Abstract
Tracheobronchial submucosal glands (SMGs) are derived from one or more multipotent glandular stem cells that coalesce to form a placode in surface airway epithelium (SAE). Wnt/β-catenin-dependent induction of lymphoid enhancer factor (Lef-1) gene expression during placode formation is an early event required for SMG morphogenesis. We discovered that Sox2 expression is repressed as Lef-1 is induced within airway SMG placodes. Deletion of Lef-1 did not activate Sox2 expression in SMG placodes, demonstrating that Lef-1 activation does not directly inhibit Sox2 expression. Repression of Sox2 protein in SMG placodes occurred posttranscriptionally, since the activity of its endogenous promoter remained unchanged in SMG placodes. Thus we hypothesized that Sox2 transcriptionally represses Lef-1 expression in the SAE and that suppression of Sox2 in SMG placodes activates Wnt/β-catenin-dependent induction of Lef-1 during SMG morphogenesis. Consistent with this hypothesis, transcriptional reporter assays, ChIP analyses, and DNA-protein binding studies revealed a functional Sox2 DNA binding site in the Lef-1 promoter that is required for suppressing β-catenin-dependent transcription. In polarized primary airway epithelium, Wnt induction enhanced Lef-1 expression while also inhibiting Sox2 expression. Conditional deletion of Sox2 also enhanced Lef-1 expression in polarized primary airway epithelium, but this induction was significantly augmented by Wnt stimulation. Our findings provide the first evidence that Sox2 acts as a repressor to directly modulate Wnt-responsive transcription of the Lef-1 gene promoter. These studies support a model whereby Wnt signals and Sox2 dynamically regulate the expression of Lef-1 in airway epithelia and potentially also during SMG development.
Keywords: Lef-1, promoter, Sox2, submucosal glands, Wnt
submucosal glands (SMGs) reside within the mesenchyme beneath the surface airway epithelium in most mammalian cartilaginous airways. In mice, airway SMGs are limited to the proximal portion of the trachea. SMGs secrete antimicrobial mucous and serous fluids that are essential to normal lung function and innate immunity. It has become increasingly recognized that dysfunctions of SMGs may contribute to many respiratory pathologies. For example, SMGs are a niche for slow cycling stem/progenitor cells (6, 8, 15) that is affected by CFTR dysfunction in cystic fibrosis (50). Additionally, many hypersecretory lung diseases such as cystic fibrosis, bronchitis, and asthma lead to expansion of gland mass through hypertrophy and/or hyperplasia (17, 22, 36). The development and maintenance of SMGs is precisely coordinated by complex reciprocal interactions between the surface epithelium and the surrounding mesenchyme. During SMG development, signal transduction events induce transcriptional changes that coordinate the maturation of glands from primordial glandular placodes (PGPs), which are morphologically distinguishable as clusters of epithelial progenitor cells basally orientated within the surface airway epithelium, into fully developed SMGs consisting of intricately branched networks of secretary acini, tubules, and ducts (8, 27, 28).
One of the key signaling pathways that mediates the maturation processes of PGPs is the Wnt/β-catenin signaling pathway, which provides a stimulatory signal to promote airway epithelial invagination, proliferation, tubulogenesis, and gland formation (10, 27). In PGPs, canonical Wnt/β-catenin signaling induces transient expression of lymphoid enhancer-binding factor 1 (Lef-1), a member of the T cell factor (TCF) family transcription factors (5, 10, 11). Lef-1 is one of the most well-characterized transcription factors downstream of the Wnt/β-catenin signaling pathway, and it is known to mediate the formation of epithelial structures, including the intestinal epithelium, hair follicles, taste buds, and mammary glands (5, 21, 28, 34, 49). Lef-1 knockout mice have abnormal development of mammary glands, teeth, and airway SMGs (5, 9, 13, 27, 46). In contrast, aberrant overexpression of Lef-1 in conjunction with hyperactive Wnt signaling has been observed in some hyperproliferative conditions, including colon cancer (1).
Lef-1 interacts with β-catenin and is recruited to specific Wnt-responsive gene promoters such as CCND1 (cyclin D1) to control proliferation (40) and genes encoding cytokeratins that may reflect changes in cellular differentiation (53). Members of the TCF family, including Lef-1, share a similar high-mobility group (HMG) DNA binding domain as well as an NH2-terminal β-catenin binding domain (1). However, Lef-1 is not redundant among other members of the TCF family in the context of Wnt activation and lineage specification during airway glandular development (10, 13, 27). Thus dissecting the mechanisms of transcriptional regulation of Lef-1 expression during SMG development will provide insights into early molecular events involved in lineage specification of glandular stem cells.
Transcriptional activation of Lef-1 within the Wnt/β-catenin pathway has been extensively investigated, yet questions still remain. Previous studies have discovered that Wnt signaling transcriptionally regulates the Lef-1 gene through a 110-bp Wnt responsive element (WRE) within the Lef-1 promoter (−879 to −769 bp), which is required for Wnt3a responsiveness in various cell lines (16). Deletion of this region results in a loss of Lef-1 promoter activity in many developing epithelial appendages in mice (11, 29). Additional studies of the Lef-1 promoter have identified several TCF factor-binding sites that function to regulate Wnt-mediated transcription (1, 16, 26, 30). These studies show that various TCF-binding sites can result in a multiplicity of inhibitory and stimulatory effects on Lef-1 transcription in a context-dependent manner (30). Furthermore, recent studies have begun to appreciate the role of Sox (SRY-related HMG box) family transcription factors in regulating Lef-1. For example, Sox17 suppresses Wnt/β-catenin-dependent transcription of Lef-1 by directly binding to multiple TCF factor binding sites (30).
Since both Sox2 and Sox17 play key roles in airway epithelial lineage specification and proliferation (18, 20, 25, 38, 45), determining whether Sox2 also regulates Lef-1 in the airway is of general importance. Sox2 plays a crucial role in maintaining adult airway epithelium. Deleting Sox2 in adult mice leads to a loss of a subset of Clara cells, ciliated cells, and basal cells, which results in a reduced ability to recover from injury (39). Sox2 is also thought to antagonize Wnt signaling during taste bud formation, lung development, and bone formation (18, 20, 32, 35). One potential mechanism to explain the ability of Sox2 to inhibit Wnt signaling is that Sox2 is capable of directly binding to β-catenin and inhibit activity of the β-catenin/TCF/Lef-1 transcriptional complex (32).
Given that Sox2 plays important roles in regulating cell phenotypes in the airway and has the ability to influence Wnt/β-catenin signaling, we hypothesized that Sox2 may directly regulate the expression of the Lef-1 gene in airway epithelia during SMG development. In support of this hypothesis, we observed that Sox2 and Lef-1 protein expression is reciprocally regulated during early stages of SMG morphogenesis when Wnt/β-catenin signaling is induced. At the early stages of PGP formation, Lef-1 expression is induced with β-catenin activation while Sox2 expression is inhibited. A similar reciprocal pattern of Sox17 and Lef-1 expression in developing PGPs has also been reported (30), suggesting that Sox2 and Sox17 may dynamically regulate Lef-1 expression during gland development. Using polarized primary airway epithelia and human lung-derived cell lines, we demonstrated that indeed Sox2 can modulate transcription of the Lef-1 gene in a manner both dependent and independent of Wnt/β-catenin signaling. DNA-protein interaction and ChIP analyses confirmed that Sox2 binds directly to the Lef-1 promoter at multiple TCF and Sox binding sites. Sox2-mediated inhibition of β-catenin-dependent activation of the Lef-1 promoter was most significantly tied to a Sox2-binding site within the WRE region. Interestingly, Sox2 binding sites within the Lef-1 promoter overlap with those previously reported for Sox17 (30). However, Sox2 appears to play a dominant role in regulating Lef-1 transcription in polarized primary airway epithelia, since deletion of Sox2, but not Sox17, significantly enhanced Lef-1 mRNA expression. In summary, our studies reveal that Sox2 can directly regulate Lef-1 transcription by binding at multiple sites within its promoter. These mechanisms likely coordinate the spatial control of Wnt/β-catenin signals important for lineage specification of glandular progenitors during early stages of airway SMG morphogenesis.
MATERIALS AND METHODS
Transgenic mice.
All studies were performed according to protocols approved by the Institutional Animal Care and Use Committee of the University of Iowa. Lef-1 knockout mice (47), Sox2 conditional knockout (Sox2COND/COND) mice (43), Sox2EGFP knock-in mice (14), Sox17 conditional knockout (Sox17COND/COND) mice (42), and Lef-1 conditional knockout (Lef-1COND/COND) mice (54) were previously described. ROSA-CreERT2 mice, B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J (stock no. 008463) (48), and β-catenin reporter mice or “BAT-gal” mice, B6.Cg-Tg(BAT-lacZ)3Picc/J (stock no. 005317)(33), were purchased from the Jackson Laboratory. ROSA-CreERT2/Sox2COND/COND double transgenic mice were achieved by mating Sox2COND/COND to ROSA-CreERT2 mice. All mouse strains were on a C57BL/6J background (at least 8 times backcrossed) with the exception of Lef-1 knockout mice, which was on a mixed C57BL/6J:ICR background.
Immunofluorescence staining.
Immunofluorescence staining was conducted on 8-μm frozen sections of mouse trachea, using a rabbit anti-mouse Lef-1 antibody (1:1,000, 2230B, Cell Signaling Technology), a goat anti-human Sox2 antibody (1:200, AF2018, R&D Systems), or a rabbit anti-mouse keratin-14 antibody (1:200, RB-2090, Lab Vision). Sections were fixed in 4% paraformaldehyde at room temperature for 20 min, followed by incubation in 0.2% Triton X-100 PBS solution at room temperature for 20 min. The sections were then incubated with blocking buffer containing 20% donkey serum in PBS at room temperature for 1 h. The mixture of primary antibodies was applied in blocking buffer with reduced donkey serum (1%) at 4°C overnight. After washing, a mixture of secondary antibodies (1:250 dilution of FITC-labeled donkey anti-goat IgG and Texas Red-labeled donkey anti-rabbit IgG; Jackson ImmunoResearch Laboratories) in buffer containing 1% donkey serum was applied at room temperature for 2 h. The sections were then washed and mounted in Vectashield Mounting Medium containing DAPI (H-1200; Vector Laboratories).
Cell lines and transient transfection experiments.
For studies involving Sox2/β-catenin responsiveness of the Lef-1 promoter, a β-catenin-S37A dominant active mutant and mouse Sox2 expression plasmid were cotransfected with the previously described Lef-1 promoter-luciferase reporter plasmid (pLF−2700/−200-Luc) (30). The Lef-1 promoter in this construct encompassed −2,700 to −200 bp of the human Lef-1 promoter relative to the translational start site of the Lef-1 protein (16). For these studies, both HEK293T and A549 cell lines were purchased from American Type Culture Collection (ATCC) and grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were grown to ∼60–70% confluency before being transfected using Lipofectamine-LTX (Invitrogen Life Technologies) according to the manufacturer's instructions. Transient transfection reporter assays were performed in six-well plates and utilized 0.2 μg of the Lef-1 promoter-luciferase reporter, 0.1, 0.2, or 0.4 μg of the β-catenin-S37A mutant and/or Sox2 expression plasmids, and 0.01 μg of the pCMV-Renilla luciferase plasmid (Promega) as an internal control for transfection efficiency. The total amount of DNA transfected in all experiments was normalized with an empty control plasmid (pcDNA3.1; Invitrogen). Transfections were carried out in serum-free medium, and cells were harvested for the firefly and Renilla luciferase assays at 48 h after transfection, as previously described (30). Protein was prepared for the assays by washing the cells in PBS, followed by lysis in 1× Passive Reporter Lysis Buffer (E1910, Promega). Protein concentrations were determined using the Bradford method, and all lysates were normalized to the same protein concentration using the lysis buffer. Ten micrograms of total cellular lysate was used for measurements of the relative luciferase activity units (RLU), for both firefly and Renilla luciferase, using the Dual Luciferase Assay kit (E1980, Promega). Transfection efficiencies were normalized by dividing the relative firefly luciferase units by the relative Renilla luciferase units. Following normalization, values were represented as RLU.
Western and Northern blots.
Western blots were performed with 100 μg of total cell lysate using goat anti-human Sox2 (1:1,000, AF2018, R&D Systems) and mouse anti-human Lef-1 (1:1,000, T100M, Exalpha Biologicals) antibodies. For Northern blots, 20 μg of total RNA was isolated from cell cultures using the TRIzol reagent (15596–018, Invitrogen) and was separated on agarose-formaldehyde gels in MOPS buffer. Gels were then transferred to Hybond-N+ membrane (RPN2250B, Amersham Biosciences). The membranes were prehybridized in prehybridization buffer [50% (vol/vol) formamide, 0.9 M NaCl, 60 mM NaH2PO4, 6 mM EDTA, 5X Denhardt's solution, 1% SDS, 200 μg/ml denatured shearing salmon sperm DNA, 10% dextran sulfate] at 42°C for 1 h prior to hybridization with 200 ng/ml of denatured biotinylated DNA probe at 42°C overnight. A full-length human Lef-1 or Sox2 cDNA was used as the DNA probe, which was biotinylated using the NEBlot Phototope Kit per manufacturer's instruction (N7550, New England Biolabs). The blots were developed by chemiluminescence using the Phototope Star Detection Kit (N7020, New England Biolabs).
Chromatin immunoprecipitation assays (ChIP).
For preparation of a single ChIP assay, three 150-mm plates of A549 cells were fixed and harvested for ChIP. Formaldehyde cross-linked DNA-protein complexes were sheared by sonication, to an average fragment length of ∼200 bp, diluted 1:10 with the ChIP dilution buffer, followed by preclearing with protein A agarose beads according to protocol of the manufacturer of the EZChIP Kit (17–371, Upstate Biologicals). Precleared DNA-protein samples were used for the ChIP assays. For immunoprecipitation, 10 μg of anti-Sox2 antibody (no. AF2018, R&D Systems), anti-β-catenin antibody (ab6302, Abcam), or nonimmune control IgG antibody was added to the ChIP reaction mixture. The recovered immunoprecipitated DNA was detected by PCR and DNA agarose gel electrophoresis using 3 μl of the final DNA precipitate. PCR was carried out for 35 cycles, each at 95°C for 15 s, 62°C for 30 s, and 72°C for 45 s. Nine primer sets were used to survey the 2.5-kb human Lef-1 promoter locus (Table 1).
Table 1.
Sequence of primer sets for ChIP assays on the 2.5-kb human Lef-1 promoter
| Amplicon | Lef-1 Promoter Position, bpa | Forward Primer | Reverse Primer |
|---|---|---|---|
| A | −2700/−2590 | 5′-GGGGATCCTTTGTAAAGTTTTAAAA-3′ | 5′-ACAATGCTGTATCTAAGTCA-3′ |
| B | −2579/−2497 | 5′-CCCCATTCAATGTTCAGCAGCCTG-3′ | 5′-CTCCATTATATAGGAATTGTCTGGC-3′ |
| C | −2417/−2327 | 5′-TGTTCTGAAGTTTTACAGATGGTTA-3′ | 5′-TTTTGTCCCTTGGTTCTCAAACCC-3′ |
| D | −2304/−2189 | 5′-GCTTCCCTTCTGCTGTAACTTTCA-3′ | 5′-AAGAC ACCAATCCGAATACAACCT-3′ |
| E | −1994/−1901 | 5′-CTCCGTACATCCCGTGGTGAGAA-3′ | 5′-TATGACGAGGAAGAAGGAACTGAAG-3′ |
| F | −1440/−1329 | 5′-TCCTTCCTTCTCTCGCCAAGTTG-3′ | 5′-CAGATCCTGTAGGACGAGTCTTGG-3′ |
| G | −1261/−1133 | 5′-CGGGCAGCCAAGGAGAGCTAG-3′ | 5′-GCAGGTGAGCGGCTGCGCCT-3′ |
| H | −1020/−902 | 5′-GCCAACTCAAGGGGCGCAGCTCTTTGCT-3′ | 5′-CCTCTTTGTTCCCGGCTCGAGTTTC-3′ |
| I | −793/−692 | 5′-CGGCAAAAACTTTATTCTTGGCA-3′ | 5′-CGTCATTTAATCTGCTAGAGAAGGAG-3′ |
Base pair position is relative to the translational ATG start site (+1) in exon 1 of the human Lef-1 gene.
Mutagenesis of Lef-1 promoter-reporter constructs.
The QuikChange MultiSite-Directed Mutagenesis kit from Stratagene was used to generate three mutated LF−2700/−200-Luciferase Lef-1 promoter activity reporter plasmids. Putative 6-bp Sox2 binding sites were mutated by changing A → C, T → G, C → A, or G → T. Mutated synthetic oligonucleotides used in the NoShift assay reflected the changes generated in the full-length promoter site-directed mutagenesis. The oligonucleotide sequences used to generate mutations within the Lef-1 promoter are listed in Table 2.
Table 2.
The sequences and positions of synthetic oligonucleotides used to generate mutants of Lef-1 promoter-luciferase constructs
| Binding Sitea | Lef-1 Promoter Position, bpb | Synthetic Oligonucleotide Sequencec |
|---|---|---|
| S1 | −2660/−2654 | 5′-GGATCCTTTGTAAAGTTTTAAAAATTGCTTCCTAGTAAcggaggATTCCCAAGAAGTAGTCGTTATTAGTAGGTGAAGTCATTAGTGC-3′ |
| S2 | −1956/−1950 | 5′-GTACATCCCGTGGTGAGAACAGAATGAAAGATATcggtggTAAAAAGCAATAATTAAAATCTAGTCTTCAGTTCCTTCTT-3′ |
| S3 | −780/−774 | 5′-GGAGTCCCCGCGCTTGCCGGCAAAAACTTTcggaggGGCAAAACTCCTCTTTCTCTTCCCCTCCTCCTCCTCGGGCC-3′ |
Three putative Sox binding sites within the human Lef-1 promoter were selected for mutation analysis.
Base pair position is relative to the translational ATG start site (+1) in exon 1 of the human Lef-1 gene.
The mutated sequences are shown in lowercase and bold.
Purification of V5/his-tagged Sox2 mouse proteins.
The mouse Sox2 cDNA was cloned in-frame into the pET151/D-TOPO vector backbone using the Champion pET Directional TOPO Expression Kit (K15101, Invitrogen), which generates an NH2-terminal V5/His-tagged fusion protein in bacteria. The QuikChange MultiSite-Directed Mutagenesis Kit (200519, Stratagene) was used to generate two mutant Sox2 variants with single amino acid changes (M49A and L97P). The oligonucleotides used to generate Sox2 variants M49A and L97P are listed in Table 3. Resultant clones were confirmed by DNA sequencing. V5/His-tagged bacterial fusion proteins were purified by HPLC using a HisTrap HP column (17–5249-01, GE Healthcare Life Sciences), and the size of the purified protein was confirmed by SDS-PAGE and Western blotting with anti-V5 antibody (46–0705, Invitrogen). Concentrations of the purified proteins were normalized using the Bradford method, and aliquots were stored at −80°C until further use.
Table 3.
The sequences of synthetic oligonucleotides used to generate Sox2 mutant bacterial expression constructs
| Mutation | Synthetic Oligonucleotide Sequencea |
|---|---|
| Sox2M49A | 5′-GCGGCCCATGAATGCCTTCgcaGTGTGGTCCCGCGGGCAGC-3′ |
| Sox2L97P | 5′-CCGTTCATCGACGAGGCTAAGCGGCCCCGAGCGccgCACATGAAGGAGCACCCG-3′ |
The mutated bases are shown in lowercase and bold.
ELISA based assay for DNA/protein interaction (NoShift assay).
To test whether Sox2 binds to putative consensus Sox DNA binding sites in the Lef-1 promoter, an ELISA based DNA-Protein interaction assay was used. Purified synthetic 5′ biotin-labeled oligonucleotides were annealed to non-biotin-tagged complementary oligonucleotides (see Table 4). The annealed products were purified using a 7.5% polyacrylamide gel in 0.5X TBE buffer, and the concentration after purification was determined by slot blot titrations using the NEB Phototope-star kit (N7020, New England Biolabs). Two nanograms of purified wild type (WT) Sox2-V5-His-tagged protein or mutated Sox2-V5-His-tagged proteins was mixed with 10 ng of the biotinylated oligonucleotides and brought to a final volume of 100 μl with binding buffer. The binding buffer contains 33 μg/μl poly(dI-dC) (P4929-25UN, Sigma), 100 ng/μl salmon-sperm DNA (15632-011, Invitrogen), 20 mM HEPES (pH 8.0), 2 mM PMSF, 5 mM MgCl2, 100 mM KCl, and 5% glycerol in nucleotide-free water. After the mixture was incubated on ice for 45 min, 100 μl was applied onto a streptavidin-labeled 96-well plate (no. 15120, Thermo) and incubated at room temperature for 2 h. After five PBS washes, the V5-His-tagged Sox2 proteins that bound to the biotinylated oligonucleotides were detected using goat anti-V5 antibodies (1:1,000, 46–0705, Invitrogen), and HRP-conjugated anti-goat secondary antibody (1:5000, Jackson ImmunoResearch Laboratories) was used to perform ELISA reactions.
Table 4.
The sequences of biotinylated oligonucleotides used for DNA/protein binding assays
| Binding Site | Lef-1 promoter position (bp)a | 5′ Biotinylated Synthetic Oligonucleotide Sequenceb |
|---|---|---|
| T1 | −2693/−2687 | Biotin-5′-GGGGATCctttgtaAAGTTTTAAAAATTGC-3′ |
| S1 | −2670/−2640 | Biotin-5′-TTCCTAGTAAattcttattCCCAAGAAGTA-3′ |
| T1/S1 | −2693/−2645 | Biotin-5′-GGGGATCctttgtaAAGTTTTAAAAATTGCTTCCTAGTAAattcttattCCCAAG-3′ |
| T2 | −2427/−2421 | Biotin-5′-CATTTCTTTATGTCctttgttTACTGTTCTG-3′ |
| T3 | −2254/−2248 | Biotin-5′-CTCCTGTctttgtaCAAAATTCATC-3′ |
| T4 | −2240/−2234 | Biotin-5′-CAAAATTCatcaaagAGACATGG-3′ |
| T3/T4 | −2254/−2234 | Biotin-5′-CTCCTGTctttgtaCAAAATTCatcaaagAGACATGG-3′ |
| S2 | −2956/−1951 | Biotin-5′-GAAAGATATattgttTAAAAAGC-3′ |
| T5 | −921/−915 | Biotin-5′-CTCGGCCGGGaacaaagAGGGGTCGG-3′ |
| S3 | −780/−760 | Biotin-5′-CAAAAActttattcttGGCA-3′ |
| Mutated T1 | −2693/−2687 | Biotin-5′-GGGGATCagggtgcAAGTTTTAAAAATTGC-3′ |
| Mutated S1 | −2670/−2640 | Biotin-5′-TTCCTAGTAAcggaggcggCCCAAGAAGTA-3′ |
| Mutated T1/S1 | −2693/−2645 | Biotin-5′- GGGGATCagggtgcAAGTTTTAAAAATTGCTTCCTAGTAAcggaggcggCCCAAG-3′ |
| Mutated T2 | −2427/−2421 | Biotin-5′-CATTTCTTTATGTCagggtggTACTGTTCTG-3′ |
| Mutated T3/T4 | −2254/−2234 | Biotin-5′-CTCCTGTagggtgcCAAAATTCcgaccctAGACATGG-3′ |
| Mutated S2 | −2956/−1951 | Biotin-5′-GAAAGATATcggtggAAAAAGC-3′ |
| Mutated T5 | −921/−915 | Biotin-5′-CTCGGCCGGGccacccctAGGGGTCGG-3′ |
| Mutated S3 | −780/−760 | Biotin-5′-CAAAAAagggcggaggGGCA-3′ |
Base pair position is relative to the translational ATG start site (+1) in exon 1 of the human Lef-1 gene.
Synthetic oligonucleotides containing the wild type or mutant sequences of putative TCF and/or Sox binding sites in the human Lef-1 promoter were labeled at the 5′ end with biotin. The potential binding sequences (wild type and mutated) are shown in lowercase and bold.
Generation of air-liquid interface (ALI) cultures of polarized mouse airway epithelia and treatment conditions.
ALI cultures were generated with primary tracheal airway epithelial cells obtained from Sox2COND/COND, Sox17COND/COND, Lef-1COND/COND, ROSA-CreERT2/Sox2COND/COND and wild-type C57BL/6J mice. Previously described procedures for isolation and culturing of mouse primary airway epithelia in ALI cultures were used (50). Briefly, pronase-dissociated cells from each experimental trachea were panned to remove fibroblasts, and the nonadherent cells were seeded onto collagen Type I-coated Millicells. Cultures were incubated for 4–6 days in BEGM medium prior to being cultured in USG supplemented differentiation medium on the basolateral side only and cultured at an ALI for an additional 2 wk. For experiments knocking down Sox2 expression, Sox2COND/COND ALI cultures were infected with Cre-EGFP-expressing recombinant adenovirus (Ad.CRE) at a multiplicity of infection (MOI) of 104 particles/cell, which typically yielded >85% of the epithelia expressed EGFP after 3 day infection (data not shown). For experiments overexpressing Sox2, wild-type C57BL/6J ALI culture were infected with Sox2-expressing recombinant adenovirus (Ad.Sox2) at MOIs of 102 or 104 particles/cell. In both experiments, equivalent titers of a virus lacking a transgene (Ad.Control, also called Ad.BglII) were used to infect control ALI cultures. ALI cultures were evaluated at 3 days following adenoviral infection in both types of experiments. For experiments deleting Sox2 in ROSA-CreERT2/Sox2COND/COND ALI culture, 4-hydroxytamoxifen (T176–50MG, Sigma) was dissolved in cell culture grade DMSO and added to BEGM growth medium at the time of seeding airway epithelial cells into Millicells. Cells were exposed to 4-hydroxytamoxifen (final concentration of 5 μM) or vehicle (DMSO) for 96 h prior to moving cells to an ALI for 2 wk, as previously described (37). For studies evaluating canonical Wnt-dependent changes in gene expression, mature ALI cultures (with or without conditional Sox2 knockout) were basolaterally exposed to a mixture of 100 ng/ml of active recombinant Wnt1 (ab84080, Abcam) and 100 ng/ml of active recombinant Wnt3a (ab81484, Abcam) for 72 h prior to the harvesting for total RNA. Since these recombinant proteins were lyophilized, they were directly dissolved in USG differentiation medium at the final concentration. Control Millicells were fed fresh USG medium without Wnts.
RT-PCR on ALI cultures of polarized mouse airway epithelia.
ALI cultures were harvested for total RNA preparation using TRIzol reagent (SKU10296–010, Invitrogen), and RNA was retrieved using RNeasy minikit (74104, Qiangen). Total RNA was used for cDNA synthesis with the reverse transcription kit (4368813, AB Applied Biosystems). The resultant cDNA was used for quantitative RT-PCR. Primer sets used to detect Sox2, Sox17, Lef-1, and GAPDH cDNAs are listed in Table 5. The PCR protocol used was 40 repeating cycles of 95°C for 15 s, 61°C for 45 s, and 72°C for 1 min. Real-time quantitative PCR was performed using Bio-Rad IQ SYBR Green Supermix and iCycler iQ Detection System (Bio-Rad, Hercules, CA). The average Ct values for duplicate PCR runs were used to calculate the relative abundance of Sox2, Sox17, and Lef-1 transcripts in each sample using the ΔCt method [relative abundance of transcript of interests = 2−(average gene of interest Ct − average GAPDH Ct)]. The ΔΔCt method was used to calculate the fold change in Sox2, Sox17, and/or Lef-1 mRNA expression in response to a specific treatment condition (i.e., Ad.CRE vs Ad.Control infection; Ad.Sox2 vs Ad.Control infection; and 4-hydroxytamoxifen vs vehicle treatment). In these cases, the fold change in gene expression was calculated as 2−(experimental ΔCt − Control ΔCt).
Table 5.
Primer sequences used for real-time PCR in mouse airway epithelia
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Sox2 | 5′-GGCAGCTACAGCATGATGCAGGAGC-3′ | 5′-CTGGTCATGGAGTTGTACTGCAGG-3′ |
| Sox17 | 5′-CCAGAAACTGCAGACCAGAAG-3′ | 5′-GGAGGTGCTGCTCATTGTATC-3′ |
| Lef-1 | 5′-ACTCTGCGCCACCGATGAGATGAT-3′ | 5′-TTGCTGGCTGGGATGATTTCGGACT-3′ |
| GAPDH | 5′-ATGGTGAAGGTCGGTGTGAA-3′ | 5′-GTCGTTGATGGCAACAATCTCC-3′ |
Statistical analysis.
Statistical significance for all comparisons was assessed using a 1-way ANOVA followed by either a Tukey's multiple comparisons test or Mann-Whitney test as described in the figure legends. Two-way ANOVA was used to assess significant interactions between Sox2 and dominant-active mutant β-catenin-S37A expression on transcription from the Lef-1 WT and Sox2 binding site mutant promoter-reporters. The resulting P values are two-tailed, and P < 0.05 was considered significant.
RESULTS
Sox2 and Lef-1 are expressed in distinct domains during airway SMG development.
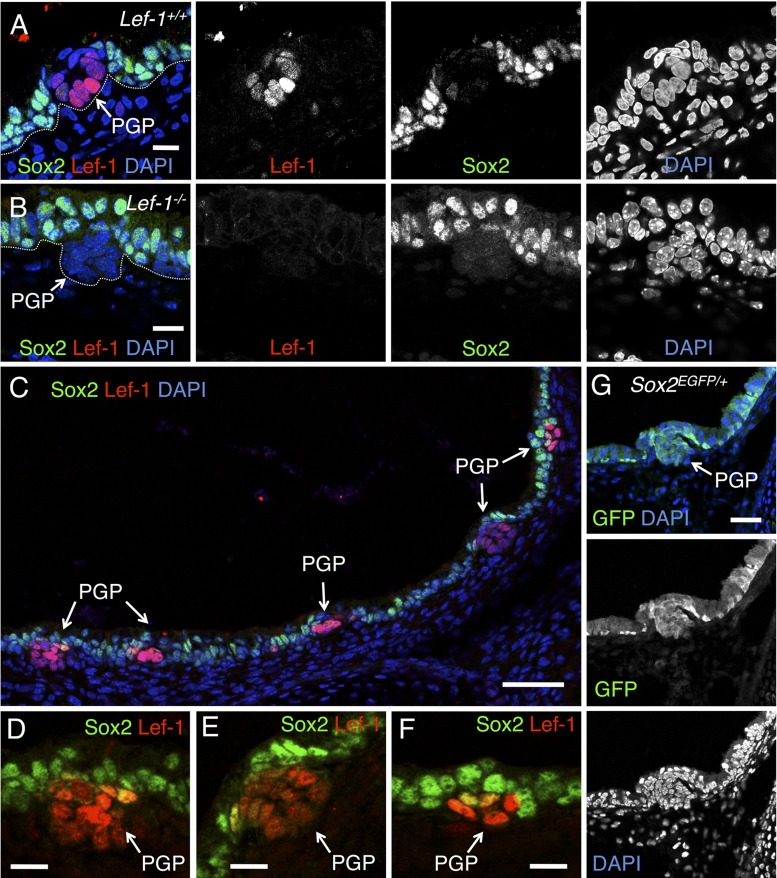
Lef-1 expression is upregulated in the PGP at both the mRNA and protein levels, and its expression is maintained in the tips of glandular tubules as the gland matures (10, 12). Previous studies have demonstrated that Sox2 is expressed in the surface airway epithelium (SAE) of the developing (20) and adult mouse airway (39, 45). However, the role of Sox2 in airway submucosal gland (SMG) development has not been thoroughly investigated. Immunolocalization of Sox2 in newborn mouse tracheal tissues demonstrated that Sox2 is highly expressed in nuclei of cells in the SAE, but significantly downregulated in cells of the PGP showing upregulated levels of Lef-1 protein (Fig. 1, A, C–F). Sox2 expression is reactivated as tubules mature and Lef-1 expression is again repressed (data not shown).
Fig. 1.
Sox2 is posttranscriptionally downregulated in Lef-1 expressing primordial gland placodes (PGPs). A and B: frozen sections of postnatal day 1 tracheas from wild-type (A) and Lef-1-knockout (KO) mice (B) stained with antibodies specific for Sox2 (green) and Lef-1 (red). DAPI (blue) was used to mark nuclei. Individual immunofluorescence channels are shown to the right of the combined channels. Dotted white line marks the basal lamina of the epithelium. C–F: collage of a postnatal day 1 wild-type trachea stained with Sox2 (green) and Lef-1 (red). D–F are higher magnification views of three PGPs shown in C. G: immunostaining for GFP (green) in a postnatal day 1 trachea from a Sox2EGFP/+ knock-in mouse. In this line, the endogenous Sox2 promoter drives EGFP expression in both surface airway epithelium (SAE) and PGP. Scale bar: A, B, D–F = 25 μm; C = 100 μm; G = 50 μm.
Based on the mutually exclusive expression patterns of Lef-1 and Sox2 in the newborn mouse trachea, we hypothesized that a direct regulatory relationship may exist between Sox2 and Lef-1 expression in PGPs. For example, Sox2 may repress Lef-1 expression within the SAE, and initiation of PGP formation may involve direct repression of Sox2 expression, thus allowing for Wnt-mediated activation of Lef-1. Alternatively, Wnt-activated Lef-1 may inhibit the expression of Sox2 within PGPs. To test whether the transcription of Sox2 is repressed in the PGP, we evaluated EGFP expression in the PGP from Sox2EGFP knock-in mice that express EGFP under the direction of the endogenous Sox2 promoter. Findings from this study demonstrated that the Sox2 promoter is active in the PGP (Fig. 1G). Additional studies evaluating Sox2 expression in the PGP of Lef-1 knockout animals confirmed that Lef-1 activation in the PGP does not participate in repression of Sox2; Lef-1 knockout mice maintained repression of Sox2 expression in the PGP as observed in wild-type mice (Fig. 1, A and B). Taken together, these findings demonstrate that repression of Sox2 protein expression in the PGP is posttranscriptional and unlikely to be directly regulated by Lef-1. Additionally, they are consistent with a model where Sox2 expression in the SAE acts to repress Lef-1 expression.
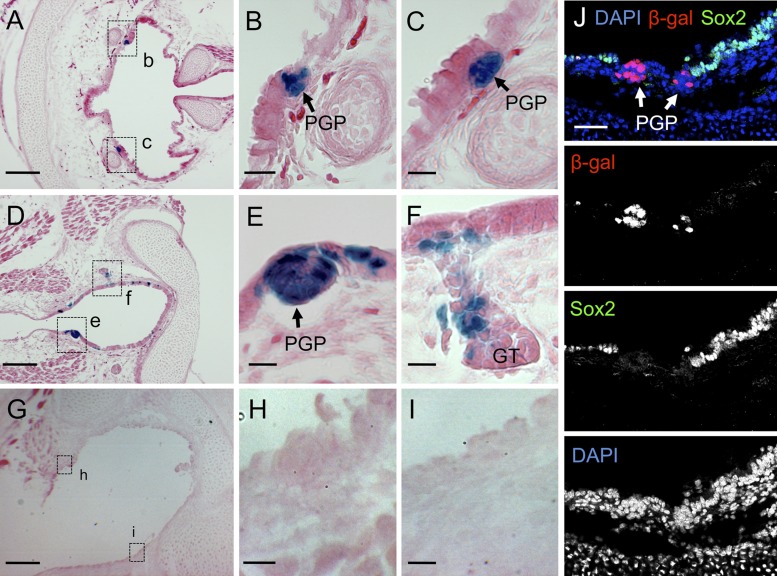
Given previous results demonstrating that PGPs fail to mature in Lef-1 knockout mice and that the Wnt3A pathway regulates activation of the Lef-1 promoter in PGPs (10), we hypothesized that the activation of Lef-1 gene expression during SMG development must lie downstream to changes in Sox2 expression. We further investigated the relationship between Sox2 and canonical Wnt/β-catenin signaling during PGP formation using BAT-gal reporter mice (33). TCF/β-catenin-dependent β-galactosidase reporter expression was strongly observed in PGPs compared with a lack of β-galactosidase expression throughout the SAE (Fig. 2, A–I). As anticipated, Sox2 protein expression was also lacking in cells of the PGP that expressed the TCF/β-catenin-dependent LacZ reporter (Fig. 2J). This result suggests that canonical Wnt/β-catenin activation in PGPs may coordinate both the posttranslational regulation of Sox2 protein levels and the activation of Lef-1 expression.
Fig. 2.
β-Catenin/T cell factor (TCF) pathway is activated in developing airway PGPs and portions of developing submucosal glands (SMGs). A–J: tracheas were dissected from newborn BAT-gal transgene positive (A–F and J) and transgene negative (G–I) mice. The BAT-gal transgene is a canonical Wnt signaling reporter that expresses nuclear-targeted β-galactosidase under the direction of a β-catenin/TCF-responsive promoter. Histological 10-μm frozen sections were stained in Xgal. Tracheal primordial gland placodes (PGPs) and glandular tubules (GT) are shown in the boxed regions of A, D, and G. B and C are enlarged regions boxed in A (labeled b and c). E and F are enlarged regions boxed in D (labeled e and f). H and I are enlarged regions boxed in G (labeled h and i). J shows an immunofluorescently stained section with antibodies specific for Sox2 (green) and β-galactosidase (red). DAPI (blue) was used to mark nuclei. The individual immunofluorescence channels are shown beneath the combined channels. Scale bars: A, D, and G = 100 μm; J = 50 μm; B, C, E, F, H, and I = 25 μm.
Wnt stimulation and Sox2 deletion synergistically enhance Lef-1 expression in polarized mouse tracheal airway epithelium.
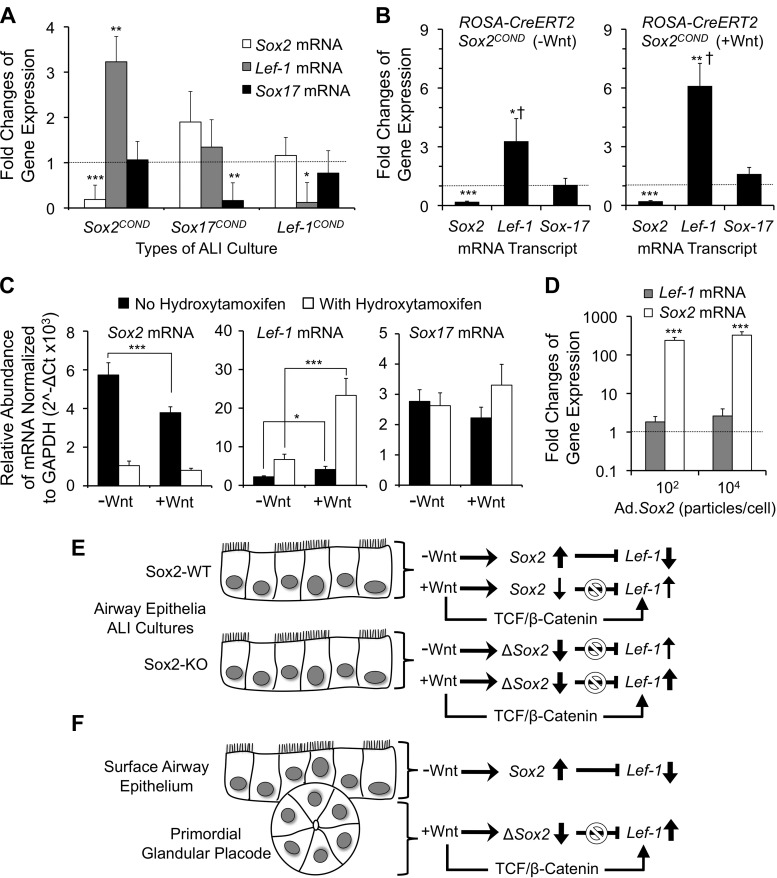
Studies have shown that Sox2 and Sox17 can bind directly to β-catenin to form a transcriptional inhibitor complex that represses the expression of Wnt responsive genes (32, 55). Furthermore, Sox17 has been shown to inhibit Wnt-mediated transcriptional activation of the Lef-1 promoter (30). Both Sox2 and Sox17 are downregulated in airway PGPs (Fig. 1) (30); thus it remained unclear if repression of both Sox2 and Sox17 is necessary for the induction of Lef-1 expression in developing PGPs. To approach this question, we generated polarized tracheal epithelia from homozygous Sox2 and Sox17 conditional knockout (Sox2COND/COND and Sox17COND/COND) mice as air-liquid interface (ALI) cultures. Cultures were then infected with Cre-expressing recombinant adenovirus (Ad.CRE) or a control adenovirus lacking a transgene insert (Ad.Control). As a control to confirm that Lef-1 gene expression did not directly influence expression of Sox2 or Sox17, we also evaluated ALI cultures from Lef-1 conditional knockout (Lef-1COND/COND) mice. Three days following infection with Ad.CRE, Sox2, Sox17, and Lef-1 transcripts were reduced ∼5-fold on the respective conditional backgrounds, compared with samples infected with Ad.Control virus (Fig. 3A), confirming efficient knockdown. Knockdown of Sox2 significantly increased Lef-1 mRNA levels 3.2-fold, while knockdown of Sox17 had no significant effect on Lef-1 mRNA expression (Fig. 3A). Furthermore, knockdown of Lef-1 had no significant effect on either Sox2 or Sox17 mRNA levels.
Fig. 3.
Sox2 deletion and Wnt stimulation synergistically enhances Lef-1 mRNA expression in polarized mouse tracheal airway epithelia grown at an air liquid interface (ALI). A: tracheal airway epithelial cells were harvested from Sox2COND/COND, Sox17COND/COND, and Lef-1COND/COND mice and used to generate ALI cultures of polarized airway epithelia. The cultures were then infected with either CRE-expressing recombinant adenovirus (Ad.CRE) or a control virus lacking a transgene (Ad.Control) at multiplicities of infection (MOI) of 104 particles/per cell. At 3 days postinfection, the cultures were harvested and total RNA was generated. Quantitative PCR for Sox2, Lef-1, Sox17, and GAPDH transcripts was performed using reverse transcribed cDNA. The graph shows the fold change in mRNA expression for Sox2, Lef-1, and Sox17 genes (normalized to GAPDH) after Ad.CRE infection for each type of ALI cultures. Fold changes in expression were calculated using the ΔΔCT method comparing Ad.CRE to Ad.Control conditions. B: primary tracheal epithelial cells were harvested from ROSA-CreERT2/Sox2COND/COND mice and used to generate ALI cultures. During the first 96 h of seeding onto filters, cultures were treated with 4-hydroxytamoxifen or vehicle (DMSO). Cultures were then allowed to develop an ALI for ∼2 wk and subsequently refed basolaterally with USG medium lacking (left panel, −Wnt) or containing (right panel, +Wnt) a mixture of Wnt1 and Wnt3a for 72 h prior to harvesting for total RNA. Quantitative PCR for Sox2, Lef-1, Sox17, and GAPDH transcripts was then performed using reverse-transcribed cDNA. The graph shows the fold change in mRNA expression for Sox2, Lef-1, and Sox17 genes (normalized to GAPDH) following treatment with 4-hydroxytamoxifen. Fold changes in expression were calculated using the ΔΔCT method comparing 4-hydroxytamoxifen to vehicle treatment conditions in the absence of Wnt (left panel) or the presence of Wnt (right panel). C: data from B are plotted as the relative abundance of Sox2, Lef-1, and Sox17 transcripts normalized to GAPDH (2−ΔCt) in the presence and absence of Wnt and/or 4-hydroxytamoxifen. D: tracheal ALI cultures were generated from wild-type C57B6 mice and then infected with either Sox2-expressing recombinant adenovirus (Ad.Sox2) or a control virus lacking a transgene (Ad.Control) at MOIs of 102 and 104 particles/per cell. At 3 days postinfection, the cultures were harvested, total RNA generated, and quantitative PCR for Sox2, Lef-1, and GAPDH transcripts was performed using reverse-transcribed cDNA. The graph shows the fold changes in mRNA expression for Sox2 and Lef-1 genes (normalized to GAPDH) after Ad.Sox2 infection at the indicated MOIs. Fold changes in expression were calculated using the ΔΔCT method comparing Ad.Sox2 to Ad.Control infection conditions. The values in all panels depict the means ± SE for the following number of independent ALI minicells for each data point (A: n = 6; B: n = 12 left panel, n = 6 right panel; and D: n = 6). Dashed lines in A, B, and D mark the threshold value [equal to 1 for Ad.Control infected samples (A and D) and without 4-hydroxytamoxifen treatment (B)] for changes in gene expression. Differences between marked groups and this threshold value (or marked comparisons in C) were significant by the nonparametric Mann-Whitney test (*P < 0.01, **P < 0.005, ***P < 0.001). B: Lef-1 mRNA levels in between +Wnt- and −Wnt-treated samples were also significantly different by nonparametric Mann-Whitney test (†P < 0.008). All data points not marked were not significantly different from threshold values. E: summary of functional studies in Sox2COND/COND polarized mouse airway epithelia demonstrating Sox2 represses Lef-1 expression while Wnt stimulation activates Lef-1 expression to a greater extent in the absence of Sox2. F: working model for Sox2 regulation of Lef-1 gene transcription during submucosal gland development. In the surface airway epithelium, the absence of a Wnt signal maintains high levels of Sox2 expression leading to inhibition of the Lef-1 promoter. In the PGP, local Wnt/β-catenin signals lead to posttranscriptional repression of Sox2 mRNA and/or Sox2 protein. In the absence of Sox2 expression, Wnt/TCF/β-catenin signals can directly activate Lef-1 expression and glandular morphogenesis.
We next used an alternative approach to conditionally delete the Sox2 gene in ALI cultures generated from ROSA-CreERT2/Sox2COND/COND mice and assess changes in the level of Sox2, Sox17, and Lef-1 transcript in the presence and absence of Wnt stimulation and/or Sox2 gene deletion. Findings from these studies (Fig. 3B) confirmed results with Ad.CRE mediated deletion of Sox2. In the absence of Wnt stimulation, Sox2 deletion with 4-hydroxytamoxifen induced Lef-1 transcript levels 3.3-fold while leaving Sox17 expression unchanged (Fig. 3B, left panel). Furthermore, in the presence of Wnt simulation, Sox2 deletion led to greater induction of Lef-1 transcripts (6.1-fold), which was significantly greater (P < 0.008) than that observed in the absence of Wnt (Fig. 3B, right panel). Interestingly, in the presence of Sox2, Wnt stimulation significantly reduced Sox2 transcripts by 34%, while significantly stimulating Lef-1 transcription 1.8-fold (Fig. 3C). These findings demonstrate that Wnt signals can influence Sox2 mRNA levels in airway epithelia. The most significant induction in Lef-1 transcripts (10.4-fold, P < 0.0001) was observed between Sox2-expressing (−Wnt) and Sox2-deleted (+Wnt) ALI cultures (Fig. 3C). In contrast to Sox2 deletion, ectopic expression of Sox2 using a recombinant adenovirus did not significantly change Lef-1 transcript levels despite a >100-fold increase in Sox2 expression (Fig. 3D). However, under this condition Lef-1 transcript levels nonsignificantly increased 1.8- to 2.6-fold in a vector dose-dependent manner above cultures infected with a control adenovirus lacking a transgene.
Cumulatively, these findings support the hypotheses that Sox2 represses transcription of the Lef-1 gene in the surface tracheal airway epithelium and that Wnt signals enhance Lef-1 transcription while also partially reducing the abundance of Sox2 transcripts (Fig. 3E). The combined deletion of Sox2 with Wnt stimulation provided the maximal induction of Lef-1 in airway epithelia. Combining these findings with those in Figs. 1 and 2, we propose a working model for understanding Lef-1 regulation in the PGP (Fig. 3F). In the absence of a Wnt signal, the SAE maintains high levels of Sox2, which in turn represses Lef-1 transcription. Downregulation of Sox2 in PGPs, by activated Wnt signals and likely other Wnt-controlled factors specific to the PGP, leads to derepression of the Lef-1 promoter and promotes Wnt/β-catenin-mediated activation of transcription.
Sox2 inhibits β-catenin-dependent activation of the Lef-1 promoter.
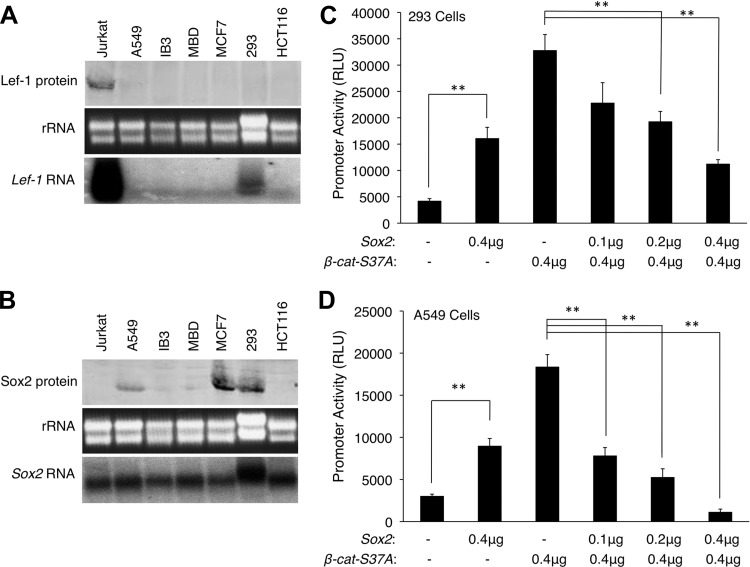
In previous studies, we generated transgenic mice that express LacZ or GFP from a 2.5-kb human Lef-1 promoter (10, 11, 29). These reporter mice recapitulate endogenous Lef-1 immunolocalization patterns in tracheal PGPs, nasal PGPs, and developing hair follicles, and demonstrated that the induction of Lef-1 expression in PGPs is controlled at the transcriptional level. To test our hypothesis that Sox2 inhibits transcription of the Lef-1 promoter (Fig. 3, E and F), we utilized this 2.5-kb human Lef-1 promoter driving firefly luciferase (30) in transient transfection assays to evaluate how Sox2 modulates β-catenin-dependent activation of the Lef-1 promoter. We utilized two human cell lines (A549 and HEK293) that were chosen following a screen of six cell lines for endogenous Sox2 and Lef-1 protein and mRNA expression (Fig. 4, A and B). The A549 lung carcinoma cell line did not express Lef-1 protein or mRNA but did express moderate levels of Sox2 protein and mRNA. The 293 kidney cell line expressed low levels of Lef-1 mRNA, but lacked Lef-1 protein, and also expressed higher levels of Sox2 protein and mRNA compared with A549 cells. Thus A549 cells are more similar to the in vivo expression patterns of Sox2 and Lef-1 in the tracheal surface airway epithelial cells of neonatal mice. It should be noted that both Lef-1 mRNA (12) and protein (Fig. 1) expression are limited to PGPs and not expressed in the surface airway epithelium.
Fig. 4.
Sox2 inhibits β-catenin-dependent activation of the Lef-1 promoter. A and B: endogenous protein and mRNA expression patterns for Lef-1 (A) and Sox2 (B) were evaluated in 6 easily transfectable cell lines (A549, IB3, MBD, MCF7, 293, and HCT116 cells). Jurkat cells were used as a control for high-level Lef-1 expression. Protein levels were determined by Western blotting of 100 μg protein with anti-Lef-1 and anti-Sox2 antibodies. mRNA levels were determined by Northern blotting of 20 μg of total mRNA. rRNA banding patterns are shown in the ethidium bromide-stained agarose gel prior to transfer. From these studies, 293 and A549 cells were chosen for expression analysis of the Lef-1 promoter. C and D: a human 2.5-kb Lef-1 promoter-luciferase reporter (Lef-1-Luciferase) construct was used to evaluate promoter activation in the presence of dominant-active β-catenin-S37A and/or Sox2. Cells were Lipofectamine transfected with a constant amount pLef-1-Luciferase (0.2 μg) together with the indicated plasmid amount of β-catenin-S37A and/or Sox2 expression plasmid. All transfections also contained 0.01 μg of a pCMV-Renilla luciferase plasmid for normalization of transfection efficiency. For all transfections, the total DNA concentration was identical and normalized by adding empty-vector control plasmid (pcDNA). Relative activity of the Lef-1 promoter is plotted as relative light units (RLU) for experiments in 293 (C) and A549 cells (D). Values represent means ± SE for n = 6 independent transfections. RLU values were normalized to Renilla luciferase expression for each transfection. Differences between marked (**) comparisons were significant by the nonparametric Mann-Whitney test (P < 0.002).
Since transcription of the Lef-1 promoter has been shown to respond to canonical Wnt/β-catenin signaling, we overexpressed a dominant-active mutant β-catenin-S37A in combination with various dosages of Sox2 expression plasmid. Transcriptional activity of the Lef-1 promoter increased significantly (3.8-fold in 293 cells and 3-fold in A549 cells) when Sox2 alone was overexpressed (Fig. 4, C and D). These findings are similar to those in primary ALI cultures with forced expression of Sox2 (Fig. 3D), although the induction of Lef-1 transcripts was not significant in this case. Studies on both A549 cells and 293 cells confirmed that the dominant-active β-catenin dramatically induces the Lef-1 promoter by 6.2- and 7.7-fold, respectively (Fig. 4, C and D). Overexpressing Sox2 inhibited this induction in a dose-dependent manner (Fig. 4, C and D). Indeed, these observations are consistent with previous findings that Sox2 can modestly induce the TCF-responsive Topflash reporter, while more significantly inhibiting β-catenin-dependent activation of the Topflash reporter (32). Consistent with some level of cell-type specificity in the regulation of the Lef-1 promoter, A549 cells demonstrated a 13.4-fold reduction in β-catenin-dependent transcription following Sox2 overexpression (Fig. 4D), while in 293 cells Sox2-mediated inhibition was only 3.0-fold (Fig. 4C). This cell-type-specific difference is perhaps not surprising given that 293 cells are able to maintain endogenous Lef-1 mRNA expression in the setting of elevated Sox2 protein (Fig. 4, A and B). This contextual biology suggests that other transcription factors likely influence Sox2 action on the Lef-1 promoter in both a positive and negative fashion, and in a manner that is controlled by the extent of β-catenin activation.
Sox2 binds both SOX and TCF family binding sites on the endogenous Lef-1 promoter.
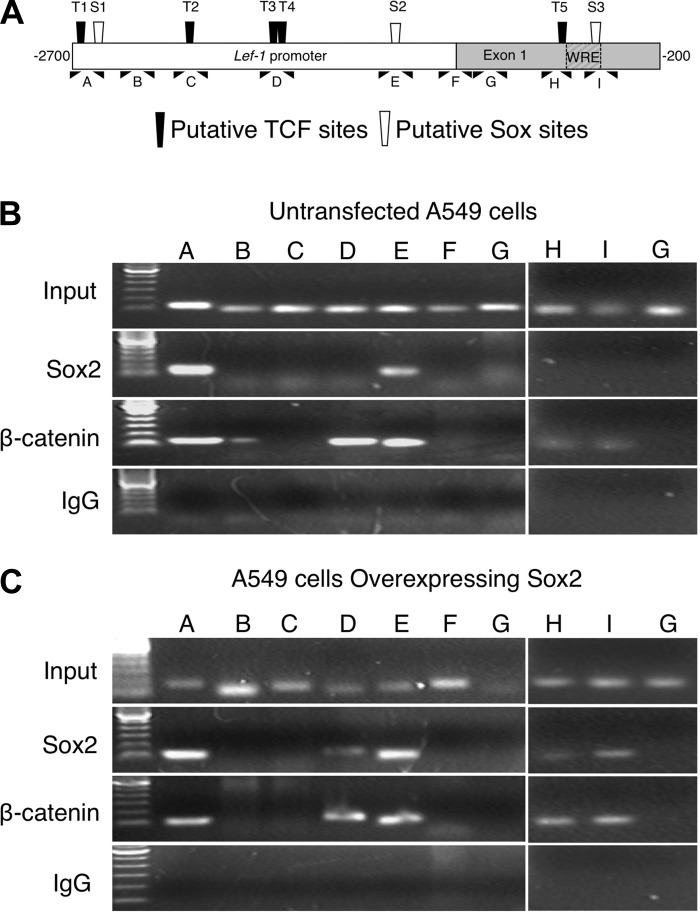
To confirm that Sox2 binds to the Lef-1 promoter we performed ChIP analysis in A549 cells in the presence and absence of ectopic Sox2 overexpression. A549 cells were chosen for this analysis since they most closely modeled the expression patterns of Sox2 and Lef-1 in mouse tracheal surface airway epithelia. Immunoprecipitation of Sox2 and β-catenin cross-linked DNA was assayed by PCR using primer sets spanning nine different segments of the 2.5-kb Lef-1 promoter sequence, including three putative SOX family binding sites (S1, S2, and S3) and five T-cell factor (TCF) family binding sites (T1, T2, T3, T4, and T5) (Table 1 and Fig. 5A). In A549 cells, endogenous Sox2 strongly associated with two assessed segments (T1/S1 and S2), which contain putative SOX family binding sites (Fig. 5, A and B). Sites “T1” and “S1” were separated by only 30 bp and thus were indistinguishable by ChIP; therefore this approach could not determine if endogenous Sox2 associates with one or both of these sites in this region. Interestingly, all of the regions within the Lef-1 promoter that bound endogenous Sox2 also immunoprecipitated with β-catenin, which is consistent with previously reported data indicating that Sox proteins can form complexes with β-catenin (30, 32). Three additional sites (T3/T4, T5, and S3) also selectively bound β-catenin, but not Sox2; these sites were previously shown to bind Sox17 and/or TCF4 (30). Moreover, control IgG failed to immunoprecipitate any Lef-1 promoter DNA segments, demonstrating specificity of the assay. Furthermore, segments of the Lef-1 promoter that did not contain putative binding sites showed no PCR amplification (Fig. 5B).
Fig. 5.
ChIP analysis of Sox2 and β-catenin binding sites in the endogenous Lef-1 promoter of A549 cells. A: displayed is a schematic representation of 9 PCR fragments used to survey the Lef-1 promoter by ChIP. Each PCR fragment covered ∼100 bp of the promoter. The location of TCF and Sox consensus sites are marked. B and C: ChIP was performed on untransfected A549 cells (B) or A549 cells transiently transfected with a Sox2 expression construct (C), using Sox2, β-catenin, or nonimmunogenic IgG as the immunoprecipitation antibody. Agarose gels of the PCR products are shown. Lane labels correspond to the PCR region shown in A. Data are representative of 3 independent experiments.
To model how Sox2 levels influence interaction with the Lef-1 promoter, ChIP analysis was performed in A549 cells transfected with a Sox2-expressing plasmid. Indeed, we found that forced expression of Sox2 led to binding within additional regions of the Lef-1 promoter. When overexpressed, Sox2 associated with three additional TCF family binding sites (T3/T4, and T5) and one additional SOX site (S3), which is within the WRE of the Lef-1 promoter. Again these sites were also associated with β-catenin (Fig. 5C). Taken together, these findings demonstrate that both Sox2 and β-catenin can physically interact with multiple sites in the Lef-1 promoter in a dose-dependent fashion and suggest a mechanism for the inhibition of β-catenin-dependent induction of the Lef-1 promoter in cells that express elevated Sox2, such as the surface airway epithelium in the developing mouse tracheal airway.
Confirmation of putative Sox2 binding sites in the Lef-1 promoter by NoShift assay.
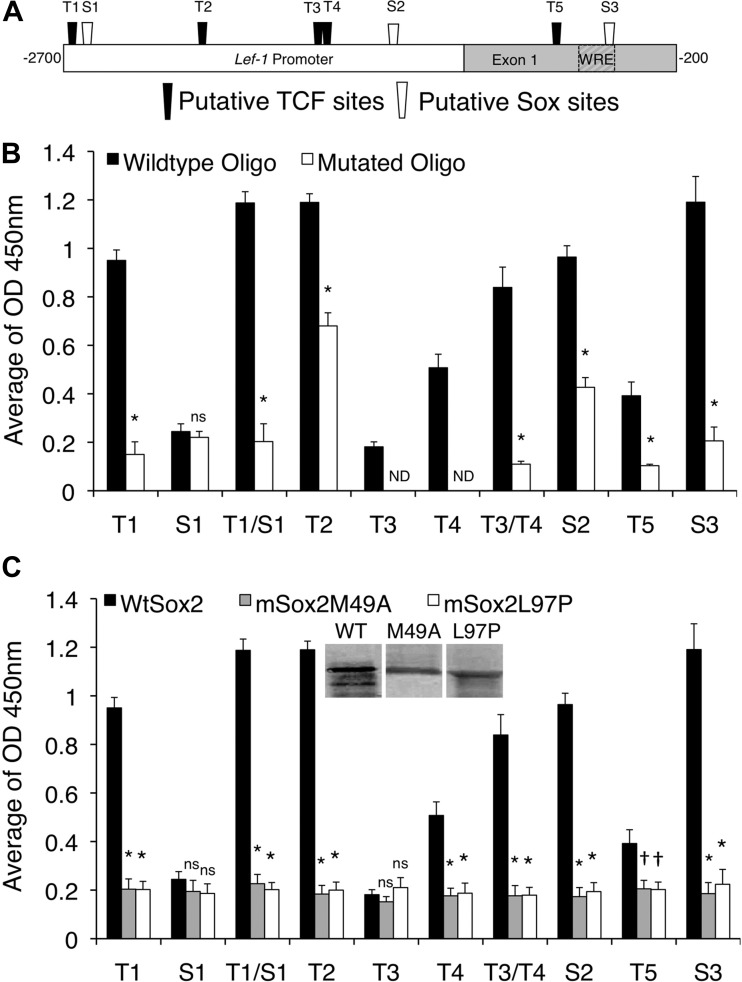
We next assessed the affinity of Sox2 binding to defined short segments of the Lef-1 promoter, focusing our analysis on the TCF (T1–T5) and SOX (S1–S3) consensus binding sites we identified by ChIP analysis. Notably, some of the TCF consensus sequences share most of their essential nucleic acids with SRY/Sox consensus sequences (CWTTGWW) (19, 30). We first engineered a recombinant, His-V5-tagged, full-length, wild-type (WT) Sox2 protein. Biotin-conjugated, double-stranded oligonucleotides, containing each of the potential WT or mutated TCF and Sox binding sequences from the Lef-1 promoter (Table 4 and Fig. 6A), were incubated with purified Sox2 protein and bound to streptavidin-coated plates. The amount of bound Sox2 was then detected using an anti-V5 antibody. Results from these studies confirmed that more WT-Sox2 protein bound to the WT oligonucleotides, compared with the mutated oligonucleotides, containing putative sites S2 and S3, as well as TCF family binding sites T1, T2, T3/T4, and T5 (Fig. 6B). Surprisingly, the S1 putative SOX family binding site did not associate with Sox2 in isolation, suggesting that Sox2 binding to the T1/S1 regions seen by ChIP was most likely facilitated through the T1 site. However, the combined T1/S1 oligo bound Sox2 more efficiently than T1 alone (P < 0.0024, by Mann-Whitney test), suggesting that the S1 site may either bind Sox2 in a T1-context-dependent manner or enhance the binding of Sox2 to T1.
Fig. 6.
Sox2 binds to several putative Sox and TCF family binding sites within the Lef-1 promoter. We analyzed the affinity of bacterially generated Sox2 to eight transcription factor-binding motifs within the Lef-1 promoter using an in vitro NoShift assay. A: schematic of the Lef-1 promoter with five TCF and three Sox family consensus binding sites. Eight wild-type and mutated biotin labeled oligonucleotides were generated to survey the Sox2 binding affinity on these binding motifs (Table 4). B: relative Sox2 binding affinity to wild-type and mutant oligos for the indicated sites. Data represent means ± SE for n = 12 samples. Significant differences using the Mann-Whitney test compared with the wild-type oligo and mutated oligo for each site: *P < 0.001; ns, nonsignificant. C: relative binding of wild type Sox2, Sox2M49A, and Sox2L97P to the wild-type oligo for the indicated sites. Inset shows Western blot of His-tagged Sox2 proteins isolated from bacteria. Data represent means ± SE for n = 12 samples. Significant differences using the Mann-Whitney test compared the wild-type Sox2 vs. each of the mutant Sox2 forms: †P < 0.05; *P < 0.001.
Since we observed variable binding to mutated oligonucleotides in the above studies, two Sox2 mutants (Sox2-L97P and Sox2-M49A) that are defective for DNA binding were used as additional negative controls (Fig. 6C). The L97P mutation was previously reported to disrupt DNA binding activity by Sox2 (23) and the M49A mutation is an analogous mutation in the HMG domain known to disrupt Sox17 DNA binding (30, 41). These mutant Sox2 constructs were used as negative controls to test the binding specificity of purified Sox2 protein. These studies confirmed that significantly greater WT-Sox2 protein bound to oligonucleotides containing T1, T1/S1, T2, T4, T3/T4, T5, S2, and S3 sites, compared with mutant-Sox2 proteins (Fig. 6C). Interestingly, although the T3 site did not bind WT-Sox2 in isolation, inclusion of this site in the T3/T4 oligo also significantly enhanced Sox2 binding over that of T4 alone (P < 0.0120, by Mann-Whitney test), suggesting that the affinity of Sox2 at T3/T4 is synergistically enhanced by the two adjacent TCF sites. Overall, these binding studies demonstrate that Sox2 binds to numerous sites in the Lef-1 promoter and that these sites overlap with previously identified TCF binding sites.
Mutations of Sox2 binding sites in the Lef-1 promoter alter Sox2- and β-catenin-dependent transcriptional changes.
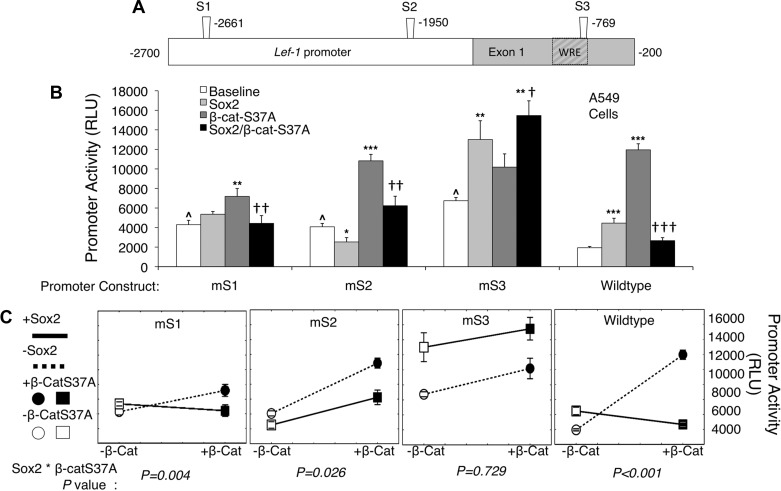
To evaluate the functional importance of Sox2 binding sites in the Lef-1 promoter on the repression of β-catenin-dependent transcription, we generated three luciferase reporter plasmids harboring mutations at S1, S2, or S3 within the Lef-1 promoter (Fig. 7A) (30). We assessed transcriptional activities of these mutated Lef-1 promoters in A549 cells expressing dominant active β-catenin (β-catenin-S37A) and/or Sox2. Notably, all of these mutations significantly elevated the baseline activity of the Lef-1 promoter, suggesting that these sites may have a repressive role in Lef-1 expression (Fig. 7B). Sox2-dependent induction of the Lef-1 promoter was significantly reduced by S1 and S2 mutations, but not the S3 mutation. All mutations retained significant induction in the presence of active β-catenin overexpression, but the level of induction was reduced compared with the WT Lef-1 promoter primarily due to the elevated baseline expression of the S1, S2, and S3 mutant promoters in the absence of active β-catenin. In this regard the level of expression of the WT vs. S2 and S3 mutant promoters was not significantly different in the presence of active β-catenin expression. The marginal, but significant, reduction in luciferase expression of the S1 mutant in the presence of activated β-catenin may reflect its close proximity to the T1 TCF-binding site that may play a role in β-catenin/TCF-mediated transcriptional activation of the promoter. Importantly, the ability of Sox2 overexpression to inhibit β-catenin-dependent activation of the Lef-1 promoter was most significantly blocked by the mutation at the S3 site (Fig. 7B). Analysis by two-way ANOVA, considering Sox2 and dominant-active β-catenin as independent variables, also demonstrated a significant interaction between Sox2 and β-catenin-S37A on the activity of all Lef-1 promoter constructs except the S3 mutant (Fig. 7C); this alternative analysis tests whether Sox2 changes the effect of β-catenin-S37A. The relationship between β-catenin-S37A and Sox2 on the activation of the Lef-1 promoter is visually apparent with each set of lines connecting the promoter activity (for each mutant and WT) in the presence or absence of overexpressed β-catenin-S37A and/or Sox2 (Fig. 7C). Promoter constructs mS1, mS2, and WT demonstrate a statistically significant relationship between β-catenin-S37A and Sox2. However, the relationship is no longer significant in construct mS3 (Fig. 7C), which suggests that S3 is the primary Sox site where interaction between β-catenin and Sox2 influence Lef-1 promoter transcription. Cumulatively, these data suggest that Sox2 regulates Lef-1 transcription independently of β-catenin at the S1 and S2 Sox2-binding sites within the Lef-1 promoter, while at the S3 binding site the inhibition of the Lef-1 promoter by Sox2 is β-catenin-dependent.
Fig. 7.
Sox2 binding-domains in the Lef-1 promoter are inhibitory in nature within A549 cells. A: schematic representation of the 2.5-kb human Lef-1 promoter with the position of candidate Sox2 protein binding sites (S1, S2, and S3) determined by ChIP and NoShift assays. Each Sox2 site was mutated by changing A to C, T to G, C to A, and G to T, to generate mutant Lef-1 promoter luciferase expression constructs (called mS1, mS2, and mS3). B: each of the four Lef-1 promoter luciferase expression constructs (wild type, mS1, mS2, and mS3) were evaluated following transient transfection of 0.2 μg DNA into A549 cells under four conditions: 1) transfection of the Lef-1-luciferase reporter alone (baseline), 2) cotransfection with 0.4 μg Sox2 expression plasmid, 3) cotransfection with 0.4 μg β-catenin-S37A expression plasmid, or 4) cotransfection with both Sox2 and β-catenin-S37A expression plasmids at 0.4 μg each. For all transfections, the total DNA concentration was identical (1.0 μg) and normalized by adding empty-vector control plasmid (pcDNA). Background luciferase activity was determined following transfection of 1.0 μg empty plasmid pcDNA, and these values were subtracted from experimental values. All transfections also contained 0.01 μg of a pCMV-Renilla luciferase plasmid for normalization of transfection efficiency for each data point. Values represent means ± SE of relative Light Units (RLU) at 48 h posttransfection for n = 9 independent transfections. RLU values were normalized to Renilla luciferase expression for each transfection. Differences between marked comparisons were significant by the nonparametric Mann-Whitney test (comparisons to baseline expression within each group: *P < 0.01, **P < 0.005, ***P < 0.001; comparison of expression in the presence of β-catenin-S37A vs. Sox2/β-catenin-S37A within each group: †P < 0.05, ††P < 0.01; †††P < 0.001; comparisons of baseline expression across groups to wild type Lef-1 promoter: ^P < 0.001). C: alternative representation of the data in B to emphasize differences in the interactions between Sox2 and β-catenin-S37A on expression from the various Lef-1 promoter constructs. Dashed lines indicate the changes in promoter activity (RLU) when β-catenin-S37A is added in the absence of Sox2. Solid lines indicate the changes in promoter activity (RLU) when β-catenin-S37A is added in the presence of Sox2. Significant differences were assessed by 2-way ANOVA using Sox2 and β-catenin-S37A as independent variables. P values shown below each dataset evaluate interaction between Sox2 and β-catenin-S37A (Sox2 * β-catenin-S37A).
DISCUSSION
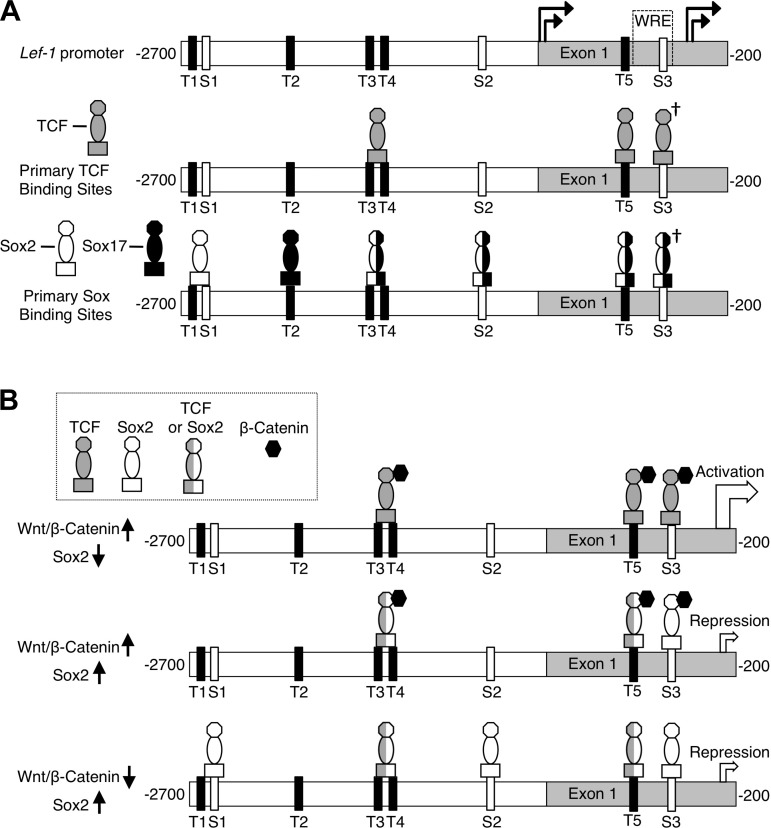
Wnt/β-catenin and Sox signaling play important roles in regulating lung development and airway stem/progenitor cell phenotypes following injury/repair (4, 7, 8, 18, 31, 52). It is clear from previous studies that Wnt signaling is required for the formation of airway SMGs in mice (10) and that the induction of Lef-1 expression by Wnt/β-catenin is critical for this process (10–13, 16). Sox family members play important roles in regulating Wnt/β-catenin signals by acting as both antagonists and agonists (24, 30, 32, 41). In the present study, we demonstrate that Sox2 represses Lef-1 expression in polarized airway epithelia, directly binds to five sites in the Lef-1 promoter, and can modulate transcription from the Lef-1 promoter through direct binding to at least three sites in the promoter (Fig. 8A). Interestingly, Sox2 has both inductive and repressive transcriptional effects on Lef-1 promoter activities that depend on the presence of β-catenin activation. These findings support a role for Sox2-dependent transcriptional control of the Lef-1 promoter in the specification of glandular progenitors during both early and late morphogenic events involved in SMG development.
Fig. 8.
Schematic summary of Sox2 binding to the Lef-1 promoter and potential mechanisms of Wnt/β-Catenin activation and Sox2 repression of Lef-1 transcription in airway epithelia. A, top: TCF (T) and Sox (S) consensus candidate binding sites in the Lef-1 promoter and positions of the four transcriptional start sites (16). A, middle: primary binding sites for TCF1 and TCF4 as previously reported by others to be important in Wnt/β-catenin activation of the Lef-1 promoter (2, 3, 16, 26, 30). A, bottom: Sox2 and Sox17 binding sites in the Lef-1 promoter demonstrated in the present study for Sox2 by ChIP and oligonucleotide binding and a previous study for Sox17 using similar assays (30). Sites that have been shown to bind both Sox2 and Sox17 are marked by half-white and half-black icons. The † marking denotes that the previous study (30) evaluating Sox17 and TCF4 binding by ChIP did not distinguish between binding at the T5 and S3 site, since the probe spanned both binding sites. However, data in Fig. 5 demonstrating that β-catenin associated with both T5 and S3 sites at steady state in the absence of Sox2 binding (i.e., without forced expression of Sox2) suggest that Sox17 and/or TCF4 likely bind to T5 and S3. B: model of proposed TCF/Sox2 occupancy of the Lef-1 promoter under various states of Wnt/β-catenin activation and Sox2 expression. B, top: when Wnt/β-catenin activation is high and Sox2 expression is low (i.e., in the primordial glandular placode), TCFs bound to β-catenin activate Lef-1 promoter transcription. B, middle: when Sox2 expression is reactivate under conditions of sustained Wnt/β-catenin activation (i.e., during later stages of glandular tubulogenesis), the Lef-1 promoter is primarily inhibited by binding of Sox2 to the S3 site based on A549 Lef-1 promoter expression studies with the S3 binding site mutant (Fig. 7). The TCF/Sox2 occupancy of T3/T4 and T5 sites under these conditions remains unclear, but Sox2 may also compete for TCF binding at these sites. B, bottom: when Wnt/β-catenin activation is low and Sox2 expression is high (i.e., as seen in the surface airway epithelium), Sox2 maintains low expression of the Lef-1 promoter by binding to S1, S2, and S3. Mutations at each of these sites elevates baseline activity of the Lef-1 promoter in A549 cells (Fig. 7). The TCF/Sox2 occupancy of T3/T4 and T5 sites under these conditions remains unclear, but Sox2 may also compete for TCF binding at these sites. This model does not attempt to predict alternative scenarios by which Sox2 overexpression can activate the Lef-1 promoter in A549 cells through binding to the S1 and S2 sites. Mutations at either of these sites blocks Sox2-mediated activation of the Lef-1 promoter in A549 cells (Fig. 7).
Localization of Sox2 and Lef-1 in the developing tracheal airway demonstrates distinct domains of expression; Lef-1 is selectively expressed in PGPs while Sox2 expression is restricted to the SAE. Cells of the PGP that induce Lef-1 and lack Sox2 expression also demonstrated significant Wnt activation. Consistent with the positive influence of Wnt signals on the induction of Lef-1, Wnt stimulation of tracheal ALI cultures induced Lef-1 while suppressing Sox2 expression. Wnt-mediated stimulation of Lef-1 expression in tracheal ALI cultures was also dramatically enhanced by Sox2 deletion, consistent with the expression patterns of Lef-1, Sox2, and Wnt signals in the PGP. Since deletion of Sox2 in polarized airway epithelia induces Lef-1 mRNA expression, but deletion of Lef-1 does not alter Sox2 expression in tracheal epithelium, downregulation of Sox2 in PGP likely precedes the induction of Lef-1. Interestingly, Sox17 is also downregulated in airway PGPs and can directly regulate Lef-1 promoter activity in cell lines when overexpressed (30). However, deletion of Sox17 in polarized airway epithelia did not significantly enhance Lef-1 mRNA expression in polarized airway epithelia. Thus, in differentiated airway epithelia, Sox2 appears to play a dominant role in suppressing Lef-1 expression. Nonetheless, it is interesting that Sox2 and Sox17 binding overlaps at three sites in the Lef-1 promoter (T3/4, S2, T5, and S3) (30), while each transcription factor also retains a unique binding site (T1/S1 and T2) (Fig. 8A). Thus it remains possible that dynamic changes in PGP expression of multiple Sox genes may regulate Lef-1 expression. Since no single cell line can duplicate the transcriptional program and environment of PGPs in vivo, these dynamic changes in Sox gene expression by PGPs are important when interpreting results from cell line studies evaluating the mechanism of Sox2 regulation of the Lef-1 promoter.
The initiating factor(s) that drives changes in Sox/Lef-1 expression within PGPs is currently unknown; however, several lines of evidence support the hypothesis that Wnt/β-catenin signals coordinate these changes in expression. First, previous studies using Lef-1 promoter reporter transgenic mice crossed to Wnt3A knockout mice have demonstrated that Wnt3A signals are required for activation of the Lef-1 promoter in PGPs (10). Second, our studies using BAT-gal mice demonstrate that β-catenin-dependent TCF/Lef-1 transcription is selectively activated in Sox2-negative cells of the PGPs. Third, Wnt stimulation of mouse tracheal airway epithelium represses Sox2 while enhancing Lef-1 mRNA expression. Last, deletion of Sox2 in ALI mouse tracheal airway epithelium induces Lef-1 mRNA expression in a manner synergistically enhanced by Wnt stimulation. Taken together, these findings demonstrate that Wnt/β-catenin signals direct changes in Sox2 and Lef-1 expression within airway epithelium and thus are candidate pathways for controlling Lef-1 gene transcription in the PGP.
To clarify whether Wnt/β-catenin signals are required for Sox2-mediated repression of the Lef-1 promoter, we performed in vitro reporter studies in transfected 293 and A549 cells. As hypothesized, Sox2 overexpression inhibited β-catenin-dependent activation of the Lef-1 promoter in a dose-dependent manner. This finding is consistent with selective activation of Lef-1 expression in PGPs demonstrating activated β-catenin-dependent transcription and low levels of Sox2 expression. However, we also observed that Sox2 expression alone induced transcription from the Lef-1 promoter in both 293 and A549 cells. Similarly, forced expression of Sox2 in polarized airway epithelium also demonstrated a trend to induction of Lef-1 mRNA. The latter findings are inconsistent with studies in polarized airway epithelia demonstrating that deletion of Sox2 induces Lef-1 mRNA expression in the absence of applied Wnts. One explanation for these contradictory findings could be that polarized airway epithelia maintain some level of β-catenin activation required for the induction of Lef-1 mRNA following deletion of Sox2. Indeed, Xgal staining of polarized airway epithelia generated from BAT-gal mice demonstrates low levels of LacZ expression that appears clonal in origin (data not shown), suggesting that a specific cellular phenotype in these epithelia maintain β-catenin activation. Thus we conclude that in vitro polarized airway epithelia maintain a subset of cells that are responsive to Sox2-dependent repression of β-catenin-dependent Lef-1 expression. Alternatively, the ability of Sox2 to activate or repress the Lef-1 promoter in cell lines, and Lef-1 mRNA expression in primary airway epithelia, could suggest that the function of Sox2 is dependent on the intensity of β-catenin signals.
β-Catenin-dependent activation of the Lef-1 promoter is controlled by two or more primary TCF1 and TCF4 binding sites (T3/4, T5, and/or S3) (2, 3, 16, 26, 30) and a WRE that functions to repress transcription in the absence of a Wnt signal (Fig. 8A) (16). Several of the TCF sites within the Lef-1 promoter have also been shown to bind Sox17 (30). Thus we assessed the extent of Sox2 binding to all known TCF and Sox binding sites within the Lef-1 promoter. Utilizing ChIP assays, we identified five Sox2 binding regions in the promoter that contained TCF and/or Sox binding consensus sequences (T1/S1, S2, T3/T4, T5, and S3). In vitro oligonucleotide binding experiments confirm the position and sequences of these Sox2 binding sites. Native low levels of Sox2 in A549 cells bound T1/S1 and S2 sites by ChIP; these sites also bound β-catenin. Interestingly, ChIP Sox2 binding at three of the five sites was observed only when Sox2 was overexpressed by transient transfection (T3/T4, T5, and S3), suggesting that the level of Sox2 expression can control binding at these sites. These three sites (T3/T4, T5, and S3) all bound β-catenin in the absence of Sox2 overexpression, and based on previous studies, β-catenin binding at these sites is likely mediated through TCFs (30). Since increased expression of Sox2 leads to binding at these sites, it is plausible that these sites compete for TCF/β-catenin and Sox2/β-catenin binding (Fig. 8B).
Our findings on Sox2/β-catenin binding to the Lef-1 promoter support a model whereby Sox2 could directly regulate Wnt-dependent transcription of the promoter in airway epithelia (Fig. 8, B). The mechanism of suppression of Sox2 in the PGP remains unclear, but given that the Sox2 promoter is not downregulated in the PGP (Fig. 1G), this mechanism likely involves either posttranscriptional regulation of Sox2 mRNA or posttranslational regulation of the Sox2 protein. Our studies demonstrating that Wnt treatment of polarized mouse tracheal epithelia significantly reduces Sox2 mRNA (albeit by only 34%) lend support for a posttranscriptional level of Sox2 mRNA regulation. MicroRNA-mediated regulation of Sox2 has indeed been previously reported in embryonic stem cells (44, 51). Alternatively, Sox17 promotes proteasome-dependent degradation of both β-catenin and TCF proteins (41). Whether similar mechanisms for Sox2 protein degradation exist remains to be determined. Such potential mechanisms of Sox2 regulation in the PGP warrant further investigation. However, it is most plausible that Wnt signals at sites of PGP formation signal activation of Lef-1 transcription, while these same Wnt signals simultaneously promote the degradation of Sox2 protein and/or Sox2 transcripts (Fig. 3F). Functional mutagenesis studies of the three Sox consensus binding sites in the promoter demonstrated that mutagenesis of S3 within the WRE most significantly impairs the ability of Sox2 to repress β-catenin-dependent Lef-1 promoter transcription. These findings are consistent with previous findings demonstrating the WRE serves as a repressor of Lef-1 promoter transcription, while also being required for Wnt-mediated induction (16). By contrast, S1 and S2 mutations most significantly impact the ability of Sox2 to induce the Lef-1 promoter in the absence of overexpressed dominant active β-catenin, but retain partial Sox2-mediated repression of β-catenin-dependent transcription. Last, S1, S2, and S3 sites all serve to repress the Lef-1 promoter in the absence of forced Sox2 expression and β-catenin signals, given that baseline expression of the promoter was significantly enhanced by S1, S2, and S3 mutations. Thus we propose a model whereby the level of Sox2 and the intensity of β-catenin signals dynamically control Wnt-dependent transcription of the Lef-1 promoter by binding to unique sites and sites that overlap with TCF/β-catenin binding (Fig. 8B).
In summary, our studies have demonstrated that Sox2 binds to multiple sites in the Lef-1 promoter to dynamically regulate transcription. The ability of Sox2 to repress transcription from the Lef-1 promoter in the presence of Wnt/β-catenin signals may be an important mechanism to coordinate spatial and temporal regulation of Lef-1 during PGP formation and glandular morphogenesis. Such findings provide new insights into how Sox family proteins contribute to the context-dependent control of Wnt/β-catenin signaling.
DISCLOSURES
No conflicts of interest, financial or otherwise are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: W.X., T.J.L., X.L., M.J.G., K.R.P., J.M.W., H.-H.X., L.H.P., and J.F.E. conception and design of research; W.X., T.J.L., X.L., S.Y., X.Z., M.L., D.M.K., X.S., Y.Y., and Y.Z. performed experiments; W.X., T.J.L., X.L., S.R.T., M.L., and J.F.E. analyzed data; W.X., T.J.L., X.L., S.R.T., M.L., M.J.G., K.R.P., and J.F.E. interpreted results of experiments; W.X., T.J.L., S.R.T., and J.F.E. prepared figures; W.X., T.J.L., and J.F.E. drafted manuscript; W.X., T.J.L., X.L., S.R.T., J.M.W., and J.F.E. edited and revised manuscript; W.X., T.J.L., X.L., S.R.T., S.Y., X.Z., M.L., D.M.K., X.S., Y.Y., Y.Z., M.J.G., K.R.P., J.M.W., H.-H.X., L.H.P., and J.F.E. approved final version of manuscript.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) Grants DK-047967 (to J. F. Engelhardt) and K08-HL-114725 (to K. R. Parekh), and an NIH grant to the University of Iowa Center for Gene Therapy (DK-54759).
REFERENCES
- 1. Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene 25: 7492–7504, 2006 [DOI] [PubMed] [Google Scholar]
- 2. Atcha FA, Munguia JE, Li TW, Hovanes K, Waterman ML. A new beta-catenin-dependent activation domain in T cell factor. J Biol Chem 278: 16169–16175, 2003 [DOI] [PubMed] [Google Scholar]
- 3. Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol 27: 8352–8363, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beers MF, Morrisey EE. The three R's of lung health and disease: repair, remodeling, and regeneration. J Clin Invest 121: 2065–2073, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol 295: 219–231, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, Randell SH. Evidence for stem-cell niches in the tracheal epithelium. Am J Respir Cell Mol Biol 24: 662–670, 2001 [DOI] [PubMed] [Google Scholar]
- 7. Cardoso WV, Lu J. Regulation of early lung morphogenesis: questions, facts and controversies. Development 133: 1611–1624, 2006 [DOI] [PubMed] [Google Scholar]
- 8. De Langhe SP, Reynolds SD. Wnt signaling in lung organogenesis. Organogenesis 4: 100–108, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 136: 2815–2823, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Driskell RR, Goodheart M, Neff T, Liu X, Luo M, Moothart C, Sigmund CD, Hosokawa R, Chai Y, Engelhardt JF. Wnt3a regulates Lef-1 expression during airway submucosal gland morphogenesis. Dev Biol 305: 90–102, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Driskell RR, Liu X, Luo M, Filali M, Zhou W, Abbott D, Cheng N, Moothart C, Sigmund CD, Engelhardt JF. Wnt-responsive element controls Lef-1 promoter expression during submucosal gland morphogenesis. Am J Physiol Lung Cell Mol Physiol 287: L752–L763, 2004 [DOI] [PubMed] [Google Scholar]
- 12. Duan D, Sehgal A, Yao J, Engelhardt JF. Lef1 transcription factor expression defines airway progenitor cell targets for in utero gene therapy of submucosal gland in cystic fibrosis. Am J Respir Cell Mol Biol 18: 750–758, 1998 [DOI] [PubMed] [Google Scholar]
- 13. Duan D, Yue Y, Zhou W, Labed B, Ritchie TC, Grosschedl R, Engelhardt JF. Submucosal gland development in the airway is controlled by lymphoid enhancer binding factor 1 (LEF1). Development 126: 4441–4453, 1999 [DOI] [PubMed] [Google Scholar]
- 14. Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci 26: 148–165, 2004 [DOI] [PubMed] [Google Scholar]
- 15. Engelhardt JF. Stem cell niches in the mouse airway. Am J Respir Cell Mol Biol 24: 649–652, 2001 [DOI] [PubMed] [Google Scholar]
- 16. Filali M, Cheng N, Abbott D, Leontiev V, Engelhardt JF. Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter. J Biol Chem 277: 33398–33410, 2002 [DOI] [PubMed] [Google Scholar]
- 17. Finkbeiner WE. Physiology and pathology of tracheobronchial glands. Respir Physiol 118: 77–83, 1999 [DOI] [PubMed] [Google Scholar]
- 18. Gontan C, de Munck A, Vermeij M, Grosveld F, Tibboel D, Rottier R. Sox2 is important for two crucial processes in lung development: branching morphogenesis and epithelial cell differentiation. Dev Biol 317: 296–309, 2008 [DOI] [PubMed] [Google Scholar]
- 19. Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res 22: 1500–1501, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hashimoto S, Chen H, Que J, Brockway BL, Drake JA, Snyder JC, Randell SH, Stripp BR. beta-Catenin-SOX2 signaling regulates the fate of developing airway epithelium. J Cell Sci 125: 932–942, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iwatsuki K, Liu HX, Gronder A, Singer MA, Lane TF, Grosschedl R, Mistretta CM, Margolskee RF. Wnt signaling interacts with Shh to regulate taste papilla development. Proc Natl Acad Sci USA 104: 2253–2258, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeffery PK. Remodeling in asthma and chronic obstructive lung disease. Am J Respir Crit Care Med 164: S28–S38, 2001 [DOI] [PubMed] [Google Scholar]
- 23. Kelberman D, Rizzoti K, Avilion A, Bitner-Glindzicz M, Cianfarani S, Collins J, Chong WK, Kirk JM, Achermann JC, Ross R, Carmignac D, Lovell-Badge R, Robinson IC, Dattani MT. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J Clin Invest 116: 2442–2455, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kormish JD, Sinner D, Zorn AM. Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Dev Dyn 239: 56–68, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lange AW, Keiser AR, Wells JM, Zorn AM, Whitsett JA. Sox17 promotes cell cycle progression and inhibits TGF-beta/Smad3 signaling to initiate progenitor cell behavior in the respiratory epithelium. PLoS One 4: e5711, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li TW, Ting JH, Yokoyama NN, Bernstein A, van de Wetering M, Waterman ML. Wnt activation and alternative promoter repression of LEF1 in colon cancer. Mol Cell Biol 26: 5284–5299, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu X, Driskell RR, Engelhardt JF. Airway glandular development and stem cells. Curr Top Dev Biol 64: 33–56, 2004 [DOI] [PubMed] [Google Scholar]
- 28. Liu X, Driskell RR, Engelhardt JF. Stem cells in the lung. Methods Enzymol 419: 285–321, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu X, Driskell RR, Luo M, Abbott D, Filali M, Cheng N, Sigmund CD, Engelhardt JF. Characterization of Lef-1 promoter segments that facilitate inductive developmental expression in skin. J Invest Dermatol 123: 264–274, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu X, Luo M, Xie W, Wells JM, Goodheart MJ, Engelhardt JF. Sox17 modulates Wnt3A/beta-catenin-mediated transcriptional activation of the Lef-1 promoter. Am J Physiol Lung Cell Mol Physiol 299: L694–L710, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev 87: 219–244, 2007 [DOI] [PubMed] [Google Scholar]
- 32. Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol 168: 1065–1076, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci USA 100: 3299–3304, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev 15: 1688–1705, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Okubo T, Pevny LH, Hogan BL. Sox2 is required for development of taste bud sensory cells. Genes Dev 20: 2654–2659, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oppenheimer EH, Esterly JR. Pathology of cystic fibrosis review of the literature and comparison with 146 autopsied cases. Perspect Pediatr Pathol 2: 241–278, 1975 [PubMed] [Google Scholar]
- 37. Ostrowski LE, Yin W, Rogers TD, Busalacchi KB, Chua M, O'Neal WK, Grubb BR. Conditional deletion of dnaic1 in a murine model of primary ciliary dyskinesia causes chronic rhinosinusitis. Am J Respir Cell Mol Biol 43: 55–63, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park KS, Wells JM, Zorn AM, Wert SE, Laubach VE, Fernandez LG, Whitsett JA. Transdifferentiation of ciliated cells during repair of the respiratory epithelium. Am J Respir Cell Mol Biol 34: 151–157, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development 136: 1899–1907, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA 96: 5522–5527, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol 27: 7802–7815, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, Wells JM. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell 17: 62–74, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev 20: 1187–1202, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128, 2008 [DOI] [PubMed] [Google Scholar]
- 45. Tompkins DH, Besnard V, Lange AW, Keiser AR, Wert SE, Bruno MD, Whitsett JA. Sox2 activates cell proliferation and differentiation in the respiratory epithelium. Am J Respir Cell Mol Biol 45: 101–110, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. PITX2, beta-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J Cell Sci 118: 1129–1137, 2005 [DOI] [PubMed] [Google Scholar]
- 47. van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev 8: 2691–2703, 1994 [DOI] [PubMed] [Google Scholar]
- 48. Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661–665, 2007 [DOI] [PubMed] [Google Scholar]
- 49. Wong MH, Huelsken J, Birchmeier W, Gordon JI. Selection of multipotent stem cells during morphogenesis of small intestinal crypts of Lieberkuhn is perturbed by stimulation of Lef-1/beta-catenin signaling. J Biol Chem 277: 15843–15850, 2002 [DOI] [PubMed] [Google Scholar]
- 50. Xie W, Fisher JT, Lynch TJ, Luo M, Evans TI, Neff TL, Zhou W, Zhang Y, Ou Y, Bunnett NW, Russo AF, Goodheart MJ, Parekh KR, Liu X, Engelhardt JF. CGRP induction in cystic fibrosis airways alters the submucosal gland progenitor cell niche in mice. J Clin Invest 121: 3144–3158, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137: 647–658, 2009 [DOI] [PubMed] [Google Scholar]
- 52. Zhang Y, Goss AM, Cohen ED, Kadzik R, Lepore JJ, Muthukumaraswamy K, Yang J, DeMayo FJ, Whitsett JA, Parmacek MS, Morrisey EE. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet 40: 862–870, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev 9: 700–713, 1995 [DOI] [PubMed] [Google Scholar]
- 54. Zhou X, Xue HH. Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J Immunol 189: 2722–2726, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, Varmus HE. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol Cell 4: 487–498, 1999 [DOI] [PubMed] [Google Scholar]