Abstract
Ionotropic glutamate receptors comprise two conformationally different A/C and B/D subunit pairs. Closed channels exhibit 4-fold radial symmetry in the transmembrane domain (TMD) but transition to 2-fold dimer-of-dimers symmetry for extracellular ligand binding and N-terminal domains. Here, to evaluate symmetry in open pores we analyzed interaction between the Q/R editing site near the pore loop apex and the transmembrane M3 helix of kainate receptor subunit GluK2. Chimaeric subunits that combined the GluK2 TMD with extracellular segments from NMDA receptors, which are obligate heteromers, yielded channels made up of A/C and B/D subunit pairs with distinct substitutions along M3 and/or Q/R site editing status, in an otherwise identical homotetrameric TMD. Our results indicate that Q/R site interaction with M3 occurs within individual subunits and is essentially the same for both A/C and B/D subunit conformations, suggesting that 4-fold pore symmetry persists in the open state.
Keywords: pore loop, docosahexaenoic acid, NMDA receptor, kainate receptor
Introduction
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate excitatory synaptic transmission throughout the central nervous system. Aberrant activation of these receptors is implicated in a number of pathological situations, which has motivated intense effort to understand their operation and to devise therapeutic interventions that might allow for regulation of iGluR activity1. The three main iGluR subtypes, named for the agonists N-methyl-D-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and kainate (KA), are made up of distinct sets of homologous subunits that combine as homo or heteromeric tetramers to form an ion conducting pathway through the membrane1. X-ray crystallography of subunit sub-domains2 and of an intact homomeric AMPA receptor in the closed state3, has revealed a modular structure with 2-fold symmetry in the extracellular amino terminal and ligand binding domains (ATD and LBD) and apparent 4-fold symmetry in the channel-forming transmembrane domain (TMD) (Fig. 1). The iGluR channels exhibit a pore-helix and selectivity filter flanked by two transmembrane helices as originally described for the KcsA channel4, but with inverted topology. In addition, all eukaryotic iGluR subunits include an additional transmembrane helix (M4) that is essential for channel function5-7.
Figure 1. Glutamate receptor domain structure.
(a) Chimaeric glutamate receptor homology model based on the homomeric GluA2 AMPA receptor closed state x-ray structure (Sobolevsky et al., 2009) illustrating the amino terminal domain (ATD), ligand-binding domain (LBD) and transmembrane domain (TMD). The cytoplasmic carboxy terminal domain (CTD) was not resolved or modeled. Each subunit is shown in a different color. In wild type NMDA receptors GluN1 and GluN2B subunits are in the A/C and B/D conformations, respectively. (b) Diagram of the four domains in 2 chimaeric subunits of a heteromeric receptor highlighting the pore loop and M1-M4 alpha helices in the TMD derived from KA receptor subunit GluK2 (white roman numerals I, II, III, IV). Left hand subunit is unedited (Q) at the apex of the pore loop and wild type L614 in M3 at the level of the central cavity. Right hand subunit is edited (R) and mutated (L614A) in M3. Subunits with N1 and N2B extracellular domains are presumed to be adjacent in functional tetramers (below), but for clarity are positioned opposite each other in the diagram. (c) Enlarged side and axial views of the homology model showing the lower portion of the LBD in faint teal (GluN1) and red (GluN2B), as well as the TMD and linkers from GluK2 in full color. The view down the central axis from the extracellular domains illustrates differences in the A/C and B/D linkers. (d) Sequence alignment for chimaeric GluN1 and GluN2B with GluK2. Green triangles indicate the position of joints in the chimaeric subunits. The purple triangle indicates an additional joint for subunits that only included the TMD from GluK2. Numbering is for the mature wild type proteins. Secondary structure is shown as cylinders (α-helices), arrows (β-strands) and lines (loops) in grey above the sequence. * indicates the Q/R/N site in the pore loop and the GluK2 L614A mutation site in M3. ◇ Indicates the M3 Ala at the middle of the bundle crossing. The W in M2 of GluN1 and the S in M3 of GluN2B that interact with each other (Siegler Retchless et al., 2012) are underlined.
In contrast to potassium channels and most other members of this ion channel superfamily, the pore loop in iGluRs is on the cytoplasmic face of the channel and the inner helix bundle crossing, believed to form the gate to ion passage, faces the extracellular side and connects to the LBD via short linkers. Motion of the LBD coincident with agonist binding is thought to pull on the linkers, inducing a conformational change in the TMD that opens the channel. Once open, iGluRs conduct monovalent cations and in some cases calcium ions1.
A striking feature of the intact homomeric GluA2 crystal structure3 is the presence of two distinct subunit configurations, with identical A/C and B/D subunit pairs arranged diagonally across from each other within the TMD. In the closed state structure the transition from 2-fold to 4-fold symmetry occurs within a narrow zone near the extracellular membrane surface where the linkers connect the LBD and TMD3. It remains unclear, however, whether this abrupt symmetry transition persists when channels open or desensitize. Indeed, several lines of evidence suggest that in the open state there may be asymmetries between the A/C and B/D subunit pairs, at least near the bundle crossing where the extracellular ends of the inner (M3) helix make close contact to occlude ion passage. For example, studies of heteromeric NMDA receptors with inner helix cysteine substitutions in and around the highly conserved SYTANLAAF motif have demonstrated asymmetric modification for homologous positions of GluN1 versus GluN2 subunits8, suggesting an offset of the A/C (GluN1) relative to B/D (GluN2) subunit pairs. In addition, homomeric GluA1 AMPA receptors with cysteine substitutions in this segment exhibit block by cadmium ions that is consistent with 2-fold symmetry in the open state9. More recent experiments on both AMPA10 and NMDA11 receptors have provided evidence for differences between the A/C and B/D subunit pairs in the extent and timing of gating-associated movements. It remains to be determined whether these apparent asymmetries are confined to the extracellular portions of M3, close to the linkers connecting to the LBD, or whether the entire TMD converts to 2-fold symmetry when channels open and/or desensitize12.
To begin addressing this question we have analyzed an interaction that was recently discovered between residues along the inner (M3) helix and the Q/R site near the apex of the pore loop of homomeric GluK2 KA receptors13. The Q/R site is a location where RNA editing can alter the primary amino acid sequence, replacing a polar, but uncharged glutamine (Q) encoded by genomic DNA with a positively charged arginine (R). Such editing occurs in the KA receptor GluK1 and GluK2 subunits, as well as the GluA2 subunit of AMPA receptors. The homologous position remains a Q in all other AMPA and KA receptor subunits and an asparagine in all GluN1 and GluN2 NMDA receptor subunits1. Q/R site editing controls a variety of channel properties1 including ion selectivity and unitary conductance, as well as susceptibility to inhibition by polyamines and by cis-unsaturated fatty acids such as arachidonic and docosahexaenoic acid (AA and DHA).
As shown by recent work13, the GluK2 pore loop Q/R site interacts with several residues along the inner (M3) helix, most notably with the side chain at position L614 at the level of the central cavity (Fig. 1). Moreover, exposure to DHA enhances the strength of this energetic coupling. For example, in control conditions Q/R site editing interacts with L614V or D substitutions with a coupling energy of ~2.5 kT and treatment with DHA increases the coupling in both cases by an additional 3 kT. The GluK2(R) L614A substitution mutant displays the largest change with DHA treatment. Whereas wild type homomeric edited GluK2(R) channels are more than 80% inhibited by exposure to DHA, when editing is combined with the L614A substitution DHA treatment potentiates agonist-evoked currents by ~10 fold. Although these results in homomeric channels demonstrated strong interaction between the Q/R site and M3 helix they did not reveal whether the interaction is equivalent for all 4 subunits or whether there are substantial differences between the relative contributions of the A/C and B/D subunit pairs. In addition, work to date has not determined whether the Q/R site to M3 interaction occurs within an individual subunit or across the boundary between two adjacent subunits13, 14.
In this study, to address these questions we have constructed chimaeric channels that combine the TMD and cytoplasmic carboxy terminal domain (CTD) of the GluK2 kainate receptor subunit with the extracellular ATD and LBD of NMDA receptor subunits GluN1 and GluN2B. Because NMDA receptors are obligate heteromers1 and the extracellular domains are strongly implicated in specifying the heteromeric arrangement of subunits within tetrameric channels15 we reasoned that functional channels would only be generated when both a GluN1/GluK2 and GluN2B/GluK2 chimaera were co-expressed, which turned out to be the case. In this way we were able to construct channels with A/C and B/D subunits that differed in their Q/R editing or M3 substitutions, or both. Using this approach we show that interactions between the Q/R site and M3 helix occur within each subunit, not between adjacent subunits; and, that pore loop to M3 interactions within the A/C and B/D subunit pairs make nearly equal contributions to the potentiation elicited by treatment with DHA, suggesting that the 4 fold radial symmetry observed for the closed state TMD is preserved when channels open, at least up to the level of the central cavity.
Results
Chimaeric subunits with a kainate receptor pore
Previous efforts to transplant just the M1 to M3 segment between NMDA and KA receptor subunits met with limited success16, probably because interaction of the M1 to M3 pore with the M4 transmembrane helix is now recognized to be essential for channel assembly7 and / or function5, 6. Therefore, we combined the entire KA receptor TMD including the linkers to the LBD, as well as the CTD, with the extracellular domains from NMDA receptor subunits GluN1 and GluN2B (Fig. 1).
In wild type NMDA receptors glutamate or NMDA binds to the GluN2 subunit, whereas the co-agonist glycine or D-serine binds to GluN11. In addition, recent work 17, 18 supports the assignment3 of GluN1 to the A/C configuration and GluN2 to the B/D configuration (but see19). As shown in figure 2a expression of each chimaeric subunit alone, in either the unedited (Q) or edited (R) form, failed to generate functional homomeric channels, however, co-expression of the GluN1-GluK2 pore (N1/K2) and GluN2B-GluK2 pore (N2B/K2) chimaeric subunits together resulted in channels that were activated by application of NMDA and the co-agonist glycine20, 21. As for wild type full length GluK2, when both subunits were unedited (Q) in the pore loop the resulting channels displayed bi-rectifying current-voltage relations (Fig. 2b) indicative of voltage-dependent block by cytoplasmic polyamines1. In contrast, if one or both subunits was in the edited (R) form, then polyamine block was diminished and the IV relation was linear (Fig. 2c-e), or showed slight outward rectification22. Importantly, exposure to DHA strongly inhibited channels in which all of the subunits were edited (R) in the pore loop but caused little or no change in channels with either or both subunits in the unedited (Q) form (P < 0.0001, one way ANOVA with post hoc Student-Newman-Keuls comparison) (Fig. 3), which is identical to the pattern of DHA modulation observed for wild type full length recombinant heteromeric kainate receptors23 (Fig. 3c). A similar outcome was obtained with constructs in which the CTD, as well as the ATD and LBD, came from GluN1 and GluN2B.
Figure 2. Whole-cell current and current-voltage relations for channels with kainate receptor pores.
(a) Whole-cell current (mean ± s.e.m.) evoked by 10 μM NMDA plus 10 μM glycine in HEK 293 cells transfected with edited (R) or unedited (Q) N1/K2 and N2B/K2 chimaeric subunits. Agonist application did not elicit current in cells transfected with the N1/K2(Q/R) or N2B/K2(Q/R) subunits alone. Sample size is shown for each bar. (inset) Current recorded from a cell transfected with N1/K2(Q)+N2B/K2(Q). (b-e) Whole-cell currents evoked by 10 μM NMDA and 10 μM glycine as the membrane potential was ramped from −160 to +110 mV at 0.75 mv msec−1. (b) Bi-rectification of current mediated by N1/K2(Q)+N2B/K2(Q) reflects block by endogenous polyamines, as well as 20 μM spermine added to the internal solution, with relief of block as the polyamines permeate the channel at positive potentials. Polyamine block was eliminated by Arg substitution at the Q/R site of either the N1/K2 (c) or N2B/K2 (d) subunit, or both subunits (e).
Figure 3. Selective DHA inhibition of fully edited chimaeric receptors.
Whole-cell current evoked by 10 μM NMDA and 10 μM glycine (open bars) at −80 mV before and after exposure to 30 μM DHA (solid bar) in a cell transfected with fully edited N1/K2(R) + N2B/K2(R) subunits (a) and in a cell that received N1/K2(Q)+N2B/K2(R) (b). (c) Plot of current evoked immediately after exposure to DHA as a fraction of control current before DHA (mean ± s.e.m.) for the 4 chimaeric combinations and, for comparison, wild type homomeric GluK2(Q) and (R) and heteromeric GluK2(R)+GluK5(Q). Sample size is shown for each bar. * significantly different from N1/K2(Q)+N2B/K2(Q) and homomeric GluK2(Q) (P < 0.0001, one way ANOVA with post hoc Student-Newman-Keuls comparison).
Taken together, these results demonstrate that the kainate receptor TMD is sufficient to determine the effect of DHA exposure: inhibition when all four subunits are edited (R), but neither inhibition nor potentiation when one or both subunits is unedited (Q). Thus, the NMDA receptor ATD and LBD domains effectively activate all of the edited and unedited GluK2 pore combinations, but are not able to prevent inhibition of R/R pores by DHA or to induce potentiation of Q/R, R/Q or Q/Q pores24, 25. Collectively, these results strongly suggest that AA and DHA act by partitioning into the membrane and directly or indirectly modifying conformational changes in the TMD associated with channel opening.
Q/R site interaction with the M3 helix
Recent work on homomeric GluK2 KA receptors has demonstrated that several amino acids along the inner transmembrane helix (M3) interact selectively with the Arg side chain at the Q/R site in fully edited channels13. In particular, a number of substitutions at position L614 completely reverse the effect of DHA exposure on GluK2(R), while having little or no effect on GluK2(Q). Substitution of L614 with alanine had the largest effect, converting ~90% inhibition of wild type GluK2(R) to approximately 10-fold potentiation for homomeric GluK2(R)L614A13. In order to determine the relative contribution of the A/C and B/D subunit pairs to this interaction and to test whether it occurs between adjacent subunits or within individual subunits, we made Q and R forms of both GluN1/K2 and GluN2B/K2 each bearing alanine substituted for the leucine homologous to L614 in full length GluK2 (henceforth referred to as L614A substitutions in the chimaeric subunits). As shown in figure 4, all twelve of the possible heteromeric combinations of mutant and non-mutant subunits were functional and a clear pattern emerged for modulation of channel function by DHA: First, the strongest potentiation by DHA (>10 fold) occurred when all 4 subunits were both edited (R) and mutated (L614A); second, intermediate potentiation (2 to 4 fold) was observed when one of the two chimaeric subunits was both edited (R) and mutated (L614A); third, less than 1.5 fold potentiation occurred when the L614A mutation was only present in unedited (Q) subunits (P < 0.0001, one way ANOVA with post hoc Student-Newman-Keuls comparison).
Figure 4. Intrasubunit interaction between the pore loop and M3 helix.
Whole-cell current evoked by 10 μM NMDA and 10 μM glycine (open bars) at −80 mV before and after exposure to 30 μM DHA (solid bar) in cells co-transfected with N1/K2(R)+N2B/K2(Q)L614A (a), N1/K2(R)L614A+N2B/K2(Q)L614A (b), N1/K2(Q)+N2B/K2(R)L614A (c), or N1/K2(R)L614A+N2B/K2(R)L614A (d). (e) Plot of current evoked immediately after exposure to DHA as a fraction of control current before DHA (mean ± s.e.m.) for the 12 chimaeric constructs and, for comparison, homomeric L614A mutants of GluK2(Q) and (R). Sample size is shown for each bar. Note the logarithmic scale. DHA had minimal effect when L614A and Q to R editing were not present on the same chimaeric subunit; green horizontal bar indicates the 95% confidence interval (C.I.) for N1/K2(Q)L614A +N2B/K2(Q)L614A. Significant potentiation was observed when one of the two subunits included the L614A substitution and Q to R editing (P < 0.0001, one way ANOVA with post hoc Student-Newman-Keuls comparison); 95% C.I. for N1/K2(Q)L614A +N2B/K2(R)L614A (yellow bar) Potentiation was greatest when both chimaeric subunits were edited (R) and mutated (L614A); 95% C.I. for N1/K2(R)L614A+N2B/K2(R)L614A (pink bar).
Together, the results in figure 4 provide strong evidence that the Arg side chain in edited Q/R sites interacts with the residue at position 614 within the same subunit. Channels that included an edited (R) subunit wild type at L614 and an L614A mutation on the adjacent unedited (Q) subunit displayed no significant potentiation relative to completely unedited (Q/Q) combinations. However, combinations in which both subunits were edited (R) but only one of them bore the L614A mutation exhibited stronger potentiation than combinations with one or both subunits mutated (L614A) but only one subunit edited (R)(P < 0.0001, one way ANOVA with post hoc Student-Newman-Keuls comparison)(Fig. 4e), suggesting that confinement of 4 Arg guanidinium groups within the pore enhances the interaction with M3. Thus, the effect of DHA is strongest when all four subunits are edited at the Q/R site, whether the effect is inhibition, as for wild type L614, or potentiation, with either 2 or 4 L614A substitutions.
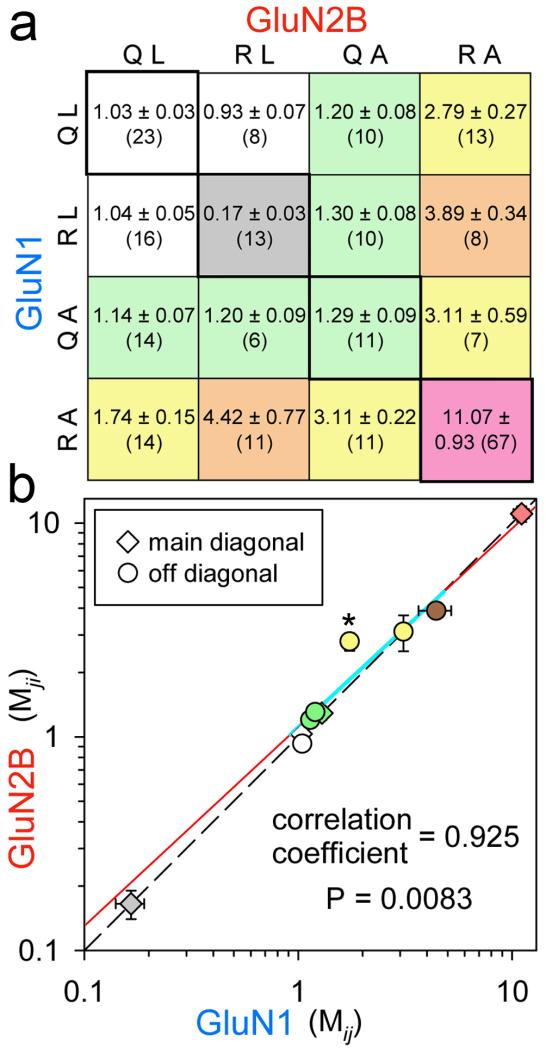
Importantly, the Q/R site interaction with M3 is likely to be equivalent in both the A/C and B/D subunit pairs because a similar level of potentiation was observed for channels that included either the N1/K2(R)L614A or the N2B/K2(R)L614A constructs (Fig. 5). The only evidence for asymmetry came from the N1/K2(R)L614A+N2B/K2(Q) combination that displayed weaker potentiation by DHA than any of the other constructs with L614A on an edited (R) subunit (P = 0.002, one-way ANOVA with post hoc Student-Newman-Keuls comparison)(Fig. 4e). Despite this difference there was strong correlation between the effects of DHA on symmetric construct pairs (Fig. 5b) (Pearson correlation coefficient = 0.925 for the 6 off-diagonal points; P = 0.0083 that the association is invalid; regression line for the off-diagonal points in cyan and extended to the axes in red). Indeed, the relation between analogous pairs of N1/K2 and N2B/K2 constructs was well described by a model assuming 4-fold radial symmetry (F test) (Fig. 5b, perfect symmetry indicated by the dashed line).
Figure 5. Radial symmetry in chimaeric receptor pores.
(a) Interaction matrix for Q/R editing and L614A substitutions to chimaeric N1/K2 and N2B/K2 subunits. Matrix elements display I DHA / I control as mean ± sem (# of cells). Color code: No A + <2R, white; No A + 2R, grey; A not with R, green; 1R with 1 or 2 A, yellow; 1A with 2R, tan; 2A with 2R, pink. (b) Diamonds plot I DHA / I control for the 4 main diagonal elements (Mji versus Mij where i = j) outlined in bold in (a). Circles plot I DHA / I control in N2B/K2 versus N1/K2 for symmetric constructs (off diagonal matrix elements, Mji versus Mij and i ≠ j). Pearson product moment correlation coefficient = 0.925 for the six off diagonal points indicates strong symmetric association (P = 0.0083 that the association is invalid). In addition, linear regression to the 6 off diagonal matrix element points (solid line, 2 parameters) was not statistically superior (F test) to the line of identity through the symmetric main diagonal elements (dashed line, no free parameters). The solid line is shown in cyan over the range of off diagonal points used for regression. It is extended to each axis in red. * Only the N1/K2(Q) +N2B/K2(R)L614A, N1/K2(R)L614A+N2B/K2(Q) coordinate pair displayed significant asymmetry (P = 0.002, one way ANOVA with post hoc Student-Newman-Keuls comparison).
Decoupling the co-agonist sites
In addition to increasing agonist-evoked currents, in some cases exposure to DHA also increased the baseline holding current (e.g. Figs. 4d, 6a). The magnitude of the change varied from cell to cell but on average was significantly larger for construct combinations that also showed the greatest potentiation of agonist-evoked current (Fig. 6b), raising the possibility that DHA was either increasing the open probability of unliganded chimaeric channels26, 27 or enhancing channel activation by trace levels of agonist that might be present in our control external solution20, 21, 28. If the change in baseline involved activation by trace agonist levels, then it should be blocked or reversed by co-application of competitive antagonists, which was indeed the case for antagonists of the GluN1 glycine binding site including ACEA 102129 and 5-fluoro-indole-2-carboxylic acid (5F-I2CA)30, whereas competitive inhibitors of the GluN2 glutamate / NMDA binding site, such as APV and CPP31, had less effect (Fig. 6c, d).
Figure 6. Changes in holding current with DHA and antagonists.
(a) Whole-cell current evoked by 10 μM NMDA and 10 μM glycine (open bars) before and after exposure to 15 μM DHA (black bars). The glycine site antagonist ACEA 1021 (1 μM) was applied together with DHA during the periods indicated by the grey bars. After the final exposure to DHA the control external solution contained 0.1% BSA. (b) Plot of current evoked immediately after exposure to DHA (mean ± s.e.m.) as a fraction of control current before DHA for the 12 chimaeric constructs in Fig. 4 versus change in holding current with DHA exposure normalized to the current evoked by NMDA and glycine. Pearson product moment correlation coefficient = 0.949 (P < 0.0001). Sample size as given in Fig. 4e and 5a. (c) Percent block of DHA induced change in holding current (mean ± s.e.m.) for N1/K2(R)L614A + N2B/K2(R)L614A by 1 μM ACEA 1021 or 1 mM 5F-I2CA (20 cells) or by 50 μM APV or 30 μM CPP. Sample size is shown for each bar. (d) Percent change in baseline holding current (mean ± s.e.m.) without DHA exposure.
Analysis of macroscopic21, 32, 33 and single channel34-36 currents suggest that efficient opening of wild-type NMDA receptor channels requires agonist occupancy of all four LBDs, with both GluN1 subunits binding glycine or D-serine and both GluN2 subunits binding glutamate or NMDA. In contrast, AMPA and kainate receptors can open with only a subset of the four agonist binding sites occupied37-39. Our initial results with chimaeric subunits showed that production of functional channels requires co-expression of N1/K2 and N2/K2 subunits (Fig. 2a) but did not determine whether simultaneous occupancy of both the glycine and NMDA sites is essential for channel activation. To test for channel activation by NMDA alone we recorded dose-response relations for NMDA in the presence of a glycine site antagonist (1 mM 5F-I2CA) and no added glycine and normalized to current evoked in the same cells by 1 mM NMDA plus 10 μM glycine and no antagonist (Fig. 7a-d). Currents were recorded both before and after exposure to DHA to see whether there was any change in apparent affinity or efficacy. For cells transfected with N1/K2(Q)+N2B/K2(Q) there was little evidence for channel activation by NMDA alone (Fig. 7a, c; n=4 cells), either before or after DHA treatment. In contrast, NMDA alone evoked almost 30% as much current as NMDA plus glycine in cells co-transfected with the M3 mutant subunits N1/K2(R)L614A+ N2B/K2(R)L614A (Fig. 7b, d; n=6 cells). Moreover, treatment with DHA produced an equivalent increase in currents evoked by NMDA alone or with glycine, such that there was no significant change in the normalized dose-response relation (Fig. 7d; F test).
Figure 7. Decoupling of coagonist sites.
(a, b) Whole-cell currents evoked by 1 mM NMDA and 10 μM glycine (solid bars), then by 1 mM, 10, 1 and 0.1 μM NMDA alone (open bars) in the presence of 1 mM 5F-I2CA, a glycine site antagonist (grey bars). N1/K2(Q) + N2B/K2(Q) (a) and N1/K2(R)L614A + N2B/K2(R)L614A (b) before (top) and after (bottom) exposure to 15 μM DHA. (c, d) Currents (mean ± sem) evoked by NMDA alone plus 1 mM 5F-I2CA (circles) normalized to 1 mM NMDA plus 10 μM glycine (diamonds), before (open) and after (filled) exposure to DHA. Smooth curve: best Hill equation fit to NMDA alone (with 5F-I2CA) before and after DHA, EC50 = 1.7 μM, n = 2.1 and Imax = 0.28 (6 cells). (e, f) Current evoked by 100 μM NMDA plus 10 μM glycine (solid bars), then by 50, 10, 2, 0.4, 0.08 and 0.016 μM glycine alone (open bars). Traces from N1/K2(Q) + N2B/K2(Q) before DHA (e) and from N1/K2(R)L614A + N2B/K2(R)L614A after DHA (f). (g, h) Currents (mean ± sem) evoked by glycine alone (circles) normalized to 10 μM NMDA plus 10 μM glycine (diamonds), before (open) and after (filled) exposure to DHA. Smooth curves: best Hill equation fit to glycine alone before and after DHA for N1/K2(Q) + N2B/K2(Q) (g) with EC50 = 0.79 μM, n = 0.82 and Imax = 0.51 (6 cells) and for N1/K2(R)L614A + N2B/K2(R)L614A (h) with EC50 = 0.57 μM, n = 0.87 and Imax = 0.38 (10 cells). Exposure to DHA did not significantly alter the concentration response relations for NMDA alone (c, d) or glycine alone (g, h) (F test).
To test for channel activation by glycine alone we recorded glycine dose-response relations in external solutions without added NMDA or glutamate (Fig. 7e-h), and in some cases including NMDA site competitive antagonists (either 50 μM APV or 30 μM CPP). Currents recorded before or after exposure to DHA were normalized to the control response evoked by 100 μM NMDA plus 10 μM glycine. In contrast to NMDA, application of glycine alone evoked substantial currents in cells expressing either the non-mutant N1/K2(Q)+N2B/K2(Q) (0.5 of control, n=6 cells) or the mutant N1/K2(R)L614A+N2B/K2(R)L614A chimaeric subunits (0.4 of control, n=10 cells)(Fig. 7e-h). Importantly, there was little difference in the half-maximal dose (EC50) for glycine between the two construct combinations (0.6 and 0.8 μM, respectively; t-test, P=0.52) and no significant change in the EC50 following treatment with DHA (F test). Collectively, these results suggest a loss of the strict requirement for all four agonist sites to be occupied in order for channels to open. Instead, chimaeric channels that include the GluK2 TMD and linkers appear to be activated by glycine alone, or by NMDA alone for the M3 mutant constructs, with only a modest reduction in efficacy compared to co-application of both agonists together. The EC50 values for activation of chimaeric constructs by glycine or NMDA alone are somewhat lower than observed for intact wild type subunits29 or neuronal NMDA receptors30, 40, which might reflect a difference in linkage from LBD to TMD41 in the chimaeric subunits, or alternatively, might be owing to the lack of negative allosteric coupling between agonist binding sites33, 40 when only one agonist is applied.
Discussion
Our results support several conclusions about the structure and operation of iGluRs and their modulation by cis-unsaturated fatty acids. Despite minimal sequence identity1, the heteromeric extracellular ATD and LBD from NMDA receptor subunits GluN1 and GluN2B were able to operate the homomeric TMD from the GluK2 kainate receptor subunit, confirming the modular arrangement of iGluRs42-44. Moreover, the GluK2 TMD embedded within the chimaeric subunits can completely reproduce the modulatory effects of DHA observed for full length homomeric wild type GluK223 and for the M3 helix L614A point mutation in homomeric GluK2 channels13. These results strongly suggest that modulation of GluK2 by DHA involves a direct effect on the TMD via the membrane. In addition, our results provide strong evidence that an edited pore loop Q/R site Arg side chain interacts with the M3 helix13 within an individual subunit rather than between adjacent subunits. Substantial potentiation by exposure to DHA was only observed when both Q to R editing and L614A substitution were present on the same chimaeric subunit. Because a similar level of potentiation was observed for L614A substitution of either the N1/K2(R) or N2B/K2(R) subunits, our results also suggest that the pore retains 4-fold radial symmetry in the open state, at least up to the level of the central cavity. Finally, our results demonstrate decoupling of the co-agonist sites in chimaeric receptors, suggesting that linkage between the LBD and TMD underlies the strict requirement of intact wild type NMDA receptors for occupation of both the agonist and co-agonist sites in order to promote channel opening (see also27).
Previous work on chimaeric iGluRs has involved transferring domains within the members of a subfamily (i.e. GluN2A and GluN2B45, 46 or GluK1 and GluK247) or between AMPA and kainate receptors43, 44, which are more closely related to each other (~30-40% identity) than to any of the NMDA receptor subunits (<20% identity)1. Chimaeric subunits with the M1-M3 segment from GluN1 transferred into GluK2 form functional homomeric channels16, however similar constructs using the M1-M3 portion of GluN2B are not functional as homomers and do not combine with the GluK2(GluN1 M1-M3) chimaera16. Moreover, homomeric GluK2/GluN1 (M1-M3) channels do not exhibit voltage-dependent block by Mg16, which is a hallmark of native NMDA receptors1. Based on increasing evidence that the M4 transmembrane helix is essential for channel function5-7, we fused the entire TMD and extracellular linkers plus the cytoplasmic C-terminal tail of GluK2 to the extracellular ATD and LBD from GluN1 and GluN2B, including from the NMDA receptor subunits both the S1 segment preceding M1 and the extracellular S2 segment between M3 and M442. Co-transfection of N1/K2 + N2B/K2 subunits resulted in functional channels activated by NMDA and/or glycine. However, transfection of individual chimaeric subunits alone failed to generate functional homomeric channels, consistent with the fact that NMDA receptors are obligate heteromers1 and that the extracellular domains appear to play a major role in heteromer assembly15. Further experiments will be needed to determine whether chimaeric subunits expressed alone fail to make it to the cell surface, or whether they assemble and transit to the surface as non functional oligomers.
Native iGluRs exhibit divergent modulation by AA and DHA. NMDA receptor-mediated currents potentiate 2 to 3 fold by an increase in channel open probability24, 25. Neuronal KA receptors48 and fully edited recombinant channels23 are strongly inhibited by AA and DHA, owing to a reduction in open probability49. Whereas, AMPA receptors and recombinant KA receptors that include unedited (Q) wild type subunits are relatively unaffected23, 50. It remains unclear whether iGluR modulation by AA and DHA involves specific interaction with hydrophobic segments in the extracellular, intracellular or transmembrane portions of each subunit or whether it reflects sensitivity to changes in bulk properties of the membrane, analogous to the effect of membrane stretch on NMDA receptor gating51. Using chimaeric constructs, wild type at M3 position L614, we found that the GluK2 TMD completely recapitulates the pattern of DHA modulation observed for full length homo and heteromeric kainate receptors: strong inhibition of fully edited (R)/(R) channels, but little effect on channels that included unedited subunits. Significantly, there was no potentiation of unedited (Q)/(Q) chimaeric channels, as might have been expected if DHA were to interact with hydrophobic regions of the NMDA receptor ATD, which are known to regulate activity of full length NMDA receptors15, 52. Although our results do not formally rule out such interactions with the ATD, they show that KA receptor modulation is completely determined by the TMD, suggesting that channel regulation likely depends on contacts within the membrane.
Recent experiments on NMDA receptors14 showed that unitary conductance, calcium permeability and the strength of magnesium block depend on an intersubunit interaction between a conserved M2 helix Trp residue in GluN1 and an M3 helix Ser (GluN2A and 2B) or Leu (GluN2C or 2D) in the adjacent GluN2 subunit (see Fig. 1d). Work on homomeric kainate receptor channels formed by GluK2 has also provided evidence for interactions between the pore loop and residues along the M3 helix13; however, our results in the present study indicate that interactions between Q/R site Arg residues and L614A at the level of the central cavity occur within an individual subunit rather than between adjacent subunits. In channels with one pair of edited (R) and one pair of unedited (Q) chimaeric subunits, DHA caused strong potentiation only when the edited (R) subunits included the L614A mutation. Alanine substitution at L614 of unedited (Q) subunits had little or no effect regardless the mutation status at L614 of the adjacent edited (R) subunits. On the other hand, channels with one pair of mutated (A) and one pair of wild type (L) subunits at position 614 exhibited stronger potentiation if both subunits were edited (R) than if the wild type L614 subunits were unedited (Q), which may reflect interaction between adjacent Q/R site arginines in fully edited channels. By far the strongest potentiation occurred when an edited Q/R site and the L614A mutation were present on both chimaeric subunit pairs, or on all 4 of the subunits in a homomeric full length GluK2(R)L614A channel13.
Agonist binding to iGluRs induces clam shell closure of the LBD53 that is thought to open channels by exerting force on the extracellular ends of the transmembrane helices via short random coil linkers3. In the process of opening, portions of the TMD might convert from 4 fold symmetry in the closed state to 2 fold symmetry in the open state10. A number of studies8, 27, 41 provide evidence for asymmetries in and around the pore of NMDA receptors, but whether these reflect local effects of the substantial differences in primary sequence between GluN1 and GluN2 subunits (Fig. 1d) or more global dissimilarities in subunit conformation within the TMD remains uncertain54. Importantly, analysis of cadmium binding to homomeric GluA1 AMPA receptors with an Ala to Cys substitution in the conserved M3 SYTANLAAF motif (indicated by an open diamond in Fig. 1d) provides evidence for 2-fold symmetry at the level of the bundle crossing9, which is thought to form the gate for ion flow. In the closed state crystal structure3, however, this conserved alanine lies right at the transition from 2-fold to 4-fold symmetry. Thus, it remains to be determined whether the sharp closed state transition in symmetry is preserved in the open state or whether local 2-fold symmetry extends further down the transmembrane helices when channels open and/or desensitize12. The present study evaluates an interaction approximately 3 turns down the M3 helix in a zone of strong 4-fold symmetry in the closed state3; and, our results suggest that 4-fold symmetry persists at this position when channels are open. This interpretation is most compatible with an alternating N1-N2-N1-N2 arrangement for chimaeric subunits imposed by the extracellular NMDA receptor segments15. Although substantial evidence supports this organization3, 17, 18, there is also some data that instead favors an adjacent N1-N1-N2-N2 arrangement5, 19, and our results cannot entirely exclude an adjacent organization. It is also notable that DHA regulates channels whether applied in the presence or absence of agonist, with kinetics of onset and recovery from modulation that are much slower than gating transitions13, 23. Thus, it seems unlikely that our evidence for radial symmetry reflects a selective interaction that is exclusive to the 4-fold symmetric closed state.
Despite the fact that formation of functional channels requires co-expression of both chimaeric N1/K2 and N2B/K2 subunits our results show that currents can be activated by glycine alone, or in some cases NMDA alone, suggesting that the LBD does not dictate the strict requirement of intact NMDA receptors for dual occupancy by both agonists. Instead, it suggests that the need for all 4 agonist sites to be occupied depends on linkage between the LBD and TMD that is unique for NMDA receptors. Previous work on NMDA receptors with point mutations in GluN1 at the bundle crossing (replacement of alanine homologous to that in GluA1 mentioned above, see Fig. 1d) also provided evidence for co-agonist site decoupling27, but mutations in this region are known to interfere with normal gating26. In contrast, L614, which was mutated to alanine in the present study, is well below the bundle crossing and co-agonist site decoupling was also observed for the N1/K2(Q) +N2B/K2(Q) combination with no M3 mutations. Importantly, both KA and AMPA receptor channels can open with less than complete agonist occupancy37-39, and our results suggest that this property can be transferred by transplantation of the GluK2 TMD together with the short linkers that extend up to the LBD. Analysis of additional chimaeric constructs with fusion joints closer to the TMD may reveal more about the structural basis for differential occupancy requirements between the iGluR family members.
Methods
cDNA constructs and cell culture
Subunit cDNAs, generously provided by Steve Heinemann, Peter Seeburg, Mark Mayer and Stefano Vicini, were all expressed from the pRK5 vector using the GluK2 signal sequence. Chimaeric subunits were generated by a restriction enzyme (RE) free PCR cloning method55 or by ligation of PCR products with novel restriction sites inserted by silent mutations. The RE-free method involves two sense and two antisense primers for each joint that create a 15 bp complementary overlap55 (underlined). S1 to M1 joint: sense Gluk2 CCCAATGGTACAAACCCAGG and CCAGGCGTCTTCTCCTTCCTG, antisense GluN1 GTTTGTACCATTGGGCTTCTTGACCAAAATGGTCAGGC and CTTCTTGACCAAAATGGTCAGGC; antisense GluN2B GTTTGTACCATTGGGGCGAGATACCATGACACTGATGC and GCGAGATACCATGACACTGATGC. M3 to S2 joint: antisense GluK2 GTCAATGGGCGACTCCATGC and CATGCGTTCCACAGTCAGAAAGGC, sense GluN1 GAGTCGCCCATTGACGGCATCAATGACCCCAGGCTC and GGCATCAATGACCCCAGGCTC, sense GluN2B GAGTCGCCCATTGACGGCCTGAGTGACAAGAAGTTCC and GGCCTGAGTGACAAGAAGTTCC. S2 to M4 joint: sense GluK2 CCGGAGGAGGAGAGCAAAGAG and AAAGAGGCCAGTGCTCTGGG, antisense GluN1 GCTCTCCTCCTCCGGGCATTCCTGATACCGAACCC and GCATTCCTGATACCGAACCC, antisense GluN2B GCTCTCCTCCTCCGGGCAAATGCCAGTGAGCCAGAG and GCAAATGCCAGTGAGCCAGAG. M4 to CTD joint: sense GluN1 TGCCCAATTGAAGCAGATGCAGCTGG and sense GluN2B GCATCAATTGATGGGTGTCTGTTCTGG primers generate Mfe I sites (bold) to match the endogenous unique site in GluK2. All constructs were sequenced by the Washington University PNACL facility. cDNAs were expressed by transient transfection in HEK 293 cells (ATCC) using lipofectamine 2000 (Invitrogen). The cells were propagated in 25 cm2 flasks with MEM plus 10% fetal bovine serum and passaged once each week with protease XXIII (Sigma). Cells used for transfection were seeded onto 12 well plates and transfected the following day. Co-expression of GFP from a second vector was used to identify transfected cells. The day after transfection, cells were plated at low density on 35 mm plates that were coated with nitrocellulose; recordings were obtained on the following two days.
Electrophysiology
Cultures were bath perfused with Tyrode’s solution (in mM): 150 NaCl, 4 KCl, 2 MgCl2, 2 CaCl2, 10 glucose, 10 HEPES, pH 7.4 with NaOH. Whole-cell electrodes were pulled from borosilicate tubing (WPI) and filled with an internal solution that contained (in mM): 140 Cs-glucuronate, 10 EGTA, 5 CsCl, 5 MgCl2, 5 ATP, 1 GTP, 0.02 spermine, and 10 HEPES, pH adjusted to 7.4 with CsOH. Agonists and antagonists were delivered in control extracellular solution (160 mM NaCl, 2 mM CaCl2 and 10 mM HEPES, pH adjusted to 7.4 with NaOH) by local perfusion from a multi-barreled pipette positioned near the recorded cell. Currents were recorded with an Axopatch 200A amplifier controlled by p-Clamp software (Molecular Devices). Current-voltage relations were generated by averaging the currents recorded during a series of 5 ascending and descending membrane potential ramps (0.75 mV msec-1)13. Concentration response relations were fit with the Hill equation: I = Imax / (1 + (EC50 / [agonist])^n). Results are presented as mean ± s.e.m. and were considered significant for P < 0.05. One-way ANOVA and t-tests were performed with SigmaStat (Systat Software). Curve fits using different numbers of parameters were evaluated by F-tests13.
Molecular modeling
Homology modeling of a receptor composed of two pair of chimaeric subunits, N1/K2(R)+N2B/K2(R), was performed using modeller56 release 9v7; available at http://salilab.org/modeller/. The GluA2 tetramer crystal structure (PDB ID: 3KG2) was used as the template with sequence alignment from Fig. S2 of reference 3 and two-fold symmetry constraints for the A/C and B/D subunit pairs.
Acknowledgements
We are grateful to S. Heinemann, P. Seeburg, M. Mayer, and S. Vicini for cDNA constructs, to W. Borschel for helpful comments on the manuscript, to A. Lu for technical assistance and to the NIH for support (NS30888, JEH and GM083914, MNL).
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Author Contributions
T.J.W, performed electrophysiology, M.N.L. and J.E.H. performed molecular biology. All authors contributed to the study design and to writing the manuscript.
References
- 1.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–96. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer ML. Emerging models of glutamate receptor ion channel structure and function. Structure. 2011;19:1370–80. doi: 10.1016/j.str.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 5.Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23:1151–8. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terhag J, Gottschling K, Hollmann M. The Transmembrane Domain C of AMPA Receptors is Critically Involved in Receptor Function and Modulation. Front Mol Neurosci. 2010;3:117. doi: 10.3389/fnmol.2010.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salussolia CL, Gan Q, Kazi R, Singh P, Allopenna J, Furukawa H, Wollmuth LP. A eukaryotic specific transmembrane segment is required for tetramerization in AMPA receptors. J Neurosci. 2013;33:9840–5. doi: 10.1523/JNEUROSCI.2626-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobolevsky AI, Rooney L, Wollmuth LP. Staggering of subunits in NMDAR channels. Biophys J. 2002;83:3304–14. doi: 10.1016/S0006-3495(02)75331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobolevsky AI, Yelshansky MV, Wollmuth LP. The outer pore of the glutamate receptor channel has 2-fold rotational symmetry. Neuron. 2004;41:367–78. doi: 10.1016/s0896-6273(04)00008-x. [DOI] [PubMed] [Google Scholar]
- 10.Dong H, Zhou HX. Atomistic mechanism for the activation and desensitization of an AMPA-subtype glutamate receptor. Nat Commun. 2011;2:354. doi: 10.1038/ncomms1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazi R, Gan Q, Talukder I, Markowitz M, Salussolia CL, Wollmuth LP. Asynchronous Movements Prior to Pore Opening in NMDA Receptors. J Neurosci. 2013;33:12052–66. doi: 10.1523/JNEUROSCI.5780-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486:135–9. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez MN, Wilding TJ, Huettner JE. Q/R site interactions with the M3 helix in GluK2 kainate receptor channels revealed by thermodynamic mutant cycles. J Gen Physiol. 2013;142:225–239. doi: 10.1085/jgp.201311000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegler Retchless B, Gao W, Johnson JW. A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nat Neurosci. 2012;15:406–13. doi: 10.1038/nn.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen KB, Furukawa H, Traynelis SF. Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol. 2010;78:535–49. doi: 10.1124/mol.110.067157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villmann C, Hoffmann J, Werner M, Kott S, Strutz-Seebohm N, Nilsson T, Hollmann M. Different structural requirements for functional ion pore transplantation suggest different gating mechanisms of NMDA and kainate receptors. J Neurochem. 2008;107:453–65. doi: 10.1111/j.1471-4159.2008.05623.x. [DOI] [PubMed] [Google Scholar]
- 17.Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. Arrangement of subunits in functional NMDA receptors. J Neurosci. 2011;31:11295–304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riou M, Stroebel D, Edwardson JM, Paoletti P. An alternating GluN1-2-1-2 subunit arrangement in mature NMDA receptors. PLoS One. 2012;7:e35134. doi: 10.1371/journal.pone.0035134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balasuriya D, Goetze TA, Barrera NP, Stewart AP, Suzuki Y, Edwardson JM. α-Amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) and N-methyl-D-aspartate (NMDA) receptors adopt different subunit arrangements. J Biol Chem. 2013;288:21987–98. doi: 10.1074/jbc.M113.469205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JW, Ascher P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 1987;325:529–31. doi: 10.1038/325529a0. [DOI] [PubMed] [Google Scholar]
- 21.Kleckner NW, Dingledine R. Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science. 1988;241:835–7. doi: 10.1126/science.2841759. [DOI] [PubMed] [Google Scholar]
- 22.Bowie D, Mayer ML. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15:453–62. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 23.Wilding TJ, Zhou Y, Huettner JE. Q/R site editing controls kainate receptor inhibition by membrane fatty acids. J Neurosci. 2005;25:9470–9478. doi: 10.1523/JNEUROSCI.2826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller B, Sarantis M, Traynelis SF, Attwell D. Potentiation of NMDA receptor currents by arachidonic acid. Nature. 1992;355:722–725. doi: 10.1038/355722a0. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa M, Kimura S, Akaike N. Facilitatory effect of docosahexaenoic acid on N-methyl-D-aspartate response in pyramidal neurones of rat cerebral cortex. J. Physiol. (London) 1994;475:83–93. doi: 10.1113/jphysiol.1994.sp020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohda K, Wang Y, Yuzaki M. Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat Neurosci. 2000;3:315–22. doi: 10.1038/73877. [DOI] [PubMed] [Google Scholar]
- 27.Blanke ML, VanDongen AM. The NR1 M3 domain mediates allosteric coupling in the N-methyl-D-aspartate receptor. Mol Pharmacol. 2008;74:454–65. doi: 10.1124/mol.107.044115. [DOI] [PubMed] [Google Scholar]
- 28.Klein RM, Howe JR. Effects of the lurcher mutation on GluR1 desensitization and activation kinetics. J Neurosci. 2004;24:4941–51. doi: 10.1523/JNEUROSCI.0660-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodward RM, Huettner JE, Guastella J, Keana JF, Weber E. In vitro pharmacology of ACEA-1021 and ACEA-1031: systemically active quinoxalinediones with high affinity and selectivity for N-methyl-D-aspartate receptor glycine sites. Mol Pharmacol. 1995;47:568–81. [PubMed] [Google Scholar]
- 30.Huettner JE. Indole-2-carboxylic acid: a competitive antagonist of potentiation by glycine at the NMDA receptor. Science. 1989;243:1611–3. doi: 10.1126/science.2467381. [DOI] [PubMed] [Google Scholar]
- 31.Watkins JC, Krogsgaard-Larsen P. Honoré, T. Structure-activity relationships in the development of excitatory amino acid receptor agonists and competitive antagonists. Trends Pharmacol Sci. 1990;11:25–33. doi: 10.1016/0165-6147(90)90038-a. [DOI] [PubMed] [Google Scholar]
- 32.Benveniste M, Mayer ML. Kinetic analysis of antagonist action at N-methyl-D-aspartic acid receptors. Two binding sites each for glutamate and glycine. Biophys J. 1991;59:560–73. doi: 10.1016/S0006-3495(91)82272-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clements JD, Westbrook GL. Activation kinetics reveal the number of glutamate and glycine binding sites on the N-methyl-D-aspartate receptor. Neuron. 1991;7:605–13. doi: 10.1016/0896-6273(91)90373-8. [DOI] [PubMed] [Google Scholar]
- 34.Banke TG, Traynelis SF. Activation of NR1/NR2B NMDA receptors. Nat Neurosci. 2003;6:144–52. doi: 10.1038/nn1000. [DOI] [PubMed] [Google Scholar]
- 35.Popescu G, Auerbach A. Modal gating of NMDA receptors and the shape of their synaptic response. Nat Neurosci. 2003;6:476–83. doi: 10.1038/nn1044. [DOI] [PubMed] [Google Scholar]
- 36.Schorge S, Elenes S, Colquhoun D. Maximum likelihood fitting of single channel NMDA activity with a mechanism composed of independent dimers of subunits. J Physiol. (London) 2005;569:395–418. doi: 10.1113/jphysiol.2005.095349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–9. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- 38.Smith TC, Howe JR. Concentration-dependent substate behavior of native AMPA receptors. Nat Neurosci. 2000;3:992–7. doi: 10.1038/79931. [DOI] [PubMed] [Google Scholar]
- 39.Fisher JL, Mott DD. Distinct functional roles of subunits within the heteromeric kainate receptor. J Neurosci. 2011;31:17113–22. doi: 10.1523/JNEUROSCI.3685-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benveniste M, Clements J, Vyklický L, Jr, Mayer ML. A kinetic analysis of the modulation of N-methyl-D-aspartic acid receptors by glycine in mouse cultured hippocampal neurones. J Physiol. (London) 1990;428:333–57. doi: 10.1113/jphysiol.1990.sp018215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murthy SE, Shogan T, Page JC, Kasperek EM, Popescu GK. Probing the activation sequence of NMDA receptors with lurcher mutations. J Gen Physiol. 2012;140:267–77. doi: 10.1085/jgp.201210786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stern-Bach Y, Bettler B, Hartley M, Sheppard PO, O'Hara PJ, Heinemann SF. Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron. 1994;13:1345–57. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 43.Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–13. doi: 10.1016/s0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 44.Carbone AL, Plested AJ. Coupled control of desensitization and gating by the ligand binding domain of glutamate receptors. Neuron. 2012;74:845–57. doi: 10.1016/j.neuron.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 45.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–7. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–58. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson GT, Gereau RW, 4th, Green T, Heinemann SF. Identification of amino acid residues that control functional behavior in GluR5 and GluR6 kainate receptors. Neuron. 1997;19:913–26. doi: 10.1016/s0896-6273(00)80972-1. [DOI] [PubMed] [Google Scholar]
- 48.Wilding TJ, Chai YH, Huettner JE. Inhibition of rat neuronal kainate receptors by cis-unsaturated fatty acids. J Physiol (Lond) 1998;513:331–9. doi: 10.1111/j.1469-7793.1998.331bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilding TJ, Fulling E, Zhou Y, Huettner JE. Amino acid substitutions in the pore helix of GluR6 control inhibition by membrane fatty acids. J Gen Physiol. 2008;132:85–99. doi: 10.1085/jgp.200810009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kovalchuk Y, Miller B, Sarantis M, Attwell D. Arachidonic acid depresses non-NMDA receptor currents. Brain Res. 1994;643:287–295. doi: 10.1016/0006-8993(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 51.Casado M, Ascher P. Opposite modulation of NMDA receptors by lysophospholipids and arachidonic acid: common features with mechanosensitivity. J Physiol. (London) 1998;513:317–30. doi: 10.1111/j.1469-7793.1998.317bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475:249–53. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–81. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 54.Dai J, Zhou HX. An NMDA receptor gating mechanism developed from MD simulations reveals molecular details underlying subunit-specific contributions. Biophys J. 2013;104:2170–81. doi: 10.1016/j.bpj.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tillett D, Neilan BA. Enzyme-free cloning: a rapid method to clone PCR products independent of vector restriction enzyme sites. Nucleic Acids Res. 1999;27:e26. doi: 10.1093/nar/27.19.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]