SUMMARY
This study examines the functional relationship between glioma cell production of hyaluronan (HA), known to play a role in glioma invasion, expression of its CD44 receptor, and glioma cell viability. Production of HA by CD44 positive mouse G26 and human U373 glioma cell lines was evaluated and compared to that of a CD44 positive mouse fibroblast-like L929 cell line. We found that both G26 and U373 MG glioma cells, but not L929 fibroblast-like cells, synthesized HA. HA synthesis by glioma cells was found during the proliferative phase as well as post-confluency, as detected by fluorophore-assisted carbohydrate electrophoresis. Eighty to ninety percent of the HA synthesized was secreted into the medium and 10–20% remained associated with the cells. To examine a possible mechanistic link between the CD44-HA interaction and endogenous HA production, glioma cells were treated with either anti-CD44 antibodies (clones KM201 or IM7) or HA oligosaccharides (hexamer oligoHA-6 or decamer oligoHA-10). We found that oligoHA-10, which was previously shown to compete effectively with the CD44-HA interaction, enhanced glioma HA synthesis by approximately 1.5-fold, without affecting cell viability. IM7 treatment of human U373 glioma cells resulted in over 50% decrease of HA production, which was associated with changes in cell size and apoptosis. Taken together, these data show that CD44 specific ligands, such as the IM7 antibody or oligoHA-10 could down-regulate or up-regulate glioma HA production, respectively. Our results suggest that interference with CD44/HA may lead to the discovery and development of new treatment modalities for glioma.
Keywords: Glioma, hyaluronan, CD44
INTRODUCTION
Malignant gliomas are locally invasive tumors that rarely metastasize, but are challenging to treat. Glioma invasion is a complex process involving interaction of tumor cells with host cells and with extracellular matrix (ECM) molecules and proteolytic enzymes [1–6]. Hyaluronan (HA), an abundant component of the ECM of brain parenchyma plays an essential role during glioma invasion through the proteolytically modified ECM in the brain [2, 5–7]. During this process, certain chondroitin sulfate (CS) proteoglycans (PGs) and HA are synthesized and released into the microenvironment [8–10]. Some ECM components and CD44 are found at the invasion front of several tumors, including glioma, at the interface with normal tissue, e.g., with brain tissue [11–16].
The ECM environment of the brain, rich in HA [17– 21], provides hydrated spaces that facilitate glioma cell migration and angiogenesis. HA, a principal ligand of the widely expressed CD44 cell adhesion molecule, is a large glycosaminoglycan (GAG) composed of repeats of the disaccharide beta-D-glucuronyl-beta-D-N-acetylglucosamine. In the central nervous system (CNS) the intracellular HA pool results from endocytic or phagocytic uptake as well as direct secretion into the cytoplasm following synthesis [18, 22, 23]. HA can be taken up by cells following its interaction with cell-surface CD44, after which it is transported into lysosomes and degraded by hyaluronidases into small oligosaccharides [24, 25]. HA localization, synthesis and breakdown are subject to dynamic regulation [18, 21]. In numerous pathologies, including tumor invasion and inflammation, an injury-associated increase in the production of HA was reported [26– 28]. The stroma of various tumors is rich in HA, with its levels being several fold higher compared to the normal parenchyma [29–31]. Both the overproduction of full-length (polymeric) HA [16] and its degradation to lower molecular weight fragments support tumor invasion and growth, as exemplified by accelerated intracerebral formation of murine astrocytoma [24, 31,32]. The diverse functions of HA are mediated in part by HA-binding proteins collectively called hyaladherins, which include cell-surface receptors such as the CD44 adhesion molecule.
CD44 is a transmembrane glycoprotein, widely expressed in many cells and found in primary brain tumors such as glioma [33–36]. CD44 serves as a surface receptor for components of the ECM such as HA and CS-PGs [1]. The standard form of CD44 (CD44s) consists of three parts, a cytoplasmic tail, a transmembrane region, and a large (90 amino acid-long) extracellular domain. The extracellular domain, including the N-terminal region, contains motifs that function as docking sites for ECM components and are responsible for binding HA [37]. Binding of HA to CD44 is influenced by variations in the CD44 polypeptide sequence, as well as by glycosylation, and oligomerization of the receptor [37, 38]. Under physiological conditions, CD44 expressed in tumor cells is cleaved by a membrane-associated metalloproteinase (MMP) at the membrane-proximal region of the ectodomain. This shedding of CD44 plays a critical role in efficient cell detachment from the HA substrate and promotes cell migration [39–44]. We found a membranous pattern of focally strong positive CD44 staining in invading G26 glioma cells and weak punctate CD44 staining in the cytoplasm [12, 45–47]. G26 cells were also shown to secrete HA and CS-PGs, and the active form of MMP-2 in vitro [8, 46]. Anti-CD44 antibody treatment of these cells in vitro resulted in decreased invasiveness but had no effect on the activity of MMP-2 [47]. The current study evaluates the HA production and viability of glioma cells treated with anti-CD44 or HA oligosaccharides, both known to compete with the CD44-HA interaction [38].
MATERIALS and METHODS
Cell lines
Two tumorigenic [mouse glioma-26 (G26) and human glioma (glioblastoma) U373-MG] and one non-tumorigenic (mouse fibroblast L929) cell lines were used in this study. L929 and U373 MG were obtained from the American Type Culture Collection (ATCC; Manassas, VA). The G26 cell line was developed in this laboratory [8, 45] using glioma tissue derived from a G26 in vivo model in C57BL/6 mice.
Reagents
The following anti-CD44 monoclonal antibodies (mAbs) were used for treatment in cell cultures: rat anti-mouse CD44 clone KM201 (Antigenix America Inc. Franklin Square, NY and from Southern Biotech, Birmingham, AL.), and rat anti-mouse/human CD44 clone IM7 [48]. Mouse MOPC-21 myeloma IgG1 was used as non-specific IgG control (Sigma-Aldrich, St. Lois, MO). All anti-CD44 mAbs used in this study are known to react with epitopes located on the extracellular portion of the CD44 molecule. The KM201 mAb recognizes an epitope in the N-terminal HA-binding region of CD44 which is distinct from the one recognized by IM7 mAb [49, 50] HA-oligosaccharides used in this study were: oligo-HA10 (decamer) and oligo-HA6 (hexamer) (gifts from Dr. Akira Asari, Japan). Mouse anti-human CD44-FITC mAb (clone L178; BD Biosciences, San Diego, CA) was used for immunostaining.
Cell Cultures
Cells were seeded at 1- 2 × 106 cells/25 cm2 tissue culture flasks and grown at 37°C in 5% CO2 in Minimum Essential Medium (MEM) growth medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) or, in selected experiments, with serum substitute (UltraCulture general purpose serum-free medium from BioWhittaker, Cambrex, Walkersville, MD). The growth medium of cell cultures designated for evaluation of HA production was changed within 48 h of culture and collected daily at 1, 2 and 3 days post-plating (time-course of HA production), or on day 3 post-plating (cumulative production of HA). The growth medium of cell cultures designated for treatments with anti-CD44 antibodies or HA-oligosaccharides was also changed within 48 hours of culture and followed by initiation of the treatments for an additional 22–24 hours.
Cell Treatment
Cells seeded in the flasks (as described above) were incubated for 22–24 hours either in fresh growth medium alone or with the addition of one of the anti-CD44 mAbs or HA-oligosaccharides. Both mAbs and the non-specific IgG1 control were used at approximately 7.5–10 µg protein/5–7×106 cells/25 cm2 flask in 3 ml culture medium. HA-oligosaccharides were used at a concentration of 100 µg/ml (total of 300 µg/culture). Following a 22–24 hour incubation of glioma cells with either antibodies or HA-oligosaccharides, endogenous HA content in cells and the medium was quantified by fluorophore-assisted carbohydrate electrophoresis (FACE). In addition, CD44 expression by the cells was evaluated by flow cytometry, cell viability by trypan blue exclusion assay and apoptosis by Annexin V staining.
Fluorophore-assisted carbohydrate electrophoresis (FACE) quantification of HA
This assay is based on detection of the enzymatically cleaved repeating disaccharide sequences of HA or chondroitin sulfate/dermatan sulfate (CS/DS) at the beta-1,4-bonds by chondroitinase ABC. This cleavage generates delta-disaccharides such as HA-derived (delta-DiHA) and CS/DS- derived disaccharides (delta-Di0S, delta-Di6S, delta-Di4S). Each lysate product contains a free reducing end that can be stoichiometrically coupled to a fluorescent tag and detected with this assay [51, 52]. The solubilization of the cell monolayers with proteinase K at 37° C for 24 h, 60°C for 5 h, and 100° C for 10 minutes is expected to lyse the cells and solubilize all proteins. HA quantification was performed in media and cell monolayers.
To evaluate HA production by the cell cultures, the growth medium was changed and either collected daily at 1, 2 and 3 days post-plating (time course of HA production), on day 3 post-plating (cumulative production of HA) or after 22–24 h following incubation of cells with anti-CD44 antibody or oligo-HA (treatments). All cultures (media and cell monolayers separately) were prepared for glycosaminoglycan (GAG) analyses as follows:
To evaluate GAG levels the culture medium was removed, combined with a 1 ml PBS wash of the cell monolayer and lyophilized. Following resuspension with 1.5 ml of water, 400 µg of proteinase K (Invitrogen, Carlsbad, CA) was added and the mixture was digested for 8 h at 60°C. Cell monolayers were digested in the flask in 1.5 ml 0.1 M sodium acetate pH 7.2 with 400 µg of proteinase K for 8 h at 60°C. Proteinase K was inactivated at 100°C for 10 min; samples were cooled on ice, and then chromatographed on G50 columns (0.5 × 5 cm) that had been preconditioned with 1 mg of bovine serum albumin (BSA) in water. Samples were eluted in water, the void volume fractions (1.5 ml) were collected and residual glucose and phenol red were removed by an additional MicroCon10 (Millipore, Billerica, MA) ultrafiltration step. GAG chains were recovered from the filters in 100 µl of 0.1 M ammonium acetate pH 7.2 and digested with 16 mU of chondroitinase ABC and 20 mU chondroitinase ACII (Seikagaku America, A Division of Associates of Cape Cod, Inc., East Falmouth, MA.) for 18 h at 37°C. Buffer was evaporated by speedvac lyophilization and samples were derivatized with aminoacridone (AMAC) and cyanoborohydride (CNBH4) and separated by FACE using monosaccharide composition gels. After separation by FACE, the relative fluorescence in each band was quantified as follows. The sample wells and the stacking gel were covered with the light impermeable tape, and the gel cassette was placed on the transilluminator light box. The gels were viewed and the images captured with a cooled, charge-coupled device (CCD) camera for analysis. The pixel densities of the bands of chondroitin lysate digestion products were determined by Gel-Pro Image analyses software. A sample containing a range of concentrations (20–300 pmole) of delta-disaccharide standards was routinely included in one lane of each FACE gel. A standard curve of pixel densities per picomole of saccharide was generated. An average pixel density per picomole of the delta-disacharide standard (based on the glucose standard) was determined. The digestion products indentified as unsaturated internal delta-disaccharides included: delta-DiHA, delta-Di0S, delta-Di6S, and delta-Di4S. The delta-disaccharide content was determined and delta-DiHA was quantified in the experimental samples. The HA levels were presented as HA µg /tissue culture flask or HA fg/single cell [51, 52].
Flow cytometry
Cells were detached from adherent cultures using either an enzymatic method with mild trypsinization or non-enzymatic method using a “cell stripper” solution (Cellgro Mediatech, Inc. Manassas, VA), washed in culture medium, counted in trypan blue, and samples of 1 × 106 cells in a 50 µl volume were reacted with CD44 antibody conjugated with fluorescein isothiocyanate (CD44-FITC), for detection of CD44 expression by flow cytometry. The mouse cells were labeled with rat anti-mouse KM201 CD44-FITC mAb, using nonspecific rat IgG1 as an isotype control (Antigenix America). The human cells were labeled with mouse anti-human CD44-FITC mAb (clone L178) using nonspecific mouse IgG1 as an isotype control (BD Biosciences, San Diego, CA). The FITC labeled cells were run in a FACSCalibur flow cytometer and analyzed using CellQuest software (BD Biosciences). Staining intensity was determined in 2-dimensional histograms of fluorescence intensity versus cell number, relative to control. (FSC) forward scatter (cell size) and (SSC) side scatter (granularity) were displayed under identical instrument settings to ensure comparability of all cell parameters.
Apoptosis Assay
Adherent U373 cell monolayers, grown in chamber slides (5 × 103 cells/well) and cells in suspension (prepared as for flow cytometry from cell monolayers) were used in this assay, employing an annexin V conjugates apoptosis detection kit (Molecular Probes, Eugene, OR). Cell monolayers were treated with IM7, KM201 or non-specific IgG for 24 h. Following treatment, cell cultures were rinsed 3 times with annexin-binding buffer and labeled with Alexa Fluor 488-conjugated annexin V. Following labeling, cells were rinsed 3 times and fixed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer. The buffer was removed, Vectashield mounting medium with DAPI was added, and the slides were coverslipped. The cells were stored at 4°C until examination and photography using a Nikon Diaphot microscope (Nikon, Melville, NY) equipped with a color CCD camera. Images were acquired and processed using software from Oncor Image (Gaithersburg, MD). The number of apoptotic cells in each experimental sample was obtained by the microscopic evaluation of multiple fields of 100 cells in five experiments. This resulted in obtaining data for a total of 15 fields (100 cells/field) for each sample in experiments using the adherent cell monolayers, and 7 fields (100 cells/field) for each sample with cells in suspension.
Statistical Analysis
Statistical analysis was performed using the Microsoft Excel Analysis Tool Pack, (Redmond, WA). Descriptive statistics (mean and SD) were obtained for the concentrations of HA and the apoptotic cells numbers. The levels of HA were expressed as either HA µg/flask or HA fg/cell, or as an average relative absorbance of the band representing delta-DiHA in FACE. The ANOVA single factor test was used for the statistical comparison of the data evaluating the effect of HA oligosaccharides on HA synthesis. The Student’s paired t-test was used for the statistical comparison of the data evaluating the effect of IM7 treatment on HA synthesis. Cell apoptosis was evaluated in a similar manner, using either the Student’s t-test or ANOVA. Differences were considered statistically significant at p<0.05.
RESULTS
HA Production
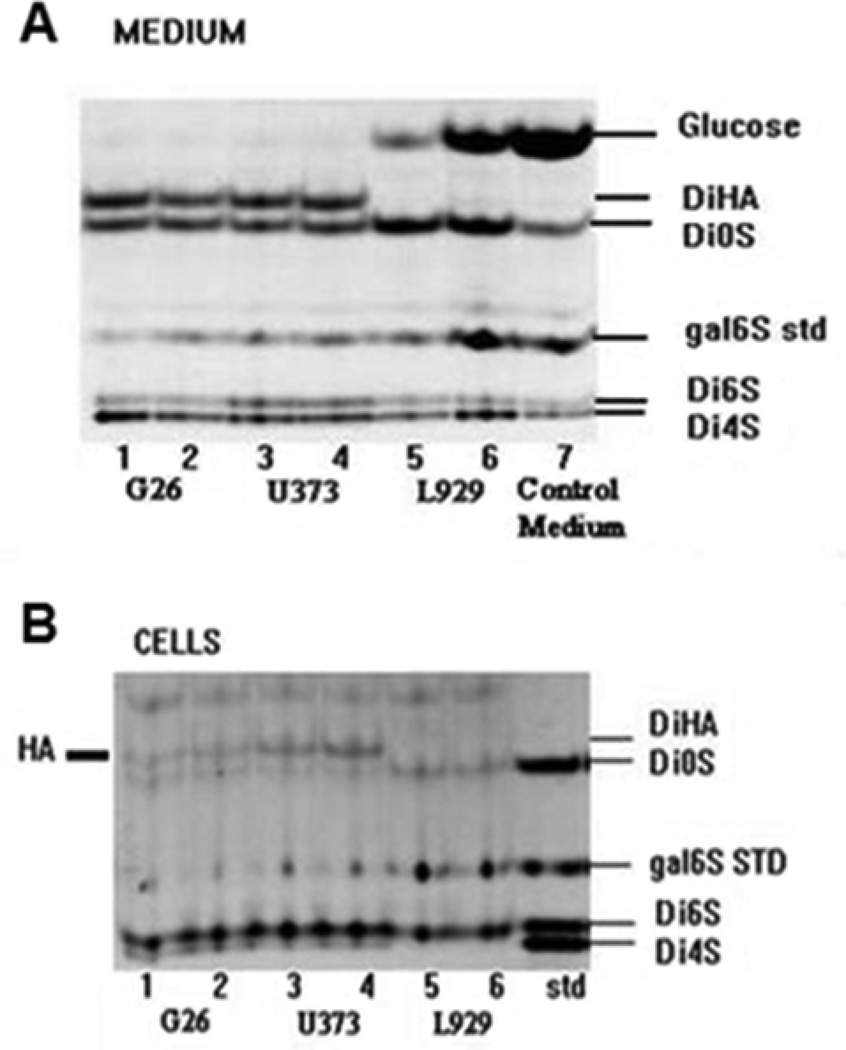
To assess the cumulative production of HA, the growth medium and cells were collected from either mouse G26 or human U373 glioma or L929 fibroblast cell cultures at 3 days post-plating, and GAG levels were evaluated (Fig. 1). Using the FACE assay, we found that HA-derived disaccharides (delta-DiHA) were present in digests from both medium (Fig. 1A) and cells (Fig. 1B) of the mouse glioma G26 (lanes 1, 2), and human glioma U373 cells (lanes 3, 4). However, they were not detectable in either fraction of the mouse fibroblast L929 cells (lanes 5, 6) (Fig. 1. A, B). Therefore, it was concluded that HA was accumulating during 3 days culture in the medium of both glioma cells but not L929 fibroblast cells. HA concentrations in the samples of medium from the cell cultures, showed a reciprocal relationship with the levels of glucose found in these samples. This suggested higher glucose “consumption” by HA-producing glioma cells than by non-HA producing L929 cells. HA was also detected at low levels in the cell pellets prepared from G26 and U373 glioma cells but not L929 cells.
Figure 1.
Cumulative secretion (A, Medium) and cellular accumulation (B, Cells) of HA by human glioma U373, mouse glioma G26, and mouse fibroblast L929 cells are represented by FACE images of chondroitin lyase digestion products from cultures harvested at the end of 3-day cultures. Shown are FACE images of HA-derived disaccharides (delta-DiHA), and CS/DS-derived disaccharides (delta-Di0S, delta-Di6S, delta-Di4S), and non-reducing hexosamine (6S galNAC) as a standard (Std). Each lane represents the products from either one or two combined 25 cm2 flasks seeded with 1- 2 × 106 cells per flask. The glucose content in medium from L929 cell culture is comparable to the control medium.
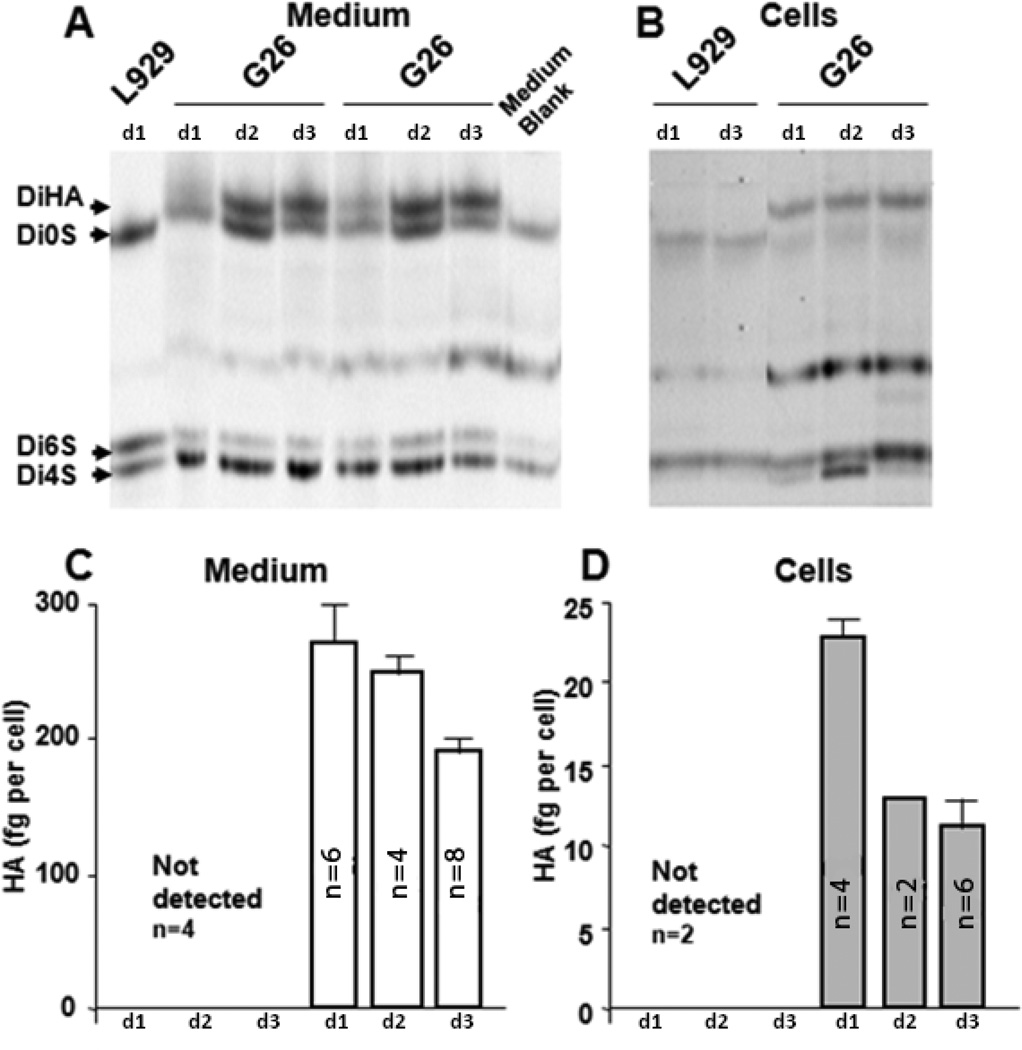
A 3-day time course of HA production was also evaluated in cell cultures seeded at 2×106 cells/25 cm2 flask in 3 ml of medium. HA secreted into the medium or associated with the cells was assayed in G26 mouse glioma and compared to HA content in L929 mouse fibroblast cultures (Fig. 2). The growth medium was collected daily (days: 0–1, 1–2, and 2–3 post-plating) and the corresponding cell cultures were trypsinized and cell counts were obtained using trypan blue exclusion. FACE quantitation of HA levels in the medium (Fig. 2A) and in the cell-associated matrix (Fig. 2B) revealed that 5–10% of HA produced by G26 glioma cells remained associated with the cells during various times of culture (Fig. 2B). HA was not detected in the cell-associated matrix in cultures of L929 cells at any of the time points (Fig. 2B). Despite the absence of HA in the L929 culture medium, other GAGs, such as chondroitin/dermatan sulfate (Di0S, Di6S and Di4S) were detected in both L929 fibroblast and G26 glioma cells cultures (Fig. 2. A, B). G26 glioma cells produced HA both during proliferation (days 0–2) and after reaching confluence (day 3) (Fig. 2. C, D). The secretion of HA into the medium at 22–24h intervals was the highest during the proliferative phase (0–48 h) of culture ,~280 fg/cell and subsequently declined by approximately 30% to ~190 fg/cell when cells had reached confluence. Furthermore, the cell associated HA concentration was also higher in proliferating cells (~22 fg/cell) compared to confluent cells (~12 fg/cell) (Fig. 2. C, D). HA detected in the cell preparation was most likely not intracellular but rather cell-bound. Although the FACE assay measures both extracellular and intracellular HA, the levels of internalized HA are expected to be minute due to extensive HA degradation occurring within the cell.
Figure 2.
Time course of HA synthesis measured in the medium (A, C) and cellular HA accumulation (B, D) by mouse glioma G26 and mouse fibroblast L929 cells. Shown are the FACE images (A,B) of chondroitin lyase digestion products (delta-DiHA, -Di0S, -Di4S, -Di6S) in media (A) and cells (B). Each lane represents the products from two 25 cm2 flasks. The quantitative data (C, D) are expressed as fg of HA/cell/22h, and are presented for medium (C) and cell-associated matrix (D) on days 1–3 (d1-d3). Shown are means + SD, and the number of cultures is indicated on the graphs. The corresponding cell numbers of G26 glioma cell cultures were as follows: Day 1 = 3.6 ×106, Day 2 = 7.5 ×106, Day 3 = 9.5 ×106 cells/flask.
HA production after cell treatment with oligo-HA-6, HA-10 or anti-CD44 antibody
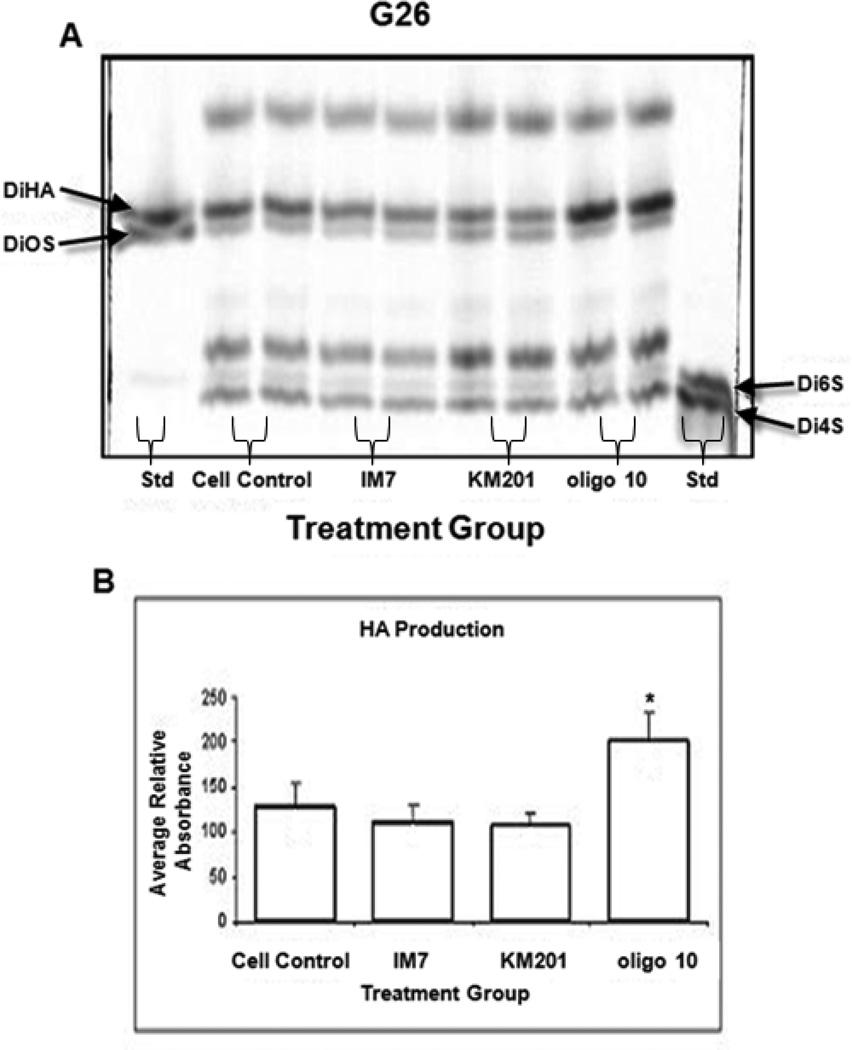
To evaluate of the relationship between CD44 engagement and HA synthesis by the glioma cells we employed 22–24 h treatment of cells with HA oligosaccharides (oligoHA-10 and HA-6) or with anti-CD44 mAbs. Using 2–3 day cell cultures, subsequently treated for additional 22–24 h with either HA-10 or HA-6, showed that they differed in their effect on HA production. When G26 glioma cells were incubated in the presence of oligo-HA-10, HA synthesis increased ~1.5 fold over the cell controls, which was statistically significant (Fig. 3). Treatment of cells with HA-6 had no effect on HA production (data not shown). Also, there was no significant effect of HA oligosaccharides on glioma cell viability or apoptosis (FlowTacs Apoptosis Detection kit, R&D Systems, data not shown).
Figure 3.
HA accumulation in mouse G26 glioma cultures, either left untreated (Cell Control) or treated with anti-CD44 mAbs (IM7 or KM201), or HA 10 oligosaccharide (HA10). Presented is the FACE image (A) of chondroitin lyase digestion products and FACE standard (Std). The corresponding graph (B) represents average relative UV absorbance. Shown are the means + SD (n=4–6; *p< 0.005).
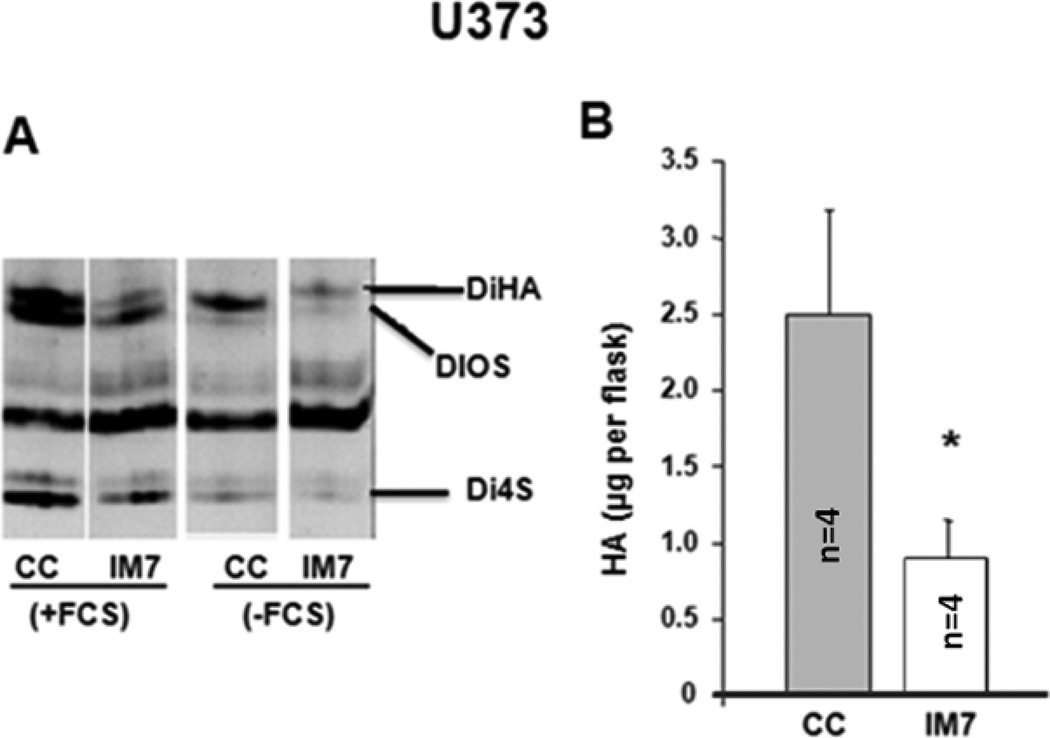
In another approach, the CD44 was targeted on glioma cells with two anti-CD44 mAbs, KM201 mAb or IM7 both of which to recognize distinct epitopes located on the extracellular domain of CD44 [53]. Furthermore, KM201 reacts only with mouse but not human CD44, whereas IM7 recognize both human and mouse CD44, and has been reported to have higher affinity for the human than for mouse CD44 [53]. Using 2–3 day cell cultures, subsequently treated for additional 22–24 h with either KM201 or IM7 showed no effect on HA production in G26 glioma cells (Fig. 3). On the other hand, when the human U373 glioma cells were cultured in the presence of IM7 (either in the medium supplemented with 10% serum or containing serum substitute), the HA content detected in the medium was significantly lower than in the medium of cell control (Fig. 4). The following decreases were observed in these cultures: from 3.007–3.145 µg/flask to 0.848–0.850 µg/flask (when cells were cultured in the medium supplemented with 10% serum), and from 2.158–1.933 µg/flask to 1.242–0.9239 µg /flask (when cells were cultured in the medium containing serum substitute). However, the cell-associated HA levels were not visibly affected by IM7 treatment (data not shown).
Figure 4.
HA accumulation in the medium (A) of human U373 glioma cells, cultured in presence of 10% FBS (+FBS) or serum substitute (-FBS) and either left untreated [cell control (CC)] or treated for 22–24 h with IM7 (IM7). Presented are the FACE image (A) of chondroitin lyase digestion products and the corresponding quantitative data (B). HA concentrations are expressed as µg/flask from 3 × 106 cells/flask/22 h). Shown are the means + SD (n=4,*p<0.05).
CD44 expression
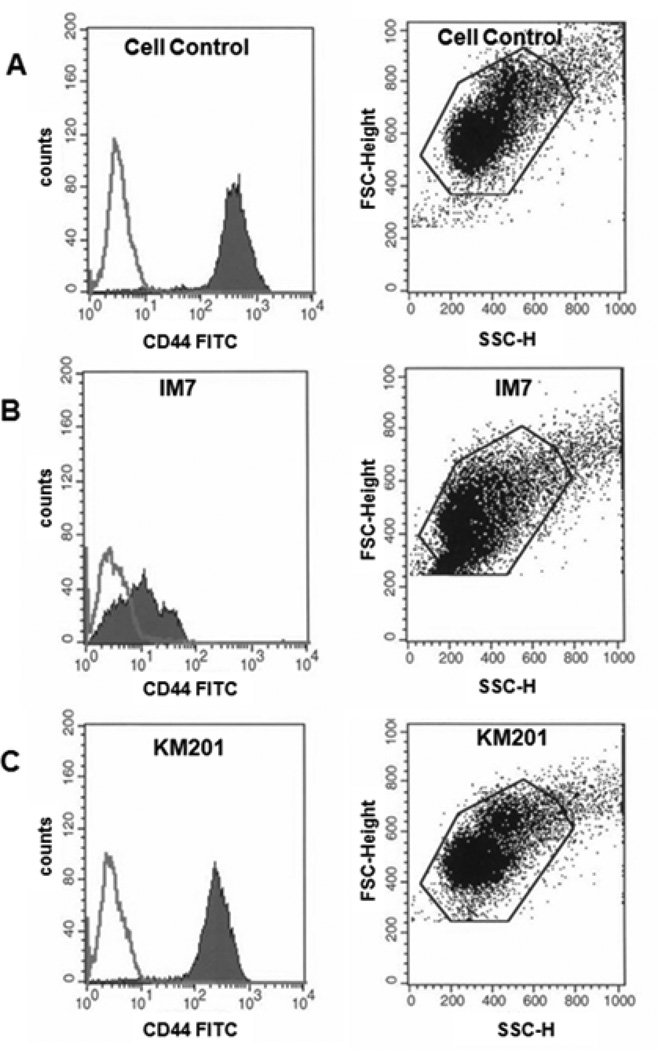
All cell lines (U373, G26 and L929) evaluated by flow cytometry showed that these cells expressed CD44 (data not shown). The cell suspension for flow cytometry was prepared either by trypsinization or non-enzymatic cell removal using “cell-stripper” solution. Both methods of cell preparation show similar levels of CD44. Treatment of mouse glioma G26 with oligoHA-6 or oligoHA-10, or with IM7 had no significant effects on CD44 fluorescence intensity. However, when compared with non-specific IgG (cell control; Fig. 5A), incubation of human glioma U373 with IM7 (Fig. 5B), but not with KM201 (Fig. 5C), resulted in decreased CD44 fluorescence intensity and a morphological change exhibited by the shift toward reduced cell size (Fig.5 B, scatter plot). The morphological alteration of IM7-treated cells detected by flow cytometry suggested the possibility of apoptosis, but morphological change was not discernible by routine light microscopic examination of the adherent U373 cells during IM7 treatment.
Figure 5.
Flow cytometric evaluation of cell-surface CD44 expression in human U373 glioma non-treated or treated with anti-CD44 Abs. The two super-imposed peaks seen in each histogram represent cells reactive with anti-CD44-FITC [L178 anti-human CD44 mAb) (filled histogram) or with nonspecific mouse IgG1 (line histogram). Representative data are presented on the left-side, showing histograms of non-treated cell cultures (A), cells treated with IM7 (B), or with KM201 (C). The Y axis of each histogram represents cell number and X-axis represents fluorescence intensity. Also, shown on the right side are the corresponding scatter dot plots of forward-scattered (FSC) and side scattered (SSC) light signals of the gated cell population, indicating a shift in the size of IM7-treated glioma cells (B) as compared to untreated (A) or KM201-treated (C) cells.
Apoptosis
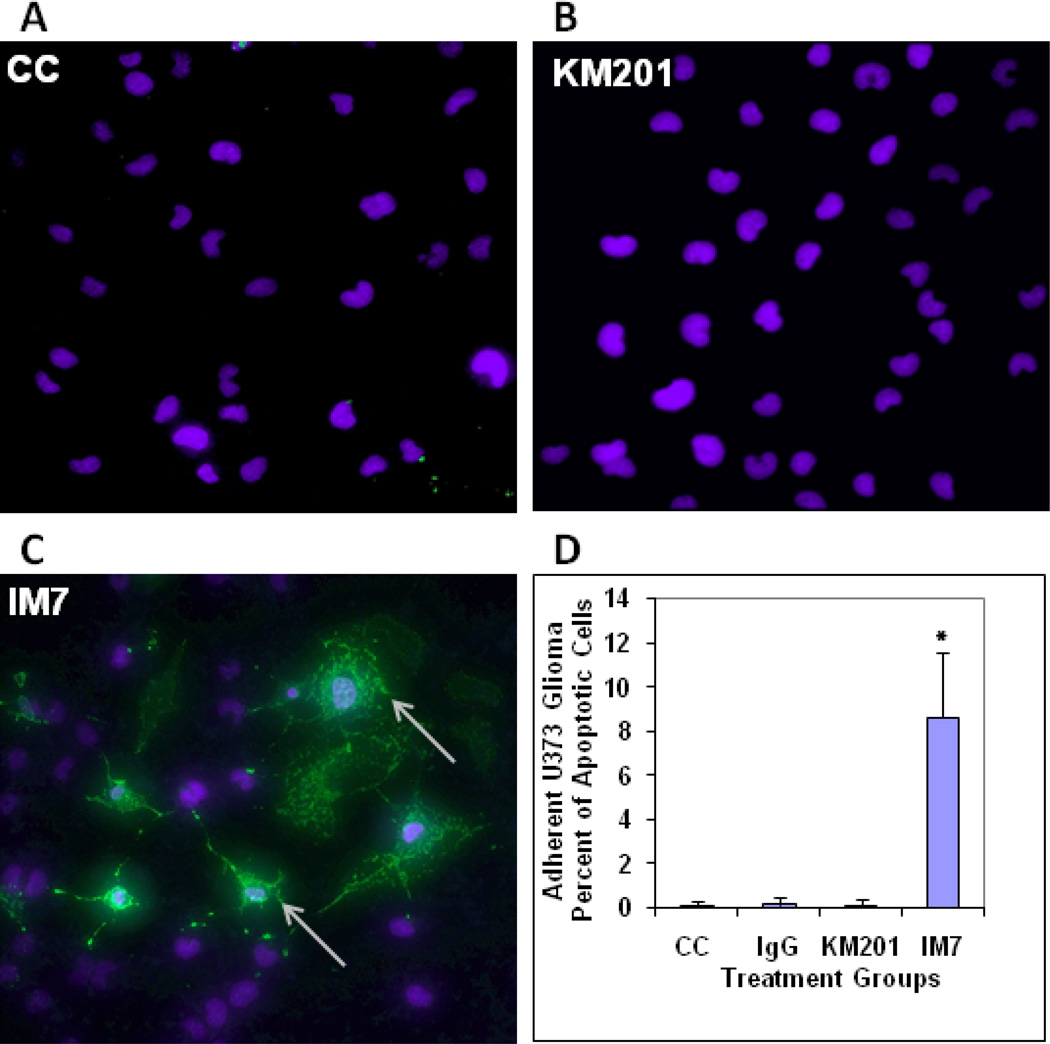
To address if apoptosis was induced by anti-CD44 mAb treatment in U373 human glioma cells, we employed annexin V staining. Exposure of adherent U373 glioma cell cultures to KM201 showed no detectable apoptosis relative to cell control (Fig. 6 A, B). However, treatment with IM7 resulted in ~9% of cells becoming Annexin V positive as evaluated by confocal microscopy (Fig. 6 C, D).
Figure 6.
Evaluation of apoptosis in adherent cultures of U373 human glioma by annexin V staining and confocal microscopy. Confocal images of U373 human glioma cells which were either non-treated (A) or treated with KM201 (B) or with IM7 (C) are shown. Exposure of U373 glioma cells to KM201 showed no detectable induction of Annexin V binding (B). Treatment with IM7 resulted in approximately 9% of the cells staining positive with annexin V (arrows in C) detected by confocal microscopy. Only nuclear (DAPI) staining is visible in the Figs. 6 A & B. Means + SD of Annexin V-positive cell numbers in untreated Cell Control (CC), IgG-treated, KM201-, and IM7- treated cultures are presented in graph (D) as percent of apoptotic cells, (n=15, *p<0.0001).
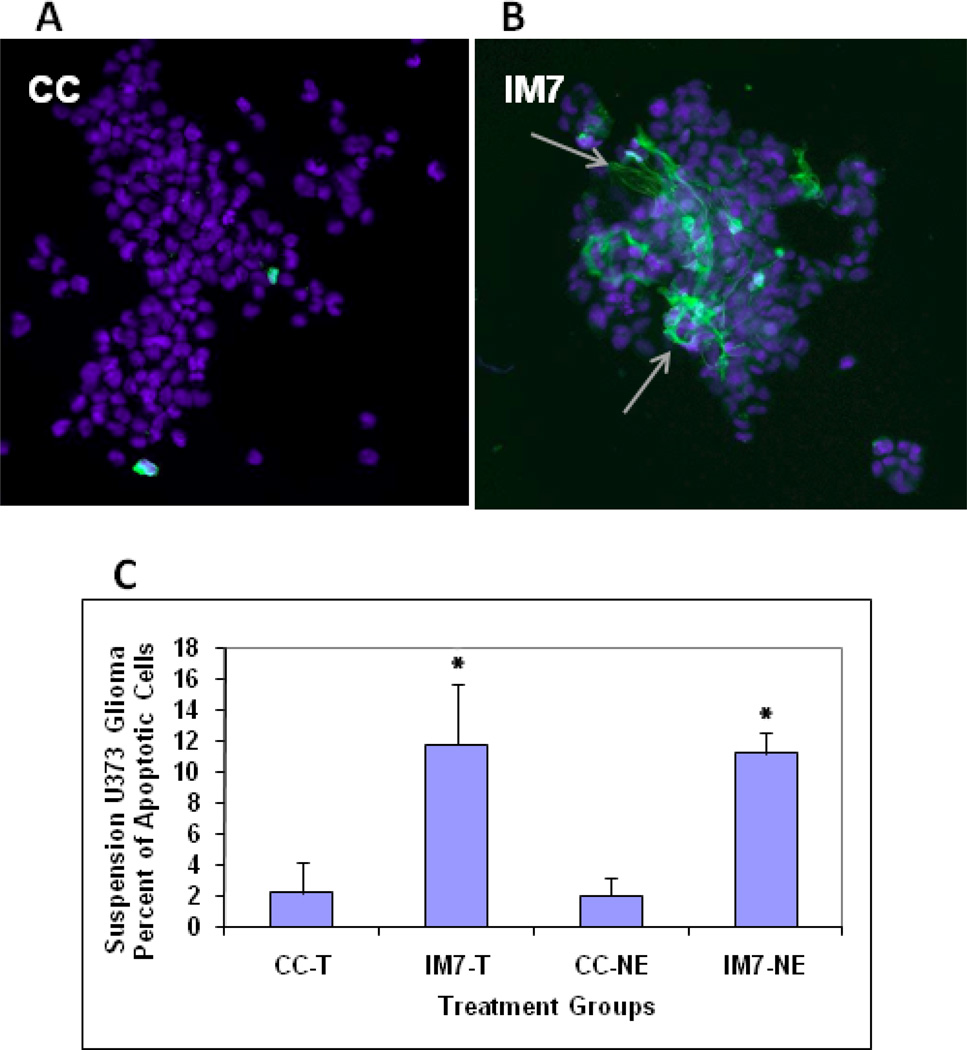
We found similar number of apoptotic cells in IM7-treated human glioma cells, which were either kept adherent throughout the apoptosis assay (~9%) (Fig 6) or used as a suspension culture after they were trypsinized (T) or non-enzymatically (NE) removed from the tissue culture (~11%) (Fig 7). Therefore, we excluded the possibility that the procedure of detaching the cells from the tissue culture dish using enzymatic or non-enzymatic method could trigger the morphological change seen upon flow cytometry, and apoptosis seen by annexin V staining.
Figure 7.
Evaluation of apoptosis in IM7-treated U373 human glioma cultures after cell detachment using enzymatic or non-enzymatic removal. Shown are confocal images of U373 human glioma cells that were either non-treated (A) or treated with IM7 (B), then detached with either trypsin or with “cellstripper”solution (non-enzymatic method) and stained with DAPI and Annexin V. Similar as in the adherent cultures (shown in Fig. 6), treatment of glioma cells with IM7 (before detachment) resulted in approximately 11% of Annexin V positive cells (arrows in B). There was no difference between cells detached with trypsinization (T) or non-enzymatic (NE) method. The graphs represent means + SD of Annexin V-positive cell numbers presented in graph (C) as percent of apoptotic cells, (n=7, *p<0.001).
DISCUSSION
Here we report that two tumorigenic CD44 positive glioma cell lines (human U373 and mouse G26) constitutively produce HA. HA was detected in the cell-associated matrix and found accumulating extracellularly, with the highest levels reached during cell proliferation. The HA-CD44 interaction has been implicated in matrix assembly, cell-matrix adhesion, tumor metastasis, cell migration and motility [5, 6, 54–57]. Increased synthesis and abnormal accumulation of HA are also known to be associated with cell migration [58–60], mitosis [61], and tumor invasion [62, 63]. For example, both U373 and G26 glioma, known to be invasive in vitro and in vivo and tumorigenic in vivo [46, 47, 64], were found here to produce and accumulate HA extracellularly.
While evaluating HA synthesis by human U373 and mouse G26 invasive glioma cell lines, a control cell line, mouse L929 fibroblast was also examined. L292 cells, unlike glioma, are known to be non-invasive, non-tumorigenic, and thus resemble the normal tissue “parenchyma”. While we found L929 cells to express CD44, these cells showed no detectable synthesis of HA. Similar observation with regard to HA production by normal and tumor cells, was made by several investigators. For example, Alaish et al [65] showed that while there was no difference in the magnitude of CD44 expression between the normal and keloid tumor derived fibroblasts, HA synthesis by keloid fibroblasts was significantly higher than in normal dermal fibroblasts in vitro and in vivo. A similar observation was made for prostate cancer tissue with regard to higher production of HA when compared to normal or benign hypertrophic prostate tissue [31]. HA synthesis by isolated plasma membranes from normal cultured human fibroblast cells was found reduced in comparison with membranes from non-metastatic melanoma cell line, and much lower than in a metastatic melanoma cell line [21]. Deposition of HA in the ECM by both mouse and human glioma cell lines investigated in this study, and the lack of HA deposition in the mouse fibroblast cell line, could result from a balance between the synthesis, release and intracellular uptake and degradation of HA.
It was previously reported that treatment of various cells with CD44-specific mAbs resulted in blocking of their HA binding [66]. We found that IM7 treatment of human glioma cells led to reduction in both HA production and CD44 expression. IM7 also induced glioma cell shrinkage and annexin V binding, both consistent with apoptosis. Shi et al [48] reported that IM7-treatment induced profound differences in the organization of filamentous actin in monocytes and fibroblasts. IM7-induced apoptosis was reported in experimental autoimmune uveoretinitis (EAU), where IM7 treatment suppressed EAU disease severity in vivo by inducing apoptosis of retina-infiltrating leukocytes 50]. In addition, an induction of apoptosis following IM7 treatment of human chondrosarcoma cell line was reported recently [67]. The IM7 mAb was shown to induce more extensive shedding of CD44 than other anti-CD44 antibodies, e.g., KM201 [48]. Annabi et al, [4] reported that CD44 shedding from the cell surface during glioma infiltration into the normal brain was a HA-mediated process, related to the up-regulation of MT1-MMP expression. Collectively, it appears that disturbance of the CD44-HA interaction from either the receptor or the ligand side may have serious biological consequences [68].
It is known that HA oligosaccharides such as HA-6 or HA-10 can displace full-length HA by competing for CD44 binding [21]. Displacement of HA can be accomplished by HA decasaccarides, but HA oligosaccharides shorter than 10-mer cannot effectively displace HA from the cell surface [38]. Ghatak et al [68] showed that mixed fractions of HA oligomers composed of 3–10 disaccharide units caused perturbation of HA-CD44 binding, leading to inhibition of anchorage–independent growth in tumor cell culture, tumor growth in vivo and apoptosis in vitro [68, 69]. A study by Deed et al [32] reported that native (large-size) HA had anti-angiogenic activity (inhibited endothelial cell proliferation and migration), but when degraded to smaller size oligosaccharide fractions (3–10 disaccharide units), this effect was lost. It has also been reported previously that a hyaluronidase, expressed by human tumors but not by normal tissues [32, 36], may be responsible for degradation of native HA leading to production of small-size HA oligosaccharides and induction of angiogenesis. Our data showing increased levels of HA produced after glioma cell treatment with HA-10, was similar to that reported by Luke and Prehm [21] who found that HA oligosaccharides (6–10-mers) stimulated HA production in human fibroblast cultures. According to Luke and Prehm [21], exogenously added HA oligosaccharides, while displacing nascent HA from the receptors, can stimulate HA synthesis. In our study, oligoHA-10 interaction with CD44 in glioma cells could have resulted in further up-regulation of HA production by these cells. HA of high molecular mass was found in normal and benign tissue while small HA fragments were found only in tumor tissue, e.g., prostate cancer [30, 31]. In addition, it was reported that low molecular-weight HA fragments and tumor-derived HA oligosaccharides strongly stimulated tumor growth, enhanced CD44 cleavage and tumor cell motility [15, 30, 70].
Further understanding of the importance of HA production and HA-CD44 interactions in glioma cell behavior and invasiveness can provide support to the development of clinical approaches targeting the HA-CD44 interaction. Information obtained from studies examining CD44-HA interactions was utilized in some therapeutic models [71]. For example, soluble recombinant CD44-HA–binding domain (CD44-HABD) was shown to act directly on endothelial cells by inhibiting their proliferation in vitro and blocking angiogenesis in vivo ,therefore, resulting in an inhibition of growth of various tumors [71]. Furthermore, HA-paclitaxel, targeted to CD44 positive ovarian carcinoma xenografts, showed antitumor efficacy, thus representing one of the first attempts to use a HA-based prodrug with anti-tumor potential [72]. Also, use of cisplatin-incorporated HA nanoparticles for antitumor drug delivery were reported recently [73].
In conclusion, our findings show that CD44 positive glioma cells produce high amounts of HA and this process parallels with the rapid growth and proliferation of these cells. Pericellular accumulation of HA creates a HA-rich microenvironment, which may support glioma cell growth. This HA-rich microenvironment may, in turn, signal through CD44, via short HA fragments e.g., oligoHA-10, to further up-regulate HA production in glioma cells. Based on the results of our study, HA production may be down-regulated by targeting a specific epitope (such as IM7 epitope) in the extracellular domain of CD44 [53], with concomitant induction of apoptosis in glioma cells. Taken together, further evaluation of CD44-HA interaction and alteration of glioma microenvironment through modulation of HA production by these cells is warranted. These results may lead to new approaches with a potential for developing specific anti-tumor therapies targeting invasive gliomas.
Statements about the novelty and impact of the paper.
Our study shows that extracellular levels of HA representing 80–90% of total HA synthesized by CD44 positive glioma cells (during the proliferative phase and post-confluence), can be modulated to affect glioma cell viability. The data obtained from treatment of glioma cells with either oligosaccharide HA-10, which enhanced HA synthesis (~ 1.5 fold), without an effect on cell viability, or with anti-CD44 antibody clone IM7, which decreased HA synthesis (>50%) of human glioma and caused apoptosis, suggest that manipulation of HA production could assist in the development of new anti-glioma therapies.
ACKNOWLEDGMENTS
Authors would like to extend their appreciation to Dr. Santo Nicosia, Chair of the Department Pathology and Cell Biology at USF, for the discussion and critique of the data, and for his insightful comments, and to Mr. Lawrence J. Stawkowski for his assistance in figure preparation. This study was supported by a grant from the American Cancer Society Florida Division.
Abbreviations
- HA
Hyaluronic acid
- oligoHA
HA oligosaccharides
- CS
chondroitin sulfate
- DS
dermatan sulfate
- PG
proteoglycan
- GAG
glycosaminoglycan
- ECM
extracellular matrix
- MMP
metalloproteinase
- MEM
Minimum Essential Mediun
- mAb
monoclonal antibody
- FBS
fetal bovine serum
- IgG
immunoglobulin G
- FITC
fluorescein isothiocyanate
- FSC
forward scatter
- SSC
side scatter
- FACE
fluorophore-assisted-carbohydrate electrophoresis
- BSA
bovine serum albumin
- AMAC
aminoacridone
- CNBH4
cyanoborohydride
- EAU
experimental autoimmune uveoretinitis
- CD44-HABD
CD44-HA-binding domain
- DiHA
HA-derived disaccharide
- CS/DS
chondroitin/dermatan sulfate
- Di0S, Di6S, Di4S
derived disaccharides
REFERENCES
- 1.Radotra B, McCormick D, Crockard A. CD44 plays a role in adhesive interactions between glioma cells and extracellular matrix components. Neuropathol Appl Neurobiol. 1994;20:399–405. doi: 10.1111/j.1365-2990.1994.tb00986.x. [DOI] [PubMed] [Google Scholar]
- 2.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046–1069. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Annabi B, Thibeault S, Moumdjian R, Beliveau R. Hyaluronan cell surface binding is induced by type I collagen and regulated by caveolae in glioma cells. J Biol Chem. 2004;279:21888–21896. doi: 10.1074/jbc.M313694200. [DOI] [PubMed] [Google Scholar]
- 4.Annabi B, Bouzeghrane M, Moumdjian R, Moghrabi A, Beliveau R. Probing the infiltrating character of brain tumors: inhibition of RhoA/ROK-mediated CD44 cell surface shedding from glioma cells by green tea catechin EGCg. J Neurochem. 2005;94:906–916. doi: 10.1111/j.1471-4159.2005.03256.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim MS, Park MJ, Kim SJ, Lee CH, Yoo H, Shin SH, Song ES, Lee SH. Emodin suppress hyaluronic acid-induced MMP-9 secretion and invasion of glioma cells. Int J Oncol. 2005;27:839–846. [PubMed] [Google Scholar]
- 6.Kim MS, Park MJ, Moon EJ, Kim SJ, Lee CH, Yoo H, Shin SH, Song ES, Lee SH. Hyaluronic acid induces osteopontin via the phosphatidylinositol 3-kinase/Akt pathway to enhance the motility of human glioma cells. Cancer Res. 2005;65:686–691. [PubMed] [Google Scholar]
- 7.Bignami A, Asher R. Some observations on the localization of hyaluronic acid in adult, newborn, and embryonal rat in brain. Int J Dev Neurosci. 1992;10:45–57. doi: 10.1016/0736-5748(92)90006-l. [DOI] [PubMed] [Google Scholar]
- 8.Wiranowska M, Naidu AK. Interferon effect on glycosaminoglycans in mouse glioma in vitro. J Neuro-Oncol. 1994;18:9–17. doi: 10.1007/BF01324598. [DOI] [PubMed] [Google Scholar]
- 9.Knott JCA, Mahesparan R, Garcia-Cabrera I, Tysnes BB, Edvardsen K, Ness GO, Mork S, Lund-Johansen M, Bjerkvig R. Stimulation of extracellular matrix components in the normal brain by invading glioma cells. Int J Cancer. 1998;75:864–872. doi: 10.1002/(sici)1097-0215(19980316)75:6<864::aid-ijc8>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 10.Mahesparan R, Read TA, Lund-Johansen M, Skaftnesmo KO, Bjerkvig R, Engebraaten O. Expression of extracellular matrix components in a highly infiltrative in vivo glioma model. Acta Neuropathol. 2003;105:49–57. doi: 10.1007/s00401-002-0610-0. [DOI] [PubMed] [Google Scholar]
- 11.Tysnes BB, Mahesparan M, Thoresen F, Haugland HK, Porwol T, Enger PO, Lund-Johansen M, Bjerkvig R. Laminin expression by glial fibrilary acidic protein positive cells in human gliomas. Int J Dev Neurosci. 1999;17:531–539. doi: 10.1016/s0736-5748(99)00055-6. [DOI] [PubMed] [Google Scholar]
- 12.Wiranowska M, Ladd S, Smith SR, Gottschall PE. CD44 adhesion molecule and neuro-glial proteoglycan NG2 as invasive markers of glioma. Brain Cell Biol. 2006;35:159–172. doi: 10.1007/s11068-007-9009-0. [DOI] [PubMed] [Google Scholar]
- 13.Rapponen K, Tammi M, Parkkinen J, Eskelinen M, Tammi R, lipponen P, Agren U, Alhava E, Kosma V-M. Tumor cell-associated hyaluronan as an unfavorable prognostic factor in colorectal cancer. Cancer Res. 1998;58:342–347. [PubMed] [Google Scholar]
- 14.Knudson W, Biswas C, Toole BP. Interaction between human tumor cells and fibroblasts stimulate hyaluronate synthesis. Proc Natl Acad Sci USA. 1984;81:6767–6771. doi: 10.1073/pnas.81.21.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujisaki T, Tanaka Y, Fujii K, Mine S, Saito K, Yamada S, Yamashita U, Irimura T, Eto S. CD44 stimulation induces integrin-mediated adhesion of colon cancer cell lines to endothelial cells by up-regulation of integrins and c-Met and activation of integrins. Cancer Res. 1999;59:4427–4434. [PubMed] [Google Scholar]
- 16.Kosaki R, Wantabe K, Yamaguchi Y. Overproduction of hyaluronan by expression of the hyaluronan synthase Has2 enhances anchorage-independent growth and tumorgenicity. Cancer Res. 1999;59:1141–1145. [PubMed] [Google Scholar]
- 17.Ruoslahti E. Brain extracellular matrix. Glycobiology. 1996;6:489–492. doi: 10.1093/glycob/6.5.489. [DOI] [PubMed] [Google Scholar]
- 18.Lynn BD, Turley EA, Nagy JI. Subcellular distribution, calmodulin interaction, and mitochondrial association of the hyaluronan-binding protein RHAMM in rat brain. J Neurosci Res. 2001;65:6–16. doi: 10.1002/jnr.1122. [DOI] [PubMed] [Google Scholar]
- 19.Wiranowska M, Plaas A. Cytokines and extracellular matrix remodeling in the central nervous system. In: Berczi I, Szentivanyi A, editors. Neuroimmune Biology, 6, Cytokines and the Brain. San Diego, CA: Elsevier Inc.; 2008. pp. 167–197. [Google Scholar]
- 20.Weigel PH, Hascall VC, Tammi M. Hyaluronan synthases. J Biol Chem. 1997;72:13997–14000. doi: 10.1074/jbc.272.22.13997. [DOI] [PubMed] [Google Scholar]
- 21.Luke H-J, Prehm P. Synthesis and shedding of hyaluronan from plasma membrane of human fibroblasts and metastatic and non-metastatic melanoma cells. Biochem J. 1999;343:71–75. [PMC free article] [PubMed] [Google Scholar]
- 22.Evanko SP, Wight TN. Intracellular localization of hyaluronan in proliferating cells. J Histochem Cytochem. 1999;47:1331–1342. doi: 10.1177/002215549904701013. [DOI] [PubMed] [Google Scholar]
- 23.Adamia S, Maxwell CA, Pilarski LM. Hyaluronan and hyaluronan synthases: potential therapeutic targets in cancer. Curr Drug Targets Cardiovasc Haematol Disord. 2005;5:3–14. doi: 10.2174/1568006053005056. [DOI] [PubMed] [Google Scholar]
- 24.Novak U, Stylli SS, Kayne AH, Lepperdinger G. Hyaluronidase-2 overexpression accelerates intracerebral but not subcutaneous tumor formation of murine astrocytoma cells. Cancer Res. 1999;59:6246–6250. [PubMed] [Google Scholar]
- 25.Culty M, Shizari M, Thompson E, Underhill C. Binding and degradation of hyaluronan by human breast cancer cell lines expressing different forms of CD44: correlation with invasive potential. J Cell Physiol. 1994;160:275–286. doi: 10.1002/jcp.1041600209. [DOI] [PubMed] [Google Scholar]
- 26.Dahl IM, Husby G. Hyaluronic acid production in vitro by synovial lining cells from normal and rheumatoid joints. Ann Rheum Dis. 1985;44:647–657. doi: 10.1136/ard.44.10.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Day AJ, de la Motte CA. Hyaluronan cross-linking: a protective mechanism in inflammation? Trends Immunol. 2005;26:637–643. doi: 10.1016/j.it.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Edward M, Fitzgerald L, Thind C, Leman J, Burden AD. Cutaneous mucinosis associated with dermatomyositis and nephrogenic fibrosing dermopathy: fibroblast hyaluronan synthesis and the effect of patient serum. Br J Dermatol. 2007;156:473–479. doi: 10.1111/j.1365-2133.2006.07652.x. [DOI] [PubMed] [Google Scholar]
- 29.Satoh C, Duff R, Rapp F, Davidson EA. Production of mucopolysaccharides by normal and transformed cells. Proc Natl Acad Sci USA. 1973;70:54–56. doi: 10.1073/pnas.70.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rooney P, Kumar S, Ponting J, Wang M. The role of hyaluronan in tumor neovascularization Int. J Cancer. 1995;60:632–636. doi: 10.1002/ijc.2910600511. [DOI] [PubMed] [Google Scholar]
- 31.Lokeshwar VB, Rubinowicz D, Schroeder GL, Forgacs E, Minna JD, Block NL, Nadji M, Lokeshwar BL. Stromal and epithelial expression of tumor markers hyaluronic acid and HYAL1 hyaluronidase in prostate cancer. J Biol Chem. 2001;276:11922–11932. doi: 10.1074/jbc.M008432200. [DOI] [PubMed] [Google Scholar]
- 32.Deed R, Rooney P, Kumar P, Norton JD, Smith J, Feemont AJ, Kumar S. Early-response gene signaling is induced by angiogenic oligosaccharides of hyaluronan in endothelial cells. Inhibition by non-angiogenic, high-molecular-weight hyaluronan. Int J Cancer. 1997;71:251–256. doi: 10.1002/(sici)1097-0215(19970410)71:2<251::aid-ijc21>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 33.Ranuncolo SM, Ladeda V, Specterman S, Varela M, Lastri J, Morandi A, Matos E, Bal de Kier Joffe E, Purcelli L, Pallota MG. CD44 expression in human gliomas. J Surg Oncol. 2002;79:30–36. doi: 10.1002/jso.10045. [DOI] [PubMed] [Google Scholar]
- 34.Pilkington GJ, Akinwunmi J, Ognjeovic N, Rogers JP. Differential binding of anti-CD44 on human gliomas in vitro. Neuroreport. 1993;4:259–262. doi: 10.1097/00001756-199303000-00008. [DOI] [PubMed] [Google Scholar]
- 35.Ariza A, Lopez D, Mate JL, Isamant M, Musulen E, Pujol M, Levy A, Navas-Palacios JJ. Role of CD44 in the invasiveness of glioblastoma multiforme and the noninvasiveness of meningioma: An immunohistochemistry study. Hum Pathol. 1995;26:1144–1147. doi: 10.1016/0046-8177(95)90278-3. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Liu J, Hofmann M. Differential CD44 expression patterns in primary brain tumors and brain metastases. Br J Cancer. 1995;72:160–163. doi: 10.1038/bjc.1995.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signaling regulators. Nature Reviews. 2003;4:33–48. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 38.Tammi R, MacCallum D, Hascall VC, Pienimaki J-P, Hyttinen M, Tammi M. Hyaluronan bound to CD44 on keratinocytes is displaced by hyaluronan decasaccharides and not hexasaccarides. J Biol Chem. 1998;273:28878–28888. doi: 10.1074/jbc.273.44.28878. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto I, Kawano Y, Tsuiki H, Sasaki J, Nakao M, Matsumoto M, Suga M, Ando M, Nakajima M, Saya H. CD44 cleavage induced by a membrane-associated metalloprotease plays a critical role in tumor cell migration. Oncogene. 1999;18:1435–1446. doi: 10.1038/sj.onc.1202447. [DOI] [PubMed] [Google Scholar]
- 40.Okomoto I, Kawano Y, Murakami D, Sasayama T, Araki N, Miki T, Wong AJ, Saya H. Proteolytic release of CD44 intracellular domain and its role in the CD44 signaling pathway. J Cell Biol. 2001;155:755–762. doi: 10.1083/jcb.200108159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami D, Okomoto I, Nagano O, Kawano Y, Tomita T, Iwatsubo T, De Strooper B, Yumoto E, Saya H. Presenilin-dependent γ-secretase activity mediates the intramembranous cleavage of CD44. Oncogene. 2003;22:1511–1516. doi: 10.1038/sj.onc.1206298. [DOI] [PubMed] [Google Scholar]
- 42.Murai T, Miyazaki Y, Nishinakamura H, Sugahara KN, Miyauchi T, Sako Y, Yanagida T, Miyasaka M. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J Biol Chem. 2004;279:4541–4550. doi: 10.1074/jbc.M307356200. [DOI] [PubMed] [Google Scholar]
- 43.Okada H, Yoshida J, Sokabe M, Wakabayashi T, Hagiwara M. Suppression of CD44 expression decreases migration and invasion of human glioma cells. Int J Cancer. 1996;66:255–260. doi: 10.1002/(SICI)1097-0215(19960410)66:2<255::AID-IJC20>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 44.Breyer R, Hussein S, Radu DL, Putz K-M, Gunia S, Hecker H, Samii M, Walter GF, Stan AC. Disruption of intracerebral progression of C6 rat glioblastoma by in vivo treatment with anti-CD44 monoclonal antibody. J Neurosurg. 2000;92:140–149. doi: 10.3171/jns.2000.92.1.0140. [DOI] [PubMed] [Google Scholar]
- 45.Wiranowska M, Gonzalvo AA, Saporta S, Gonzalez OR, Prockop LD. Evaluation of blood-brain barrier permeability and the effect of interferon in mouse glioma model. Neuro-Oncol. 1992;14:225–236. doi: 10.1007/BF00172598. [DOI] [PubMed] [Google Scholar]
- 46.Wiranowska M, Rojiani A, Gottschall P, Moscinski L, Johnson J, Saporta S. CD44 expression and MMP-2 secretionby mouse glioma cells: Effect of interferon and anti-CD44antibody. Anticancer Res. 2000;20:4301–4306. [PubMed] [Google Scholar]
- 47.Wiranowska M, Tresser N, Saporta S. The effect of interferon and anti-CD44 antibody on mouse glioma invasiveness in vitro. Anticancer Res. 1998;18:3331–3338. [PubMed] [Google Scholar]
- 48.Shi M, Dennis K, Peschon JJ, Chandrasekaran R, Mikecz K. Antibody-induced shedding of CD44 from adherent cells is linked to the assembly of the cytoskeleton. J Immunol. 2001;167:123–131. doi: 10.4049/jimmunol.167.1.123. [DOI] [PubMed] [Google Scholar]
- 49.Mikecz K, Brennan FR, Kim JH, Glant TT. Anti-CD44 treatment abrogates tissue oedema and leukocyte infiltration in murine arthritis. Nat Med. 1995;1:558–563. doi: 10.1038/nm0695-558. [DOI] [PubMed] [Google Scholar]
- 50.Xu H, Manivannan A, Liversidge J, Sharp PF, Forrester JV, Crane IJ. Involvement of CD44 in leukocyte trafficking at the blood-retinal barrier. J Leukoc Biol. 2002;72:1133–1141. [PubMed] [Google Scholar]
- 51.Plaas AHK, West L, Midura RJ, Hascall VC. Disaccharide composition of hyaluronan and chondroitin/dermatan sulfate. Analysis with Fluoro-Assisted Carbohydrate Electrophoresis. In: Iozzo RV, editor. Methods of Molecular Biology. vol 171: Proteoglycan Protocols. Totowa, N.J.: Humana Press Inc; 2000. pp. 117–128. [Google Scholar]
- 52.Calabro A, Benavides M, Tammi M, Hascall VC, Midura RJ. Microanalysis of enzyme digest of hyaluronan and chondroitin/dermatan sulfate by fluoro-assisted carbohydrate electrophoresis (FACE) Glycobiology. 2000;10:273–281. doi: 10.1093/glycob/10.3.273. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Z, Katoh S, He Q, Oritani K, Miyake K, Lesley J, Hayman R, Hamik A, Parkhouse RM, Farr AG, Kincade PW. Monoclonal antibodies to CD44 and their influence on hyaluronan recognition. J Cell Biol. 1995;130:485–495. doi: 10.1083/jcb.130.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherman L, Sleeman J, Herrlich P, Ponta H. Hyaluronate receptors: key players in growth, differentiation, migration and tumor progression. Current Opinion Cell Biol. 1994;6:726–733. doi: 10.1016/0955-0674(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 55.Mikecz KK, Dennis M, Shi M, Kim JH. Modulation of hyaluronan receptor (CD44) function in vivo in a murine model of rheumatoid arthritis. Arthritis Rheum. 1999;42:659–668. doi: 10.1002/1529-0131(199904)42:4<659::AID-ANR8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 56.Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 57.Hamilton SR, Fard SF, Paiwand FF, Tolg C, Veiseh M, Wang C, MacCarthy JB, Bissell MJ, Koropatnick J, Turley EA. The hyaluronan receptors CD44 and Rham (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J Biol Chem. 2007;282:16667–16680. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monz K, Maas-Kuck K, Schumacher U, Schulz T, Hallmann R, Schnaker EM, Schneider SW, Prehm P. Inhibition of hyaluronan export attenuates cell migration and metastasis of human melanoma. J Cell Biochem. 2008;105:1260–1266. doi: 10.1002/jcb.21925. [DOI] [PubMed] [Google Scholar]
- 59.Toole BP. Proteoglycans and hyaluronan in morphogenesis and differentiation. In: Hay ED, editor. Cell Biology of the Extracellular Matrix. New York, NY: Plenum Press; 1991. pp. 305–341. [Google Scholar]
- 60.Itano N, Atsumi F, Sawai T, Yamada Y, Miyaishi O, Senga T, Hamaguchi M, Kimata K. Abnormal accumulation of hyaluronan matrix diminishes contact inhibition of cell growth and promotes cell migration. Proc Natl Acad Sci USA. 2002;99:3609–3614. doi: 10.1073/pnas.052026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brecht M, Mayer U, Schlosser E, Prehm P. Increased hyaluronate synthesis is required for fibroblast detachment and mitosis. Biochem J. 1986;239:445–450. doi: 10.1042/bj2390445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toole BP, Biswas C, Gross J. Hyaluronate and invasiveness of the rabbit V2 carcinoma. Proc Natl Acad Sci USA. 1979;76:6299–6303. doi: 10.1073/pnas.76.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tzircotis G, Thorne RF, Isacke CM. Chemotaxis towards hyaluronan is dependent on CD44 expression and modulation by cell type variation in CD44-hyaluronan binding. J Cell Sci. 2005;118:5119–5128. doi: 10.1242/jcs.02629. [DOI] [PubMed] [Google Scholar]
- 64.deRidder LI, Laerum OD, Mork SJ, Bigner DD. Invasiveness of human glioma cell linesin vitro: Relation to tumorigenicity in athymic mice. Acta Neuropathol. 1987;72:207–213. doi: 10.1007/BF00691091. [DOI] [PubMed] [Google Scholar]
- 65.Alaish SM, Yager DR, Diegelmann RF, Cohen IK. Hyaluronic acid metabolism in keloid fibroblasts. J Pediatr Surg. 1995;30:949–952. doi: 10.1016/0022-3468(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 66.Knudson W, Biswas C, Li XQ, Nemec RE, Toole BP. The role and regulation of tumor-associated hyaluronan. Ciba Found Symp. 1989;143:150–159. doi: 10.1002/9780470513774.ch10. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida M, Yasuda T, Hiramitsu T, Ito H, Nakamura T. Induction of apoptosis by anti-CD44 antibody in human chondrosarcoma cell line SW1353. Biomed Res. 2008;29:47–52. doi: 10.2220/biomedres.29.47. [DOI] [PubMed] [Google Scholar]
- 68.Ghatak S, Misra S, Toole BP. Hyaluronan oligosaccharides inhibit anchorage-independent growth of tumor cells by suppressing the phospoinositide 3-kinase/Akt cell survival pathway. J Biol Chem. 2002;277:38013–38020. doi: 10.1074/jbc.M202404200. [DOI] [PubMed] [Google Scholar]
- 69.Zheng C, Toole BP, Kinney SD, Kuo J-W, Stamenkovic I. Inhibition of tumor growth in vivo by hyaluronan oligomers. Int J Cancer. 1998;77:369–401. doi: 10.1002/(sici)1097-0215(19980729)77:3<396::aid-ijc15>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 70.Sugahara KN, Hirata T, Hayasaka H, Stern R, Murai T, Miyasaka M. Tumor cells enhance their own CD44 cleavage and motility by generating hyaluronan fragments. J Biol Chem. 2006;281:5861–5868. doi: 10.1074/jbc.M506740200. [DOI] [PubMed] [Google Scholar]
- 71.Pall T, Gad A, Kasak L, Drews M, Stromblad S, Kogerman P. Recombinant CD44-HABD is a novel and potent direct angiogenesis inhibitor enforcing endothelial cell-specific growth inhibition independently of hyaluronic acid binding. Oncogene. 2004;23:7874–7881. doi: 10.1038/sj.onc.1208083. [DOI] [PubMed] [Google Scholar]
- 72.Auzenne E, Gosh SC, Khodadadian M, Rivera B, Farquhar D, Price RE, Ravoori M, Kundra V, Freedman RS, Klostergaard J. Hyaluronic acid-paclitaxel: antitumor efficacy against CD44(+) human ovarian carcinoma xenografts. Neoplasia. 2007;9:479–486. doi: 10.1593/neo.07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeong YI, Kim ST, Jin SG, Ryu HH, Jin YH, Jung TY, Kim IY, Jung S. Cisplatin-incorporated hyaluronic acid nanoparticles based on ion-complex formation. J Pharm Sci. 2008;97:1268–1276. doi: 10.1002/jps.21103. [DOI] [PubMed] [Google Scholar]