Abstract
We report the separation of several quadruplex species formed by ten promoter sequences by Size Exclusion Chromatography (SEC). Modification at the 5’ or 3’ ends or in loop regions of quadruplex forming sequences has become the standard technique for dealing with quadruplex polymorphism. However, conformations produced employing this method or by other means of artificially shifting the equilibrium may not represent the species that are present in vivo. This method enables an unperturbed view of the structural polymorphism inherent to quadruplex formation. Separation via SEC facilitates studies on quadruplex structure and biophysical properties without the need for sequence modification.
G-quadruplexes formed from a particular DNA sequence in solution are usually polymorphic1,2. A common technique for simplifying the ensemble of species to enable structural analysis is to make sequence modifications3,4. The presumption is that such alterations merely perturb the finely balanced equilibrium, which is in fact untested, and therefore structures produced in this manner may not represent the species that are present initially or in vivo. We have used Size Exclusion Chromatography (SEC) to analyze and separate the ensemble of quadruplex species formed by several oncogene promoter sequences. Separation via SEC facilitates studies on quadruplex structure without sequence modification or structural perturbation while potentially accessing a greater quadruplex folding space.
Guanine-rich DNA with at least four runs of two guanines can form three-dimensional structures called G-quadruplexes in the presence of certain monovalent cations such as Na+, K+, or NH4+. G-quadruplexes are made up of stacks of two or more square planer arrays of four guanines (a G-quartet) comprised of Hoogsten hydrogen bonds with coordination of a cation to the O6 of the guanines5. Although most often associated with telomeres, potential quadruplex-forming sequences have been found throughout the genome6 including in the promoter regions of many proto-oncogenes such as c-myc7, c-kit8,9, bcl-210, VEGF11, and HIF-1α12, and in the 5’-untranslated regions of mRNA of proto-oncogenes13. Quadruplex DNA is of interest because of possible roles in biological regulation14, bio/nanotechnology applications15, and as potential therapeutic drug targets15,16 or as therapeutic agents17,18 themselves.
Polymorphism is inherent in quadruplex formation for most quadruplex-forming sequences. When factors such as strand orientation, loop type and arrangement, glycosyl torsion angles, intramolecular versus intermolecular formation, ion type and concentration, DNA concentration, the presence of organic solvents and various biological molecules, and annealing profile are considered, even the relatively simple human telomeric sequence d(GGGTTA)4 can theoretically fold into more than 200 intramolecular conformations1,19,20. This intrinsic polymorphism is an issue for those who wish to investigate quadruplex structure or the thermodynamics of quadruplex formation. Structural and biophysical studies of quadruplex DNA have been hindered by our inability to isolate and study the individual quadruplex species that may occur in solution. Low resolution methods such as circular dichroism, UV-vis spectrophotometry, analytical ultracentrifugation, gel electrophoresis, or calorimetry may not be able to differentiate among conformations with similar physical properties and high resolution techniques such as NMR spectroscopy are limited for examining these complex mixtures2.
Polymorphism has been generally dealt with by altering the sequence of interest to produce an enriched configuration for study. This usually requires either adding or deleting bases at the 5’ or 3’ ends of the sequence or lengthening, shortening, or eliminating guanines from putative loop regions to ensure that a single fold is enriched3,4. Substitution of 8-aminoguanine promotes formation of tetramolecular parallel quadruplexes such as those formed by TG4T21, incorporation of 8-methylguanine or 8-bromoguanine is known to produce quadruplex structures with a syn glycosidic configuration22–24, while use of O6-methylguanine, inosine, or 6-thioguanine has been shown to destabilize quadruplex formation25–28. Modification of the sugar phosphate backbone by insertion of 5’-5’ or 3’-3’ polarity inversion has also been shown to have a remarkable effect on quadruplex formation and stability29. RNA and LNA force adoption of a syn glycosidic guanosine conformation30–33.
Reduction of quadruplex polymorphism is possible using methods other than sequence modification. Solvent conditions can greatly affect the range of configurations formed. Choice of and concentration of monovalent cation, primarily K+ or Na+, or inclusion of divalent cations, such as Mn2+, Co2+, Ni2+, Mg2+, Pb2+, or Sr2+, can stabilize or destabilize G-quadruplex structures. Potassium concentration has been shown to be essential in selecting which quadruplex conformation is formed by the human telomere sequence34,35, and inclusion of divalent cations has been shown to induce a transition from antiparallel to parallel G-quadruplex structure for the G4T4G4 sequence36,37. Finally, addition of organic or biological molecules such as PEG, acetonitrile, proteins, or polysaccharides can contribute greatly to quadruplex formation and stability38,39.
In most cases a putative quadruplex forming sequence undergoes several modifications and/or truncations before producing a supposedly single species in solution. For example, the bcl-2 promoter parent thirty nine base sequence has six runs of three or more guanines and was truncated to a twenty three base sequence centred on the four central sets of guanine. This twenty three base sequence was chosen from a set of three possible sequences10,40,41. The selection process included the use of CD data which has been shown to be unreliable for unambiguous determination of structural details of quadruplex configuration42. The sequence was further modified by substitution of thymine for two guanines, thereby forcing a loop, to produce a final sequence for study somewhat removed from the original native sequence potential folding topology. This is not unique and is a common practice as further evidenced by the sequences used to produce the solution structures of c-kit and c-myc. Production of a c-kit sequence suitable for solution structure determination took a series of five sequence substitutions for the 2KJ2 structure and a series of twelve sequence substitutions for the 2O3M structure. In the case of c-myc, the parent twenty seven base sequence containing six consecutive runs of guanines, five of which are comprised of three or more guanines, was shown to form a complex mixture of quadruplexes by NMR43. The efforts to reduce and simplify the spectra for structural study produced three different structures from two different truncated sequences selected from different areas of the parent sequence and utilized two different thymine/guanine substitutions7,43. Another difficulty is the reproducibility of reported data as these sequences can be exquisitely sensitive to solvation and annealing conditions. The substitution of two thymines in the sequence used to produce the c-myc 1XAV solution structure was stated as required as the truncated sequence which produced a suitable 1D 1H NMR spectrum for study by one research group43 produced a complex spectra when examined by a second research group7.
While these methods can yield a less complex ensemble of species and have been used to produce many NMR and crystal structures, the relevance of these results is uncertain as they represent only a small number of the species possible in solution. If the array of quadruplex conformations is at or near equilibrium then sequence modification as a method for perturbing this equilibrium could lead to unreliable and unpredictable results. Therefore, the results from such modifications do not necessarily reflect the diversity of species actually present in solution for the parent sequence, and the modified sequences likely represent a further removal from conditions found in vivo1.
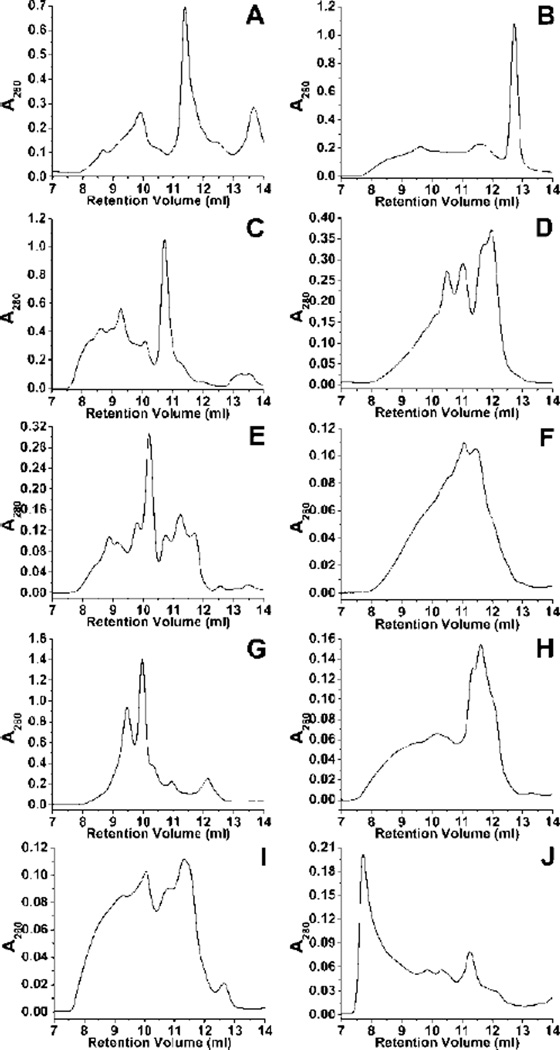
We report the separation profile of several quadruplex species formed by ten proto-oncogene promoter sequences by Size Exclusion Chromatography. The set of sequences was chosen because they have been previously studied and have either a deposited structure or a reported topology (Table 1). Each of the chosen sequences showed high polymorphism with variable degrees of separation (Figure 1). These results agree with AUC data for each of the sequences (Supplementary Figure S1) which indicates that each promoter sequence yields a mixture of conformations. It is clear that the modification of the parent sequence changes the quadruplex ensemble. This is consistent with previous reports for the human telomere3 and is dramatically shown for the two c-kit promoter studies that used significantly different sequences (Table 1) and obtained completely different structures8,9. The SEC method shows that for these two sequences the distribution of species is indeed changed (Figure 1A,B). More subtle sequence modifications, such as the inclusion of flanking bases as in the case of the HIF-1α promoter (Table 1), also have significant perturbation of the quadruplex populations as seen by SEC (Figure 1C,D). The remainder of the oncogene promoter sequences in Table 1 display significant polymorphism by SEC (Figure 1E-J). In some cases, such as with the KRAS sequence (Figure 1J), the chromatogram may also be indicative of higher order species such as g-wire. Figure 1 shows that all of the oncogene promoter sequences have a wide distribution of species that are partially resolved by SEC. Many of the sequences show 6–8 peaks via SEC. Some of the individual peaks appear to be broadened. These broad peaks could be a combination of several conformations that elute at similar elution volumes. Broadened peak shapes may also signify rapid inter-conversion or re-equilibration of quadruplex species. If the distribution of conformations is near equilibrium then such inter-conversions are possible on the HPLC time scale. In such a case, the possibility of population perturbation by the act of separation must also be considered.
Table 1.
Quadruplex-forming sequences from various proto-oncogene promoter sequences utilized in this study.
| Sequence Name: | Sequence: | Topology or PDB ID: | Reference: |
|---|---|---|---|
| c-kit 2KJ2 | CGGGCGGGCGCGAGGGAGGGT | 2KJ2 | 8, 44 |
| c-kit 2O3M | AGGGAGGGCGCTGGGAGGAGGG | 2O3M | 9 |
| HIF-1α with flanking | GCGCGGGGAGGGGAGAGGGGGCGGGAGCGCG | all-parallel propeller | 12 |
| HIF-1α without flanking | GGGGAGGGGAGAGGGGGCGGGA | all-parallel propeller | 12 |
| Retinoblastoma | CGGGGGGTTTTGGGCGGC | anti-parallel basket | 45 |
| c-myc | TGGGGAGGGTGGGGAGGGTGGGGAAGG | 1XAV | 7 |
| Her2 | AGGAGAAGGAGGAGGTGGAGGAGGAGGGC | putative quadruplex | 46 |
| bcl-2 | AGGGGCGGGCGCGGGAGGAAGGGGGCGGGAGCGGGGC | 2F8U | 10 |
| VEGF | GGGCGGGCCGGGGGCGGGGTCCCGGCGGGGCGGGAG | all-parallel propeller | 11 |
| KRAS | GGGAAGAGGGAAGAGGGGGAGG | all-parallel propeller | 47 |
Figure 1.
SEC separations for the promoter sequences. A. c-kit 2KJ2, B. c-kit 2O3M, C. HIF-1α, D. HIF-1α without flanking sequences, E. Retinoblastoma, F. c-myc, G. Her2, H. bcl-2, I. VEGF, and J. KRAS.
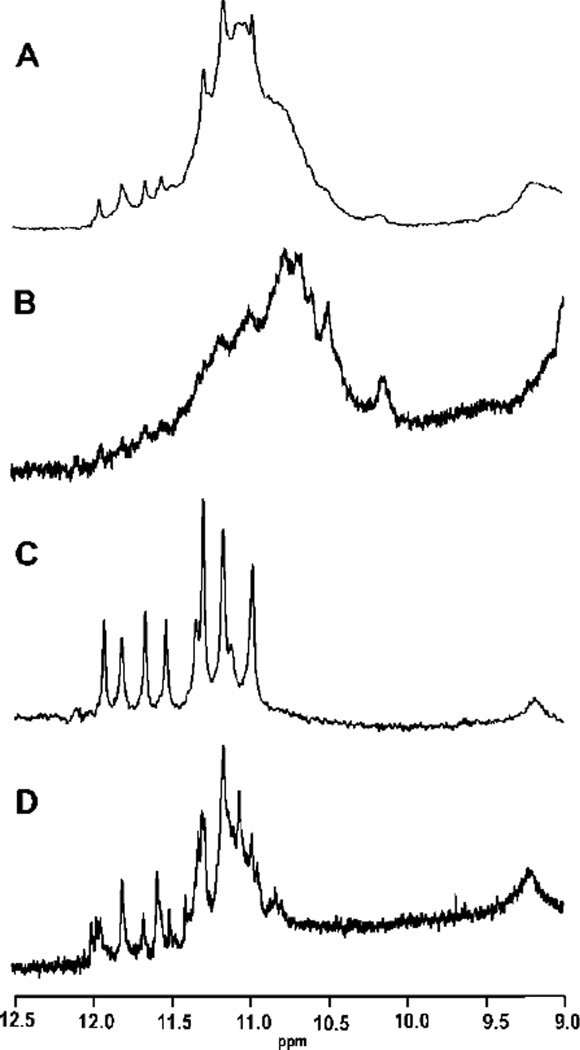
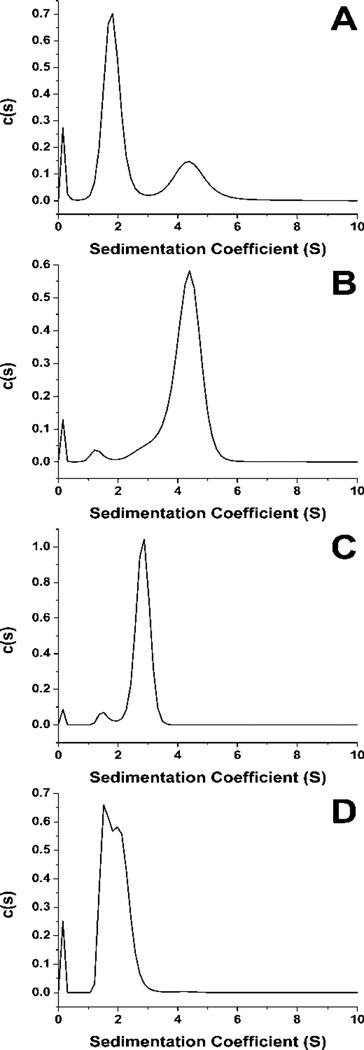
The c-kit 2KJ2 sequence was chosen for further study since this sequence has ample NMR data available8,44. The sequence was annealed and separated via SEC and fractions corresponding to the major components were collected. Material from several HPLC runs were combined. The 1D 1H NMR spectra of the combined fractions and of the un-separated material were recorded (see Supplementary Materials for Methods). The un-separated material produced an NMR spectrum indicative of a mixture of several quadruplex conformations. The spectrum displays many overlapping imino proton resonances with chemical shifts characteristic of quadruplex formation in K+ buffer48,49 (Figure 2A). AUC for the parent mixture (Figure 3A) supports the presence of multiple species in solution. The first purified fraction, corresponding to a retention volume of 9.90 ml, also displayed a broad envelope of overlapping imino proton signals (Figure 2B). This result could be indicative of either a complex mixture unresolvable by SEC or of a structure consisting of multiple strands of DNA such as a g-wire. The AUC data for this fraction supports the presence of a species that is at least a tetramer (Figure 3B). In contrast, the 1D 1H NMR data for the purified fraction 2, corresponding to a retention volume of 11.38, indicates only one quadruplex species in solution (Figure 2C). AUC data for this fraction indicates that fraction 2 is at least a dimeric species of quadruplex (Figure 3C). Finally, the third fraction collected yields a complex mixture of imino signals Figure 2D). AUC data provides support that fraction 3 is a mixture of components (Figure 3D). The 1D 1H NMR spectrum of the parent sequence and each fraction displays GN1H resonances between 12 to 10 ppm. The occurrence of these inimo/amino resonances is indicative of Hoogsteen hydrogen bonding characteristic of the presence of a G-quadruplex48,49.
Figure 2.
NMR of the c-kit 2JK2 sequence at 293 K. A. The spectrum of 35 the parent c-kit sequence showing an overlapping set of GN1H resonances in the quadruplex imino/amino proton region. B. The NMR spectrum of the fraction 1. C. The NMR spectrum of fraction 2. D. The NMR spectrum of fraction 3.
Figure 3.
AUC of the un separated c-kit 2JK2 sequence (A) and of each of the major fraction presented in Figure 2 (B-D).
Re-injection of each of the NMR samples shows that fraction 1 and fraction 2 matched the original elution profile, whereas fraction 3 showed significant re-equilibration (Supplementary Figure S2). UV-vis and CD data for each of the fractions is similar (Supplimentary Figure S3 and S4 respectively) and therefore little use for indicating different species. Interestingly, none the purified species or un-separated mixture matches the previously reported NMR data for this sequence8,44. In fact, the major species formed by this sequence under the conditions of this study is a dimer which is at odds with the previously reported data. Clearly formation of a particular quadruplex configuration is dependent not only on the sequence but also several other factors, including buffer conditions and annealing profile.
The data presented here, exemplified by the chromatograms of the c-kit sequences, indicates that these modified sequences are capable of forming far more than the single species reported in the literature. This disparity has been observed previously with the c-myc sequence7,43. This discrepancy could be due to differences in annealing conditions producing an altered equilibrium of quadruplex conformations. The disparity in results could also be attributed to the 13C, 15N labelling method commonly used for quadruplex structural determination. At the low enrichment levels used for unambiguous assignment of residues, typically 3-6% or less7,10,41,50,51, conformations with a low relative abundance would simply be lost in the baseline noise of the spectra. In other cases, such as the in the determination of the topology of the Retinoblastoma sequence (Figure 1E), researchers depended on low resolution methods to obtain their results45. The results in figures 1 and S1 clearly show multiple species in solution instead of a single structure.
When compared to the protein calibration curves (Supplementary Materials Figure S5) results from the separation of the c-kit quadruplex sequence (Figure 1A), at 6,634.3 g/mole with an estimated Stokes Radius (RS) of approximately 16 Å, show that the species isolated for further study elutes at an apparent molecular weight of 28-30 kDa and RS of approximately 26 Å. A facile answer to this observation would be the formation of a tetramer species, yet this material is shown to be a single, dimeric species via 1H NMR (Figure 2C) and AUC (Figure 3C). The Retinoblastoma sequence (Figure 1E) demonstrates a series of putative quadruplex species with apparent RS ranging from 25 Å to over 40 Å. Comparison to the elution profile of several human telomere sequences illustrates very different behaviour (Supplementary Materials Figure S6 and Table S1). Experimental values for the RS of these sequences also somewhat disagree with the calculated values (Supplementary Materials Table S2), however, these sequences elute in a much more consistent manner than do the oncogene promoter sequences.
There are several possible explanations for these results. First, there is the basic difference between telomeric sequences and promoter derived sequences. Telomeric sequences form structures with only one of three types of loop, double chain reversal, chair, and diagonal, and no matter how the G-quartets are stacked the result is always a very similar compact structure. More irregular sequences, typical of promoter sequences, are capable of more diverse stacking, even formation of alternate G-quartets, and greater differences in loop configuration. However, it seems unlikely that these differences in configuration could result in the separation demonstrated here and yet be undetectable by other biophysical means. Second, proteins can undergo changes in stokes radius due to buffer dependent changes in configuration52. This is unlikely for quadruplexes because of their exceptional stability. Third, protein samples can experience associative and repulsive interactions with column packing material53,54. Some negatively charged species may experience exclusion by repulsive interaction with the column matrix. The results from the promoter sequences may be from differences in these interactions based on the differences in topology.
This approach facilitates the isolation of the actual species formed by a parent sequence without the need for extensive sequence modification with possible structural perturbation. An added benefit is that purification may yield more than one isolatable species providing the opportunity for simultaneous study of several structures. The separation technique could be used for a variety of applications including identification of protein/quadruplex interactions or drug/quadruplex binding studies from the parent mixtures. This method may not be suitable for every quadruplex forming system. If the distribution of conformations at or near equilibrium for a specific quadruplex mixture then inter-conversions are possible on the HPLC time scale. The possibility of population perturbation by the act of separation must also be considered. As shown for 2KJ2 fraction 3, isolated fractions may also undergo re-equilibration and lack the stability required for further study. As such, each of these variables should be investigated thoroughly before incorporating SEC into procedures for the preparation of quadruplex DNA.
Supplementary Material
Acknowledgments
‡ This work was supported by National Institutes of Health (CA113735-01 to J.O.T), National Institutes of Healt Grant Number P20RR018733 from the NCRR, The JG Brown Foundation, and the Kentucky Challenge for Excellence. The authors would like to thank Dr. Robert Gray for useful discussions and advice. The authors would also like to thank Dr. Andrew N. Lane for assistance with NMR data collection and Robert Buscaglia for assitance with assistance with CD data collection.
Footnotes
† Electronic Supplementary Information (ESI) available: Methods, figures, and tables are available. See DOI: 10.1039/b000000x/
Notes and references
- 1.Lane AN, Chaires JB, Gray RD, Trent JO. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dailey MM, Miller MC, Bates PJ, Lane AN, Trent JO. Nucleic Acids Res. 2010;38:4877–4888. doi: 10.1093/nar/gkq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai J, Carver M, Yang D. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang D, Okamoto K. Future Med Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson JR. Annu. Rev. Biophys. Biomolec. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 6.Huppert JL, Balasubramanian S. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambrus A, Chen D, Dai JX, Jones RA, Yang DZ. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 8.Hsu STD, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S. Journal of the American Chemical Society. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. J Am Chem Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai J, Chen D, Jones RA, Hurley LH, Yang D. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun DY, Guo KX, Rusche JJ, Hurley LH. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Armond R, Wood S, Sun DY, Hurley LH, Ebbinghaus SW. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 13.Patel DJ, Phan AT, Kuryavyi V. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maizels N. Nature Structural & Molecular Biology. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 15.Huppert JL. Philos T R Soc A. 2007;365:2969–2984. doi: 10.1098/rsta.2007.0011. [DOI] [PubMed] [Google Scholar]
- 16.Hurley LH, Siddiqui-Jain A. Genet Eng News. 2005;25:26–26. [Google Scholar]
- 17.Cogoi S, Quadrifoglio F, Xodo LE. Biochemistry. 2004;43:2512–2523. doi: 10.1021/bi035754f. [DOI] [PubMed] [Google Scholar]
- 18.Ireson CR, Kelland LR. Molecular Cancer Therapeutics. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webba da Silva M. Chemistry. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 21.Gros J, Avino A, de la Osa JL, Gonzalez C, Lacroix L, Perez A, Orozco M, Eritja R, Mergny JL. Chem Commun. 2008:2926–2928. doi: 10.1039/b801221k. [DOI] [PubMed] [Google Scholar]
- 22.Virgilio A, Esposito V, Randazzo A, Mayol L, Galeone A. Nucleic Acids Res. 2005;33:6188–6195. doi: 10.1093/nar/gki924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Virgilio A, Esposito V, Randazzo A, Mayol L, Galeone A. Bioorgan Med Chem. 2005;13:1037–1044. doi: 10.1016/j.bmc.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 24.Esposito V, Randazzo A, Piccialli G, Petraccone L, Giancola C, Mayol L. Organic & Biomolecular Chemistry. 2004;2:313–318. doi: 10.1039/b314672c. [DOI] [PubMed] [Google Scholar]
- 25.Mekmaysy CS, Petraccone L, Garbett NC, Ragazzon PA, Gray R, Trent JO, Chaires JB. J Am Chem Soc. 2008;130:6710–6711. doi: 10.1021/ja801976h. [DOI] [PubMed] [Google Scholar]
- 26.Petrovic AG, Polavarapu PL. Journal of Physical Chemistry B. 2008;112:2255–2260. doi: 10.1021/jp075873v. [DOI] [PubMed] [Google Scholar]
- 27.Marathias VM, Sawicki MJ, Bolton PH. Nucleic Acids Res. 1999;27:2860–2867. doi: 10.1093/nar/27.14.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spackova N, Cubero E, Sponer J, Orozco M. Journal of the American Chemical Society. 2004;126:14642–14650. doi: 10.1021/ja0468628. [DOI] [PubMed] [Google Scholar]
- 29.Esposito V, Virgilio A, Randazzo A, Galeone A, Mayol L. Chem Commun. 2005:3953–3955. doi: 10.1039/b504455c. [DOI] [PubMed] [Google Scholar]
- 30.Bonifacio L, Church FC, Jarstfer MB. International Journal of Molecular Sciences. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar N, Maiti S. J Phys Chem B. 2007;111:12328–12337. doi: 10.1021/jp072705u. [DOI] [PubMed] [Google Scholar]
- 32.Tang CF, Shafer RH. J Am Chem Soc. 2006;128:5966–5973. doi: 10.1021/ja0603958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi J, Shafer RH. Biochemistry. 2007;46:7599–7606. doi: 10.1021/bi602392u. [DOI] [PubMed] [Google Scholar]
- 34.Gray RD, Li J, Chaires JB. Journal of Physical Chemistry B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray RD, Petraccone L, Trent JO, Chaires JB. Biochemistry. 2010;49:179–194. doi: 10.1021/bi901357r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blume SW, Guarcello V, Zacharias W, Miller DM. Nucleic Acids Res. 1997;25:617–625. doi: 10.1093/nar/25.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyoshi D, Nakao A, Sugimoto N. Nucleic Acids Res Suppl. 2001:259–260. doi: 10.1093/nass/1.1.259. [DOI] [PubMed] [Google Scholar]
- 38.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Journal of the American Chemical Society. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 39.Miller MC, Buscaglia R, Chaires JB, Lane AN, Trent JO. J Am Chem Soc. 2010 doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- 40.Dexheimer TS, Sun D, Hurley LH. Journal of the American Chemical Society. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masiero S, Trotta R, Pieraccini S, De Tito S, Perone R, Randazzo A, Spada GP. Organic & biomolecular chemistry. 2010;8:2683–2692. doi: 10.1039/c003428b. [DOI] [PubMed] [Google Scholar]
- 43.Phan AT, Modi YS, Patel DJ. J Am Chem Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuryavyi V, Phan AT, Patel DJ. Nucleic Acids Res. 2010;38:6757–6773. doi: 10.1093/nar/gkq558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Sugiyama H. Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziemba AJ, Zhilina ZV, Krotova-Khan Y, Stankova L, Ebbinghaus SW. Oligonucleotides. 2005;15:36–50. doi: 10.1089/oli.2005.15.36. [DOI] [PubMed] [Google Scholar]
- 47.Cogoi S, Xodo LE. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feigon J, Koshlap KM, Smith FW. Nuclear Magnetic Resonance and Nucleic Acids. 1995;261:225–255. doi: 10.1016/s0076-6879(95)61012-x. [DOI] [PubMed] [Google Scholar]
- 49.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martenson RE. J Biol Chem. 1978;253:8887–8893. [PubMed] [Google Scholar]
- 53.Frigon RP, Leypoldt JK, Uyeji S, Henderson LW. Analytical Chemistry. 1983;55:1349–1354. doi: 10.1021/ac00259a037. [DOI] [PubMed] [Google Scholar]
- 54.Kuo CY, Provder T. Acs Symposium Series. 1987;352:2–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.