Abstract
P63, a p53 family member, plays pivotal roles in epidermal development, aging, and tumorigenesis. Thus, understanding how p63 expression is controlled has biological and clinical importance. RBM24 is an RNA-binding protein and shares a high sequence similarity with RBM38, a critical regulator of p63. In the current study, we investigated whether RBM24 is capable of regulating p63 expression. Indeed, we found that ectopic expression of RBM24 decreased, whereas knockdown of RBM24 increased, the levels of p63 transcript and protein. To explore the underlying mechanism, we found that RBM24 was able to bind to multiple regions in the p63 3′ untranslated region and subsequently, destabilize p63 transcript. Furthermore, we showed that the 3′ untranslated region in p63 transcript and the RNA-binding domain in RBM24 were required for RBM24 to bind p63 transcript and consequently, inhibit p63 expression. Taken together, our data provide evidence that RBM24 is a novel regulator of p63 via mRNA stability.
Implications
Our study suggests that p63 is regulated by RBM24 via mRNA stability, which gives an insight into understanding how posttranscriptional regulatory mechanisms contribute to p63 expression.
Keywords: RNA-binding protein, RBM24, p63, mRNA stability, p53
Introduction
P63 is a member of the p53 family including p53, p63, and p73 (1). All three proteins are transcriptional factors and share a high sequence similarity, especially in the DNA binding domain (2). Like other p53 family members, p63 gene has complex expression patterns due to the usage of two distinct promoters and alternative splicing at the C-terminus. The usage of two promoters results in two major p63 isoforms, TAp63 and ΔNp63, and each isoform is alternatively spliced into at least five variants, a, β, γ, δ, and ε (3, 4). Importantly, TAp63 isoforms, transcribed from the upstream P1 promoter, contain a transactivation domain similar to that in p53 and thus can induce a number of p53 target genes including p21 and MDM2 (5). By contrast, ΔNp63 isoforms, transcribed from the P2 promoter in intron 3, lack the N-terminal transactivation domain and are presumably thought to be transcriptionally inactive. Interestingly, some studies showed that ΔNp63 carries a ΔN activation domain and retains transcriptional activity under certain circumstances (2, 5, 6).
The biological function of p63 is complex due to the presence of multiple isoforms with opposing functions. Studies suggest that the ΔNp63 isoforms have oncogenic potential (7, 8), whereas the TAp63 isoforms play a role in tumor suppression (9). This apparent conflict was recently addressed by generating isoform-specific p63 knockout mice models. Specifically, total p63 knockout mice have defects in skin, teeth, mammary gland, and limb, and die soon after birth (10, 11), suggesting a critical role of p63 in epidermal development. Interestingly, mice deficient in ΔNp63 isoforms largely phenocopy total p63 knockout mice (12). These mice die shortly after birth due to several developmental defects, such as truncated forelimbs and the absence of hind limbs. By contrast, mice deficient in TAp63 isoforms are born live and tumor-prone (9). In addition, these mice develop several phenotypes, including accelerated aging, obesity, insulin resistance, and glucose intolerance (13, 14). Together, these in vivo studies indicate a critical role of p63 in skin development, aging, metabolism, and tumorigenesis.
Given the biological importance of p63, studies have been carried out to elucidate how p63 expression is controlled. For instance, upon exposure to various stimuli, the level of p63 transcript is regulated by p53 and several other transcription factors (15–17). Moreover, p63 can be posttranscriptionally regulated by RNA-binding protein (RBP) RBM38 and HuR via mRNA stability and protein translation, respectively (18, 19). In addition, several microRNAs, including miR-302, miR-130b, and miR-203, are found to regulate p63 mRNA stability (20–22). Furthermore, p63 protein stability is regulated by a set of E3 ligases, such as itch, Pirh2, wwp1, and SCFβTrCP1 (23–26). Nevertheless, other regulators, which are critical in modulating p63 expression, remain to be elucidated.
Materials and Methods
Reagents
Anti-RBM24, raised in rabbit, was generated by Cocalico Biologicals (Reamstown, PA). Anti-p63, 4A4, was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-HA was purchased from Covance (San Diego, CA). Anti-actin, proteinase inhibitor cocktail, RNase A, and protein A/G beads were purchased from Sigma (St. Louis, MO). Scrambled siRNA (GGC CGA UUG UCA AAU AAU U) and siRNA against RBM24 (CAC UGG AGC UGC AUA CGC A) were purchased from Dharmacon RNA Technologies (Chicago, IL). Transfection reagent Metafectene was purchased from Biontex (Germany). Silentfect lipid was purchased from Bio-Rad Laboratories (Hercules, CA). Trizol reagent purchased from Invitrogen (Carlsbad, CA). The MMLV reverse transcriptase was purchased from Promega (Madison, WI). EST clone, containing a full length human TAp63α (clone ID 5552611), was purchased from OpenBiosystem (Huntsville, AL),
Plasmids
To generate pcDNA3-HA-RBM24, a PCR product was amplified by using cDNA samples from MCF7 as a template and then inserted into pcDNA3-HA vector via EcoRI and XhoI sites. The primers were a forward primer, 5′ GGG GAA TTC ATG CAC ACG ACC CAG AAG 3′, and a reverse primer, 5′ GGG CTC GAG CTA TTG CAT TCG GTC TGT CTG 3′. To generate pcDNA3-RBM24, a PCR product was amplified by using cDNA samples from MCF7 as a template and then inserted into pcDNA3 vector via HindIII and XhoI sites. The primers were a forward primer, 5′ AAA AAG CTT CAC CAT GAT GCA CAC GAC CCA GAA GGA CAC GAC GTA CA 3′, and a reverse primer, which is the same as the one for pcDNA3-HA-RBM24. To generate pcDNA4-RBM24 vector, a DNA fragment was digested from pcDNA3-HA-RBM24 and then inserted into pcDNA4 vector via EcoRI and XhoI sites.
To generate pcDNA3-RBM24-ΔRNP1 vector, a two-step PCR strategy was used. The first-step PCR was performed to separately amplify two DNA fragments by using RBM24 EST clone as a template. Fragment 1 was amplified with a forward primer, 5′ AAA AAG CTT CAC CAT GAT GCA CAC GAC CCA GAA GGA CAC GAC GTA CA 3′, and a reverse primer, 5′ TCG GCA GCA GCC CGG TCA GCG GAC TTG CCC GTC TGC CGG TCG GTG 3′. Fragment 2 was amplified with a forward primer, 5′ ACC GGC AGA CGG GCA AGT CCG CTG ACC GGG CTG CTG CCG AAA GGG 3′, and a reverse primer, 5′ GGG CTC GAG CTA TTG CAT TCG GTC TGT CTG 3′. The second-step PCR was performed using a mixture of fragment 1 and 2 as a template with a forward primer, 5′ AAA AAG CTT CAC CAT GAT GCA CAC GAC CCA GAA GGA CAC GAC GTA CA 3′, and a reverse primer, 5′ GGG CTC GAG CTA TTG CAT TCG GTC TGT CTG 3′. This PCR product was then inserted into pcDNA3-RBM24 vector via HindIII and XhoI sites to generate pcDNA3-RBM24-ΔRNP1. To generate pcDNA3-RBM24-ΔRNP2 vector, the same strategy was used with different primers. The primers to amplify fragment 1 were a forward primer, 5′ AAA AAG CTT CAC CAT GAT GCA CAC GAC CCA GAA GGA CAC GAC GTA CA 3′, and a reverse primer, 5′ GCG TCG GTG GTG TGG TAG GGC TTG GTG TAC GTC GTG TCC T 3′. The primers to amplify fragment 2 were a forward primer, 5′ AGG ACA CGA CGT ACA CCA AGC CCT ACC ACA CCA CCG ACG CCA GCC 3′, and a reverse primer, 5′ GGG CTC GAG CTA TTG CAT TCG GTC TGT CTG 3′. The primers for second-step PCR were a forward primer, 5′ AAA AAG CTT CAC CAT GAT GCA CAC GAC CCA GAA GGA CAC GAC GTA CA 3′, and a reverse primer, 5′ GGG CTC GAG CTA TTG CAT TCG GTC TGT CTG 3′.
To generate pGEX-5X-1-RBM24 vector, a DNA fragment was digested from pcDNA3-HA-RBM24 and then inserted into pGEX-5X-1 vector via EcoRI and XhoI sites. To generate pGEX-5X-1-RBM24-ΔRNP1, a PCR product was amplified by using pcDNA3-RBM24-ΔRNP1 as a template and then inserted into pGEX-5X-1 via EcoRI and XhoI sites. The PCR primers were a forward primer, 5′ CGG AAT TCA TGC ACA CGA CCC AGA AGG ACA CGA CGT ACA 3′, and a reverse primer, 5′ GGG CTC GAG CTA TTG CAT TCG GTC TGT CTG 3′. To generate pGEX-5X-1-RBM24-ΔRNP2, the same strategy was used except that pcDNA3-RBM24-ΔRNP2 was used as a template.
To generate pcDNA3-TAp63α expression vector, a DNA fragment were amplified by using TAp63α EST clone as a template with a forward primer, 5′ GGG GAA GCT TGC CAC CAT GAA TTT TGA AAC TTC ACG G 3′, and a reverse primer, 5′ GGG GGA TCC TCA CTC CCC CTC CTC TTT GAT G 3′. The PCR products were cloned into pcDNA3 via HindIII and BamHI sites. To generate pcDNA3-TAp63α-3′UTR expression vector, a DNA fragment containing the full-length p63 3′UTR was amplified by using TAp63α EST clone as a template and then inserted into pcDNA3-TAp63α vector via BamHI and XhoI sites. The PCR primers were a forward primer 5′ GGG GGA TCC GCC TCA CCA TGT GAG CTC TTC C 3′, and a reverse primer, 5′ GGG GCT CGA GCA ATT TCT TAA TTA GTT TTT ATT TAT TTT TTA AAT TTT ATT GCA TGT CCT GGC AAA CAA AAA GAG 3′.
Cell culture and cell line generation
HaCaT, ME180, MIA-PaCa2, MCF10A, HCT116, and MCF7 cells were cultured in DMEM supplemented with 10% fetal bovine serum as previously described (18, 27). MEFs were cultured in DMEM supplemented with 10% fetal bovine serum, 1x nonessential amino acids, and 55 μM beta-mercaptoethanol. HCT116 and MCF7 stable cell lines that can inducibly express RBM24 were generated as described previously (28). Briefly, pcDNA4-RBM24 was transfected into HCT116 and MCF7 parental cells, which express a tetracycline repressor (pcDNA6) (28). The RBM24 expressing cells were selected with zeocin (150 μg/ml) and confirmed by Western blot analysis. To induce RBM24 expression, tetracycline (1 μg/ml) was added to medium for various times.
MEF isolation
RBM38−/−;p53−/− MEFs were isolated as previously described (29). To generate TAp63−/− MEFs, mice heterozygous for TAp63 (14), a gift from Dr Elsa R. Flores’ lab, were bred. MEFs were isolated from 13.5-day-old embryos as previously described (30). All animals are housed at the University of California at Davis CLAS vivarium facility. All animals and use protocols were approved by the University of California at Davis Institution Animal Care and Use Committee.
Western blot analysis
Western blot analysis was performed as previously described (5). Briefly, cells lysates were collected and resuspended with 1x SDS sample buffer. Proteins were then resolved in an 8–12% SDS-PAGE gel and transferred to a nitrocellulose membrane, followed by ECL detection. The level of protein was quantified by densitometry. The data are representative of three independent experiments.
Recombinant protein purification, RNA probe generation, and REMSA
Bacteria BL21 was transformed with a pGEX-5X-1 vector expressing GST-tagged RBM24, ΔRNP1, or ΔRNP2 and positive clones were selected. The recombinant proteins were then purified by glutathione sepharose beads (Amersham Biosciences). RNA probes containing various regions of p63 3′UTR were generated as previously described (18). The p21 probe was generated as previously described (31). REMSA was performed as described previously (18). Briefly, 32P-labeled probes were incubated with recombinant protein in a binding buffer (10 mM HEPES-KOH at pH 7.5, 90 mM potassium acetate, 1.5 mM magnesium acetate, 2.5 mM DTT, 40 U of RNase inhibitor [Ambion]) at 30°C for 30 min. RNA-protein complexes were resolved on a 6% acrylamide gel and radioactive signals were detected by autoradiography.
RNA isolation and RT–PCR analysis
Total RNAs were isolated using TRIzol reagent. cDNA was synthesized using MMLV reverse transcriptase according to the user’s manual. The PCR program used for amplification was (i) 94°C for 5 min, (ii) 94°C for 45 s, (iii) 58°C for 45 s, (iv) 72°C for 1 min, and (v) 72°C for 10 min. From steps 2 to 4, the cycle was repeated 20 times for human and mouse actin or 30 times for RBM24 and P63. To amplify human actin, two pairs of primers were used. The first pair of primers was used for the RT-PCR analysis in Figs. 2A, 2C, 3A, and 4A whereas the second pair of primers were used for all other RT-PCR analysis.
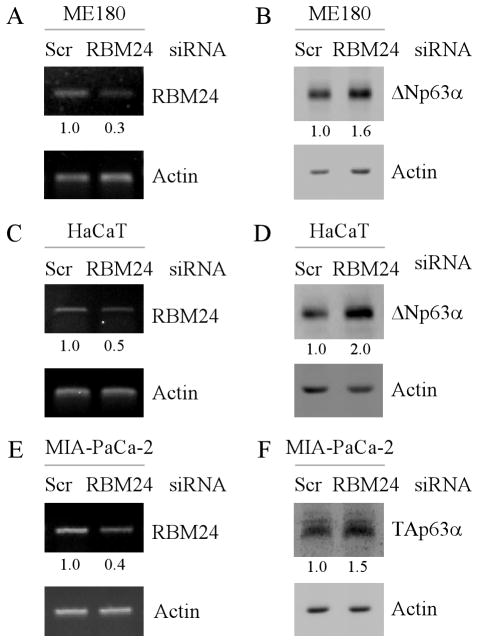
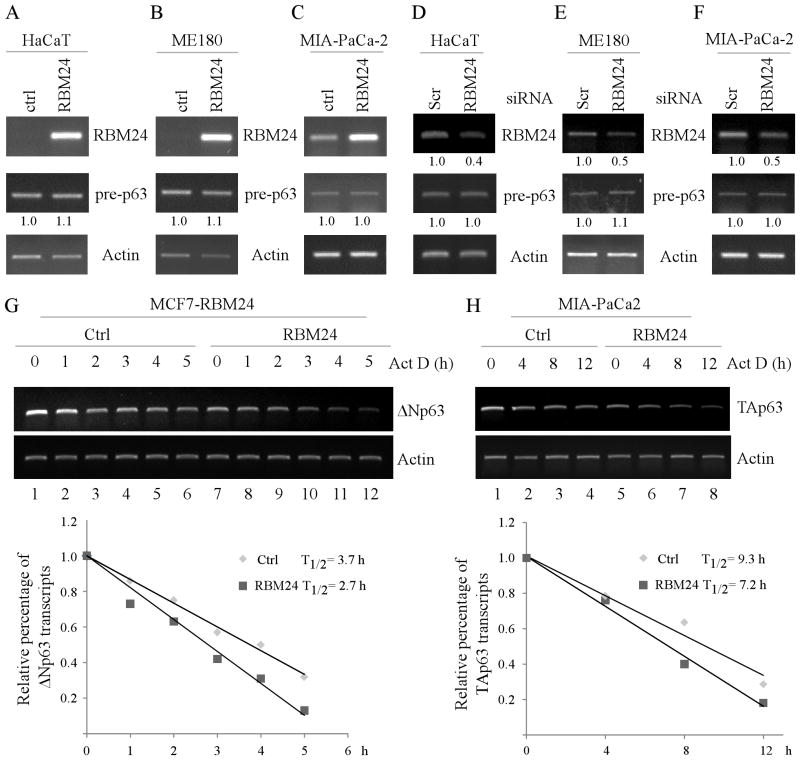
Figure 2. Knockdown of RBM24 increases p63 expression.
(A and C) ME180 (A) and HaCaT (C) cells were transiently transfected with a control or RBM24 siRNA for 72 h, followed by RT-PCR analysis to determine the level of RBM24 and actin transcripts. The level of RBM24 transcript was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane. (B and D) ME180 (B) and HaCaT (D) cells were treated as describe in (A), and the level of ΔNp63α and actin proteins was determined by Western blot analysis. The level of ΔNp63α protein was normalized to that of actin protein and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane. (E) MIA-PaCa2 cells were transiently transfected with a control or RBM24 siRNA for 72 h and the level of RBM24 and actin transcripts was determined by RT-PCR analysis. (F) RBM24 knockdown increases TAp63α expression. MIA-PaCa2 cells were treated as describe in (E), and the level of TAp63α and actin proteins was determined by Western blot analysis. The level of TAp63α protein and transcript were normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane.
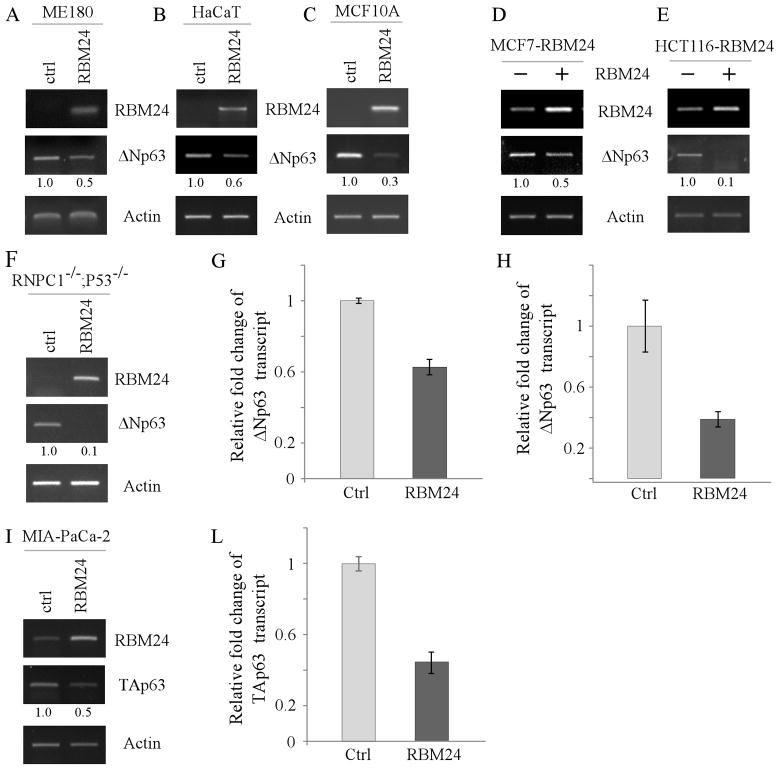
Figure 3. The level of p63 transcript is decreased by ectopic expression of RBM24.
(A–C and F) ME180 cells (A), HaCaT cells (B), MCF10A cells (C), and RBM38−/−;p53−/− MEFs (F) were transiently transfected with a control vector or a vector expressing HA-tagged RBM24 for 48 h. Total RNAs were isolated and subjected to RT-PCR analysis to determine the level of RBM24, ΔNp63, and actin transcripts. The level of ΔNp63 transcript was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane. (DE) HCT116 (D) and MCF7 (E) cells were uninduced or induced to express RBM24 for 48 h. Total RNAs were isolated and subjected to RT-PCR analysis to determine the level of RBM24, ΔNp63, and actin transcripts. (G) The level of ΔNp63 transcript in HaCaT cells, which were transfected with a control vector or Rbm24-expressing vector, was measured by quantitative RT-PCR. The level of actin mRNA was measured as an internal control. (H) The experiment was performed as in (G) except that MCF7 cells, which were uninduced or induced to express RBM24, were used. (I–L) MIA-PaCa2 cells were transiently transfected with a control vector or a vector expressing HA-tagged RBM24 for 48 h. Total RNAs were isolated and subjected to RT-PCR analysis (I) or quantitative RT-PCR (L) to determine the level of RBM24, TAp63, and actin transcripts. The level of TAp63 transcript was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane.
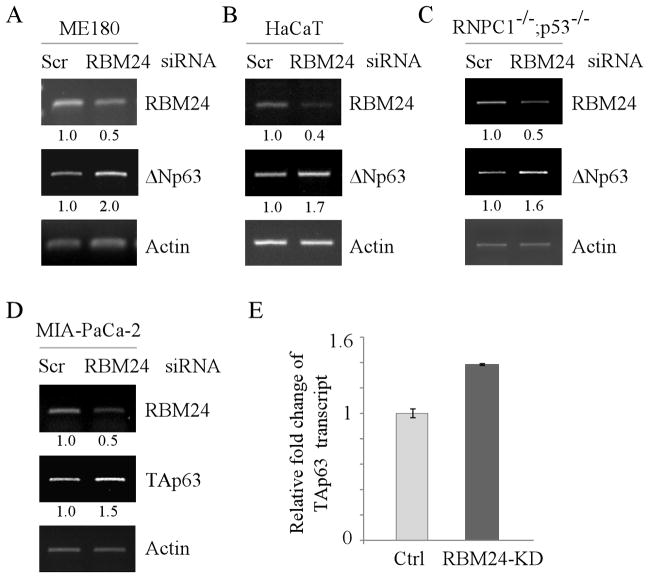
Figure 4. Knockdown of RBM24 increases the level of p63 transcript.
(A–C) ME180 cells (A), HaCaT cells (B), and RBM38−/−;p53−/− MEFs (C) were transiently transfected with a control or RBM24 siRNA for 72 h. Total RNAs were purified and subjected to RT-PCR analysis to determine the level of RBM24, ΔNp63, and actin transcripts. The level of ΔNp63 transcript was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane. (D–E) The level of TAp63 transcript is increased by knockdown of RBM24. MIA-PaCa2 cells were transiently transfected with a control or RBM24 siRNA for 72 h. Total RNAs were purified and subjected to RT-PCR (D) or quantitative RT-PCR (E) to determine the level of RBM24, TAp63, and actin transcripts. The level of TAp63 transcript was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane.
The first pair of primers were forward primer, 5′ CTG AAG TAC CCC ATC GAG CAC GGC A 3′, and reverse primer, 5′ GGA TAG CAC AGC CTG GAT AGC AAC G 3′. The second pair of primers were forward primer, 5′ AGC GCG GCT ACA GCT TCA 3′ and reverse primer, 5′ CGT AGC ACA GCT TCT CCT TAA TGT C 3′. These primers were used for the rest of RT-PCR analysis. The primers to amplify mouse actin were forward primer, 5′ CCC ATC TAC GAG GGC TAT 3′, and reverse primer, 5′ AGA AGG AAG GCT GGA AAA 3′. The primers to amplify human RBM24 were forward primer, 5′ AGC CTG CGC AAG TAC TTC G 3′, and reverse primer, 5′ CAG GCC CTT TCG GCA G 3′. The primers to amplify mouse RBM24 were forward primer, 5′ ACC CAG AAG GAC ACG ACG TA 3′, and reverse primer, 5′ TCG ATG ATG GGG TTG GGA T 3′. The primers to amplify human ΔNp63 were forward primer, 5′ TAC CTG GAA AAC AAT GCC 3′, and reverse primer, 5′ ACT GCT GGA AGG ACA CG 3′. The primers to amplify human TAp63 were forward primer, 5′ AGC CCA TTG ACT TGA ACT T 3′, and reverse primer, 5′ GGA CTG GTG GAC GAG GA 3′. The primers to amplify mouse ΔNp63 were forward primer, 5′ TAC CTG GAA AAC AAT GCC CA 3′, and reverse primer, 5′ GCT GGA AGG ACA CAT CGA A 3′. To measure the precursor mRNA, total RNAs were isolated using TRIzol reagent and then treated with Danes I to remove genomic DNA prior to cDNA synthesis. The primers for human p63 pre-mRNA were forward primer, 5′ CTT GTT GTT AAC AAC AGC ATG AG 3′, and reverse primer, 5′ AGA AAG CCT GTG CCA CTC AC 3′.
Quantitative PCR (qPCR) was performed using 2X qPCR SYBR Green Mix (ABgene, Epsom, UK) with 5 μM primers. Reactions were run on a realplex (Eppendorf, Germany) using a two-step cycling program: 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, 68°C for 30 s. A melting curve (57–95°C) was generated at the end of each run to verify the specificity. The primers for human actin were forward primer, 5′TCC ATC ATG AAG TGT GAC GT 3′, and reverse primer, 5′ TGA TCC ACA TCT GCT GGA AG 3′. The primers for human TAp63 were forward primer, 5′ AGC CCA TTG ACT TGA ACT T 3′, and reverse primer, 5′ GGA CTG GTG GAC GAG GA 3′. The primers to amplify human ΔNp63 were forward primer, 5′ TAC CTG GAA AAC AAT GCC 3′, and reverse primer, 5′ ACT GCT GGA AGG ACA CG 3′.
RNA-Chip analysis
RNA-Chip analysis was performed as previously described (32). Briefly, cells (2 × 107) were lysed with an immunoprecipitation buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes, 1 mM DTT, and 0.5% NP-40), and then incubated with 2 μg of anti-RBM24 or rabbit IgG at 4 °C overnight. The RNA-protein immunocomplexes were brought down by protein A/G beads, followed by RT-PCR analysis.
Statistics
All experiments were performed at least three times. Numerical data were expressed as mean ± standard deviation (SDs). Two group comparisons were analyzed by two-sided Student t test. P values were calculated, and a P of <0.05 was considered significant.
Results
Ectopic expression of RBM24 suppresses, whereas knockdown of RBM24 increases, p63 expression
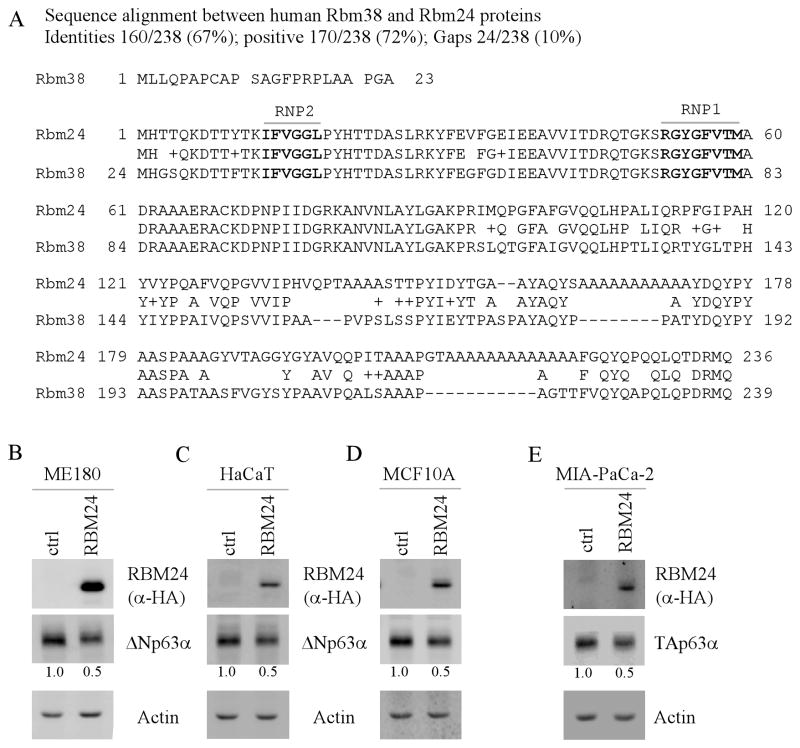
In an effort to understand the underlying mechanisms by which p63 expression is controlled, we showed previously that RBM38, also called RNPC1, is able to destabilize p63 transcript and plays a critical role in p63-mediated keratinocyte differentiation (18). Interestingly, a search of gene database revealed that RBM38 has a paralogue, named RBM24, which shares a high degree of sequence similarity with that of RBM38 (Fig. 1A). The RBM24 gene encodes 236 aa and is located on chromosome 6. Structure analysis shows that RBM24 contains one RNA-binding domain, which is composed of two submotifs, RNP1and RNP2. Most remarkably, the RNA-binding domain in RBM24 is identical to the one in RBM38 (Fig. 1A). Therefore, it is plausible that RBM24 may regulate p63 expression.
Figure 1. Ectopic expression of RBM24 suppresses p63 expression.
(A) Sequence similarity between human RBM38 and RBM24. The RNP1 and RNP2 submotifs are shown in boxes. (B–D) Ectopic expression of RBM24 inhibits ΔNp63α expression. ME180 (B), HaCaT (C), and MCF10A (D) cells were transiently transfected with a control vector or a vector expressing HA-tagged RBM24 for 48 h, and the level of HA-tagged RBM24, ΔNp63α, and actin was determined by Western blot analysis. The level of ΔNp63α protein was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane. (E) RBM24 inhibits TAp63α expression. MIA-PaCa2 cells was transiently transfected with a control vector or a vector expressing HA-tagged RBM24 for 48 h, followed by Western blot analysis to determine the level of HA-tagged RBM24, TAp63α, and actin. The level of TAp63α protein was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane.
To determine whether RBM24 regulates p63 expression, a control vector or a vector expressing HA-tagged RBM24 was transiently transfected into ME180 cells. The level of RBM24 was detectable upon transfection (Fig. 1B, RBM24 panel). Interestingly, we found that the ΔNp63α protein was markedly inhibited by RBM24 (Fig. 1B, ΔNp63α panel). Similarly, we found that RBM24 inhibited ΔNp63α expression in HaCaT and MCF10A cells (Fig. 1C–D, ΔNp63α panels). Furthermore, we tested whether RBM24 has an effect on TAp63 expression by using MIA-PaCa2 cells, in which TAp63α is highly expressed (27). We found that the level of TAp63α protein was markedly decreased by ectopic expression of RBM24 (Fig. 1E, TAp63α panel). Together, these data suggest that p63 expression is repressed by ectopic expression of RBM24.
To determine whether endogenous RBM24 regulates p63 expression. ME180 and HaCaT cells were transiently transfected with a control siRNA or a siRNA against RBM24. Again, we found that the level of RBM24 transcript was markedly reduced by RBM24, but not by control, siRNA (Fig. 2A and 2C, RBM24 panels). Importantly, we found that the level of ΔNp63α proteins was increased by RBM24 knockdown (Fig. 2B and 2D, ΔNp63α panels). Furthermore, we tested whether TAp63α expression is regulated by endogenous RBM24 and found to be increased upon RBM24 knockdown in MIA-PaCa2 cells (Fig. 2E–F). Together, these data suggest that knockdown of RBM24 increases p63 expression.
Ectopic expression of RBM24 decreases, whereas knockdown of RBM24 increases, the level of p63 transcript
RBPs are known to posttranscriptionally regulate their targets mainly through mRNA stability or protein translation. Thus, to explore how RBM24 regulates p63 expression, the level of p63 transcript was measured in ME180 cells transiently transfected with a control or RBM24 expression vector. We found that upon transient expression of RBM24, the level of ΔNp63 transcript was decreased in ME180 cells (Fig. 3A, ΔNp63 panel). Similarly, ectopic expression of RBM24 was able to reduce the level of ΔNp63 transcript in HaCaT and MCF10A cells (Fig. 3B–C, ΔNp63 panels). To verify this, HCT116 and MCF7 cells that can inducibly express RBM24 were used. We found that the level of ΔNp63 transcript was decreased upon RBM24 induction (Fig. 3D–E, ΔNp63 panels). Next, we determined whether RBM24 regulates p63 expression in the absence of p53 and RBM38. To address this, RBM38−/−;p53−/− MEFs were transiently transfected with a control or RBM24 expression vector and the level of p63 transcript was measured. We found that RBM24 was able to significantly decrease the level of p63 transcript in the absence of p53 and RBM38 (Fig. 3F, ΔNp63 panel). Consistently, qPCR analysis showed that the level of ΔNp63 transcript was decreased by ectopic expression of Rbm24 in HaCaT and MCF7 cells (Fig. 3G–H). Furthermore, we determined whether RBM24 regulates TAp63 transcript by RT-PCR and qPCR. We showed that the level of TAp63 transcript was markedly decreased by ectopic expression of RBM24 in MIA-PaCa2 cells (Fig. 3I–L). Together, these data suggest that ectopic expression of RBM24 decreases the level of p63 transcript.
Next, to determine whether endogenous RBM24 regulates p63 transcript, ME180 and HaCaT cells were transiently transfected with a control siRNA or a siRNA against RBM24. We found that the level of ΔNp63 transcript was increased by RBM24 knockdown (Fig. 4A–B, ΔNp63 panels). Likewise, knockdown of RBM24 resulted in increased levels of ΔNp63 transcript in RBM38−/−;p53−/− MEFs (Fig. 4C, ΔNp63 panel). Furthermore, we found that the level of TAp63 transcript in MIA-PaCa2 cells was increased by RBM24 knockdown (Fig. 4D–E). Together, these data suggest that the level of p63 transcript is increased by RBM24 knockdown.
RBM24 destabilizes p63 transcript
To investigate the underlying mechanism by which RBM24 regulates p63 expression, we first determined whether RBM24 regulates p63 transcription. Specifically, the level of p63 pre-mRNA was measured in HaCaT cells, transiently transfected with a control or RBM24 expression vector. We found that overexpression of RBM24 had no effect on the level of p63 pre-mRNA in HaCaT cells (Fig. 5A, pre-p63 panel). Consistent with this, the level of p63 pre-mRNA was not altered by ectopic expression of RBM24 in ME180 and MIA-PaCa2 cells (Fig. 5B–C, pre-p63 panels). Similarly, RBM24 knockdown had no effect on p63 pre-mRNA in HaCaT, ME180, and MIA-PaCa2 cells (Fig. 5D–F, pre-p63 panels). These results suggest that p63 is regulated by RBM24 via posttranscriptional mechanisms, such as mRNA stability. Thus, the half-life of ΔN and TA p63 transcripts was measured in cells treated with actinomycin D, which inhibits de-novo RNA synthesis. Specifically, MCF7 cells were uninduced or induced to express RBM24 for 48 h, followed by actinomycin D treatment for various times. Similarly, MIA-PaCA2 cells were transfected with a control vector or a vector expressing RBM24, followed by actinomycin D treatment for various times. The level of ΔN and TA p63 transcripts was determined by RT-PCR analysis and the relative half-life of ΔN and TA p63 transcripts was calculated. We showed that the half-life of ΔNp63 mRNA was deceased from 3.7 h in the control cells to 2.7 h in cells with RBM24 expression (Fig. 5G), whereas the half-life of TAp63 mRNA was deceased from 9.3 h in the control cells to 7.2 h in cells with RBM24 expression (Fig. 5H). Together, these data suggest that RBM24 shortens the half-life of p63 mRNA.
Figure 5. RBM24 destabilizes p63 transcript.
(A–C) Ectopic expression of RBM24 has no effect on the level of p63 pre-mRNA. HaCaT (A), ME180 (B), and MIA-PaCa2 (C) cells were transiently transfected with a control vector or a vector expressing RBM24 for 48 h. Total RNAs were isolated and subjected to RT-PCR analysis to determine the level of p63 pre-mRNA, RBM24, and actin mRNA. The level of p63 pre-mRNA was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane. (D–F) Knockdown of RBM24 has no effect on the level of p63 pre-mRNA. HaCaT (D), ME180 (E) and MIA-PaCa2 (F) cells were transiently transfected with a control or RBM24 siRNA for 72 h. Total RNAs were purified and subjected to RT-PCR analysis to determine the level of RBM24 and actin mRNA as well as p63 pre-mRNA. The level of p63 pre-mRNA or RBM24 mRNA was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane. (G) RBM24 shortens the half-life of ΔNp63 transcript. Upper panel: MCF7 cells were uninduced or induced to express RBM24 for 48 h, followed by treatment with 7 μg/ml actinomycin D for various times. Total RNAs were isolated and then subjected to RT-PCR analysis to determine the level of ΔNp63 and actin transcripts. Lower panel: The level of ΔNp63 transcript was normalized to that of actin and plotted along with a time course to calculate the relative half-life of ΔNp63 mRNA. (H) RBM24 shortens the half-life of TAp63 transcript. Upper panel: MIA-PaCa2 cells were transiently transfected with a control vector or RBM24-expressing vector for 48 h, followed by treatment with 7 μg/ml actinomycin D for various times. Total RNAs were isolated and then subjected to RT-PCR analysis to determine the level of TAp63 and actin transcripts. Lower panel: The level of TAp63 transcript was normalized to that of actin and plotted along with a time course to calculate the relative half-life of TAp63 mRNA.
RBM24 binds to multiple regions in the 3′UTR of p63 transcript
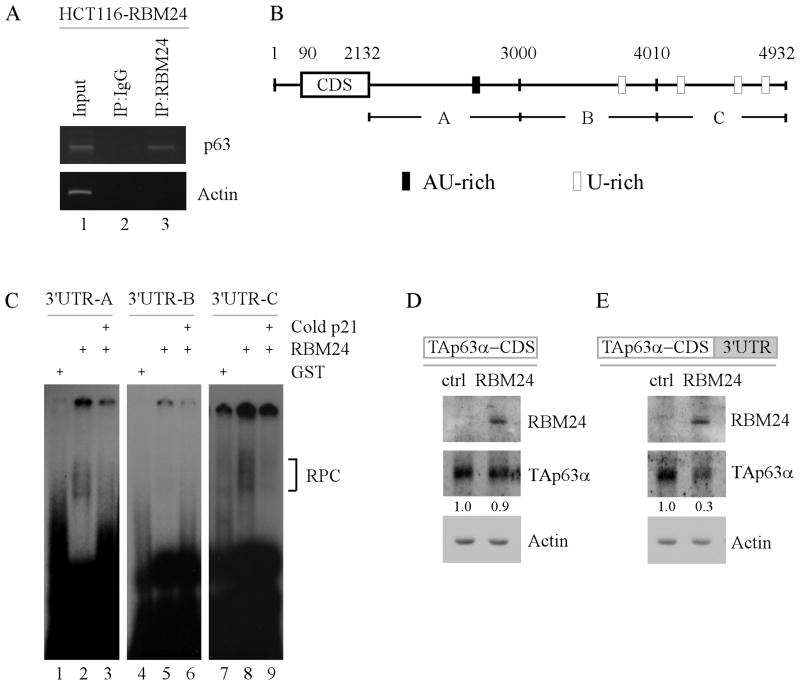
To further decipher the underlying mechanism by which RBM24 destabilizes p63 mRNA, we determined whether RBM24 associates with p63 transcript in vivo by performing an RNA-Chip analysis. We found that p63 transcript was present in RBM24, but not control IgG, immunoprecipitates (Fig. 6A, compare lane 2 with 3). As a control, RBM24 was unable bind to actin mRNA (Fig. 6A, lane 3). Next, the binding site(s) of RBM24 in p63 transcript was mapped by performing RNA electrophoretic mobility assay (REMSA). Specifically, radiolabeled probes (A–C), spanning the entire p63 3′ UTR (Fig. 6B), were incubated with recombinant GST or GST-tagged RBM24 protein, followed by electrophoresis. We found that the recombinant GST-tagged RBM24, but not GST protein, formed a complex with probes A and C, but not probe B (Fig. 6C, compare lanes 1, 4 and 7 with 2, 5 and 8, respectively). Importantly, these RNA-RBM24 complexes were further disrupted by cold probe derived from p21 3′UTR (Fig. 6C, lanes 3, 6 and 9), which is known to carry an AU-rich element.
Figure 6. RBM24 binds to multiple regions in the 3′UTR of p63 transcript.
(A) RBM24 associates with p63 transcript in vivo. RBM24-expressing HCT116 cell extracts were immunoprecipitated with a control IgG or RBM24 antibody to bring down the protein-RNA complex. Total RNAs were isolated and subjected to RT-PCR analysis to measure the level of p63 and actin transcripts. Five percent of cell lysate was used as input. (B) Schematic representation of p63 transcript and the location of probes used for REMSA. The AU- or U-rich elements were shown in shaded boxes. (C) RBM24 binds to multiple regions in p63 3′UTR. REMSA was performed by mixing recombinant GST or GST-fused RBM24 protein with 32P-labeled probe A, B, or C. The bracket indicates RNA-protein complexes. For competition assay, unlabeled p21 probe was added to the reaction mix prior to incubation with the 32P-labeled probe A, B, or C. (D-E) The p63 3′UTR is required for RBM24 to inhibit TAp63α expression. TAp63−/− MEF cells were co-transfected with a control or RBM24-expressing vector along with a TAp63α expression vector that contains the coding region alone (D) or in combination with a full-length p63 3′UTR (E). Cell lysates were collected and the level of RBM24, TAp63α, and actin proteins was determined by Western blot analysis. The relative level of TAp63α was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane.
To further verify that the p63 3′UTR is required for RBM24 to inhibit p63 expression, we generated a TAp63α expression vector that contains TAp63α coding sequence alone or together with a full length p63 3′UTR. Next, these TAp63α expression vectors were transiently transfected into TAp63−/− MEFs along with a control or RBM24 expression vector. We found that RBM24 had no effect on TAp63α expression from an expression vector that only contains TAp63α coding sequence (Fig. 6D). By contrast, TAp63α expression was significantly inhibited by RBM24 when the TAp63α expression vector contains a full length p63 3′UTR (Fig. 6E). Together, these results suggest that the p63 3′UTR is necessary for RBM24 to repress p63 expression.
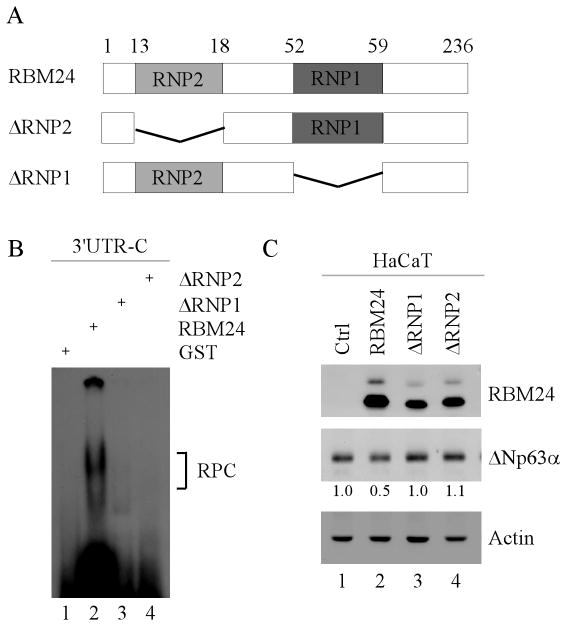
The RNA-binding domain is required for RBM24 to inhibit p63 expression
The RNA-binding domain in RBM24 is composed of two RNA recognition submotifs, RNP1 and RNP2 (Fig. 7A). Thus, to determine whether the RNA-binding domain is required for RBM24 to inhibit p63 expression, we generated two RBM24 deletion mutants, which lack RNP1(ΔRNP1) and RNP2 (ΔRNP2), respectively (Fig. 7A). Next, the ability of ΔRNP1 or ΔRNP2 to bind to p63 3′UTR was determined by REMSA. We found that neither ΔRNP1 nor ΔRNP2 were capable of binding to the p63 3′UTR (Fig. 7B). Furthermore, we found that unlike wild-type RBM24, neither ΔRNP1 nor ΔRNP2 were capable of inhibiting ΔNp63α expression in HaCaT cells (Fig. 7C). Taken together, these data suggest that the RNA-binding domain is required for RBM24 to bind p63 transcript and consequently, inhibits p63 expression.
Figure 7. The RNA-binding domain is required for RBM24 to inhibit p63 expression.
(A) Schematic representation of wild-type RBM24, ΔRNP1, and ΔRNP2. (B) Neither ΔRNP1 nor ΔRNP2 are able to bind to the p63 3′UTR. REMSA was performed by incubating 32P-labeled probe C with recombinant GST or GST-tagged RBM24, ΔRNP1, or ΔRNP2. The bracket indicates the RNA-protein complexes. (C) The RNA-binding domain is required for RBM24 to inhibit p63 expression. HaCaT cells were transiently transfected with a control vector or a vector expressing RBM24, ΔRNP1, or ΔRNP2 for 48 h. The relative level of ΔNp63α was normalized to that of actin and arbitrary set as 1.0 in control cells. The relative fold changes were shown below each lane.
Discussion
Although regulation of p63 expression has been extensively studied, very little is known about the posttranscriptional regulation of p63 by either RBPs or microRNAs. RBPs are key regulators in post-transcriptional control of RNAs and altered expression of RBPs is implicated in several kinds of human diseases including cancer (33–36). In the current study, we identified RNA-binding protein RBM24 as a novel regulator of p63 via mRNA stability. Specifically, we showed that the levels of p63 protein and transcript are decreased by ectopic expression of RBM24. Consistent with this, knockdown of endogenous RBM24 increases the levels of p63 transcript and protein. Moreover, we showed that RBM24 inhibited p63 expression by reducing the half-life of p63 transcript. Consistently, RBM24 is able to bind to multiple regions in the 3′UTR of p63 transcript and subsequently, destabilize p63 transcript. Furthermore, we showed that both the 3′UTR in p63 transcript and the RNA-binding domain in RBM24 are required for regulating p63 expression.
The biological function of RBM24 and its downstream targets remain largely unknown. To date, RBM24 is suggested to be involved in skeletal muscle differentiation by regulating MyoD (37) and myogenin (38). More recently, RBM24 is found to be involved in sarcomeric assembly and cardiac contractility (39, 40), suggesting a critical role of RBM24 in heart development. However, as a RBM24 paralogue, RBM38 was found to be critical in tumorigenesis (41, 42). Therefore, it is likely that RBM24 and RBM38 have their own distinct functions although both proteins share high degree of sequence similarity. In our study, we found that p63 is a novel downstream target of RBM24. Although the biological significance of this regulation remains unknown, it is likely that RBM24 participates in the p63 network by regulating p63 expression via various pathways. For example, RBM24 may play a role in TAp63-mediated tumor suppression or in ΔNp63-mediated epidermal development. Moreover, RBM38 is a target of the p53 family and forms a feedback or feed-forward regulatory loop with the p53 family proteins (18, 29, 43, 44). As a RBM38 closely related protein, RBM24 may participate in the p53 family-RBM38 autoregulatory loop, including regulation of p53 and p73 expression. Thus, future studies to address these questions will help us better understand the biological function of RBM24.
We have previously reported that RBM38 is able to regulate p63 mRNA stability by binding to the 3′UTR of p63 transcript (18). In the current study, we found that like RBM38, RBM24 was able to bind to the p63 3′UTR and destabilize p63 transcript. Of note, the binding sites of RBM24 to p63 transcripts are located in the same regions as that of RBM38 [Fig. 6 and (18)]. We postulate that the similar regulation to p63 by RBM38 and RBM24 is due to their high degree of sequence similarity (Fig. 1A). Interestingly, RBM24 can regulate p63 expression in the absence of RBM38 (Figs. 3F and 4C), suggesting that the function of these two proteins may not be redundant. Nevertheless, it still remains to be elucidated whether RBM24 and RBM38 cooperatively or antagonistically regulate p63 mRNA stability. First, RBM24 and RBM38 may compete to bind to p63 transcript. Second, it is likely that RBM24 enhances the RNA-binding activity of RBM38 to p63 transcript and vice versa, resulting in destabilized p63 transcript. Third, due to their high degree of sequence similarity, RBM24 and RBM38 may form a heterodimer and negatively regulate p63 mRNA stability. These issues need to be addressed in the future studies.
Of note, several micro RNAs, including miR-130b (21), miR-302 (20), and miR-203 (22), are found to posttranscriptionally regulate p63 expression. In addition, it is now well accepted that RBPs work closely with microRNA to either positively or negatively modulate their target expression. In support of this idea, RBM38 was found to modulate the ability of several microRNAs to bind to their targets (45). Therefore, it will be interesting to determine whether RBM24 alone or together with RBM38 is able to modulate the ability of microRNAs to bind to p63 transcripts and subsequently, affect p63 activity. By addressing these questions, it will help us further understand how posttranscriptional regulatory mechanisms contribute to p63 expression and consequently, affect the p63 network.
Acknowledgments
We thank Dr. Elsa R. Flores for providing the TAp63 knockout mice.
Financial Support: This work is supported in part by a National Institutes of Health grant (CA102188).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 2.Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61:822–42. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangiulli M, Valletti A, Caratozzolo MF, Tullo A, Sbisa E, Pesole G, et al. Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res. 2009;37:6092–104. doi: 10.1093/nar/gkp674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanbokhoven H, Melino G, Candi E, Declercq W. p63, a story of mice and men. The Journal of investigative dermatology. 2011;131:1196–207. doi: 10.1038/jid.2011.84. [DOI] [PubMed] [Google Scholar]
- 5.Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20:3193–205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- 6.Helton ES, Zhu J, Chen X. The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J Biol Chem. 2006;281:2533–42. doi: 10.1074/jbc.M507964200. [DOI] [PubMed] [Google Scholar]
- 7.Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M, et al. TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle. 2007;6:274–85. doi: 10.4161/cc.6.3.3797. [DOI] [PubMed] [Google Scholar]
- 8.Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci U S A. 2000;97:5462–7. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–90. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 11.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 12.Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, et al. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–82. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su X, Gi YJ, Chakravarti D, Chan IL, Zhang A, Xia X, et al. TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell metabolism. 2012;16:511–25. doi: 10.1016/j.cmet.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell stem cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmes DC, Bresnick E, Lubin EA, Watson JK, Heim KE, Curtin JC, et al. Positive and negative regulation of deltaN-p63 promoter activity by p53 and deltaN-p63-alpha contributes to differential regulation of p53 target genes. Oncogene. 2003;22:7607–16. doi: 10.1038/sj.onc.1207129. [DOI] [PubMed] [Google Scholar]
- 16.Qian Y, Jung YS, Chen X. DeltaNp63, a target of DEC1 and histone deacetylase 2, modulates the efficacy of histone deacetylase inhibitors in growth suppression and keratinocyte differentiation. The Journal of biological chemistry. 2011;286:12033–41. doi: 10.1074/jbc.M110.207241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herfs M, Hubert P, Suarez-Carmona M, Reschner A, Saussez S, Berx G, et al. Regulation of p63 isoforms by snail and slug transcription factors in human squamous cell carcinoma. The American journal of pathology. 2010;176:1941–9. doi: 10.2353/ajpath.2010.090804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Jun Cho S, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc Natl Acad Sci U S A. 2010;107:9614–9. doi: 10.1073/pnas.0912594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan W, Zhang Y, Zhang J, Cho SJ, Chen X. HuR is necessary for mammary epithelial cell proliferation and polarity at least in part via DeltaNp63. PLoS One. 2012;7:e45336. doi: 10.1371/journal.pone.0045336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheel AH, Beyer U, Agami R, Dobbelstein M. Immunofluorescence-based screening identifies germ cell associated microRNA 302 as an antagonist to p63 expression. Cell Cycle. 2009;8:1426–32. doi: 10.4161/cc.8.9.8324. [DOI] [PubMed] [Google Scholar]
- 21.Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A. 2012;109:1133–8. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–95. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 23.Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci U S A. 2006;103:12753–8. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung YS, Qian Y, Yan W, Chen X. Pirh2 E3 ubiquitin ligase modulates keratinocyte differentiation through p63. J Invest Dermatol. 2013;133:1178–87. doi: 10.1038/jid.2012.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008;15:1941–51. doi: 10.1038/cdd.2008.134. [DOI] [PubMed] [Google Scholar]
- 26.Gallegos JR, Litersky J, Lee H, Sun Y, Nakayama K, Lu H. SCF TrCP1 activates and ubiquitylates TAp63gamma. The Journal of biological chemistry. 2008;283:66–75. doi: 10.1074/jbc.M704686200. [DOI] [PubMed] [Google Scholar]
- 27.Yan W, Chen X, Zhang Y, Zhang J, Jung YS. Arsenic suppresses cell survival via Pirh2-mediated proteasomal degradation of DeltaNp63 protein. The Journal of biological chemistry. 2013;288:2907–13. doi: 10.1074/jbc.M112.428607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harms KL, Chen X. Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer research. 2007;67:3145–52. doi: 10.1158/0008-5472.CAN-06-4397. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Cho SJ, Shu L, Yan W, Guerrero T, Kent M, et al. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 2011;25:1528–43. doi: 10.1101/gad.2069311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scoumanne A, Cho SJ, Zhang J, Chen X. The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic acids research. 2011;39:213–24. doi: 10.1093/nar/gkq778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 2010;38:2256–67. doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, et al. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–80. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 33.Heinonen M, Bono P, Narko K, Chang SH, Lundin J, Joensuu H, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer research. 2005;65:2157–61. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 34.Denkert C, Koch I, von Keyserlingk N, Noske A, Niesporek S, Dietel M, et al. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2006;19:1261–9. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- 35.Chen AJ, Paik JH, Zhang H, Shukla SA, Mortensen R, Hu J, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes & development. 2012;26:1459–72. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang XY, Penalva LO, Yuan H, Linnoila RI, Lu J, Okano H, et al. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Molecular cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li HY, Bourdelas A, Carron C, Shi DL. The RNA-binding protein Seb4/RBM24 is a direct target of MyoD and is required for myogenesis during Xenopus early development. Mechanisms of development. 2010;127:281–91. doi: 10.1016/j.mod.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Jin D, Hidaka K, Shirai M, Morisaki T. RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation. Genes to cells : devoted to molecular & cellular mechanisms. 2010;15:1158–67. doi: 10.1111/j.1365-2443.2010.01446.x. [DOI] [PubMed] [Google Scholar]
- 39.Poon KL, Tan KT, Wei YY, Ng CP, Colman A, Korzh V, et al. RNA-binding protein RBM24 is required for sarcomere assembly and heart contractility. Cardiovascular research. 2012;94:418–27. doi: 10.1093/cvr/cvs095. [DOI] [PubMed] [Google Scholar]
- 40.Maragh S, Miller RA, Bessling SL, McGaughey DM, Wessels MW, de Graaf B, et al. Identification of RNA binding motif proteins essential for cardiovascular development. BMC developmental biology. 2011;11:62. doi: 10.1186/1471-213X-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–19. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- 42.Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 43.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–72. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan W, Zhang J, Zhang Y, Jung YS, Chen X. p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability. Molecular and cellular biology. 2012;32:2336–48. doi: 10.1128/MCB.00215-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nature communications. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]