Abstract
During early development, GATA factors have been shown to be important for key events of coronary vasculogenesis, including formation of the epicardium. Myocardial GATA factors are required for coronary vascular (CV) formation; however, the role of epicardial localized GATAs in this process has not been addressed. The current study was conducted to investigate the molecular mechanisms by which the epicardium controls coronary vasculogenesis, focusing on the role of epicardial GATAs in establishing the endothelial plexus during early coronary vasculogenesis. To address the role of epicardial GATAs, we ablated GATA4 and GATA6 transcription factors specifically from the mouse epicardium and found that the number of endothelial cells in the sub-epicardium was drastically reduced, and concomitant coronary vascular plexus formation was significantly compromised. Here we present evidence for a novel role for epicardial GATA factors in controlling plexus formation by recruiting endothelial cells to the sub-epicardium.
Keywords: GATA4, GATA6, Epicardial signaling, Coronary vascular development
Introduction
The origin of coronary vascular (CV) endothelial cells has been a longstanding topic of interest and investigation. It was initially thought that coronary endothelial cells arose from the heart itself (Manasek, 1969). Based on cell lineage tracing studies in avian systems, an extra-cardiac structure derived from the septum transversum termed the proepicardium (PE) was subsequently proposed as the origin of CV endothelial cells (Mikawa and Gourdie, 1996). These findings led to a model in which endothelial precursor cells, along with other CV precursor cells, migrate from the PE to the myocardium where they form the epicardium. A subset of epicardial cells then invades the sub-epicardial space to differentiate into the coronary endothelial cells, coronary smooth muscle cells, and cardiac fibroblasts that form the coronary vasculature (Dettman et al., 1998; Merki et al., 2005; Mikawa and Gourdie, 1996). The observation that blocking PE attachment to the myocardium halted CV development supported the idea that all CV cells are derived from the PE, including vascular endothelial cells, or at least, that the PE is required for differentiation of endothelial cells that make up the coronary vasculature (Manner, 1993; Olivey et al., 2004). Recent murine studies indicate that coronary endothelial cells have multiple sources of origin and derive largely from the sinus venosus, the PE, and the endocardium (Katz et al., 2012; Red-Horse et al., 2010).
The coronary vascular system consists of a network of coronary arteries and coronary veins. The coronary arteries are generally located within the myocardium, while the coronary veins are typically located in the sub-epicardial space between the epicardium and myocardium (Lavine and Ornitz, 2008). It was thought that endothelial cells that contribute to the coronary arteries and veins derived from separate sources within the developing heart (Wu et al., 2012). However, recent work indicates that endothelial cells, which contribute to both the coronary veins and the coronary arteries, derive from the sub-epicardial endothelial cells (SEECs) (Red-Horse et al., 2010; Tian et al., 2013). These findings place the appearance of endothelial cells in the sub-epicardium as a critical early step in coronary vascular formation.
Multiple studies indicate that formation of the endothelial CV plexus involves complex reciprocal signaling between cells in the epicardium and myocardial cells of the sub-epicardium (Lavine and Ornitz, 2008; Olivey and Svensson, 2010; Tomanek et al., 2010). Plexus formation has typically been used as the benchmark in many of these studies; therefore, less has been done to distinguish regulatory control mechanisms that are operating prior to plexus formation. Formation of the coronary vascular plexus can be envisaged as having at least three steps: 1-migration and appearance of endothelial cells or precursors in the sub-epicardium; 2-coalescenceof vascular endothelial cells; and 3-assembly of the initial plexus. Overall, plexus formation has been found to be dependent on epicardial–myocardial reciprocal signaling that centers on an angiogenic FGF-SHH-VEGF/Ang-2 signaling axis (Lavine and Ornitz, 2009). The role of this axis in recruiting endothelial cells, mobilizing resident hemangioblasts, or assembly of the initial plexus from endothelial precursors is the topic of significant investigation. In the studies here, we present evidence that addresses GATA-dependent regulatory control of these earliest steps and suggests a model in which the epicardium is the source of signals that regulate the appearance of SEECs.
The GATA family of transcription factors are important regulators of vascular development (Lepore et al., 2005; Watt et al., 2004; Xin et al., 2006; Zhao et al., 2005). The six members of the GATA family are divided into two sub-groups based on their expression profiles and roles in various developmental processes. GATA1/2/3 are involved in hematopoietic development, while GATA4/5/6 regulate cardiac and endodermal formation (Molkentin, 2000). Global knockout of either Gata4 or Gata6 results in failure of extra-embryonic tissues leading to embryonic lethality during gastrulation, while Gata5 knockouts were found to be viable (Koutsourakis et al., 1999; Molkentin et al., 1997, 2000). Rescue of extra-embryonic tissue failure by tetraploid complementation revealed that Gata4 knockouts are unable to form a PE, demonstrating a central role for GATA4 in CV development (Watt et al., 2004). Little is known concerning the role of GATA6 in CV development; however, compound heterozygous Gata4+/− Gata6+/− mice die at embryonic day 13.5 (E13.5) due to vascular defects (Xin et al., 2006). This finding raises the possibility that GATA4 and GATA6 are required for CV development. Consistent with this idea, in vitro work has shown that GATA6 can promote angiogenesis via suppression of anti-angiogenic, autocrine signaling (Froese et al., 2011). Moreover, friend of GATA-2 (FOG-2) a well-known GATA cofactor, has been found to be crucial for the formation of the vascular plexus (Tevosian et al., 2000). These findings suggest roles for GATA factors in coronary vascular development and a more specific role for epicardial GATAs.
Here we address the role of GATA4 and GATA6 in epicardial-mediated formation of the coronary plexus. Our results show that GATA4 and GATA6 are expressed in the epicardium and required for plexus formation. Using a Cre-lox system to conditionally knock out both Gata4 and Gata6 (dcKO) in an epicardial-specific manner, we found that the loss of epicardial GATAs resulted in a drastic loss of coronary plexus formation. These results suggest a model for formation of both coronary veins and arteries in which epicardial GATAs regulate the number of endothelial cells in the sub-epicardium.
Materials and methods
Animals
All animal experiments were approved by the Institutional Animal Care and Use committee (IACUC) at the Medical College of Wisconsin. The WT1(RP23-8C14)-Cre mouse line contains a BAC expression construct in which the Cre recombinase gene was inserted in the 5′ UTR of the first exon within the Wilms Tumor-1 gene. This construct was designed to target the epicardium and epicardial-derived cells. The WT1(RP23-8C14)-Cre line has been maintained on a C57B16/J background and was obtained as a generous gift from Dr. John Burch. The Gata4f/f/ Gata6f/f mice were generated by crossing the previously described Gata4f/f and Gata6f/f mouse lines (Watt et al., 2004; Sodhi et al., 2006). The Gata4f/f line contains loxP sites flanking exons 3–5, which contain the nuclear localization and DNA binding domains. The Gata6f/f line contains loxP sites flanking exon 2, which contains most of the GATA6 sequence. Tie2-Cre, Rosa26R-eYFP, and Rosa26R -βgal mice have been previously described (Kisanuki et al., 2001; Soriano, 1999; Srinivas et al., 2001).
Embryos were generated by timed matings designating E0.5 as noon on the day a vaginal plug was observed. Genotyping was performed with PCR by standard protocols using genomic DNA isolated from embryonic tail tissue.
Primers used are as follows:
| Cre | F: GTT CGC AAG AAC CTG ATG GAC A R: CTA GAG CCT GTT TTG CAC GTT C |
| Gata4f/f | F: CCC AGT AAA GAA GTC AGC ACA AGG AA R: AGA CTA TTG ATC CCG GAG TGA ACA TT |
| Gata6f/f | F: GTG GTT GTA AGG CGG TTT GT R: ACG CGA GCT CCA GAA AAA GT |
Histology and immunofluorescence
Embryos were harvested from timed pregnant females, photographed, and fixed overnight in zinc buffered formalin. Embryos were then embedded in paraffin wax, and sectioned at 5 µm according to standard protocols. Sections were then subject to antigen retrieval with 10 mmol/L citric acid buffer (pH 6.0) at 100 °C for 10 min. Sections were then blocked for 45 min and stained overnight with primary antibody in blocking solution. Sections were then washed three times with PBS and incubated with secondary antibody in blocking solution for 2 h. Finally, sections were washed three times with PBS and incubated in 100 mmol/L glycine with DAPI for 1 h.
Sections were immunofluorescently labeled according to standard protocols using anti-PECAM1 (1:100, BD Biosciences, clone MEC13.3), anti-ERG (1:500, Santa Cruz Biotechnology, clone C-20), anti-GFP (1:500, Invitrogen), anti-sarcomeric myosin (1:200, DSHB, clone MF-20), anti-smooth muscle myosin heavy chain (1:200, Biomedical Technologies BT-562), anti-GATA4 (1:100, Santa Cruz Biotechnology, sc-9053), anti-GATA6 (1:5000, a generous gift from Dr. Xiang-Xi Xu), anti-WT1 (1:100, DAKO, 6F-H2) and anti-SHH (1:100, DSHB, clone 5E-1) primary antibodies and corresponding fluorescent secondary antibodies from Invitrogen.
Images were captured using a Nikon Eclipse 80i microscope equipped with a Nikon Digital Sight DS-2MBW monochrome camera and NIS Elements-D imaging software. Photo processing was done with Photoshop.
Whole-mount immunohistochemistry
Whole-mount PECAM staining was conducted as previously described (Lavine et al., 2005). Isolated E12.5 hearts were fixed overnight in 4% paraformaldehyde. Following fixation, hearts were dehydrated with methanol and incubated in 6% hydrogen peroxide for 2 h. Hearts were then rehydrated, blocked in 5% goat serum/1% Triton X-100/PBS, and incubated overnight with PECAM primary antibody (1:100, BD Biosciences, clone MEC13.3). Following 5 × 1 h washes in blocking solution, hearts were incubated overnight with biotinylated goat anti-rat IgG secondary antibody (1:200, Vector). Following 5 × 1 h washes in blocking solution, hearts were incubated overnight in ABC reagent (Vectastain). After a final set of washes, the hearts were then exposed to DAB and imaged. PECAM stained hearts were then embedded, sectioned, lightly counterstained with hematoxylin, and imaged for quantification of vessels per section. Three sections from at least three wild type and three dcKO hearts were used for quantification.
Lac-Z staining
Embryos were fixed in 4% paraformaldehyde for 30 min on ice. They were rinsed in wash buffer (0.1 mol/L phosphate buffer (pH 7.3), 0.01% deoxycholate, 0.02% Nonidet-P40, 2 mmol/L MgCl2) for 15 min at room temperature. They were then incubated overnight in β-galactosidase substrate buffer (wash buffer plus 1 mg/mL X-gal, 5 mmol/L potassium ferrocyanide, 5 mmol/L potassium ferricyanide). Embryos were then rinsed in wash buffer for 15 min at room temperature and then processed as listed above.
BrdU proliferation assay
Timed pregnant females were intraperitoneally injected 4 h prior to harvest with 1 mL/100 g of BrdU labeling reagent (Invitrogen, 00-0103). Embryos were isolated, processed, and sectioned as listed above. Sections were immunofluorescently labeled according to standard protocols using anti-BrdU (1:500, Invitrogen, 03-3900), Sarcomeric Myosin (1:200, DSHB, clone MF-20), and Nkx2.5 (1:500, Santa Cruz Biotechnology, sc-8697) primary antibodies and corresponding fluorescent secondary antibodies. Sections were counterstained with DAPI to label nuclei. For the myocardial proliferation assay, four fields in the myocardium consisting of fifty Nkx2.5 expressing cells were counted for each image. Three images were analyzed for each heart and three hearts were quantified for each genotype.
Results
WT1(RP23-8C14)-Cre is expressed in the epicardium and epicardial-derived cells
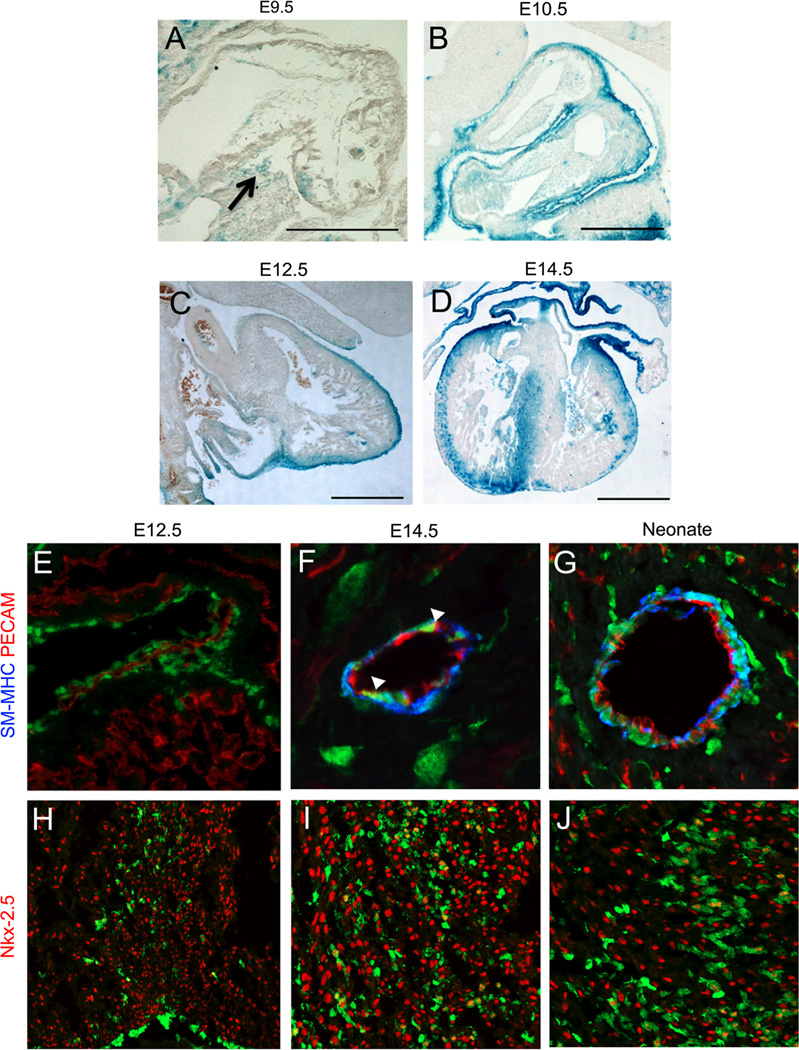
To target the knockout of Gata4 and Gata6 in the epicardium and epicardial derivatives, we utilized a WT1(RP23-8C14)-Cre mouse transgenic line. Two other similar WT1 -Cre lines have been published, the WT280Cre YAC line and the WT1(RP23-266M16)-Cre BAC line (Norden et al., 2010; Wilm et al., 2005). These previously published WT1-Cre lines show Cre expression in the epicardium, coronary smooth muscle cells, and a subset of adult coronary endothelial cells (Norden et al., 2010; Wilm et al., 2005). To characterize the expression of the WT1(RP23-8C14)-Cre used in this study, WT1(RP23-8C14)-Cre mice were crossed with either the Rosa26R -βgal reporter mice or the Rosa26R-eYFP reporter mice. We observed that the WT1(RP23-8C14)-Cre was expressed in a pattern similar to the previously published WT1-Cre lines (Norden et al., 2010; Wilm et al., 2005). At E9.5, reporter expression was seen in the proepicardium (Fig. 1A). At E10.5 epicardial expression of the reporter was observed, and this continued through E14.5 (Fig. 1B and D). Reporter-expressing cells were observed migrating into the myocardium at E12.5 (Fig. 1C). At E14.5 we saw extensive reporter expression in the sub-epicardium and septum of the developing myocardium (Fig. 1D). To determine the contribution of eYFP-positive cells to coronary vascular cell types, we evaluated expression of WT1(RP23-8C14)-Cre by co-staining with antibodies against platelet endothelial cell adhesion molecule (PECAM) to label coronary endothelial cells, and smooth muscle myosin heavy chain (SM-MHC) to label coronary smooth muscle cells. At E12.5 before the appearance of coronary smooth muscle cells, we saw no expression of the eYFP reporter in coronary endothelial cells (Fig. 1E). At E14.5 we saw eYFP expression in coronary smooth muscle cells and a few coronary endothelial cells (Fig. 1F). In neonate hearts, we see continued expression of the reporter in coronary smooth muscle cells and only occasional expression in coronary endothelial cells (Fig. 1G). Additionally, we immunofluorescently stained for eYFP and with an antibody against WT1, and found 96% (± 0.5%) of the WT1 marked cells were eYFP + at E12.5 (Supplemental Fig. 1). The expression pattern we observe is in agreement with the expression pattern of other WT1 derived Cre lines that showed reliable epicardial labeling and very rare expression in endothelial cells (Wilm et al., 2005; Zhou et al., 2009). These results are also consistent with previous observations in other studies, which indicate that cells from the epicardium largely give rise to coronary smooth muscle cells and fibroblasts and only occasionally to endothelial cells in mammals.
Fig. 1.
WT1(RP23-8C14)-Cre is expressed in the epicardium and epicardial derived lineages. The fate of cells expressing WT1(RP23-8C14)-Cre was determined by crossing with Rosa26R-βgal (A)–(D) and Rosa26R-eYFP (E)–(J) mice. At E9.5 WT1(RP23-8C14)-Cre+ cells are seen in the proepicardium ((A), arrow). At E10.5 (B) reporter expression is observed in the epicardium. At 12.5 (C) WT1(RP23-8C14)-Cre continues to label the epicardium and sub-epicardial mesenchymal cells. At 14.5 (D) WT1(RP23-8C14)-Cre labels cells of the epicardium, the sub-epicardium, and the myocardium. Co-staining with Rosa26R.eYFP (Green, (E)–(J)) found the majority of vascular eYPF+ cells co-stained with the smooth muscle cell marker, smooth muscle myosin heavy chain (Blue, (E)–(G)) and occasional co-expression (arrowheads) with the endothelial cell marker, PECAM (Red, (E)–(G)). Co-staining with the cardiomyocyte marker, Nkx2.5 (Red, (H)–(J)), found WT1(RP23-8C14)-Cre does express in a small population of cardiomyoctyes from E12.5 ((H)–(J)) on. Scale bars in (A)–(D) represent 500 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
It has been suggested that a small percentage of cardiomyocytes are derived from the epicardium, and previous reports with other WT1 derived Cre′s have shown reporter expression in cardiomyoctyes (Zhou et al., 2008). Therefore to further characterize the WT1(RP23-8C14)-Cre, we also evaluated expression in cardiomyocytes. We used Nkx2.5 as a marker for cardiomyocytes and co-stained for eYFP (Fig. 1H and J). Consistent with other WT1 derived Cre′s, the reporter was expressed in a sub-set of cardiomyocytes from E12.5 to later times (Fig. 1H and J) (Rudat and Kispert, 2012). At E14.5 we found eYFP labeled 14.03% (± 3.96%) of cardiomyocytes in the septum and 4.96% (± 2.19%) of cardiomyocytes in the ventricular walls. This suggests a small number of cardiomyocytes may derive from the epicardium, or alternatively that the WT1(RP23-8C14)-Cre could exhibit sporadic cardiomyocyte expression. The results in Fig. 1demonstrate that the expression pattern of WT1(RP23-8C14)-Cre is similar to other WT1 derived Cre lines (Wilm et al., 2005; Zhou et al., 2008) and that this pattern of expression replicates the known expression of endogenous WT1 in mesothelium-derived tissues (Rudat and Kispert, 2012).
WT1(RP23-8C14)-Cre efficiently disrupts expression of GATA4 and GATA6 in the epicardium
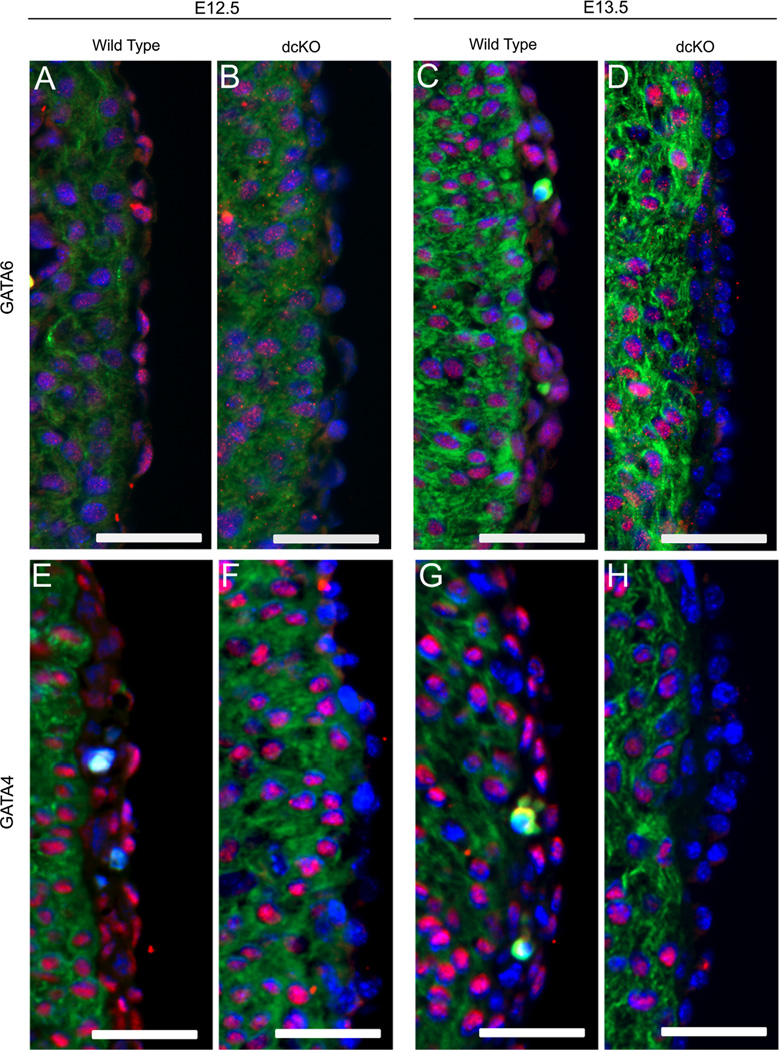
Previous work has shown GATA4 is highly expressed in the epicardium and proepicardium (Watt et al., 2004), but the expression pattern of GATA6 in the epicardium was unknown. To determine if GATA6 is expressed in the epicardium, we immunohistochemically stained E12.5 and E13.5 wild type hearts with an antibody against GATA6. We observed enriched epicardial expression of GATA6 along with comparably lower levels of myocardial expression (Fig. 2A and C). This, along with observations from Watt et al., demonstrates that both GATA6 and GATA4 are expressed in the epicardium at times critical for establishing the coronary vasculature.
Fig. 2.
WT1(RP23-8C14)-Cre disrupts expression of GATA6 and GATA4 in the epicardial and sub-epicardial cells. The epicardial expression of GATA6 and GATA4 was visualized with immunofluorescence for GATA6 (A)–(D) and for GATA4 (Red, (E)–(H)). GATA6 (A) and (C) and GATA4 (E) and (G) were found to be expressed in epicardial and sub-epicardial cells of wild type hearts. In dcKO hearts, there is was an observable decrease in the epicardial expression of GATA6 (B) and (D) and GATA4 (F) and (H) when compared to the wild type (A), (C), (E) and (G). Hearts were co-stained against sarcomeric myosin (Green) to visualize the myocardium and DAPI (Blue) to visualize nuclei. Scale bars represent 50 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To determine if WT1(RP23-8C14)-Cre disrupts expression of GATA4 and GATA6 in the epicardium, we crossed WT1(RP23-8C14)-Cre+/− Gata4f/+ Gata6f/+ males with Gata4f/f Gata6f/f females to generate WT1(RP23-8C14)-Cre+/− Gata4f/f Gata6f/f (dcKO) mice. We then visualized GATA expression in the hearts using antibodies against GATA4 or GATA6.We observed a consistent decrease in the epicardial expression of both factors by E12.5 (Fig. 2B and F). To confirm the loss of GATA4 and GATA6 persisted in epicardial derived cells migrating into the myocardium, we crossed WT1(RP23-8C14)-Cre+/− Gata4f/+ Gata6f/+ males with Gata4f/f Gata6f/f Rosa26R-eYFPf/f females generating dcKO mice expressing the eYFP reporter. We stained E14.5 hearts and observed a loss of GATA4 and GATA6 in eYFP expressing cells derived from the epicardium (Supplemental Fig. 2). These results showed WT1(RP23-8C14)-Cre is able to ablate expression of GATA4 and GATA6 in the epicardium and epicardial derived cells. This then allowed us to investigate the role of epicardial GATA4 and GATA6 in coronary vascular development.
Deletion of Gata4 and Gata6 in WT1(RP23-8C14)-Cre mice results in embryonic lethality by E15.5
To address the effects of WT1(RP23-8C14)-Cre -driven conditional knockout of Gata4 and Gata6 during development, we crossed WT1(RP23-8C14)-Cre+/− Gata4f/+ Gata6f/+ males with Gata4f/f Gata6f/f females to generate litters aged from E10.5 to weanling. Genotyping revealed a normal Mendelian ratio of dcKO mice through E14.5 with the observation that dcKO embryos exhibited hind limb polydactyly (Supplemental Fig. 3). By E15.5 all dcKOs observed were in various stages of necrosis, and no dcKO mice were found at weaning (Table 1). In contrast, WT1(RP23-8C14)-Cre+/− Gata4f/+ Gata6f/f and WT1(RP23-8C14)-Cre+/− Gata4f/f Gata6f/+ mice were viable at E15.5, with a small percentage observable at weaning. This indicates WT1(RP23-8C14)-Cre -driven loss of both Gata4 and Gata6 results in mid-gestational-lethality. In addition to epicardial expression, WT1(RP23-8C14)-Cre shows some expression in other mesothelial tissues and a small subset of cardiomyocytes (Fig. 1); therefore, embryonic lethality cannot be unequivocally ascribed to the epicardial loss of GATA4 and GATA6. However, ~E15 is a common age of lethality among phenotypes with deficient coronary vascular formation, and our observations are consistent with a loss of coronary vascularity as a primary cause of lethality (Conway et al., 2003). Significantly, however, survival of dcKO embryos to E14.5 allowed us to study the role of epicardial GATAs in plexus formation during early coronary vascular development.
Table 1.
dcKO is embryonic lethal by E15.5.
| Age | Total | Expected | dcKO | G6 KO | G4 KO |
|---|---|---|---|---|---|
| E10.5 | 41 | 5 | 6 | 5 | 6 |
| E11.5 | 77 | 10 | 7 | 8 | 8 |
| E12.5 | 51 | 6 | 5 | 3 | 5 |
| E13.5 | 81 | 10 | 11 | 10 | 7 |
| E14.5 | 59 | 7 | 11 | 9 | 4 |
| E15.5 | 41 | 5 | 0 (4) | 4 | 4 |
| Weaning | 48 | 6 | 0 | 2 | 1 |
Genotypic analysis of mouse embryos generated by crossing.
WT1(RP23-8C14)-Cre+/−/GATA-4f/+/GATA-6f/+ males with GATA-4f/f/GATA-6f/f. Values represent observed viable embryos at each embryonic day. (X)=necrotic embryos; dcKO = WT1(RP23-8C14)-Cre+/−/GATA-4f/f/ GATA-6f/f; G6 KO=WT1(RP23-8C14)-Cre+/−/GATA-4f/+/GATA-6f/f; G4 KO=WT1(RP23-8C14)-Cre+/−/GATA-4f/f/GATA-6f/+.
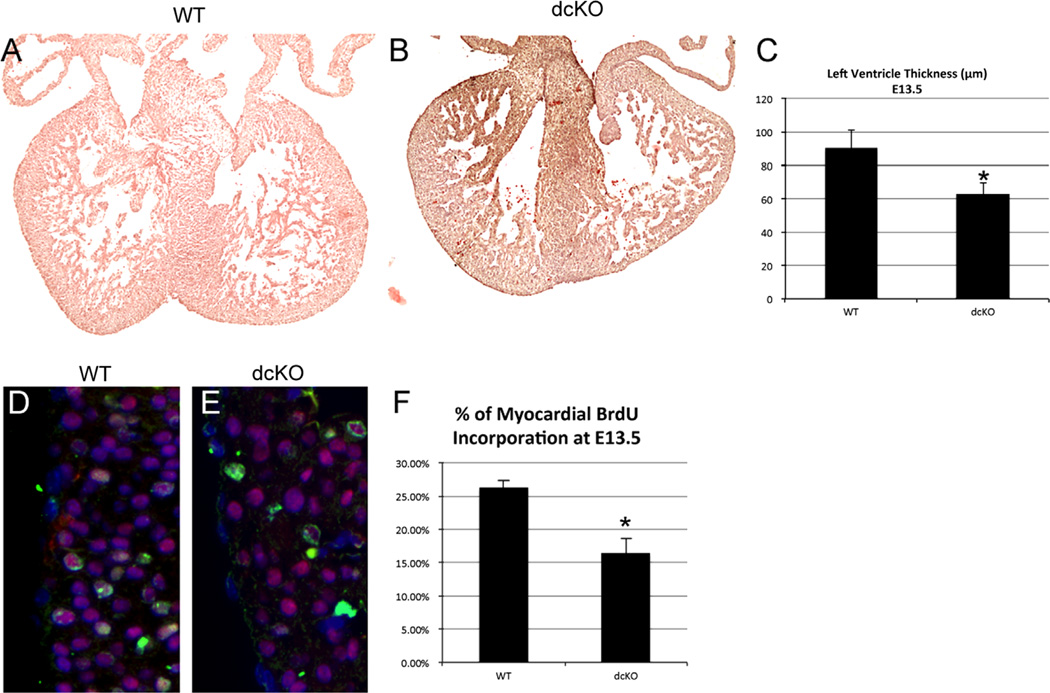
Loss of epicardial GATAs inhibits cellular proliferation in the heart
Epicardial function is crucial for maturation of the developing myocardium (Lavine et al., 2005). Because we targeted the knockout of GATA4 and GATA6 to the epicardium, we assessed the effect on the myocardium. In 33% of E13.5 dcKO hearts, we found myocardial thinning greater than 2 standard deviations from the mean of the wild type. Ventricular thickness in dcKO hearts that displayed thinning was significantly decreased compared to wild type (p < 0.05, Fig. 3C). This suggests incomplete penetrance of the thinning phenotype (Fig. 3A and B) possibly due to inherent variations in the Cre-loxP system (Rajewsky et al., 1996). Myocardial thinning during development is commonly associated with a decrease in myocardial proliferation (Stuckmann et al., 2003). To test if this was the case in the dcKO hearts, we utilized a BrdU incorporation assay. In hearts that showed thinning, we found a statistically significant decrease in the percentage of BrdU positive myocardial cells when compared to the wild type hearts (Fig. 3F). Moreover, we did not observe an increase in myocardial apoptosis (data not shown). Ultimately, these data suggest that loss of epicardial GATAs can result in some loss of myocardial cell proliferation, which may manifest as myocardial thinning. Since there is minimal expression of the Cre in cardiomyocytes, these observations are consistent with the observed thinning being due to either defective epicardial signaling or plexus disruption.
Fig. 3.
dcKO hearts display myocardial thinning and decreased proliferation. Hematoxylin and eosin staining was used to visualize the morphology of wild type (A) and dcKO (B) hearts at E13.5. We observed myocardial thinning in dcKO hearts (B) when compared to wild type hearts (A). Measurement of left ventricle thickness revealed a significant decrease between dcKO hearts that displayed thinning and wild type hearts (* = p < 0.05, (C)). To measure myocardial proliferation, BrdU incorporation (Green) was quantified in Nkx2.5 (Red) expressing cardiomyocytes (D)–(E).We found a statistically significant decrease (* = p < 0.005) in the percentage of BrdU positive cells within the myocardium of dcKO hearts when compared to wild type hearts (F). Images were counterstained with DAPI (Blue) to label nuclei. Error bars represent standard deviation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Epicardial GATAs are required for the formation of the coronary plexus
Defects in myocardial growth are often associated with deficient coronary vascular development. GATA4 and GATA6 have been reported to be important for vascular development. GATA4 is required for the formation of the proepicardium, and GATA6 is required in vascular smooth muscle cells (Lepore et al., 2005; Watt et al., 2004). Moreover, global compound heterozygosity of both Gata4 and Gata6 results in severe vascular defects (Xin et al., 2006). To address whether epicardial GATA4 and GATA6 may be important for coronary development, we assessed the effect of their loss on coronary vascular formation.
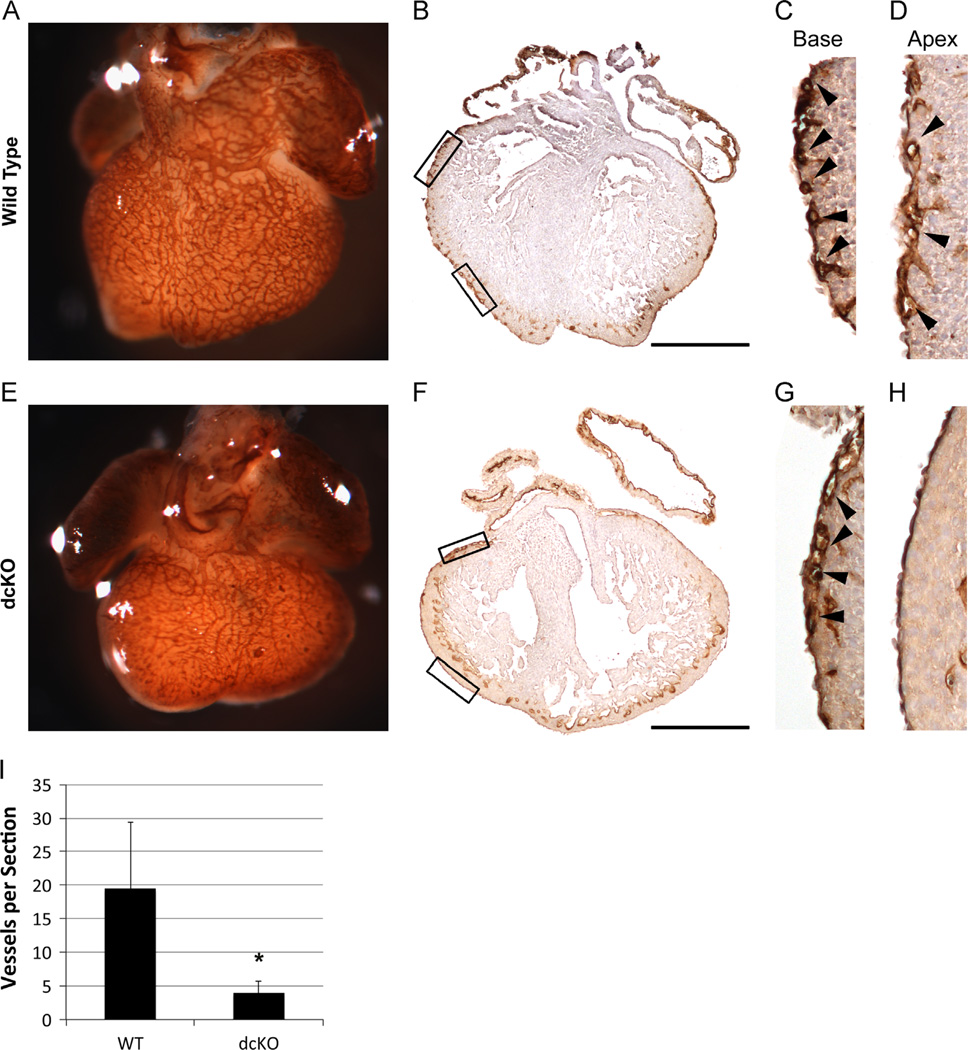
During normal CV development, coronary endothelial cells appear first in the sub-epicardium near the base of the heart to form the initial vascular plexus at ~E11. They then spread towards the apex and migrate covering the heart by E14.5 (Olivey et al., 2004). Therefore, we initially assessed plexus formation in dcKO mice by whole mount staining of E13.5 hearts using an antibody against platelet endothelial cell adhesion molecule (PECAM). Wild type hearts demonstrated an organized plexus covering the majority of the heart (Fig. 4A). In marked contrast, the dcKO hearts show limited plexus formation near the base of the heart but spreading towards the apex is severely diminished (Fig. 4E). Sections of whole mount PECAM stained hearts further confirmed decreased plexus coverage of the heart due to the loss of GATA4 and GATA6 (Fig. 4B, D and F–H). Moreover, quantification confirmed an 80% decrease in the number of vessels per section in the dcKO (p < 0.05, Fig. 4I). These results demonstrate that epicardial GATA4 and GATA6 are crucial for proper formation of the coronary plexus.
Fig. 4.
dcKO hearts exhibit decreased coronary plexus formation. Whole mount PECAM staining was used to visualize the formation of the coronary plexus at E13.5 (A) and (E). dcKO hearts (E) display decreased coverage of the ventricle when compared to wild type hearts (A). Sections prepared from whole mount PECAM stained hearts were used to visualize the formation of the coronary plexus at E13.5 (B)–(D), (F)–(H). Wild type hearts (B) display extensive sub-epicardial vessel formation, while dcKO hearts (F) show very few vessels. Higher magnification of outlined boxes in whole section images show vessels at the base of both wild type (C) and dcKO hearts (G); however, the spreading of vessels towards the apex of the wild type heart (D) is not replicated in the dcKO (H). Black arrowheads (C), (D), (G) and (H) mark sub-epicardial vessels. Decreased PECAM signal in endocardium (B)–(D) and (F)–(G) is due to limited penetrance of whole mount staining. (I) Quantification of vessels per section revealed a statistically significant (* = p < 0.05) decrease in dcKO hearts when compared to wild type hearts. Error bars represent standard deviation. Scale bars represent 500 µm.
Epicardial GATA4 and GATA6 are required for the appearance of subepicardial endothelial cells
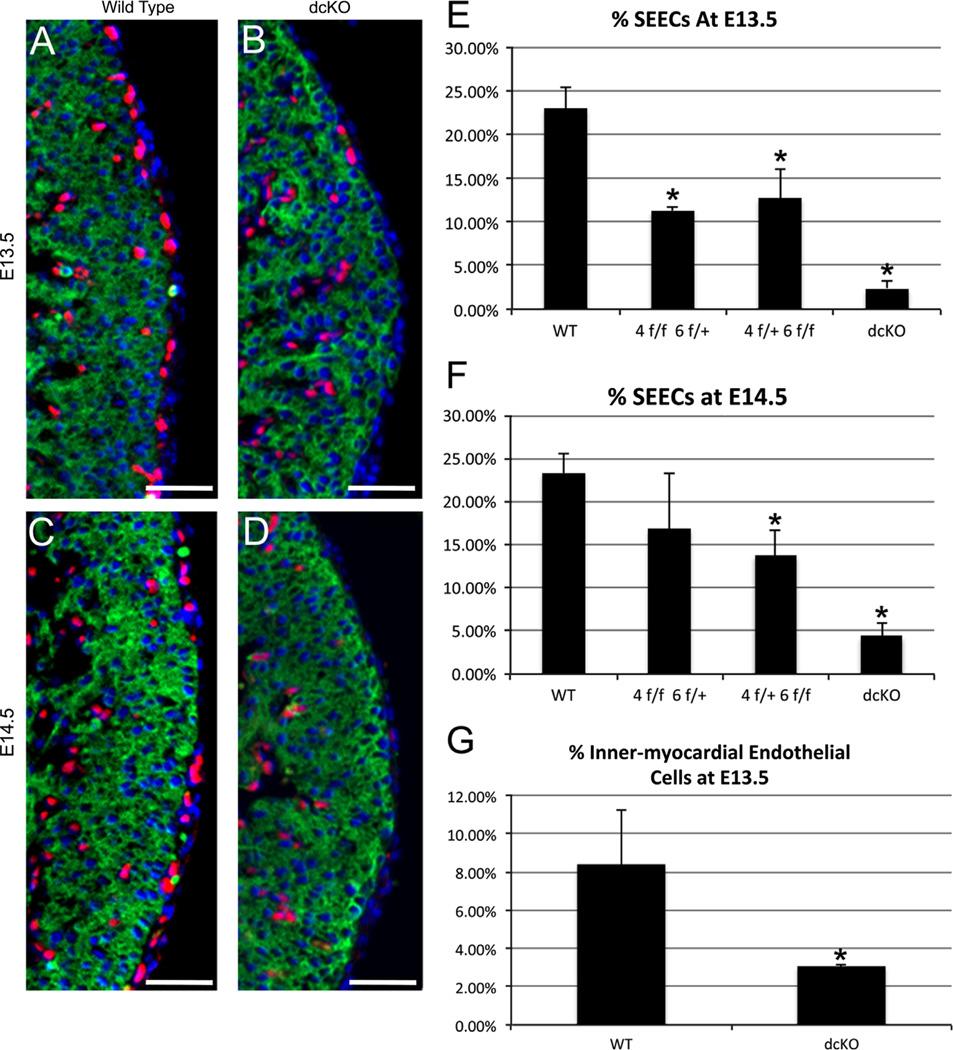
We reasoned that this decrease in plexus formation might be due to a decrease in the number of sub-epicardial endothelial cells (SEECs) that directly contribute to formation of the plexus. To quantify the changes in the number of SEECs, we visualized endothelial cells by immunofluorescent detection of ETS-related gene (ERG), which labels the nuclei of endothelial cells (Fig. 5A and D, Supplemental Fig. 4showing ERG co-expression with both PECAM and Tie2, well-established markers of endothelial differentiation) (Nikolova-Krstevski et al., 2009). To distinguish cells of the sub-epicardium and epicardium, we labeled the myocardium with MF20. Sub-epicardial and epicardial cells were identified as cells exterior to themyocardium. We determined the percentage of SEECs by dividing the number of ERG+ cells in the sub-epicardium by the total number of epicardial and sub-epicardial cells. Compared to wild type, we observed an 80–90% decrease in the percentage of SEECs at E13.5 and E14.5 in dcKO hearts (Fig. 5E and F), consistent with the decrease seen in plexus formation. We also assessed the potential loss of SEECs in the WT1(RP23-8C14)-Cre+/− Gata4f/+ Gata6f/f and WT1(RP23-8C14)-Cre+/− Gata4f/f Gata6f/+ hearts and found a statistically significant decrease in both at E13.5 (Fig. 5E). However, the loss of SEECs was not as severe as observed in the dcKO. Together, the results in Figs. 4 and 5, demonstrate epicardial expression of both GATA4 and GATA6 is required for proper formation of the endothelial coronary plexus, and further suggest that this occurs through a mechanism that involves a decrease in the number of SEECs.
Fig. 5.
dcKO hearts exhibit decreased SEECs and inner-myocardial endothelial cells. The presence of SEECs was observed by immunofluorescent labeling of endothelial cells with an anti-ERG antibody (Red, (A)–(D)).We determined the percentage of SEECs by counting the number of SEECs and dividing that value by the total number of epicardial and sub-epicardial cells. We observed a significant decrease in the percentage of sub-epicardial endothelial cells in the dcKO hearts (B) and (D) when compared to the wild type hearts (A) and (C). This decrease was found to be statistically significant (* = p < 0.005) at both E13.5 (E) and E14.5 (F). Additionally, we found decreases in the percentage of SEECs in WT1(RP23-8C14)-Cre+/− Gata4f/f Gata6f/+ (4f/f 6f/+) and WT1(RP23-8C14)-Cre+/− Gata4f/+ Gata6f/f hearts(4f/+ 6f/f, (E)–(F), * = p < 0.005). (G) dcKO hearts display a statistically significant (* = p < 0.05) decrease in the number of inner-myocardial endothelial cells at E13.5. Hearts were co-stained against sarcomeric myosin (MF20, Green) to visualize the myocardium and DAPI to label nuclei. All scale bars represent 50 µm. Error bars represent standard deviation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The loss of SEECs disrupts the appearance of inner-myocardial endothelial cells
Recent findings have shown that SEECs are major contributors to the endothelium of the coronary arteries within the myocardium wall (Tian et al., 2013). Loss of SEECs should, therefore, result in a decrease of inner-myocardial endothelial cells. To test this hypothesis, we labeled the inner-myocardial endothelial cells using antibodies against the endothelial specific transcription factor ERG (Nikolova-Krstevski et al., 2009). The percentage of inner-myocardial endothelial cells was then determined by dividing the number of ERG labeled nuclei in the basal half of the compact zone by the total number of nuclei in same region. We observed a ~ 3 fold decrease in the number of inner-myocardial endothelial cells in the dcKOs when compared to the wild type hearts (Fig. 5G). This result confirms that the loss of epicardial GATA4 and GATA6 not only affects the appearance of SEECs, but also the appearance of inner-myocardial endothelial cells.
The direct loss of GATA4 and GATA6 in endothelial cells
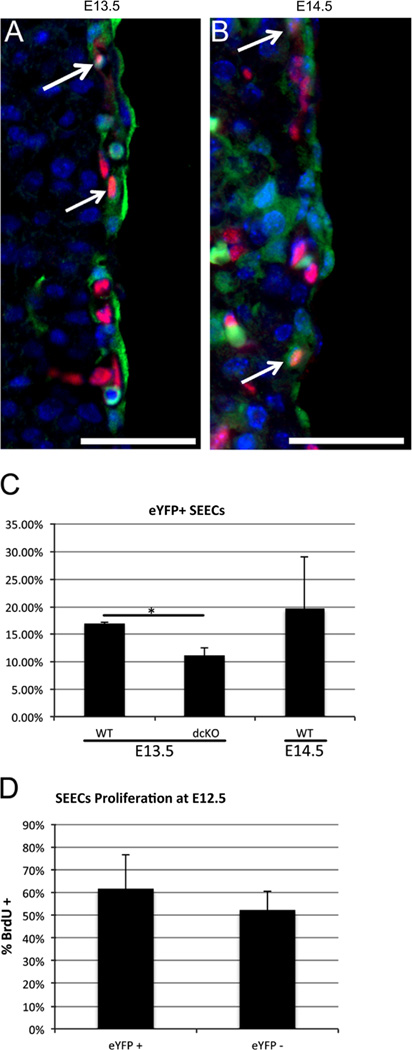
Loss of GATA4 and GATA6 in our experiments is largely confined to the epicardium, suggesting that the resultant decrease in SEEC numbers is due to signaling defects between the epicardium and surrounding cells. However, Fig. 1also shows that loss of GATAs occurs in a small subset of SEECs. This raises the possibility that direct effects on SEECs may account for the decrease in the total number of SEECs observed in the dcKO. To begin to address this, we quantified the percentage of SEECs expressing the WT1(RP23-8C14)-Cre reporter. At E13.5 and E14.5, we observed approximately 20% of SEECs were eYFP+ (Fig. 6A and C), whereas dcKO animals showed an 80–90% loss of SEECs. These results indicate that GATA loss is observed in only a small number of SEECs, the WT1(RP23-8C14)-Cre expressing subset, and therefore is unlikely to fully account for the observed loss of total SEECs. We also assessed the percentage of SEECs that were labeled with the eYFP reporter in the dcKO and found it was decreased ~30% (Fig. 6C), significantly less than the total 80–90% loss of SEECs. Together these observations indicate that GATA4 and GATA6 may be autonomously required by SEECs. However, the direct knockout of GATAs in a subset of SEECs does not fully account for the total loss of SEECs observed in the dcKO animals, supporting a model in which epicardial cells indirectly control SEEC number.
Fig. 6.
WT1(RP23-8C14)-Cre expression is limited in SEECs. Embryos generated from mating WT1(RP23-8C14)-Cre mice with Rosa26R-eYFP reporter mice were stained with antibodies against eYFP (Green) and ERG (Red) to label endothelial cells. At E13.5 (A) and E14.5 (B) we observed eYFP negative SEECs along with a few eYFP positive, SEECs (arrows). The percentage of eYFP+ SEECs was found to be 16.89% at E13.5 and 19.58% at E14.5 (C). At E13.5, we found a statistically significant decrease in the percentage of SEECs in dcKO hearts when compared to wild type hearts (* = p < 0.005, (C)). We found no significant difference in the proliferation rates between eYFP+ and eYFP− SEECs at E12.5 (D). All scale bars represent 50 µm. Error bars represent standard deviation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To address whether the WT1(RP23-8C14)-Cre expressing subset of SEECs may disproportionately contribute to plexus formation, we examined the proportions of WT1(RP23-8C14)-Cre expressing SEECs at E13.5 and found that they contributed at most 20% of the population. We also examined whether this WT1(RP23-8C14)-Cre expressing population may be more proliferative than other SEECS. We measured the proliferation rates of the WT1(RP23-8C14)-Cre expressing and non-expressing populations at E12.5 and found no significant change in the proliferation rates (Fig. 6D). Additionally, we did not observe an increase in apoptosis of SEECs (data not shown). This data, along with the significant loss of GATAs in the epicardium but only in a small sub-set of SEECs, is consistent with a model in which epicardial GATA4 and GATA6 may regulate epicardial signaling pathways needed for the appearance, or an increase in number, of sub-epicardial endothelial cells, and thereby the formation of the early vascular plexus.
dcKO does not alter the number of epicardial cells
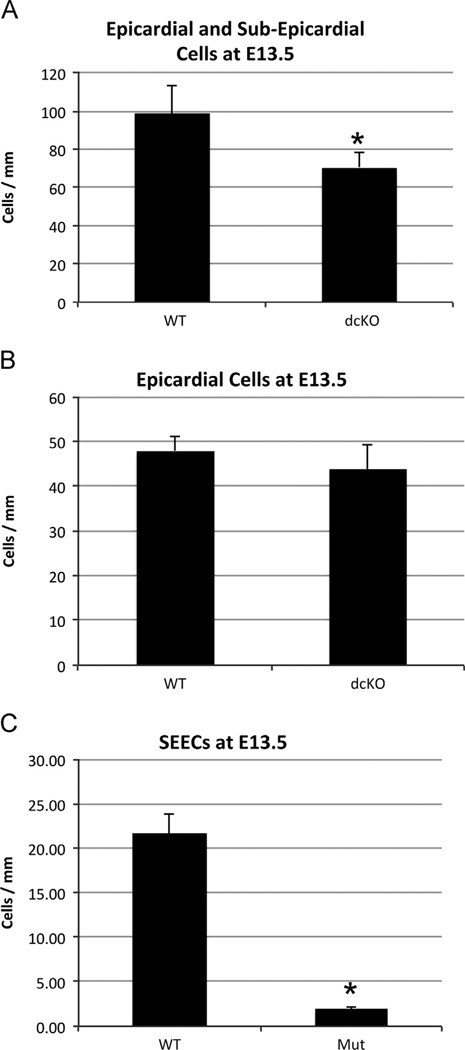
To address whether loss of epicardial GATA4 and GATA6 may result in loss of epicardial derived endothelial precursors and that this may account for the drastic loss of SEECs, we counted the total number of epicardial and sub-epicardial cells in the dcKO animals. We first counted the number epicardial and sub-epicardial cells per mm of the epicardium. We observed a significant decrease (~ 25 cells/mm) in the number of epicardial and sub-epicardial cells of dcKOs (Fig. 7A). We then analyzed if the decease in epicardial and sub-epicardial cells was due to a loss of epicardial cells or SEECs. We used the WT1(RP23-8C14)-Cre driven reporter to label epicardial cells in the wild type and dcKO cells and found no difference in the number of epicardial cells per mm (Fig. 7B). We then counted the number of SEECs per mm and found a significant decrease (~ 20 cells/mm, Fig. 7C). This agrees with previous data in Fig. 5showing a decrease in the percentage of SEECs. Together, these data indicated that even though loss of GATAs occurred primarily in epicardial cells, there was not significant change in the number of epicardial cells. There is, however, a significant loss in the number of SEECs. This further supports the hypothesis that loss of GATA4 and GATA6 in epicardial cells disrupts an epicardial signaling mechanism that may be required for accumulation and/or recruitment of SEECs.
Fig. 7.
dcKO does not affect the number of epicardial cells. The number of epicardial and sub-epicardial cells was quantified by counting the number of nuclei per mm exterior to the myocardium this revealed a significant decrease in the dcKO hearts when compared to the wild type hearts ((A), * = p < 0.005). Epicardial cells were labeled with the WT1(RP23-8C14)-Cre driven Rosa26R-eYFP reporter and counted per mm to determine no significant difference between the wild type and dcKO mice at E13.5 (B). SEECs were labeled with ERG and counted per mm to determine a significant decrease between the wild type and dcKO hearts ((C), * p < 0.005). Error bars represent standard deviation.
Discussion
In this report, we investigated the role of epicardial GATA4 and GATA6 during early CV development. Previous work has shown that GATAs are required for the formation of the epicardium, and that myocardial GATA factors are important in CV formation; however, no role for GATAs in epicardium function had been determined (Watt et al., 2004). Our results show that epicardial GATA4 and GATA6 play a critical role in CV formation, and that they are required for the formation of the coronary plexus. It is interesting that the CV phenotype is most severe upon epicardial loss of both GATA4 and GATA6. This is consistent with a model in which GATA4 and GATA6 can functionally compensate for each other (Molkentin et al., 1997). GATA4 and GATA6 are similar factors that share a very high degree of sequence conservation, can interact with a shared binding motif, and can regulate similar target genes (Molkentin et al., 1997; Nemer and Nemer, 2003; Pikkarainen et al., 2004). Moreover, whole embryo knockouts of Gata4 demonstrated elevated levels of GATA6, suggesting an in vivo compensatory response (Molkentin et al., 1997). This redundancy has been shown in multiple developmental systems, and our results add coronary vasculogenesis to this roster (Park et al., 2009; Xin et al., 2006).
While the origin of CV endothelial cells is still not clear, recent models have described predominantly non-epicardial sources for SEECs (Katz et al., 2012; Red-Horse et al., 2010). SEECs were originally thought to arise from the myocardium and endocardium (Viragh and Challice, 1981). Subsequent lineage-tracing studies in the chick suggested the epicardium as a source for coronary endothelial cells (Mikawa and Gourdie, 1996). More recent mouse studies suggest the sinus venosus, endocardium, and proepicardium as source tissue, and that each contribute in varying degrees to the coronary endothelial population with only a very minor contribution from the epicardium (Red-Horse et al., 2010; Cossette and Misra, 2011; Katz et al., 2012). In our experiments we observed that the WT1(RP23-8C14)-Cre -driven reporter reliably labeled the entire epicardium but only 20% of SEECs (Fig. 5). Further, expression of the Cre reporter used here is only seen in a small subset of endothelial cells in developed vessels (Fig. 1). The observation that the preponderance of epicardial cells express the Cre reporter while only a small subset of the coronary endothelial cells express the reporter is consistent with models in which the majority of the coronary endothelial cells are not derived from the epicardium (Katz et al., 2012; Red-Horse et al., 2010; Wilm et al., 2005; Zhou et al., 2008). Our observations, however, are consistent with a model in which the epicardium generates GATA dependent signals to recruit endothelial cells from the sinus venosus and/or PE to the sub-epicardium.
The epicardium is a source of many signaling factors required for cardiac and CV development. Studies directly inhibiting epicardial signaling have found defects in epicardial adhesion, myocardial formation, and CV formation (Olivey and Svensson, 2010). Based on factors known to be secreted from the epicardium, we hypothesized that epicardial sonic hedgehog (SHH) may be affected in the dcKO (Lavine et al., 2006). We analyzed hedgehog expression using a monoclonal antibody raised against the N-terminus of SHH and observed a loss of SHH in the sub-epicardium of dcKO hearts (Ericson et al., 1996) (Supplemental Fig. 5A and B), suggesting that GATA-dependent epicardial hedgehog signaling pathways control SEEC numbers. Consistent with an important role of SHH, we found that we could rescue the loss of SEECs in an ex vivo assay by supplementation with exogenous SHH, thereby rescuing the phenotype (Supplemental Fig. 5E and F). While these findings strongly support the hypothesis that epicardial GATAs recruit endothelial cells in to the sub-epicardial space via SHH, we were unable to find a significant difference in the levels of shh mRNA between the wild type and dcKO (Supplement Fig. 4G). The reason for this discrepancy is unclear. GATAs may not directly regulate transcription of the shh gene, but rather may be involved in post-transcriptional regulation of shh. Alternatively, the SHH antibody may be cross-reacting with other epitopes. However, rescue experiments using exogenous SHH support a central role for SHH and are in agreement with the loss of SHH seen in the antibody results (Supplemental Fig. 5C and F). Alternatively, the SHH antibody we used does detect the other members of the hedgehog family, Indian hedgehog and Desert hedgehog (Ericson et al., 1996), and loss of antibody signal in the dcKO could be due to changes in the expression of non-sonic hedgehogs. However, SHH is thought to be the only significant hedgehog in coronary vascular development, and Indian and Desert hedgehog are known to be expressed at very low levels (Lavine et al., 2006). Future work will have to address more carefully the potential relationship between epicardial GATAs and SHH signaling, or other signaling pathways that may control SEEC accumulation and early plexus formation.
In summary, we have established for the first time in this study that GATA4 and GATA6 are expressed in the epicardium during coronary vascular development. We have also presented evidence that epicardial GATAs play a critical role in the establishment of the coronary vasculature by regulating the number of SEECs during early plexus formation. In addition, based on the model proposed in Lavine et al., 2006, in which epicardial SHH plays a key signaling role in establishing the coronary plexus, results presented in Supplemental Fig. 5support a role for GATA4 and GATA6 in regulating epicardial SHH signaling. However, it is important to point out that our data does not directly validate this pathway, which should be verified by future research.
Supplementary Material
Acknowledgements
The authors would like to thank Harold Olivey for assistance with the whole heart culture assay. We thank Eric Svensson for helpful discussions. We greatly appreciate Dr. Xiang-Xi Xu for supplying the anti-GATA6 antibody. We thank Dr. John Burch for supplying the WT1-Cre mice.
Sources of funding
This work was supported by a Predoctoral Fellowship from the American Heart Association Midwest Affiliate (S.M.C.); the Advancing a Healthier Wisconsin Fund, the Marcus Family, the Phoebe R. and John D. Lewis Foundation, the Sophia Wolf Quadracci Memorial Fund, the Dr. James Guhl Memorial Fund, PO1 HL089471, and RO1 DK55743 (S.A.D.); and R01HL084636 (R.P.M.).
Footnotes
Appendix A. Supplementary materials
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2013.12.033.
References
- Conway SJ, Kruzynska-Frejtag A, Kneer PL, Machnicki M, Koushik SV. What cardiovascular defect does my prenatal mouse mutant have, and why. Genesis. 2003;35:1–21. doi: 10.1002/gene.10152. [DOI] [PubMed] [Google Scholar]
- Cossette S, Misra R. The identification of different endothelial cell populations within the mouse proepicardium. Dev. Dyn. 2011;240:2344–2353. doi: 10.1002/dvdy.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman RW, Denetclaw WJ, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev. Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- Froese N, Kattih B, Breitbart A, Grund A, Geffers R, Molkentin JD, Kispert A, Wollert KC, Drexler H, Heineke J. GATA6 promotes angiogenic function and survival in endothelial cells by suppression of autocrine transforming growth factor beta/activin receptor-like kinase 5 signaling. J. Biol. Chem. 2011;286:5680–5690. doi: 10.1074/jbc.M110.176925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz T, Johnson RL, Epstein JA, Tabin CJ. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev. Cell. 2012 doi: 10.1016/j.devcel.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Lavine KJ, Ornitz DM. Fibroblast growth factors and Hedgehogs: at the heart of the epicardial signaling center. Trends Genet. 2008;24:33–40. doi: 10.1016/j.tig.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Ornitz DM. Shared circuitry: developmental signaling cascades regulate both embryonic and adult coronary vasculature. Circ. Res. 2009;104:159–169. doi: 10.1161/CIRCRESAHA.108.191239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lepore JJ, Cappola TP, Mericko PA, Morrisey EE, Parmacek MS. GATA-6 regulates genes promoting synthetic functions in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2005;25:309–314. doi: 10.1161/01.ATV.0000152725.76020.3c. [DOI] [PubMed] [Google Scholar]
- Manasek FJ. Embryonic development of the heart. J. Embry. Exp. Morphol. 1969;22:333–348. [PubMed] [Google Scholar]
- Manner J. Experimental study on the formation of the epicardium in chick embryos. Anat. Embryol. (Berlin) 1993;187:281–289. doi: 10.1007/BF00195766. [DOI] [PubMed] [Google Scholar]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc. Nat. Acad. Sci. U.S.A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev. Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Tymitz KM, Richardson JA, Olson EN. Abnormalities of the genitourinary tract in female mice lacking GATA5. Mol. Cell Biol. 2000;20:5256–5260. doi: 10.1128/mcb.20.14.5256-5260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemer G, Nemer M. Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and -6. Dev. Biol. 2003;254:131–148. doi: 10.1016/s0012-1606(02)00026-x. [DOI] [PubMed] [Google Scholar]
- Nikolova-Krstevski V, Yuan L, Le Bras A, Vijayaraj P, Kondo M, Gebauer I, Bhasin M, Carman CV, Oettgen P. ERG is required for the differentiation of embryonic stem cells along the endothelial lineage. BMC Dev. Biol. 2009;9:72. doi: 10.1186/1471-213X-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden J, Grieskamp T, Lausch E, van Wijk B, van den Hoff MJ, Englert C, Petry M, Mommersteeg MT, Christoffels VM, Niederreither K, Kispert A. Wt1 and retinoic acid signaling in the subcoelomic mesenchyme control the development of the pleuropericardial membranes and the sinus horns. Circ. Res. 2010;106:1212–1220. doi: 10.1161/CIRCRESAHA.110.217455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivey HE, Compton LA, Barnett JV. Coronary vessel development: the epicardium delivers. Trends Cardiovascul Med. 2004;14:247–251. doi: 10.1016/j.tcm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ. Res. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Kim HS, Kim JD, Seo J, Chung KS, Lee HS, Huh TL, Jo I, Kim YO. Isolation of a ventricle-specific promoter for the zebrafish ventricular myosin heavy chain (vmhc) gene and its regulation by GATA factors during embryonic heart development. Dev. Dyn. 2009;238:1574–1581. doi: 10.1002/dvdy.21964. [DOI] [PubMed] [Google Scholar]
- Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc. Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Rajewsky K, Gu H, Kuhn R, Betz UA, Muller W, Roes J, Schwenk F. Conditional gene targeting. J. Clin. Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudat C, Kispert A. Wt1 and epicardial fate mapping. Circ. Res. 2012;111:165–169. doi: 10.1161/CIRCRESAHA.112.273946. [DOI] [PubMed] [Google Scholar]
- Sodhi CP, Li J, Duncan SA. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev. Biol. 2006;6:19. doi: 10.1186/1471-213X-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC. Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev. Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. i > FOG-2, a cofactor for <i> 4 GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, Yu W, He L, Yang Z, Zhang Z, Zhong TP, Yang X, Yang Z, Yan Y, Baldini A, Sun Y, Lu J, Schwartz RJ, Evans SM, Gittenberger-de Groot AC, Red-Horse K, Zhou B. Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res. 2013 doi: 10.1038/cr.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanek RJ, Christensen LP, Simons M, Murakami M, Zheng W, Schatteman GC. Embryonic coronary vasculogenesis and angiogenesis are regulated by interactions between multiple FGFs and VEGF and are influenced by mesenchymal stem cells. Dev. Dyn. 2010;239:3182–3191. doi: 10.1002/dvdy.22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat. Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc. Nat. Acad. Sci. U.S.A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez RA, Markwald RR, O’Rourke BP, Sharp DJ, Zheng D, Lenz J, Baldwin HS, Chang CP, Zhou B. Endocardial cells form the coronary arteries by angiogenesis through myocardial–endocardial VEGF signaling. Cell. 2012;151:1083–1096. doi: 10.1016/j.cell.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Davis CA, Molkentin JD, Lien CL, Duncan SA, Richardson JA, Olson EN. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc. Nat. Acad. Sci. U.S.A. 2006;103:11189–11194. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol. Cell Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Kong SW, Hu Y, Campbell PH, McGowan FX, Ackerman KG, Wu B, Zhou B, Tevosian SG, Pu WT. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J. Clin. Invest. 2009;119:1462–1476. doi: 10.1172/JCI38723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.