Abstract
In recent years, new prospects for the use of nucleic acids as anticancer drugs have been discovered. Aptamers for intracellular targets can regulate cellular functions and cause cell death or proliferation. However, intracellular aptamers have limited use for therapeutic applications due to their low bioavailability. In this work, we selected DNA aptamers to cell organelles and nucleus of cancer cells, and showed that an aptamer NAS-24 binds to vimentin and causes apoptosis of mouse ascites adenocarcinoma cells in vitro and in vivo. To deliver the aptamer NAS-24 inside cells, natural polysaccharide arabinogalactan was used as a carrier reagent. The mixture of arabinogalactan and NAS-24 was injected intraperitonealy for 5 days into mice with adenocarcinoma and inhibited adenocarcinoma growth more effectively than free arabinogalactan or the aptamer alone. The use of aptamers to intracellular targets together with arabinogalactan becomes a promising approach for anticancer therapy.
Introduction
Aptamers are single-stranded DNA (ssDNA) or RNA (ssRNA) oligonucleotides that fold to form well-defined tertiary structures, allowing them to bind with high affinity and specificity to their targets (Ellington et al., 1990; Tuerk et al., 1990). They are promising agents for therapeutics due to their nonimmunogenic and nontoxic properties (ULRICH, 2006). Aptamers have been selected against many different cancer cell lines and demonstrated targeted antitumor activity alone, or coupled with toxins (Huang et al, 2009; Keefe et al., 2010; Ray et al., 2010). Since their emergence, aptamers have become increasingly popular as possible therapeutics (Meyer et al., 2011). They are selected from large random-sequence nucleic acid libraries against different targets (Codrea et al., 2010; Labib et al, 2012; Muharemagic et al., 2012; Kolovskaya et al., 2013). It has been shown that aptamers selected for intracellular targets have the potential to influence receptors and effectively regulate cellular functions, causing various cellular events such as apoptosis (Jian et al., 2009) and proliferation (Li et al., 2011). Intracellular, targets are usually inaccessible to DNA oligonucleotides in live cells and this problem becomes even more challenging when these aptamers need to be delivered into cells in vivo. A lot of efforts have been made in order to deliver therapeutic DNA and RNA but no solution is currently available. Most of the methods are based on protocols used for small interfering RNA (siRNA) transfection (Juliano et al., 2008; Whitehead et al., 2009) which is used for activating, silencing, introducing, or knocking out genes both in vitro and in vivo. Gene delivery is used for corrective or cytotoxic gene therapy (Stribley et al., 2002; Al-Hendy et al., 2006). Nonviral in vivo DNA delivery procedure involves: (1) direct injection of naked DNA and artificial chromosomes into targeted organs to avoid its rapid degradation upon systemic administration (Suda et al., 2008; Kazuki et al., 2011); (2) encapsulation of DNA into liposomes (Sellins et al., 2005), synthetic polymers (Eliyahu et al., 2005; Li et al., 2012), carbohydrate-based nanogels (Ahmed et al., 2012), and chimeric capsid proteins (Choi et al., 2012), with further transfer into cells; and (3) self-formed complexes of DNA and natural polymer ligands containing oligosaccharides, useful due to specific recognition by sugar-binding protein receptors (lectins) on the surface of many animal cells (Gabor et al., 2004).
In recent years, natural polysaccharides have been widely used in pharmaceutical technology for production of macromolecular pro-drugs, which can exhibit favorable biopharmaceutical properties and enforce therapeutic performance of parent drugs. Natural polysaccharide-based molecules can interact with living cells displaying antioxidant, antimicrobial, anti-mutating, anticoagulating, immunostimulating, and cell-differentiating biological properties. In addition, they can provide high biocompatibility, biodegradability and physicochemical properties that can be used in drug delivery (Lehr et al., 2004) leading to synergistic therapeutic effects.
Arabinogalactan (AG) is a polysaccharide with a branched structure (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat) comprising of a backbone of 1,3-linked galactopyranose connected by 1,3-glycosidic linkages, comprised of 3,4,6-; 3,6-; 3,4-; and 3-linked residues. Larch AG is a biologically active substance with a low toxicity (LD50>5g/kg) and a wide spectrum of biological effects. It was reported that AG administered in rats was extensively accumulated in liver parenchymal cells by specific interactions with the asialoglycoprotein receptor (ASGPR) (Groman et al., 1994). Therefore, AG can be used for delivery of small or large, single- or double-stranded DNA in any ASGPR-positive cells.
In this study, we describe a procedure for aptamer selection to intracellular targets, as well as a characterization of their biological effect and identification of their protein targets by mass spectrometry. Finally, we used arabinogalactan, a natural polysaccharide in order to enhance in vivo delivery of antivimentin aptamer and observed tumor growth suppression.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institute of Health. The protocol was approved by the Local Committee on the Ethics of Animal Experiments of the Krasnoyarsk State Medical University. All surgery was performed under anesthesia and all efforts were made to minimize suffering of the animals.
Cell lines and animals
The HuH-7 cell line (well-differentiated hepatocyte from a liver tumor with high expression of ASGPR) was kindly provided by John P. Pezacki (National Research Council, Ottawa, Canada) and Vero cell line (kidney epithelial cells extracted from an African green monkey lacking ASGPR) was provided by Dr. John Bell (Ottawa Hospital Research Institute, Ottawa, Canada). Mouse Ehrlich ascites adenocarcinoma cell line was kindly provided by Evgeny Inzhevatkin (Krasnoyarsk Research Center, Siberian Branch, Russian Academy of Science, Krasnoyarsk, Russia). Plasmid enhanced green fluorescent protein (pEGFP)-N1 vector was purchased from Clontech. White 6-week-old 25-g Imprinting Control Region (ICF) mice were provided by State Research Center of Virology and Biotechnology “Vector” (Koltsovo, Novosibirsk, Russia).
Cell culture
Primary mouse ascites cells and HuH-7 cells were cultured in 75-mm flasks in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich), supplemented with 100 U/mL penicillin, 100 U/mL streptomycin, and 10% (v/v) fetal bovine serum (FBS) in a humidified atmosphere containing 5% CO2 at 37°C. Cells were subcultured regularly using trypsin/EDTA (StemCell Technologies Inc.). Vero cells were grown using the same conditions but without antibiotics. Prior to use, cell cultures were trypsinized (37°C, 5 minutes) and the reaction was stopped by adding DMEM medium with 10% FBS.
Purification of arabinogalactan
Arabinogalactan was obtained from small wood pieces, 2–5 mm of Larix sibírica; briefly, a metal reactor was filled with 10 g of chopped larch wood and 80 mL of distilled water and subjected to shock-acoustic effects at room temperature for 1 minute. Filtered sawdust extracts were then evaporated, cooled and poured with stirring into 120 mL of ethanol. Precipitated arabinogalactan was filtered, separated from ethanol, and dried. The output of arabinogalactan was 24%–26% of wood weight. The recovery of arabinogalactan reached 80%–90%.
Preparation of arabinogalactan–ssDNA complexes
AG–ssDNA complex was prepared by mixing 200 μL of 200 nM ssDNA with 200 μL of 2 mg/mL of arabinogalactan [each component was diluted in colorless Hank's balanced solution, phosphate-buffered saline (PBS), or DMEM medium] and allowed to incubate for 25–30 minutes at room temperature.
Aptamer selection
Detailed aptamer selection and affinity analyses are described in the Supplementary Data. Briefly, ascites cell organelles were isolated from cells taken from the abdominal cavity of ICR mice on day nine after tumor transplantation and aptamers selection was based on a SELEX (systematic evolution of ligands by exponential enrichment) scheme. A total of 11 rounds of selection to ascites cells were performed at room temperature using the protocol schematically presented in Supplementary Data (Supplementary Fig. S2b). The aptamer pool with the highest affinity to cell organelles was cloned and sequenced according to protocols described in Supporting Information. Four aptamer clones were chemically synthesized. The 80-nucleotide ssDNA library which consists of a randomized region of 40 nucleotides (A:T:C:G=25%:25%:25%:25%) flanked by two constant sites (5′-CTC CTC TGA CTG TAA CCA CG-N40-GC ATA GGT AGT CCA GAA GCC-3′) with a fluorescent label FAM at the 5′ end, as well as FAM-labeled and nonlabeled aptamers, were purchased from Integrated DNA Technologies.
In vitro studies
In vitro arabinogalactan-mediated ssDNA delivery
Mouse Ehrlich ascites carcinoma cells were used for in vitro experiments. In order to prevent nonspecific binding to plasma membranes, cells were preincubated in DPBS with 0.1 mg/mL of masking DNA from salmon sperm for 20 minutes at room temperature. All cell samples were pelleted, 106 cells were resuspended in 250 μL of DMEM with AG-ssDNA complex, and incubated for 2 hours in 5% CO2 at 37°C. Control samples were prepared the same way, but without arabinogalactan. After incubation, cells were washed three times with colorless Hanks balanced solution to remove excess of AG and ssDNA. The level of fluorescence was determined using a FC-500 flow cytometer (Beckman Coulter), fluorescence microscope CX41 with imaging system Cell F (Olympus) and FLUOVIEW FV10i confocal microscope (Olympus). The experiment was repeated independently two times, and each one was done in triplicate.
In vitro arabinogalactan-mediated EGFP gene delivery
Two million ascites cells were isolated from peritoneal cavity of mice with Ehrilch adenocarcinoma and cultured in colorless high glucose DMEM medium with 2.5 μM of pEGFP-N1 vector in the presence of AG (final concentration of 1 mg/mL) for 26 hours at 37°C. Control cells were cultured with 2.5 μM of pEGFP-N1 vector without AG. The success of delivery was confirmed by EGFP fluorescence in treated cells using a flow cytometer FC-500 (Beckman Coulter).
In vitro competitive analysis of asialofetuin-mediated inhibition of ssDNA delivery
HuH-7 cells (106 cells in total) were preincubated for 2 hours with 18 mg/mL of alternative splicing factor (ASF) in DMEM in 5% CO2 at 37°C, then mixed with AG-ssDNA complex and incubated for an additional 2 hours under the same conditions and washed three times with PBS. The following controls were made: HuH-7 cells with ssDNA, cells with 6 mg/mL of ASF mixed with 200 nM ssDNA in DPBS, cells with AG–ssDNA complex. Competitive analysis with 18, 9 and 6 mg mL1 of ASF was performed using fluorescein isothiocyanate (FITC)-labeled AG. The competitive effect of asialofetuin was demonstrated using a flow cytometer FC-500 (Beckman Coulter,).
In vitro antitumor activity of aptamer–arabinogalactan complex
Two million ascites cells were isolated from peritoneal cavity of mice with Ehrilch adenocarcinoma and cultured in colorless high glucose DMEM medium with 1 μM of aptamer NAS-24 in presence of AG (final concentration of 1 mg/mL) for 24 hours at 37°C in a humidified atmosphere containing 5% CO2. Control cells were cultured with 1 μM ssDNA library with AG or aptamer alone. Antitumor effect was estimated by cell proliferation (CellTrace™ Far Red DDAO-SE, Invitrogen Corporation) and cell viability (Annexin V Alexa Fluor® 488, Invitrogen Corporation; propidium iodide, Sigma-Aldrich) by flow cytometry, according to manufacturer's protocols.
Aptamer-mediated protein target identification
Aptamer mediated protein discovery was performed on the basis of the aptamer-facilitated biomarker discovery (AptaBiD) procedure (Berezovski et al., 2008), optimized for a reliable identification of the target for a single aptamer clone. Cell organelles were isolated from whole live cells, and 106 cells were incubated for half an hour at room temperature in 2 mL of 24 μM of “soft” anionic detergent sodium deoxycholate (Thermo Scientific), which is suitable for mass spectrometric analyses. Cells were then gently homogenized by pipetting and the mixture of organelles and membranes was centrifuged at 5,000 g for 10 minutes at 4°C. The supernatant was discarded and the pellet was resuspended in DPBS and washed two times using the same conditions. In order to suppress nonspecific aptamer binding, cellular organelles and membranes were preincubated in 1 μM of masking DNA from salmon sperm and 100 nM of unlabeled native ssDNA library in DPBS. Afterwards, the incubation was continued with 25 nM of biotinylated aptamer clones or 25 nM of biotinylated native ssDNA-library as a control in total volume of 100 μL. This concentration of a high-affinity aptamer was enough to displace the library from the protein target. Another negative control was done without using nonspecific ssDNA oligonucleotides in order to evaluate the washing procedure and to exclude nonspecifically absorbed proteins. The samples were then mixed and incubated on a shaker for another 30 minutes with 1 mg of Streptavidin MagneSphere Paramagnetic Particles (Promega Corporation) preincubated at 25°C with masking DNA from salmon sperm. The organelle–aptamer–magnesphere complexes were removed from the mixture using a magnetic stand and washed five times with DPBS buffer. Vials and tips were changed after each washing in order to remove proteins and DNA nonspecifically bound to plastic. Afterwards, the pellet was suspended in 200 μL of cell lysing buffer in PBS with 0.1% (v/v) sodium deoxycholate and incubated for 30 minutes at 25°C. Magnetic particles with attached proteins were washed again three to five times with DPBS to remove detergent residues, cell debris, and unspecific proteins. Vials and tips were also changed after each washing. Protein targets were dissociated from aptamer-coated beads by adding 30 μL of 8 M urea solution and incubated for 30 minutes at 4°C. Magnetic particles were retained using a magnetic stand, and the supernatant was removed and stored at −20°C. Protein concentration in samples was measured using NanoVue plus spectrophotometer (GE Healthcare, Life Sciences) and varied from 0.4 to 1.1 mg/mL, depending on the type of the probe.
All water solutions were prepared using deionized water from Millipore ICW-3000 water purification system (EMD Millipore Corporation). A fraction of 10 μL of denatured proteins was diluted with 15 μL of 50 mM ammonium bicarbonate (Sigma-Aldrich) and 1.5 μL of 100 mM dithiothreitol (Sigma-Aldrich), incubated for one hour at 37°C, mixed with 3 μL of 100 mM iodoacetamide (Pierce Biotechnology), and incubated for 30 minutes at 25°C in the dark. The samples were diluted with 15 μL of 50 mM ammonium bicarbonate to get 4 mM of urea. Proteins in each sample were digested with porcine trypsin (Sigma-Aldrich) for 16 hours at 37°C by adding 2 μL of 100 ng/μL trypsin. The peptide mixture was desalted using ready-to-go pipette tips filled with C18 spherical silica reversed phase material (ZipTipC18, Millipore), according to manufacturer's protocol. Peptides were eluted with 20 μL of 80% acetonitrile and 0.1% trifluoroacetic acid, evaporated with ScanVac VacSafe 15 (LaboGene ApS), and resuspended with 0.1% formic acid. Shot-gun mass spectromic analysis of 10 μL of protein-digest sample was performed by nanoflow ultra high pressure liquid chromatography (Easy-nLC 1000, Thermo Scientific) and tandem mass spectrometry with an Orbitrap Velos Pro mass spectrometer (Thermo Scientific). Full scan spectra were obtained using Fourier-transform mass analyzer with a resolution of 60,000. Fragment spectra of top 15 precursors, activated by collision-induced dissociation, were obtained by mass analyzer of ion trap. At the beginning and the end of experimental sequences, bovine serum albumin standard digest analyses were performed to estimate the quality of evolved results. Accuracy deviations for precursors were less than 10 ppm, which is appropriate for the external calibration standards. Database searches were done with Proteome Discoverer 1.3 software, Sequest search engine and SwissProt database, and the label-free quantitative analyses were performed using MaxQuant 1.3.0.5 proteomic software (Cox et al., 2008). Each experiment was made in triplicate. Experiments were repeated two times independently. Results from the independent experiments were analyzed separately.
Animal experiments
In vivo arabinogalactan-mediated GFP plasmid delivery
Six-week-old, 25-g ICR male mice were used in this study, 6 animals per group. One million Ehrlich ascites adenocarcinoma cells were transplanted into the abdominal cavity of each mouse. On the third day, mice received intraperitoneal injection of sterile PBS (control 1), 20 μg free plasmid DNA (pEGFP-N1 vector) in PBS without AG (control 2), or plasmid-AG complex [20 μg pEGFP-N1 vector with 400 μg (100 μL 2mg mL−1) of AG] in PBS. Total injection volume was 100 μL. Twenty-six hours after injection, ascites cells, blood cells, and internal organs (brain, liver, kidneys, and spleen) were isolated from the mice. Organs were frozen slowly to −80°C and sliced with Microm HM650V (Thermo Scientific) and observed under a fluorescence microscope. Liver, kidney, and spleen tissues were minced into 2–3 mm pieces, washed with Hank's balanced solution, and dissociated with collagenase (200 units in Hank's balanced solution) during 4 hours of incubation at 37°C, followed by filtration through a sterile steel needle syringe; obtained cell suspension was washed twice with Hanks balanced solution. EGFP expression was evaluated with a fluorescence microscope CX41, FC-500 flow cytometer.
In vivo antitumor activity of aptamer–arabinogalactan self-forming complex
Six-week-old, 25-g ICR male mice were used in this study, five animals per group. One million Ehrlich ascites adenocarcinoma cells were transplanted into the abdominal cavity of each mouse. Each day, starting from day four after tumor transplantation until day nine, animals were treated with aptamer AG complex, free AG or aptamer, and just PBS. Daily injections for the four groups (5 days total) were as follows:
Group 1: Injection of PBS (100 μL).
Group 2: Injection of AG (100μL 1 mg/mL1).
Group 3: Injection of AG (100μL 1 mg/mL) preincubated with aptamer NAS-24 (0.8 nmole per 25-g mouse (or 0.8 μg/kg).
Group 4: Injection of aptamer NAS-24 (0.8 nmole per 25-g mouse (or 0.8 μg/kg) in 100 μL of PBS.
After 5 days of treatment, all ascites cells were isolated and washed out from the abdominal cavity of mice. Treated cells were washed three times with Hanks balanced solution and cell concentrations were counted using Scepter 2.0 Handheld Automated Cell Counter (Millipore Corporation). Smaller particles such as blood cells and debris were excluded from calculations. The total number of cells in each animal was calculated according to the total volume of ascites cells in ascites liquid. Cell viability was determined using Annexin V Alexa Fluor® 488, (Invitrogen Corporation) and propidium iodide (Sigma-Aldrich), according to manufactures protocols on the FC-500 flow cytometer.
Statistical analysis
Data were expressed as means of six (plasmid EGFP delivery), five (aptamer AG treatment), or separate experimental groups. The differencsbetween groups were assessed using unpaired two-tailed Student's t-test, p<0.05 was considered statistically significant.
Results
Aptamer selection for cell organelles an nucleus
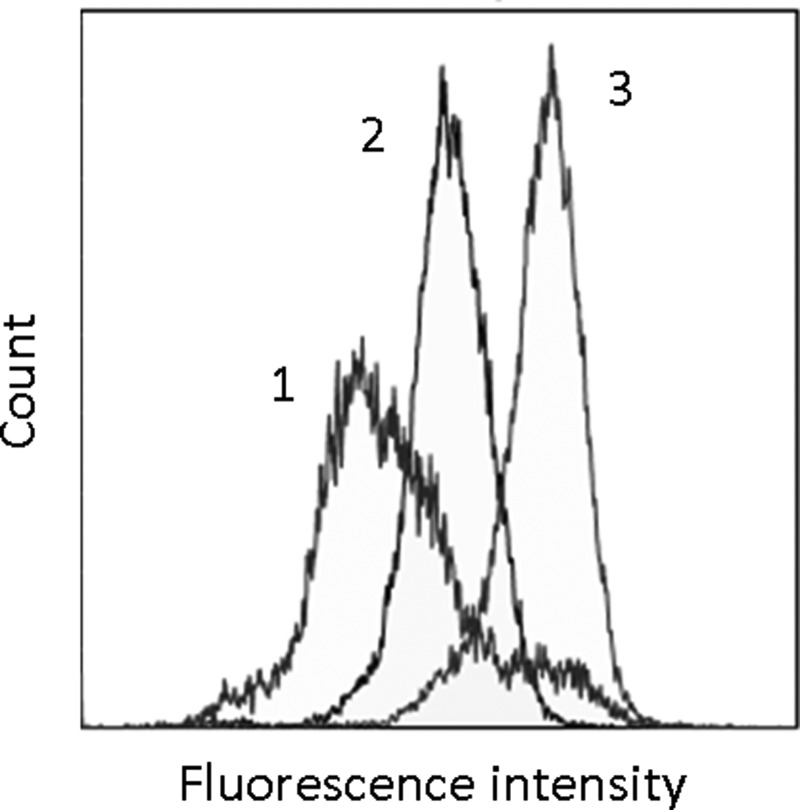
Cell organelles were isolated from whole live cells using the protocol presented in Supplementary Fig. S2a. Aptamers for cell organelles and nucleus were selected based on a SELEX technique: an in vitro selection technique of nucleic acid binders to molecular targets from a random ssDNA library. A total of 13 rounds of selection were performed at room temperature using the protocol described in detail in Supplementary Materials and Methods and presented in Supplementary Fig. S2b. Selection included negative steps to whole mouse blood and intact ascites cells to exclude sequences binding to the cell membrane. In general, the selection consisted from the following steps: (1) incubation with “negative” cells, (2) centrifugation and collection of a supernatant with unbound DNA, (3) incubation of supernatant with “positive” organelles and nucleus, (4) washing of unbound ssDNA, (5) extraction of bound ssDNA, and (6) DNA amplification by symmetric and asymmetric polymerase chain reaction (PCR). We added salmon sperm DNA (0.1 mg/L) as masking DNA in each positive selection step to eliminate nonspecific DNA binding to increase the stringency of aptamer selection. Aptamer affinity to ascites cell organelles was analyzed using flow cytometry. The gating strategy for cell organelles is represented in Supplementary Data (Supplementary Fig. S3). Relative binding analysis of DNA pools revealed that the affinity started to increase after the fourth round and reached its maximum in the eleventh round (Fig. 1). This 11th pool was cloned and sequenced. Four clones with the best affinity were synthesized. The aptamer NAS-24 with the following sequence CTC CTC TGA CTG TAA CCA CGC CTG GGA CAG CCA CAC AGA AGT GTA GAC CTC GCG GAA TCG GCA TAG GTA GTC CAG AAG CC was used for further experiments because it showed the ability to cause apoptosis in ascites when it was delivered inside the cell by arabinogalactan.
FIG. 1.
Flow cytometry binding analysis of the fluorescein (FAM)-labeled single-stranded DNA (ssDNA) to organelles and nucleus from ascites cells. Histograms of ssDNA library and 11th pool binding to organelles, where (1) curve is for intact organelles, (2) is for ssDNA library with organelles, and (3) is for the best pool binding to organelles.
Aptamer protein target identification
Affinity purification of a molecular target for NAS-24 aptamer was performed by using aptamer coated magnetic particles according to the protocol described in “Materials and Methods.” The key point of this procedure was the avoiding binding to nonspecific targets, such as cellular membranes, plastic vials, tips, and magnetic particles. To achieve this, we used several techniques such as incubating with 200 nM of scrambled DNA and 200 nM of nonlabeled naïve library to cover the organelles and mask the proteins. Afterward, we added 25 nM of biotinylated NAS-24 aptamer with high selectivity and affinity in order to replace the nonspecific DNA from the target by linking the protein of interest through biotin-labeled aptamer with streptavidin magnetic particles. We used biotinylated library instead of the aptamer as a negative control to exclude false positives of random nonspecific DNA-binding proteins. Another important procedure was exclusion of proteins bound to plastic. This was minimized by changing vials and tips during the washing; additional control group without adding any DNA was included in the experiment. All experiments were repeated twice, each one was done in triplicate. Results from independent experiments were analyzed separately. Protein sequence searches were performed using Proteome Discoverer 1.3 software, Sequest search machine, and SwissProt database. Quantitative analyses were performed using MaxQuant 1.3.0.5 proteomics software.
All proteins found in aptamer and library experiments were grouped in one table. First, the contaminant proteins according to the contaminant database included in the MaxQuant software package were excluded from further analyses. Only proteins with high confidence (proteins from the aptamer group identified by five or more unique peptides) were considered. The final list consisted of 15 and 14 protein hits for aptamer and library experimental groups, respectively. The DNA-free experiment was used to control plastic-binding proteins; the results did not influence the protein list as all hits from this experiment were already identified as contaminants. Finally, we compared and quantified aptamer and control (the native library instead of an aptamer) results using MaxQuant.
We analyzed quantitatively the protein distributions for each sample. Proteins that were present in the negative control groups (biotin-labeled naïve library instead of an aptamer and DNA-free group), in same or larger amount than in aptamer samples, were excluded from the list. Finally, three proteins were left in each list from two separate experiments. Only one protein, vimentin, appeared in both cases. Furthermore, this protein was the leading candidate; it had the highest confidence percentage and the lowest posterior error probability of identification (that functions as a p-value). Summed up extracted ion current of all isotopic clusters for the identified sequence ]label-free quantitation (LFQ) intensity] and the number of MS/MS spectra recorded for the peptide (MS/MS counts) were at least 10 times higher for vimentin than the other protein hits. LFQ intensities and MS/MS counts for all repetitions from each experimental group were summarized. The intensity and the number of MS/MS counts for vimentin in the aptamer group are ten and seven times higher than the library group. Therefore, vimentin was identified as the most probable target for NAS-24 aptamer.
Vimentin is an intermediate filament protein that can be attached to the nucleus, endoplasmic reticulum or mitochondria (Challa et al., 2011). It is overexpressed in various human epithelial cancers, including breast cancer (Korsching et al., 2005; Vora et al., 2009), gastrointestinal tumors (Fuyuhiro et al., 2010), prostate cancer (Zhao et al., 2008; Sethi et al., 2010), lung cancer (Rho et al., 2009), malignant melanoma (Li et al., 2010), and other cancer types (Hu et al., 2004; Williams et al., 2009). Furthermore, its high level of expression is associated with invasion (Gilles et al., 2003) and poor prognosis (Jin et al., 2010; Satelli et al., 2011).
Vimentin is involved in various signaling pathways, including cell adhesion, migration and apoptosis (Ivaska et al., 2007; Lahat et al., 2010; Challa et al., 2011). Interestingly, several studies have showed that vimentin has the potential of being a target for anticancer therapeutics (Satelli et al., 2011).
In vitro arabinogalactan-mediated delivery of aptamers and a plasmid containing an EGFP gene
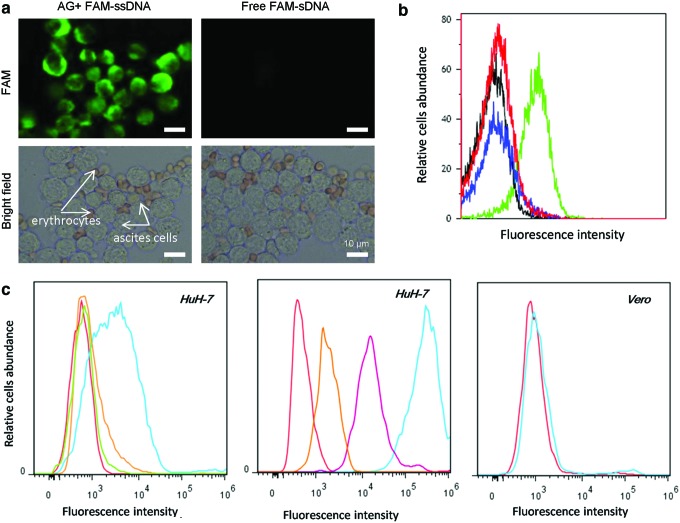
FAM-labeled 80-nucleotide single-stranded aptamers were delivered in vitro to Ehrlich ascites adenocarcinoma cells isolated from mice with arabinogalactan. In order to prevent nonspecific binding to plasma membranes, cells were preincubated in DPBS with masking DNA obtained from salmon sperm and then washed once to remove free DNA. After incubation with either AG–aptamer self-forming complex, free AG, or free aptamer, the cells were washed tree times with PBS without calcium and magnesium in order to remove nonspecific aptamer binding to the cell membrane. AG demonstrated strong delivery of FAM-labeled ssDNA aptamers into ascites cells (Fig. 2a) when compared to control experiments without AG. We did not observe the delivery in erythrocytes, which is likely attributed to the lack of asialoglycoprotein receptor (ASGPR) on erythrocytes. This does represent a limitation on the system, as the DNA complex can be only delivered to ASGPR positive cells.
FIG. 2.
In vitro delivery of plasmid ssDNA and plasmid enhanced green fluorescent protein (pEGFP)-N1 vector by means of arabinogalactan. (a) Brightfield and fluorescence images of ascites cells and erythrocytes loaded by arabinogalactan (AG)–ssDNA complex (500 nM FAM-labeled 80-nt ssDNA with AG 2 mg/mL). Only ascites cells exhibit strong fluorescence at 488 nm excitation. (b) Flow cytometry histogram of EGFP expressed in Ehrlich ascites cells at 26 hours of their incubation with plasmid pEGFP-N1 vector and arabinogalactan complex (20 μg of plasmid and 500 μg arabinogalactan) (green curve), free plasmid pEGFP-N1 vector (20 μg) (blue curve), free arabinogalactan (black curve), and intact cells (red curve). (c) Flow cytometry analysis of AG-mediated delivery of FAM-labeled ssDNA library in the presence of alternative splicing factor (ASF). (Left) AG-mediated delivery of ssDNA into HuH-7 cells. (Center) fluorescein isothiocyanate (FITC)-labeled AG internalization into HuH-7 is suppressed by asialofetuin. (Right) AG-mediated delivery of the ssDNA library does not happen in Vero cells that have the lack of asialoglycoprotein receptor (ASGPR), where red curve represents control (intact cells); green, cells with 6 mg/mL of ASF and 200nM ssDNA library; orange, cells with 18 mg/mL of ASF 2 mg/mL of AG and 200nM ssDNA library; dark blue, cells with 9 mg/mL of ASF 2 mg/mL of AG and 200nM ssDNA library; purple, cells with 6 mg/mL of ASF 2 mg/mL of AG and 200 nM ssDNA library; and light blue represents cells with 2 mg/mL of AG and 200 nM ssDNA library. Color images available online at www.liebertpub.com/nat
We performed delivery experiments with a DNA plasmid carrying the EGFP gene. The plasmid was incubated with ascites cells with and without AG. After 26-hour incubation of plasmid and AG with ascites cells, we detected strong green fluorescence of EGFP in cells (Fig. 2b). This fluorescence was not observed in samples without AG. Thus, AG facilitates the delivery of the plasmid into cells. We hypothesize that AG-DNA delivery is facilitated by receptor-mediated endocytosis. It was reported that AG administered in rats was extensively accumulated in liver parenchymal cells by specific interactions with the ASGPR (Groman et al., 1994). We tested both small, 80-nt aptamers and EGFP-carrying plasmid, one can note that the delivery mediated by AG is not DNA-size dependent. Thus, this method has a dual utility, as AG can be used as a mediator for aptamer therapeutics and selection to intracellular targets, or for gene delivery into cells.
Inhibition of AG-facilitated DNA delivery by asialofetuin
Oligosaccharide DNA delivery is mostly governed by the recognition of DNA–oligosaccharide complex with ASGPR on cell membranes and following endocytosis (Groman et al., 1994), ASGPR is a 95–150 kDa integral plasma membrane multimeric protein composed of two subunits (ASGPR1 and ASGPR2). Each subunit contains four functional domains: cytosolic, transmembrane, stalk, and a carbohydrate-recognition domain. In humans, ASGPR is found mainly in hepatocytes, immature dendritic, and some tumor cells (Valladeau et al., 2001; Li et al., 2008). Polysaccharides, together with DNA, form a complex with ASGPR, which is internalized into cells; the receptor and ligand dissociate and ASGPR then returns to the cell membrane (Supplementary Fig. S4). To confirm that AG-mediated delivery is associated with ASGPR, we used a competitive ligand for ASGPR, asialofetuin (ASF) from fetal calf serum. ASF is a galactose-terminated glycoprotein with three N-linked glycan and tri-antennary sugar chains (Cawley et al., 1981). Flow cytometry experiments showed that ASF inhibited AG-mediated delivery of ssDNA and FITC-labeled AG into HuH-7 hepatocarcinoma cells (Fig. 2c). Interestingly, ASF alone did not transport ssDNA into HuH-7 cells indicating that a unique complex forms between AG and DNA (Fig. 2c). Additionally, we did not observe the delivery of ssDNA into asialoglycoprotein receptor (ASPGR)-negative Vero cells, further indicating the dependence of ASPGR (Fig. 2c). Thus, DNA uptake is ASGPR cell-specific and AG is a key mediator, as molecules binding to the same receptor cannot facilitate this uptake.
Biological effect of anti-vimentin aptamers in vitro
FAM-labeled NAS-24 was only delivered inside the cell in combination with AG (Fig. 3a). Aptamer NAS-24 with a binding constant (Kd) of 5.9 nM showed the ability to cause cell apoptosis and did not significantly influence cell proliferation when delivered by AG in vitro (Fig. 3b,c). The level of necrosis in AG-aptamer treated cells did not increase, which suggests that the complex did not cause inflammation in vivo, only programmed cell death. Interestingly, when AG was administered alone, cells had a lower apoptotic rate.
FIG. 3.
In vitro delivery by AG of NAS-24 aptamer to intracellular targets and its effects on ascites cells. (a) Fluorescence and brightfield images of ascites cells loaded by free 100 nM FAM-labeled aptamer, or AG-aptamer complex. (b) Effect of AG-aptamer complex, free AG and free aptamer on cell proliferation (determined by using CellTrace™ Far Red DDAO-SE). (c) Effect of AG-aptamer complex, free AG, and free aptamer on apoptosis (translocation phosphatidylserine determined by using Annexin V Alexa Fluor 488) and necrosis (determined by using propidium iodide). Color images available online at www.liebertpub.com/nat
Plasmid delivery in mice
We found that aptamer NAS-24 could promote apoptosis in ascites cell cultures when delivered into the cell by arabinogalactan. To ensure that this complex can suppress tumor growth in vivo, we tested whether AG could deliver DNA inside cells in mice. A plasmid (20 μg of pEGFP-N1 vector in 200 μL PBS) coding for EGFP was injected intraperitoneally with 0.5 mg of AG or without AG in mice (n=6 per group) containing Ehrlich ascites adenocarcinoma (on day three after tumor transplantation). After 26 hours, we detected EGFP expression in ascites cells, liver, kidneys, spleen, brain, and blood cells as seen in Fig. 4. Maximum EGFP fluorescence was observed in liver and kidney. This is likely the result of an increased expression of ASGPR in these tissues. In spleen, ascites cells and blood EGFP fluorescence was significantly lower. Injection of the plasmid without AG did not result in any significant EGFP fluorescence in all tested cell types. In order to estimate the distribution of EGFP, we imaged whole organ slices and observed EGFP fluorescence (Fig. 4). This was localized mostly on the periphery of the brain, liver, and kidney, whereas in spleen EGFP was distributed evenly. We observed that AG was able to carry in vivo the plasmid coding for EGFP inside the cells from different organs, leading to the expression of this protein.
FIG. 4.
(a) Flow cytometry histogram of EGFP expressed in cells derived from different tissues at 26 hours of intraperitoneal injection of plasmid pEGFP-N1 vector and arabinogalactan complex (green curve), free plasmid pEGFP-N1 vector (blue curve), free arabinogalactan (black curve), and intact cells (red curve). (b) Fluorescent imaging of organ slices: brain, liver, kidney, and spleen after intraperitoneal injection of the mixture of pEGFP-N1 and AG in PBS and free pEGFP-N1 in PBS. Color images available online at www.liebertpub.com/nat
In vivo antitumor activity of aptamer–arabinogalactan complex
In vivo antitumor efficacy of NAS-24-AG self-forming complex was evaluated using Ehrlich ascites adenocarcinoma mouse model. AG-NAS-24 complex (1.6 μg/kg, 1:1), free NAS-24, free AG and PBS for control groups were injected daily intratumorally for 5 days. Number of tumor cells washed out from the abdominal cavity and the apparent necrosis and apoptosis levels on day five after starting the treatment are presented in Fig. 5. On day nine, after the 5-day treatment, carcinoma growth of mice treated with AG and NAS-24 mixture was effectively suppressed when compared with free aptamers, free AG or just PBS. Aptamer delivered by AG caused apoptosis in ascites cells. This study illustrates biological and pharmacological use of an aptamer delivered into the cell by natural polysaccharide arabinogalactan and targets vimentin, initiating signaling events that lead to cellular apoptosis.
FIG. 5.
In vivo antitumor activity of ssDNA–aptamer arabinogalactan complex in comparison with free aptamer, free arabinogalactan, and PBS after 5 days of treatment. (a) Total amount of cells in ascites tumor. (b) The level of necrosis (determined by using propidium iodide) and apoptosis (translocation phosphatidylserine determined using Annexin V Alexa Fluor 488) in ascites cells.
Discussion
One of the largest limitations for the use of nucleotides for research and therapeutic purposes is the difficulty of transporting them into intracellular space. For DNA-based therapeutics to be viable, a delivery system that is both simple and nontoxic is required. Here we show that arabinogalactan obtained from the larch tree is an efficient delivery agent for small (80 nt) and large (4,733 bp) DNA molecules into cells in vitro and in vivo. High biocompatibility, biodegradability of this natural DNA carrier together with its antioxidant, antimicrobic, antimutant, and immunostimulanting properties make arabinogalactan substantial for oligonicleic acid-based therapeutics. AG-mediated delivery of the plasmid with EGFP gene in mice via ASGPR-mediated endocytosis causes strong expression of EGF protein not only in a common target for AG like liver but in other organs as well, such as kidney, spleen, and adenocarcinoma cells. We applied this delivery technique to ssDNA aptamers specially selected to intracellular targets. Coupled with AG, aptamer NAS-24 was delivered into cytoplasm and caused apoptosis in Ehrlich ascites adenocarcinoma cells in vitro and in vivo; free AG or a free aptamer did not demonstrate these properties. We used mass spectrometry for the identification of the aptamer's protein target with proapoptotic properties. Vimentin was found as the most probable protein target for the aptamer NAS-24 to Ehrilch adenocarcinoma cells, which are originated from breast cancer. This protein is also known as a biomarker for several cancer types. Vimentin is over-expressed in a large number of cancers. Significantly higher level of activated soluble vimentin in tumor vs. normal cells has been reported. Also there is a difference in subcellular localization of vimentin patterns in cancer and nontransformed cells (Lahat et al., 2010). Therefore, anti-vimentin drugs should be selective to vimentin-expressing cancer cells and have only minimal effect on normal mesenchymal cells. The potential use of this protein as an anti-cancer target was highlighted by findings demonstrating proapoptotic effects induced by vimentin cleavage. It is known that vimentin cleavage results in an irreversible disruption of vimentin filaments, causes cytoskeleton reorganization, and apoptosis. Furthermore, caspase cleavage of vimentin amplifies the cell death signal by generating proteolytic products that activate more caspases by creating a positive feedback loop (Byun et al., 2001). For example, withaferin-A binding to vimentin elicits its degradation and caspase activation resulted in apoptosis in vitro and in vivo (Lahat et al., 2010). Probably NAS-24 aptamer also promoted vimentin degradation that caused in the morphological manifestations and apoptosis mostly in cancer cells with higher vimentin content. Therefore, vimentin could be considered as an attractive target for cancer therapy. This study opens up new possibilities for the development of novel promising therapeutic agents based on DNA aptamers and natural polysaccharides as their carriers.
Supplementary Material
Acknowledgments
The authors thank Dr. Evgeny Inzhevatkin for Ehrlich ascites adenocarcinoma cell line; Dr. John P. Pezacki for HuH-7 cell line; Dr. John C. Bell for Vero cell line; and Dr. Roman A. Zubarev, Dr. Denis Rogosin, Dr. Maria Trusova, and Dr. Valentina A. Kratasyuk for inspiration and constant support. This work was granted in parts by Krasnoyarsk Regional Science Foundation No. 09/12 and by Siberian Branch of Russian Academy of Science (Interdisciplinary Integration Project No. 26).
Author Disclosure Statement
No competing financial interests exist.
References
- AHMED M., and NARAIN R. (2012). Intracellular delivery of DNA and enzyme in active form using degradable carbohydrate-based nanogels. Mol. Pharm. 9,3160–3170 DOI: 10.1021/mp300255p [DOI] [PubMed] [Google Scholar]
- AL-HENDY A., and SALAMA S. (2006). Gene therapy and uterine leiomyoma: a review. Hum. Reprod. Update 12,385–400 [DOI] [PubMed] [Google Scholar]
- BEREZOVSKI M.V., LECHMANN M., MUSHEEV M.U., MAK T.W., and KRYLOV S.N. (2008). Aptamer-facilitated biomarker discovery (AptaBiD). J. Am. Chem. Soc. 130,9137–9143 [DOI] [PubMed] [Google Scholar]
- BYUN Y., CHEN F., CHANG R., TRIVEDI M., GREEN K.J., CRYNS V.L. (2001). Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 8,443–450 [DOI] [PubMed] [Google Scholar]
- CAWLEY D.B., SIMPSON D.L., and HERSCHMAN H.R. (1981). Asialoglycoprotein receptor mediates the toxic effects of an asialofetuin-diphtheria toxin fragment A conjugate on cultured rat hepatocytes. Proc. Nat. Acad. Sci. U. S. A. 78,3383–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHALLA A.A., and STEFANOVIC B. (2011). A novel role of vimentin filaments: binding and stabilization of collagen mRNAs. Mol. Cell Biol. 31,3773–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOI K., KIM K., KWON I.C., KIM I.S., and AHN H.J. (2012). Systemic delivery of siRNA by chimeric capsid protein: tumor targeting and RNAi activity in vivo. Mol. Pharm. DOI: 10.1021/mp300211a [DOI] [PubMed] [Google Scholar]
- CODREA V., HAYNER M, HALL B., JHAVERI S., and ELLINGTON A. (2010). In vitro selection of RNA aptamers to a small molecule target. Curr. Prot. Nucleic Acid Chem. DOI: 10.1002/0471142700.nc0905s40 [DOI] [PubMed] [Google Scholar]
- COX J., and MANN M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26,1367–1372 [DOI] [PubMed] [Google Scholar]
- ELIYAHU H., BARENHOLZ Y., and DOMB A.J. (2005). Polymers for DNA delivery. Molecules 10,34–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLINGTON A., and SZOSTAK J. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346,818–822 [DOI] [PubMed] [Google Scholar]
- FUYUHIRO Y., YASHIRO M., NODA S., KASHIWAGI S., MATSUOKA J., DOI Y., KATO Y., KUBO N., OHIRA M., and HIRAKAWA K. (2010). Clinical significance of vimentin-positive gastric cancer cells. Anticancer Res. 30,239–243 [PubMed] [Google Scholar]
- GABOR F. B.OGNER E. W.EISSENBOECK A., and WIRTH M. (2004). The lectin-cell interaction and its implications to intestinal lectin-mediated drug delivery. Adv. Drug Deliv. Rev. 56,459–480 [DOI] [PubMed] [Google Scholar]
- GILLES C., POLETTE M., MESTDAGT M., NAWROCKI-RABY B., RUGGERI P., BIREMBAUT P., and FOIDART J.M. (2003). Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res. 63,2658–2664 [PubMed] [Google Scholar]
- GROMAN E.V., ENRIQUEZ P.M., JUNG C., and JOSEPHSON L. (1994). Arabinogalactan for hepatic drug delivery. Bioconjug. Chem. 5,547–556 [DOI] [PubMed] [Google Scholar]
- HU L., LAU S.H., TZANG C.H., WEN J.M., WANG W., XIE D., HUANG M., WANG Y., WU M.C., HUANG J.F., et al. (2004). Association of Vimentin overexpression and hepatocellular carcinoma metastasis. Oncogene 23,298–302 [DOI] [PubMed] [Google Scholar]
- HUANG Y.F., SHANGGUAN D., LIU H., PHILLIPS J.A., ZHANG X., CHEN Y., and TAN W. (2009). Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. Chembiochem 10,862–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IVASKA J., PALLARI H.M., NEVO J., and ERIKSSON J.E. (2007). Novel functions of vimentin in cell adhesion, migration, and signaling. Exp. Cell Res. 313,2050–2062 [DOI] [PubMed] [Google Scholar]
- JIAN Y, GAO Z, SUN J, SHEN FENG QF.JING Y, YANG C. (2009). RNA aptamers interfering with nucleophosmin oligomerization induce apoptosis of cancer cells. Oncogene 28,4201–4211 [DOI] [PubMed] [Google Scholar]
- JIN H., MOROHASHI S., SATO F., et al. (2010). Vimentin expression of esophageal squamous cell carcinoma and its aggressive potential for lymph node metastasis. Biomed. Res. 31, 105–112 [DOI] [PubMed] [Google Scholar]
- JULIANO R., ALAM M.R., DIXIT V., KANG H. (2008). Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides. Nucleic Acids Res. 36,4158–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAZUKI Y., HOSHIYA H., TAKIGUCHI M., ABE S., IIDA Y., OSAKI M., KATOH M., HIRATSUKA M., SHIRAYOSHI Y., HIRAMATSU K., et al. (2011). Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther. 18,384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KEEFE A.D., PAI S., and ELLINGTON A. (2010). Aptamers as therapeutics. Nat. Rev. Drug Discov. 9,537–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOLOVSKAYA O.S., SAVITSKAYA A.G., ZAMAY T.N., RESHETNEVA I.T., ZAMAY G.S., et al. (2013). Development of bacteriostatic DNA aptamers for salmonella. J. Med. Chem. 56,1564–1572 [DOI] [PubMed] [Google Scholar]
- KORSCHING E., PACKEISEN J., LIEDTKE C., HUNGERMANN D., WULFING P., VAN DIEST P.J., BRANDT B., BOECKER W., and BUERGER H. (2005). The origin of vimentin expression in invasive breast cancer: epithelial-mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential. J. Pathol. 206,451–457 [DOI] [PubMed] [Google Scholar]
- LABIB M., ZAMAY A.S., MUHAREMAGIC D., CHECHIK A., BELL J.C., and BEREZOVSKI M.V. (2012). Electrochemical differentiation of epitope-specific aptamers. Anal. Chem. 84,2548–2556 [DOI] [PubMed] [Google Scholar]
- LAHAT G., ZHU Q.S., HUANG K.L., WANG S., BOLSHAKOVA S., LIU J., TORRES K., LANGLEY R.R., LAZAR A.J., HUNG M.C. and LEV D. (2010). Vimentin is a novel anti-cancer therapeutic target; insights from in vitro and in vivo mice xenograft studies. PLoS One 5,e10105 DOI: 10.1371/journal.pone.001010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- LEHR C.M., and GABOR F. (2004). Lectins and glycoconjugates in drug delivery and targeting. Adv. Drug Deliv. Rev. 56,419–420 [DOI] [PubMed] [Google Scholar]
- LI H., SUN X.ZHAO D., and ZHANG Z. (2012). A cell-specific poly(ethylene glycol) derivative with a wheat-like structure for efficient gene delivery. Mol. Pharm. 9,2974–2985 DOI: 10.1021/mp300321n [DOI] [PubMed] [Google Scholar]
- LI M., ZHANG B., SUN B., WANG X., BAN X., SUN T., LIU Z., and ZHAO X. (2010). A novel function for vimentin: the potential biomarker for predicting melanoma hematogenous metastasis. J. Exp. Clin. Cancer Res. 29, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI N., NGUYEN H.H., BYROM M., and ELLINGTON A.D. (2011). Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS One 6,e20299, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI Y., HUANG G., DIAKUR J., and WIEBE L.I. (2008). Targeted delivery of macromolecular drugs: asialoglycoprotein receptor (ASGPR) expression by selected hepatoma cell lines used in antiviral drug development. Curr. Drug Deliv. 5,299–302 [DOI] [PubMed] [Google Scholar]
- MEYER C., HAHN U., and RENTMEISTER A. (2011). Cell-specific aptamers as emerging therapeutics. J. Nucleic Acids 2011,904750 DOI: 10.4061/2011/904750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUHAREMAGIC D., LABIB M., GHOBADLOO S.M., ZAMAY A.S., BELL J.C., and BEREZOVSKI M.V. (2012). Anti-fab aptamers for shielding virus from neutralizing antibodies. J. Am. Chem. Soc. 134,17168–17177 [DOI] [PubMed] [Google Scholar]
- RAY P., and WHITE R.R. (2010). Aptamers for targeted drug delivery. Pharmaceuticals 3,1761–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RHO J.H., ROEHRL M.H., and WANG J.Y. (2009). Glycoproteomic analysis of human lung adenocarcinomas using glycoarrays and tandem mass spectrometry: differential expression and glycosylation patterns of vimentin and fetuin A isoforms. Protein J. 28,148–160 [DOI] [PubMed] [Google Scholar]
- SATELLI A, LI S. (2011). Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol. Life Sci. 68,3033–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SELLINS K., FRADKIN L., LIGGITT D., and DOW S. (2005). Type I interferons potently suppress gene expression following gene delivery using liposome(-)DNA complexes. Mol. Ther. 12,451–459 [DOI] [PubMed] [Google Scholar]
- SETHI S., MACOSKA J., CHEN W., and SARKAR F.H. (2010). Molecular signature of epithelial-mesenchymal transition (EMT) in human prostate cancer bone metastasis. Am. J. Transl. Res. 3,90–99 [PMC free article] [PubMed] [Google Scholar]
- STRIBLEY J.M., REHMAN K.S., NIU H., and CHRISTMAN G.M. (2002). Gene therapy and reproductive medicine. Fertil. Steril. 77,645–657 [DOI] [PubMed] [Google Scholar]
- SUDA T., SUDA K., and LIU D. (2008). Computer-assisted hydrodynamic gene delivery. Mol. Ther. 16,1098–1104 [DOI] [PubMed] [Google Scholar]
- TUERK C., and GOLD L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249,505–510 [DOI] [PubMed] [Google Scholar]
- ULRICH H. (2006). RNA aptamers: from basic science towards therapy. Handb. Exp. Pharmacol. 173,305–326 [DOI] [PubMed] [Google Scholar]
- VALLADEAU J., DUVERT-FRANCES .V, PIN J.J., KLEIJMEER M.J., AIT-YAHIA S., RAVEL O., VINCENT C., VEGA F., HELMS A., GORMAN D., et al. (2001). Immature human dendritic cells express asialoglycoprotein receptor isoforms for efficient receptor-mediated endocytosis. J. Immunol. 167,5767–5774 [DOI] [PubMed] [Google Scholar]
- VORA H.H., PATEL N.A., RAJVIK K.N., MEHTA S.V., BRAHMBHATT B.V., SHAH M.J., SHUKLA S.N., and SHAH P.M. (2009). Cytokeratin and vimentin expression in breast cancer. Int. J. Biol. Markers. 24,38–46 [DOI] [PubMed] [Google Scholar]
- WHITEHEAD K.A., LANGER R., and ANDERSON D.G. (2009). Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 8,129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS A.A., HIGGINS J.P., ZHAO H., LJUNBERG B., and BROOKS J.D. (2009). CD 9 and vimentin distinguish clear cell from chromophobe renal cell carcinoma. BMC Clin. Pathol. 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHAO Y., YAN Q., LONG X., CHEN X., and WANG Y. (2008). Vimentin affects the mobility and invasiveness of prostate cancer cells. Cell Biochem. Funct. 26,571–577 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.