One of the most striking facts about the elements is their unequal distribution and occurrence in nature.
—C. R. Hammond
free calcium is highly controlled in biological tissues, and much like the physical chemistry of elements (5), its concentration is unequal in distribution (for recent reviews on calcium signaling see Ref. 2). However, how much free calcium exists within various subcellular domains, and how these concentrations shift in response to environmental cues, is still poorly understood. In this issue of American Journal of Physiology-Cell Physiology, Subedi and colleagues (12) report on the development of a research tool that enables discrimination between calcium signals at the plasma membrane, in the bulk cytosol, and in the nucleoplasm. These tools open new opportunities for researchers to pair regionally restricted calcium signals with physiological outcomes.
Free calcium approaches 2 mM concentrations in blood and interstitial fluids in animals, yet its concentration is approximately 20,000 times lower inside the cytosolic compartment of cells—approximately 100 nM. Several cellular mechanisms maintain low cytosolic calcium concentrations (2). Calcium pumps, including the sarcoplasmic, endoplasmic reticulum calcium ATPase and plasma membrane calcium ATPase, utilize ATP to remove calcium from the cytosol. In addition, many exchangers (i.e., the sodium/calcium exchanger), uniporters, and channels contribute to maintaining low cytosolic calcium concentrations. Calcium is complexed within the cytosol by both high- and low-affinity calcium buffers, and this buffering assists in lowering the free calcium concentrations. These mechanisms combine to create a physiological environment where relatively small elevations in cytosolic calcium are used as a signal to initiate changes in effector protein function; hence, calcium serves as a response signal (i.e., second messenger) critical to cell/organism survival. The cell response to calcium transitions can be acute, such as calcium stimulation of myosin light chain kinase that initiates increased cellular tension, or it can be prolonged, such as calcium activation of transcription factors that promote gene transcription.
The lumens of intracellular organelles possess free calcium pools that are different from the cytosolic compartment. Determining the calcium concentration within various organelles has been challenging, owing to the inexact nature of available techniques. Endoplasmic reticulum calcium concentration is estimated to be between 250 and 600 μM (3), while mitochondrial free calcium concentration is about 100 nM (7). Nuclear free calcium is estimated to be approximately 100 nM (1). Each of these examples represents estimates of basal free calcium, yet the organelle and cytosolic calcium concentrations are not fixed, and they can change abruptly.
Calcium within the endoplasmic reticulum lumen is mobilized by inositol 1,4,5-trisphosphate (InsP3) (2). InsP3 binding to its receptor on the endoplasmic reticulum membrane causes opening of the receptor channel, resulting in calcium release from the organelle into the cytosol. Calcium release from the endoplasmic reticulum transiently “depletes” free calcium in the lumen and increases free calcium in the cytosol. The cytosolic calcium response is short-lived, lasting only tens of seconds in nonexcitable cells before it is recaptured out of the cytosol and into the endoplasmic reticulum. An intimate association between the endoplasmic reticulum and mitochondria allows for privileged calcium uptake, where calcium released through the InsP3 receptor can enter the mitochondria through a calcium uniporter (and potentially other channels) (10). InsP3 receptors are also found on the nucleoplasmic reticulum surface, which is continuous with the endoplasmic reticulum and nuclear envelope (4, 6). These calcium signals do not appear to be generated merely from fluctuations in cytosolic calcium; the nucleoplasmic reticulum calcium signals may be generated by InsP3 receptor-mediated release directly into the nucleoplasm and localized subnuclear regions, or by diffusion from other calcium sources through nuclear pores (4, 6). How much of the calcium that is released through the InsP3 receptor reaches the mitochondria and/or nucleus, and the fate of this calcium, remains incompletely understood.
Depletion of stored calcium by InsP3 results in opening of plasmalemmal calcium channels that allows for calcium influx into the cell (i.e., store-operated calcium entry; see Refs. 8, 9). Diffusion of free calcium is then restricted by buffering, by physical barriers, and by sequestration/extrusion mechanisms (2). Because of this diffusion limitation, elevations in calcium concentration are greater in the immediate vicinity of the channel pore as compared with the bulk cytosol. Many different theoretical and experimental models have been used to calculate the size and duration of calcium microdomains; however, precise measurement is still a technological challenge. Numerical simulations based on optical single-channel recording by Shuai and Parker (11) determined that the calcium concentration at the mouth of a channel exceeds 15 μM, more than 150 times greater than basal cytosolic levels. They further established that the microdomain does not extend beyond 200 nm and collapses rapidly within a few milliseconds of channel closure. Exactly how much calcium escapes the microdomain, and how far it reaches within the cell, remain unanswered questions.
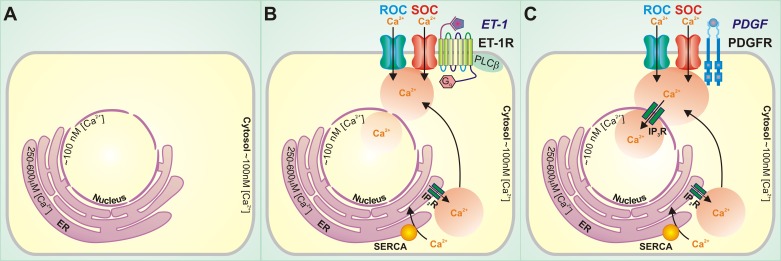
Here, Subedi et al. develop new technology that allows improved accuracy for measurement of free calcium at the plasma membrane, in the bulk cytosol, and in the nucleoplasm. Their studies relied on use of a genetically encoded cameleon, adapted to contain plasma membrane (Lyn) and nucleoplasm (nuclear localization sequence) targeting sequences. Careful calibration within each compartment enabled accurate time and concentration measurements. The Gq agonists endothelin-1 and angiotensin II elicited calcium signals most prominently at the plasma membrane domain, with relatively little calcium reaching the nucleoplasm (Fig. 1). In stark contrast, platelet-derived growth factor induced a calcium response of similar magnitude in both plasma membrane and nuclear compartments. These data vet the use of probes that enable accurate measurement of highly compartmentalized calcium signals and demonstrate the distinct signals that arise in response to different modes of environmental stimuli. This lays the groundwork for significant future progress, where investigators can begin to map the fate of calcium signals that move between subcellular compartments. Subcellular calcium transitions can be mapped alongside important physiological readouts, ultimately pairing calcium signals and protein effectors with temporal and geographic precision. Perhaps such insight will provide answers as to why one signal, such as endothelin-1, primarily promotes contraction, while another signal, such as platelet-derived growth factor, principally induces cell growth.
Fig. 1.
Endothelin-1 and platelet-derived growth factor induce distinctive subcellular calcium signatures. A: free calcium concentrations ([Ca2+])vary within subcellular compartments, where basal cytosolic calcium is approximately 100 nM, nucleoplasmic calcium is approximately 100 nM, and endoplasmic reticulum (ER) calcium is 250–600 μM. B: endothelin-1 (ET-1) stimulation of its receptor (ET-1R) in smooth muscle cells elicits an abrupt increase in membrane-associated calcium that accesses the bulk cytosol but does not readily access the nuclear compartment. C: platelet-derived growth factor (PDGF) stimulation of its receptor (PDGFR) in smooth muscle cells increases membrane-associated and cytosolic calcium, and this signal is conveyed into the nucleus. ROC, receptor-operated calcium entry channel(s); SOC, store-operated calcium entry channel(s); PLCβ, phospholipase C-β; SERCA, sarcoplasmic, endoplasmic reticulum calcium ATPase; IP3R, inositol 1,4,5-trisphosphate receptor.
GRANTS
The authors are supported by National Heart, Lung, and Blood Institute Grants HL-66299 (to T. Stevens), HL-60024 (to T. Stevens, D. L. Cioffi, and M. Francis), and HL-107778-01A1 (to D. L. Cioffi), and an American Heart Association predoctoral fellowship (to N. Xu).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
N.X., M.F., D.L.C., and T.S. drafted manuscript; N.X., M.F., D.L.C., and T.S. edited and revised manuscript; N.X., M.F., D.L.C., and T.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Drs. Ivan F. McMurtry, Thomas Rich, and Mary Townsley for review and discussion of this editorial. A special appreciation is extended to Ruby A. Fernandez (Univ. of Illinois at Chicago) for assistance in generating Fig. 1.
REFERENCES
- 1.Allbritton NL, Oancea E, Kuhn MA, Meyer T. Source of nuclear calcium signals. Proc Natl Acad Sci U S A 91: 12458–12462, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berridge MJ. Calcium signalling remodelling and disease. Biochem Soc Trans 40: 297–309, 2012 [DOI] [PubMed] [Google Scholar]
- 3.Demaurex N, Frieden M. Measurements of the free luminal ER Ca2+ concentration with targeted “cameleon” fluorescent proteins. Cell Calcium 34: 109–119, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat Cell Biol 5: 440–446, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond CR. The elements. In: Handbook of Chemistry and Physics (81st ed.), Boca Raton, FL: CRC Press, 2000, p. 4–1 [Google Scholar]
- 6.Leite MF, Thrower EC, Echevarria W, Koulen P, Hirata K, Bennett AM, Ehrlich BE, Nathanson MH. Nuclear and cytosolic calcium are regulated independently. Proc Natl Acad Sci USA 100: 2975–2980, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyata H, Silverman HS, Sollott SJ, Lakatta EG, Stern MD, Hansford RG. Measurement of mitochondrial free Ca2+ concentration in living single rat cardiac myocytes. Am J Physiol Heart Circ Physiol 261: H1123–H1134, 1991 [DOI] [PubMed] [Google Scholar]
- 8.Putney JW. Capacitative calcium entry: from concept to molecules. Immunol Rev 231: 10–22, 2009 [DOI] [PubMed] [Google Scholar]
- 9.Putney JW. Origins of the concept of store-operated calcium entry. Front Biosci (Schol Ed) 3: 980–984, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280: 1763–1766, 1998 [DOI] [PubMed] [Google Scholar]
- 11.Shuai J, Parker I. Optical single-channel recording by imaging Ca2+ flux through individual ion channels: theoretical considerations and limits to resolution. Cell Calcium 37: 283–299, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Subedi KP, Paudel O, Sham JS. Detection of differentially regulated subsarcolemmal calcium signals activated by vasoactive agonists in rat pulmonary artery smooth muscle cells. Am J Physiol Cell Physiol (December 18, 2013). 10.1152/ajpcell.00341.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]