: (1) average island promoter methylation with a minimum absolute change of 30%; and (2) a P value < 0.05. Additionally, a false discovery rate (FDR) corrected q-value estimate was calculated and is reported. After identifying covariate-associated DNA methylation patterns within CpG islands (see Supplemental Materials), differential methylation was also examined according to sample type (maternal vs. newborn), higher vs. lower Cd-exposed, and cotinine-exposed vs. cotinine-unexposed (see Supplemental Materials). The same statistical requirements were applied to all analyses.
Gene ontology/pathway enrichment analysis
Gene ontology/pathway enrichment analysis was performed using two independent methodologies. The differentially methylated genes were analyzed in the context of interacting networks using Ingenuity Pathway Analysis Software (Ingenuity Systems, Inc.) and functional clustering using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (available online: http://david.abcc.ncifcrf.gov/tools.jsp).
Enriched DNA motif identification
CpG island sequences were retrieved from the UCSC genome browser website64 for each exposure-associated gene set. These sequences are representative of CpG islands within promoter regions of the genome. Position-specific letter probability matrices, also known as motifs, were identified using Multiple EM for Motif Elicitation (MEME) version 4.8.1.65 A first order Hidden Markov Model and negative position specific priors were calculated from a background set of 200 randomly selected CpG island sequences that were not differentially methylated. Additional parameter specifications are reported in the Supplemental Materials. The motif with the highest statistical significance (P value) for each group of differentially methylated genes was compared with known transcription factor binding sites using TOMTOM.65
Validation of MIRA results
For gene-specific analysis, DNA from 15 newborn leukocyte samples was selected. DNA was bisulfite converted using the Zymo EZ DNA Methylation-Lightening kit (Zymo Research) according to the manufacturer’s instructions. Methylation was assessed using the EpiTect MethyLight Assay (Qiagen). Methylation-independent sequence-specific primers were designed for the apoptosis-associated gene PRR13 with forward primer sequence: (5′ GGTTTGGGTG ATTAGGAAGAGT 3′) and reverse primer sequence: (5′ AAAATCCAAATACCCCATCAC 3′). A methyl-specific reporter probe for quantification was also designed (5′ 6FAM-GCCGCCTAAACTTACTACGT 3′). The amplified region of PRR13represents promoter region on chromosome 12, position 52121562–52121835, based upon HG18. Amplicons were assessed using qPCR in technical triplicate. qPCR methylation abundance was determined as 100/(1 + cycle threshold (CT)) for each sample, averaged across triplicates. A Spearman rank correlation and corresponding p-value were calculated to compare the methylation abundances between the qPCR and MIRA assays.
Genome-wide comparison was performed on two maternal DNA samples stratified by maternal blood Cd levels using the 0.2 μg/L exposure cutoff and matched on maternal race, age, and insurance status. Bisulfite conversion was performed using the Zymo EZ DNA Methylation kit (Zymo Research) according to the manufacturer’s instructions. Methylation was assessed at 485 577 CpGs in maternal DNA using the Illumina Infinium HumanMethylation450 BeadChip (Illumina Inc.). BeadChip processing was performed at Expression Analysis Inc. (www.expressionanalysis.com) and processed with Illumina’s GenomeStudio Methylation module Version 1.8 (Illumina Inc.). The proportion of methylation (β) for each CpG was calculated as the ratio of methylated signal intensity divided by the sum of both methylated and unmethylated signals. For quality control, probes with detection P value < 0.0001 were required. β values were excluded from analysis for probes which did not meet this minimum threshold for detection. The data were further filtered for probes positioned within CpG islands for comparison with the MIRA assay. For statistical comparison, average β values were calculated and a ratio of β Cd-exposed/ β Cd-unexposed determined for each gene. A Spearman rank correlation was calculated to compare the β ratio to the MIRA FC for all comparable genes (n = 11 347), the focused set of 92 Cd-associated genes in maternal DNA, and the subset of 20 Cd-associated apoptosis-related genes.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was funded in part by grants from the NIEHS (T32-ES007018, P42-ES005948, ES019315, and P30-ES010126), the USEPA (RD83329301), and in part by the North Carolina Translation and Clinical Sciences Institute (NC TRaCS) grants UL1RR025747, KL2RR025746, and TLRR025745 from the NIH National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health. We acknowledge researchers at Expression Analysis in Durham, NC for their assistance with the processing of the Illumina 450K microarrays.
Glossary
Abbreviations:
- 5mC
5-methylcytosine
- Cd
Cadmium
- CpG
Cytosine-phosphate-guanine
- CEHI
Children’s Environmental Health Initiative
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DL
Detection limit
- DNA
Deoxyribonucleic acid
- FDR
false discovery rate
- ICP-MS
Inductively Coupled Plasma-Mass Spectrometry
- MEME
Multiple EM for Motif Elicitation
- MTF-1
Metal-responsive transcription factor-1
- MIRA
methylation CpG island recovery assay
- NHANES IV
Fourth National Report on Human Exposure to Environmental Chemicals
- PRR13
Proline rich 13
- qPCR
Quantitative Polymerase Chain Reaction
- SCEDDBO
Southern Center on Environmentally-Driven Disparities in Birth Outcomes
- SRF
Serum response factor
- TCF7L1
Transcription factor 7-like 1 (also known as TCF3)
- TCFAP2E
Transcription factor AP-2-eplison
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/26798
References
- 1.ATSDR. Priority List of Hazardous Substances. Online at: http://wwwatsdrcdcgov/spl/ 2011.
- 2.ATSDR. Toxicological Profile for Cadmium. Atlanta, GA: Centers for Diseaes Control, 2008. [Google Scholar]
- 3.Arita A, Costa M. Epigenetics in metal carcinogenesis: nickel, arsenic, chromium and cadmium. Metallomics. 2009;1:222–8. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waalkes MP. Cadmium carcinogenesis in review. J Inorg Biochem. 2000;79:241–4. doi: 10.1016/S0162-0134(00)00009-X. [DOI] [PubMed] [Google Scholar]
- 5.IARC Volume 58. Cadmium 1993:119-238 [Google Scholar]
- 6.Fréry N, Nessmann C, Girard F, Lafond J, Moreau T, Blot P, Lellouch J, Huel G. Environmental exposure to cadmium and human birthweight. Toxicology. 1993;79:109–18. doi: 10.1016/0300-483X(93)90124-B. [DOI] [PubMed] [Google Scholar]
- 7.Lin CM, Doyle P, Wang DL, Hwang YH, Chen PC. Does prenatal cadmium exposure affect fetal and child growth? Occup Environ Med. 2011;68:641–6. doi: 10.1136/oem.2010.059758. [DOI] [PubMed] [Google Scholar]
- 8.Nishijo M, Tawara K, Honda R, Nakagawa H, Tanebe K, Saito S. Relationship between newborn size and mother’s blood cadmium levels, Toyama, Japan. Arch Environ Health. 2004;59:22–5. doi: 10.3200/AEOH.59.1.22-25. [DOI] [PubMed] [Google Scholar]
- 9.Salpietro CD, Gangemi S, Minciullo PL, Briuglia S, Merlino MV, Stelitano A, Cristani M, Trombetta D, Saija A. Cadmium concentration in maternal and cord blood and infant birth weight: a study on healthy non-smoking women. J Perinat Med. 2002;30:395–9. doi: 10.1515/JPM.2002.061. [DOI] [PubMed] [Google Scholar]
- 10.Ronco AM, Arguello G, Muñoz L, Gras N, Llanos M. Metals content in placentas from moderate cigarette consumers: correlation with newborn birth weight. Biometals. 2005;18:233–41. doi: 10.1007/s10534-005-0583-2. [DOI] [PubMed] [Google Scholar]
- 11.Llanos MN, Ronco AM. Fetal growth restriction is related to placental levels of cadmium, lead and arsenic but not with antioxidant activities. Reprod Toxicol. 2009;27:88–92. doi: 10.1016/j.reprotox.2008.11.057. [DOI] [PubMed] [Google Scholar]
- 12.Tian LL, Zhao YC, Wang XC, Gu JL, Sun ZJ, Zhang YL, Wang JX. Effects of gestational cadmium exposure on pregnancy outcome and development in the offspring at age 4.5 years. Biol Trace Elem Res. 2009;132:51–9. doi: 10.1007/s12011-009-8391-0. [DOI] [PubMed] [Google Scholar]
- 13.Shirai S, Suzuki Y, Yoshinaga J, Mizumoto Y. Maternal exposure to low-level heavy metals during pregnancy and birth size. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2010;45:1468–74. doi: 10.1080/10934529.2010.500942. [DOI] [PubMed] [Google Scholar]
- 14.Bonithon-Kopp C, Huel G, Moreau T, Wendling R. Prenatal exposure to lead and cadmium and psychomotor development of the child at 6 years. Neurobehav Toxicol Teratol. 1986;8:307–10. [PubMed] [Google Scholar]
- 15.Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–7. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodruff TJ, Zota AR, Schwartz JM. Environmental chemicals in pregnant women in the United States: NHANES 2003-2004. Environ Health Perspect. 2011;119:878–85. doi: 10.1289/ehp.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Järup L, Akesson A. Current status of cadmium as an environmental health problem. Toxicol Appl Pharmacol. 2009;238:201–8. doi: 10.1016/j.taap.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Yu XD, Yan CH, Shen XM, Tian Y, Cao LL, Yu XG, Zhao L, Liu JX. Prenatal exposure to multiple toxic heavy metals and neonatal neurobehavioral development in Shanghai, China. Neurotoxicol Teratol. 2011;33:437–43. doi: 10.1016/j.ntt.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Godfrey K, Walker-Bone K, Robinson S, Taylor P, Shore S, Wheeler T, Cooper C. Neonatal bone mass: influence of parental birthweight, maternal smoking, body composition, and activity during pregnancy. J Bone Miner Res. 2001;16:1694–703. doi: 10.1359/jbmr.2001.16.9.1694. [DOI] [PubMed] [Google Scholar]
- 20.Klebanoff MA, Levine RJ, Clemens JD, DerSimonian R, Wilkins DG. Serum cotinine concentration and self-reported smoking during pregnancy. Am J Epidemiol. 1998;148:259–62. doi: 10.1093/oxfordjournals.aje.a009633. [DOI] [PubMed] [Google Scholar]
- 21.Jarvis MJ, Russell MA, Benowitz NL, Feyerabend C. Elimination of cotinine from body fluids: implications for noninvasive measurement of tobacco smoke exposure. Am J Public Health. 1988;78:696–8. doi: 10.2105/AJPH.78.6.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takiguchi M, Achanzar WE, Qu W, Li GY, Waalkes MP. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res. 2003;286:355–65. doi: 10.1016/S0014-4827(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 23.Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79(Suppl):1514–9. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Zamudio R, Ha HC. Environmental epigenetics in metal exposure. Epigenetics. 2011;6:820–7. doi: 10.4161/epi.6.7.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benbrahim-Tallaa L, Waterland RA, Dill AL, Webber MM, Waalkes MP. Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase. Environ Health Perspect. 2007;115:1454–9. doi: 10.1289/ehp.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang DJ, Zhang YM, Qi YM, Chen C, Ji WH. Global DNA hypomethylation, rather than reactive oxygen species (ROS), a potential facilitator of cadmium-stimulated K562 cell proliferation. Toxicol Lett. 2008;179:43–7. doi: 10.1016/j.toxlet.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Benbrahim-Tallaa L, Tokar EJ, Diwan BA, Dill AL, Coppin JF, Waalkes MP. Cadmium malignantly transforms normal human breast epithelial cells into a basal-like phenotype. Environ Health Perspect. 2009;117:1847–52. doi: 10.1289/ehp.0900999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doi T, Puri P, McCann A, Bannigan J, Thompson J. Epigenetic effect of cadmium on global de novo DNA hypomethylation in the cadmium-induced ventral body wall defect (VBWD) in the chick model. Toxicol Sci. 2011;120:475–80. doi: 10.1093/toxsci/kfr022. [DOI] [PubMed] [Google Scholar]
- 29.Jiang GF, Xu L, Song SZ, Zhu CC, Wu Q, Zhang L, Wu L. Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology. 2008;244:49–55. doi: 10.1016/j.tox.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 30.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suter M, Ma J, Harris AS, Patterson L, Brown KA, Shope C, Showalter L, Abramovici A, Aagaard-Tillery KM. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6:1284–94. doi: 10.4161/epi.6.11.17819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun Ø, Cupul-Uicab LA, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120:1425–31. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smeester L, Rager JE, Bailey KA, Guan XJ, Smith N, García-Vargas G, Del Razo LM, Drobná Z, Kelkar H, Stýblo M, et al. Epigenetic changes in individuals with arsenicosis. Chem Res Toxicol. 2011;24:165–7. doi: 10.1021/tx1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, García-Vargas G, Del Razo LM, Drobná Z, Stýblo M, Fry RC. Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. J Biochem Mol Toxicol. 2013;27:106–15. doi: 10.1002/jbt.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bollati V, Baccarelli A. Environmental epigenetics. Heredity (Edinb) 2010;105:105–12. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waisberg M, Joseph P, Hale B, Beyersmann D. Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology. 2003;192:95–117. doi: 10.1016/S0300-483X(03)00305-6. [DOI] [PubMed] [Google Scholar]
- 38.Lih CJ, Wei W, Cohen SN. Txr1: a transcriptional regulator of thrombospondin-1 that modulates cellular sensitivity to taxanes. Genes Dev. 2006;20:2082–95. doi: 10.1101/gad.1441306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benton MA, Rager JE, Smeester L, Fry RC. Comparative genomic analyses identify common molecular pathways modulated upon exposure to low doses of arsenic and cadmium. BMC Genomics. 2011;12:173. doi: 10.1186/1471-2164-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol. 2000;59:95–104. doi: 10.1016/S0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- 41.Shin HJ, Park KK, Lee BH, Moon CK, Lee MO. Identification of genes that are induced after cadmium exposure by suppression subtractive hybridization. Toxicology. 2003;191:121–31. doi: 10.1016/S0300-483X(03)00210-5. [DOI] [PubMed] [Google Scholar]
- 42.Robinson JF, Yu XZ, Moreira EG, Hong SW, Faustman EM. Arsenic- and cadmium-induced toxicogenomic response in mouse embryos undergoing neurulation. Toxicol Appl Pharmacol. 2011;250:117–29. doi: 10.1016/j.taap.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson JF, Yu X, Hong S, Griffith WC, Beyer R, Kim E, Faustman EM. Cadmium-induced differential toxicogenomic response in resistant and sensitive mouse strains undergoing neurulation. Toxicol Sci. 2009;107:206–19. doi: 10.1093/toxsci/kfn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–8. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 45.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–92. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 46.Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–7. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 47.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 48.Selvaraj A, Balamurugan K, Yepiskoposyan H, Zhou H, Egli D, Georgiev O, Thiele DJ, Schaffner W. Metal-responsive transcription factor (MTF-1) handles both extremes, copper load and copper starvation, by activating different genes. Genes Dev. 2005;19:891–6. doi: 10.1101/gad.1301805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews GK. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals. 2001;14:223–37. doi: 10.1023/A:1012932712483. [DOI] [PubMed] [Google Scholar]
- 50.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–55. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winger Q, Huang J, Auman HJ, Lewandoski M, Williams T. Analysis of transcription factor AP-2 expression and function during mouse preimplantation development. Biol Reprod. 2006;75:324–33. doi: 10.1095/biolreprod.106.052407. [DOI] [PubMed] [Google Scholar]
- 53.Nelson TJ, Balza R, Jr., Xiao Q, Misra RP. SRF-dependent gene expression in isolated cardiomyocytes: regulation of genes involved in cardiac hypertrophy. J Mol Cell Cardiol. 2005;39:479–89. doi: 10.1016/j.yjmcc.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 54.Anglim PP, Alonzo TA, Laird-Offringa IA. DNA methylation-based biomarkers for early detection of non-small cell lung cancer: an update. Mol Cancer. 2008;7:81. doi: 10.1186/1476-4598-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinnaeve PR, Donahue MP, Grass P, Seo D, Vonderscher J, Chibout SD, Kraus WE, Sketch M, Jr., Nelson C, Ginsburg GS, et al. Gene expression patterns in peripheral blood correlate with the extent of coronary artery disease. PLoS One. 2009;4:e7037. doi: 10.1371/journal.pone.0007037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–86. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pilsner JR, Hu H, Ettinger A, Sánchez BN, Wright RO, Cantonwine D, Lazarus A, Lamadrid-Figueroa H, Mercado-García A, Téllez-Rojo MM, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117:1466–71. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perera F, Tang WY, Herbstman J, Tang DL, Levin L, Miller R, Ho SM. Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One. 2009;4:e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miranda ML, Edwards S, Maxson PJ. Mercury levels in an urban pregnant population in Durham County, North Carolina. Int J Environ Res Public Health. 2011;8:698–712. doi: 10.3390/ijerph8030698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Psychiatric Epigenetics Data KCL. [http://epigenetics.iop.kcl.ac.uk/brain%5D
- 63.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32(Database issue):D493–6. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202-8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.