Abstract
Myofibrillar protein turnover is a key component of muscle growth and degeneration, requiring proteolytic enzymes to degrade the skeletal muscle proteins. The objective of this study was to investigate the role of the calpain proteolytic system in muscle growth development using μ-calpain knockout (KO) mice in comparison with control wild-type (WT) mice, and evaluate the subsequent effects of silencing this gene on other proteolytic systems. No differences in muscle development between genotypes were observed during the early stages of growth due to the up regulation of other proteolytic systems. The KO mice showed significantly greater m-calpain protein abundance (P < 0.01) and activity (P < 0.001), and greater caspase 3/7 activity (P < 0.05). At 30 wk of age, KO mice showed increased protein:DNA (P < 0.05) and RNA:DNA ratios (P < 0.01), greater protein content (P < 0.01) at the expense of lipid deposition (P < 0.05), and an increase in size and number of fast-twitch glycolytic muscle fibers (P < 0.05), suggesting that KO mice exhibit an increased capacity to accumulate and maintain protein in their skeletal muscle. Also, expression of proteins associated with muscle regeneration (neural cell adhesion molecule and myoD) were both reduced in the mature KO mice (P < 0.05 and P < 0.01, respectively), indicating less muscle regeneration and, therefore, less muscle damage. These findings indicate the concerted action of proteolytic systems to ensure muscle protein homeostasis in vivo. Furthermore, these data contribute to the existing evidence of the importance of the calpain system’s involvement in muscle growth, development, and atrophy. Collectively, these data suggest that there are opportunities to target the calpain system to promote the growth and/or restoration of skeletal muscle mass.
Keywords: body composition, muscle fiber type, proteases
INTRODUCTION
Protein turnover in skeletal muscle is unique due to the continuous structure of the myofibrils within the muscle cell, and, therefore, turnover must be accomplished without disrupting this structure (Goll et al., 2008). Dayton et al. (1976) proposed that myofibrillar proteins are turned over by releasing filaments from the surface of the fibril and must be targeted and degraded by a protease system. The μ- and m-calpains are comprised of a large 80 kDa subunit, unique to each, and a smaller 28 kDa subunit, which is common to both and encoded by the Capn4 gene (Sorimachi et al., 1989). Silencing the Capn4 gene is embryonically lethal, as is silencing the large subunit of m-calpain, but knocking out the large subunit of μ-calpain is not. These animals are both viable and fertile (Azam et al., 2001). Calpains have been identified to be involved in muscular dystrophies (Badalamente and Stracher, 2000; Briguet et al., 2008) and wasting conditions associated with muscle disuse (Dargelos et al., 2008; Brule et al., 2010). The μ-calpain knockout (KO) studies have shown severe attenuation of proteolysis of skeletal muscle proteins (Geesink et al., 2006). Furthermore, calpain activity is essential for myoblast fusion and proliferation, and cell growth (Dedieu et al., 2004; Moyen et al., 2004).
It is evident that μ-calpain has important roles, both in normal postnatal muscle growth and in muscle wasting. Consequently, understanding how muscle proteins are turned over metabolically and how this turnover is regulated has important implications in muscle growth and loss. However, the effect of silencing the large subunit of μ-calpain mice during the entirety of development has not been studied. Thus, the objective of this study was to investigate the role of the calpain proteolytic system in muscle growth development, using μ-calpain KO mice in comparison with control wild-type mice, and evaluate the subsequent effects of silencing this gene on other proteolytic systems.
MATERIAL AND METHODS
The United States Meat Animal Research Center Animal Care and Use Committee approved the use of animals in this study.
Generation of μ-calpain Knockout Mice and Sample Collection
The μ-calpain KO (Capn1 inactivation) mice were generated in the laboratory of A. H. Chishti, as described by Azam et al. (2001). Heterozygous μ-calpain± C57BL/6J mice were used to generate male and female KO and wild-type (WT) mice for this study. At 3, 5, 10, 20, and 30 wk of age (n = 6 per genotype per sex) mice were sacrificed by cervical dislocation. The gastrocnemius, quadriceps, and extensor digitorum longus (EDL) muscles from both hind limbs, and the tibialis anterior (TA) and soleus from the left hind limb were dissected, snap frozen in liquid nitrogen, weighed, pooled together, and stored at −80°C for subsequent analysis. The TA and soleus muscles from the right hind limb were quantitatively dissected and frozen in ice-cold isopentane for histology analysis. The heart, liver, and spleen were removed and weighed.
Total protein, water, fat, and ash body composition were evaluated (n = 6 per genotype per gender per age), and accretion rates calculated (proximate analysis; AOAC, 1997). Six groups of newborn mice per genotype (n = 35 to 39 per group) were sacrificed for initial body composition analysis.
Quantification of DNA, RNA, and Protein
Quantitative determination of total DNA from 30 mg of pooled crushed muscle samples was performed using a standard salt extraction procedure. Total RNA was extracted from 10 mg of muscle samples using TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA and DNA concentrations were quantified using a ND-1000 spectrophotometer (Nano-Drop Technologies, Wilmington, DE). Protein concentrations were determined using the micro-BCA assay (Pierce, Rockford, IL).
Sample Preparation
Fifty milligrams of pooled crushed muscle samples were homogenized in 10 vol of extraction buffer (100 mM Tris, 5 mM EDTA, pH 8.3). One-fourth of the total homogenate was removed and prepared for SDS-PAGE by the addition of 2X protein denaturing buffer (PDB; 125 mM Tris, 4% SDS, 20% glycerol, 10% β-mercaptoethanol, and 0.01% bromophenol blue, pH 6.8). The remaining three-fourths was prepared for either analysis of proteins in the soluble fraction (Wheeler and Koohmaraie, 1999), casein zymography (Veiseth et al., 2001), or calpastatin activity (Shackelford et al., 1994).
SDS-PAGE and Immunoblotting
For electrophoresis, 30 μg of protein was loaded onto SDS-PAGE gels. Twelve percent resolving gels were used for calpastatin, myogenin, and myoD, 10% for PTP1B and neural cell adhesion molecule (NCAM), and 7.5% for m-calpain and calpain 3, with 4% stacking gels. Gels were run at a constant of 200 V for 45 min. The separated proteins were transferred onto Hybond-P polyvinylidene difluoride (PVDF) membrane (GE Healthcare, Piscataway, NJ) by Western blotting, at a constant current of 200 mA for 1 h at 4°C. Western blots were immunoprobed overnight at 4°C with either anti-m-calpain antibody diluted 1:2,500 (Ab39165); anti-calpastatin, 1:5,000 (Ab28252); anti-myogenin, 1:600 (Ab82843); anti-myoD, 1:1,000 (Ab64159); anti-PTP1B, 1:7,500 (Ab52650; Abcam, Cambridge, MA); anti-calpain 3, 1:50 (CALP-12A2; Novacastra, Bannockburn, IL); or anti-NCAM, 1:50 (MAB310; Millipore, Billerica, MA); and detected with SuperSignal Chemiluminescence substrate (Pierce). Protein band intensities were quantified using ChemiImager 5500 digital imaging analysis system (Alpha Innotech, San Leandro, CA).
Zymography and Nondenaturing PAGE
Casein zymography was performed according to the procedure of Veiseth et al. (2001). Polyacrylamide gels (12.5%) were loaded with 100 μg of protein and electrophoresed at 125 V for 6 h at 4°C before incubation and staining. Protein band intensities were quantified using ChemiImager 5500 digital imaging analysis system (Alpha Innotech).
Calpastatin, Caspase 3/7, and Proteasome Activity
Calpastatin activity was determined using the BODIPY-labeled casein assay based on the method of Thompson et al. (2000), with some modifications. In brief, 25 μL of sample was combined with 750 ng of semi-purified ovine m-calpain and brought to a final volume of 100 μL with dilution buffer (20 mM Tris, 1 mM EDTA, 100 mM KCl, 0.1% β-mercaptoethanol, pH 7.5). The reaction was initiated with 100 μL of BODIPY-casein (5 μg BODIPY-labeled casein/mL) in dilution buffer containing 6 mM CaCl2, incubated at 37°C for 30 min and end-point fluorescence measured.
Caspase 3/7 activity was measured in muscle samples using Apo-One Homogenous Caspase-3/7 Assay (Promega, Madison, WI), adapted for tissue samples (Wagner et al., 2003). The assay was incubated at room temperature for 3 h and end-point fluorescence measured.
Proteasome activity was measured in muscle samples using Proteasome-Glo Assay (Promega), adapted for tissue samples (Strucksberg et al., 2010). The assay detects the 3 proteolytic activities of the proteasome: chymotrypsin-like, trypsin-like, and caspase-like activity, using the luciferin-labeled substrates Suc-LLVY, Z-LRR, and Z-nLPnLD, respectively. The assay was incubated at room temperature for 20 min and end-point luminescence measured.
Optimal incubation times for the protease activity assays were determined empirically and all samples were assayed in duplicate. Luminescence and fluorescence were measured on a Wallac 1420 Victor2 multilabel counter (EG&G Wallac, Turku, Finland). Fluorescence was measured at an excitation wavelength of 485 ± 20 nm and an emission wavelength of 530 ± 25 nm. The amount of luminescence or fluorescence generated is directly proportional to the amount of activity present in the sample. Protein concentrations of sample supernatants used in the activity assays were determined using micro-BCA assay (Pierce).
Fiber Typing
Soleus and TA muscles were transverse sectioned 10-μm thick with a cryostat. Sections were stained for NADH and myofibrillar ATPase activities, a simultaneous combination staining procedure, with an acid preincubation solution (pH 4.52 for TA and pH 4.20 for soleus), as described by Solomon and Dunn (1988). A minimum of 200 fibers per animal were classified as slow-twitch red oxidative Type I fibers (dark purple), fast-twitch intermediate oxidative Type IIA fibers (purple), or fast-twitch white glycolytic Type IIB fibers (light purple), according to the classification of Ashmore and Doerr (1971).
Statistical Analysis
Data were analyzed using the PROC MIXED procedure with an autoregressive covariance structure (SAS Inst. Inc., Cary, NC). The model tested fixed effects of genotype, age, and gender, and all interactions. When no gender differences were observed, gender data were combined. Least squares means were generated for all interactions and main effects. Least squares means for significant interactions and main effects not involved in higher order interactions were separated, using the Diff option (pairwise t tests). A predetermined significance level of 0.05 was used for all determinations of statistical significance. P < 0.10 was considered a trend towards significance.
RESULTS
Growth
Body, organ, and muscle weights increased with age (P < 0.001), and were heavier in males than females (P < 0.001), with the exception of the spleen (P = 0.51; Table 1). At 30 wk, WT mice were significantly heavier than KO mice (genotype × age, P = 0.046; Table 1). Liver weights were affected by genotype, with WT mice having significantly larger livers (P = 0.04) than KO mice. Genotype did not affect heart or spleen weights (P = 0.16, P = 0.43, respectively). Muscle weights of the soleus, TA, gastrocnemius, quadriceps, and EDL were also unaffected by genotype (P = 0.12, P = 0.22, P = 0.68, P = 0.91, P = 0.42, correspondingly). Other than BW at 30 wk, no other phenotypical differences were observed between genotypes.
Table 1.
Least squares means for animal organ and muscle weights from wild type (WT) and μ-calpain knockout (KO) mice
| Main Effect | BW, g | Liver, mg | Heart, mg | Spleen, mg | Quads,1 mg | Gastro,1 mg | TA,1 mg | Soleus, mg | EDL,1 mg |
|---|---|---|---|---|---|---|---|---|---|
| Genotype | |||||||||
| WT (n = 30) | 19.95 | 1,082.98 | 107.87 | 77.42 | 97.06 | 78.63 | 29.18 | 6.12 | 6.06 |
| KO (n = 30) | 19.54 | 1,021.43 | 104.40 | 74.78 | 96.73 | 79.48 | 28.15 | 5.73 | 5.83 |
| SEM | 0.18 | 21.02 | 1.71 | 2.32 | 2.03 | 1.44 | 0.62 | 0.17 | 0.20 |
| P-value | 0.106 | 0.042 | 0.155 | 0.425 | 0.907 | 0.679 | 0.244 | 0.122 | 0.417 |
| Gender | |||||||||
| Male (n = 30) | 21.28 | 1,129.55 | 113.42 | 75.02 | 105.85 | 86.47 | 31.45 | 6.35 | 6.43 |
| Female (n = 30) | 18.21 | 974.87 | 98.85 | 77.18 | 87.93 | 71.63 | 25.88 | 5.50 | 5.47 |
| SEM | 0.14 | 26.26 | 2.73 | 2.90 | 2.87 | 2.05 | 0.87 | 0.24 | 0.28 |
| P-value | <0.0001 | <0.0001 | <0.0001 | 0.511 | <0.0001 | <0.0001 | <0.0001 | <0.001 | <0.001 |
| Age | |||||||||
| 3 wk (n = 12) | 9.85 | 444.96 | 68.71 | 58.08 | 39.71 | 34.31 | 13.21 | 2.92 | 3.06 |
| 5 wk (n = 12) | 16.08 | 848.75 | 88.54 | 73.83 | 63.79 | 50.56 | 21.46 | 4.21 | 4.54 |
| 10 wk (n = 12) | 21.10 | 1,231.25 | 118.08 | 71.17 | 105.38 | 86.04 | 28.67 | 6.33 | 5.83 |
| 20 wk (n = 12) | 25.02 | 1,302.75 | 126.08 | 79.67 | 131.58 | 112.71 | 40.17 | 7.67 | 8.04 |
| 30 wk (n = 12) | 26.67 | 1,433.33 | 129.25 | 97.75 | 144.00 | 111.63 | 39.83 | 8.50 | 8.25 |
| SEM | 0.21 | 43.87 | 2.96 | 2.05 | 4.64 | 3.69 | 1.25 | 0.26 | 0.26 |
| P-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Interaction2, G × A at 30 wk | 0.0462 | 0.472 | 0.337 | 0.089 | 0.445 | 0.935 | 0.366 | 0.279 | 0.904 |
Quads = quadriceps; Gastro = gastrocnemius; TA = tibialis anterior; EDL = extensor digitorum longus.
G × A = genotype × age interaction.
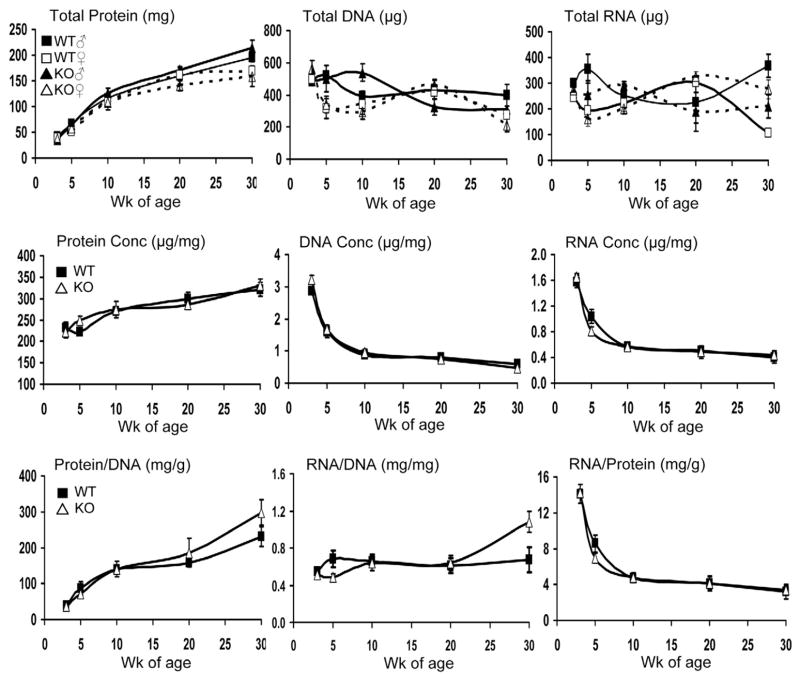
Total DNA, RNA, and protein content, and their concentrations were unaffected (P > 0.71) by genotype (Fig. 1). Protein content (P < 0.001) and concentration (P = 0.03) increased with age, indicating protein accumulation. DNA content and concentration decreased with age (P < 0.001). Although total RNA fluctuated among age groups (P = 0.38), RNA concentration decreased with age (P < 0.05). As expected, males had greater total DNA, RNA, and protein content than females (P < 0.05), reflecting increased muscle mass. Concentrations of RNA, DNA, and protein were unaffected (P > 0.86) by gender.
Figure 1.
Total RNA, DNA, and protein content, concentrations (Conc), and ratios of skeletal muscle from wild-type (WT) and μ-calpain knockout (KO) mice at 3, 5, 10, 20, and 30 wk of age. The top panel shows mean content of protein, DNA, and RNA ± SEM. Values were not affected by genotype; males were greater than females (P < 0.05). Age significantly affected protein and DNA content (P < 0.0001). The middle panel represents protein, DNA, and RNA concentrations. For all parameters, age was a significant source of variation (P < 0.05); there was no effect of sex or genotype. The bottom panel shows the protein:DNA, RNA:DNA, and RNA:protein ratios. Age had a significant effect on protein:DNA and RNA:protein (P < 0.001), but not RNA:DNA. Genotype effects were present at 30 wk, with KO mice having greater protein:DNA and RNA:DNA ratios (P < 0.05 and P < 0.01, respectively).
Protein:DNA increased with age (Fig. 1; P < 0.001), whereas RNA:protein decreased (P < 0.001). Ratios were unaffected (P > 0.56) by gender. The RNA:DNA ratio did not differ (P > 0.68) among age groups in WT mice, whereas in KO mice there was a significant increase at 30 wk (P < 0.01) and a genotype × age interaction observed (P < 0.01). At 30 wk, the protein:DNA ratio was significantly greater in the KO mice (P = 0.02). Genotype did not affect (P > 0.78) RNA:protein ratio.
Body Composition
Overall, KO mice showed greater protein percentage than WT animals (Table 2; P = 0.04). Protein content increased up to 10 wk in WT mice, whereas in KO mice protein continued to accumulate until 20 wk, before decreasing. The KO mice showed significantly greater protein percentages than WT at 20 and 30 wk (genotype × age interaction; P = 0.02 and P = 0.01, respectively), at the expense of lipid deposition at 30 wk (P = 0.03). Genotype differences were independent of sex (P > 0.44). Water and ash percentages decreased and increased with age, respectively (P < 0.001), autonomous of sex and genotype.
Table 2.
Protein, lipid, ash, and water body composition percentage in wild type (WT) and μ-calpain knockout (KO) mice at 3, 5, 10, 20, and 30 wk of age1
| Item | Protein, % | Lipid, % | Ash, % | Water, % |
|---|---|---|---|---|
| WT | ||||
| NB2 | 12.4 ± 0.1 | 2.1 ± 0.1 | 2.1 ± 0.1 | 83.4 ± 0.3 |
| 3 wk | 17.3 ± 0.2 | 9.8 ± 0.6 | 4.6 ± 0.1 | 68.6 ± 1.3 |
| 5 wk | 19.5 ± 0.2 | 6.9 ± 0.5 | 4.8 ± 0.1 | 68.8 ± 1.4 |
| 10 wk | 20.2 ± 0.4 | 8.1 ± 0.5 | 5.2 ± 0.1 | 66.6 ± 1.2 |
| 20 wk | 19.9 ± 0.3* | 8.9 ± 0.4 | 5.3 ± 0.2 | 65.1 ± 1.3 |
| 30 wk | 18.7 ± 0.5* | 14.8 ± 1.7* | 5.7 ± 0.3 | 61.0 ± 1.5 |
| KO | ||||
| NB | 12.8 ± 0.1 | 1.9 ± 0.1 | 2.1 ± 0.1 | 83.2 ± 0.1 |
| 3 wk | 17.4 ± 0.3 | 9.6 ± 0.3 | 4.7 ± 0.1 | 68.6 ± 0.3 |
| 5 wk | 19.2 ± 0.3 | 8.4 ± 0.4 | 4.8 ± 0.2 | 68.1 ± 0.4 |
| 10 wk | 20.0 ± 0.2 | 7.5 ± 0.9 | 4.9 ± 0.2 | 67.7 ± 0.9 |
| 20 wk | 21.3 ± 0.6* | 8.3 ± 1.1 | 5.5 ± 0.3 | 65.1 ± 1.2 |
| 30 wk | 20.3 ± 0.8* | 11.9 ± 1.4* | 5.8 ± 0.3 | 62.7 ± 1.4 |
Values are least squares means ± SEM; n = 6. Means within age group between genotypes are significantly different (*P ≤ 0.05).
NB = newborn.
Protein and water accretion rates were unaffected (P > 0.10) by the main effects of genotype and gender (Table 3). During the 3- to 5-wk growth period, KO mice had significantly greater lipid accretion (genotype × age; P = 0.01) than WT mice. However, this observation was reversed during the 5- to 10-wk period, with WT mice having increased lipid accretion (genotype × age; P = 0.05) and a tendency for increased ash accretion (P = 0.08).
Table 3.
Whole body protein, lipid, ash, and water accretion rates in wild type (WT) and μ-calpain knockout (KO) mice at 3, 5, 10, 20, and 30 wk of age1
| Item | Protein, mg/d | Lipid, mg/d | Ash, mg/d | Water, mg/d |
|---|---|---|---|---|
| WT | ||||
| NB2 to 3wk | 75.3 ± 2.7 | 46.8 ± 5.0 | 21.0 ± 1.2 | 280.0 ± 12.3 |
| 3 to 5 wk | 90.8 ± 12.2 | 3.3 ± 8.4** | 18.9 ± 1.5 | 263.1 ± 34.3 |
| 5 to 10 wk | 34.3 ± 8.4 | 18.2 ± 4.2* | 9.5 ± 1.2† | 92.1 ± 20.1 |
| 10 to 20 wk | 10.6 ± 1.8 | 7.7 ± 2.8 | 3.5 ± 0.6 | 32.7 ± 4.7 |
| 20 to 30 wk | 2.0 ± 3.3 | 26.8 ± 9.5 | 3.6 ± 1.8 | 7.1 ± 8.8 |
| NB to 30 wk | 23.5 ± 0.7 | 19.4 ± 2.9 | 7.4 ± 0.5 | 74. 2 ± 2.1 |
| KO | ||||
| NB to 3 wk | 75.5 ± 1.7 | 44.7 ± 1.9 | 21.2 ± 0.4 | 276.61 ± 5. 2 |
| 3 to 5 wk | 98.4 ± 7.7 | 27.8 ± 5.7** | 21.7 ± 1.5 | 297.4 ± 16.4 |
| 5 to 10 wk | 27.6 ± 2.4 | 5.1 ± 5.1* | 6.4 ± 0.8† | 80.5 ± 8.1 |
| 10 to 20 wk | 17.5 ± 3.6 | 7.9 ± 6.0 | 5.2 ± 1.4 | 33.6 ± 8.9 |
| 20 to 30 wk | −0.1 ± 5.3 | 14.9 ± 7.3 | 2.2 ± 1.4 | 1.0 ± 7.3 |
| NB to 30 wk | 24.5 ± 2.0 | 14.9 ± 7.3 | 7.1 ± 0.4 | 72.44 ± 3.0 |
Values are least squares means ± SEM; n = 6. Means within age group between genotypes are significantly different (†P ≤ 0.1, *P ≤ 0.05, **P ≤ 0.01).
NB = newborn.
Protease Systems Activity and Protein Expression
Casein zymography confirmed no μ-calpain activity in the KO mice. In WT mice, μ-calpain activity was affected by age (P < 0.001), with activity peaking at 10 wk (Table 4). The m-calpain activity and protein expression was assessed using casein zymography and Western blotting, respectively. The m-calpain was detected in both KO and WT mice, and was significantly affected by age (P < 0.0001; Table 4). The m-calpain activity was greater in KO mice (P < 0.0001), with significant genotype effects at 3, 5, and 10 wk (genotype × age; P = 0.01, P = 0.0001, P = 0.001, respectively; Table 4). However, as mice reached maturity, this up regulation of m-calpain became less apparent with no genotype effect detected (P > 0.14) at 20 or 30 wk. Protein abundance of m-calpain followed a similar pattern as activity (Table 4), with greater m-calpain expression in KO mice than WT (P < 0.01) and a significant genotype × age interaction at 5 wk (P < 0.001). In agreement with activity, genotype differences in m-calpain protein abundance were less evident as the animals aged. Age, therefore, was a significant source of variation (P < 0.001). Although age had a significant effect on calpain 3 protein abundance (P < 0.001), with expression at 5 wk greater than all other age groups, silencing the μ-calpain gene had no effect (P > 0.22) on calpain 3 expression (Table 4).
Table 4.
Protease expression and activity ± SEM in wild type (WT) and μ-Calpain knockout (KO) mice at 3, 5, 10, 20, and 30 wk of age1
| Item | μ-Calpain activity2 | m-Calpain activity2 | m-Calpain protein3 | Calpain 3 protein3 | Calpastatin protein3 | Calpastatin activity4 | Caspase 3/7 activity4 | Proteasome activity5 |
|---|---|---|---|---|---|---|---|---|
| WT | ||||||||
| 3 wk | 1.01 ± 0.1 | 0.87 ± 0.1* | 0.58 ± 0.1 | 1.03 ± 0.1 | 0.23 ± 0.1 | 56.3 ± 4.0* | 1,155.6 ± 128.5 | 28.2 ± 2.5 |
| 5 wk | 0.95 ± 0.1 | 0.98 ± 0.1*** | 0.63 ± 0.0*** | 1.22 ± 0.2 | 0.35 ± 0.1* | 47.3 ± 2.2 | 484.3 ± 38.0 | 30.9 ± 3.3* |
| 10 wk | 1.27 ± 0.1 | 1.05 ± 0.1** | 0.46 ± 0.0 | 0.85 ± 0.1 | 0.88 ± 0.2† | 39.4 ± 2.2 | 340.3 ± 28.3 | 23.0 ± 2.1 |
| 20 wk | 1.13 ± 0.0 | 0.91 ± 0.0 | 0.34 ± 0.0 | 0.47 ± 0.0 | 0.32 ± 0.1 | 35.8 ± 0.6 | 311.4 ± 25.9 | 22.6 ± 2.3 |
| 30 wk | 0.73 ± 0.0 | 0.79 ± 0.0 | 0.27 ± 0.0 | 0.80 ± 0.1 | 0.25 ± 0.1 | 38.6 ± 1.2 | 301.0 ± 39.3 | 15.0 ± 1.1 |
| KO | ||||||||
| 3 wk | ND6 | 1.14 ± 0.1* | 0.69 ± 0.1 | 0.97 ± 0.1 | 0.43 ± 0.1 | 49.7 ± 1.8* | 1,188.1 ± 161.7 | 26.7 ± 1.9 |
| 5 wk | ND | 1.45 ± 0.1*** | 0.92 ± 0.1*** | 1.20 ± 0.1 | 0.68 ± 0.1* | 44.3 ± 1.8 | 684.9 ± 111.9* | 22.4 ± 1.8* |
| 10 wk | ND | 1.34 ± 0.1** | 0.51 ± 0.1 | 0.90 ± 0.1 | 1.15 ± 0.1† | 38.1 ± 0.9 | 477.5 ± 58.3 | 17.5 ± 2.9 |
| 20 wk | ND | 1.06 ± 0.1 | 0.36 ± 0.0 | 0.64 ± 0.1 | 0.35 ± 0.1 | 34.7 ± 0.6 | 454.3 ± 44.2 | 23.9 ± 1.8 |
| 30 wk | ND | 0.97 ± 0.0 | 0.29 ± 0.0 | 0.68 ± 0.1 | 0.51 ± 0.1 | 38.5 ± 2.3 | 322.3 ± 19.8 | 16.0 ± 2.8 |
Values are least squares means ± SEM; n = 6. Means within age group between genotypes are significantly different (†P ≤ 0.1, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
The μ- and m-calpain activity, as detected by casein zymography, are expressed as mean arbitrary units of densitometry.
Protein expression, as detected by Western blotting, is expressed as mean arbitrary units of densitometry.
Activity expressed as arbitrary units of fluorescence/milligrams protein.
Activity expressed as arbitrary units of luminescence/milligrams protein.
ND = not detectable.
Protein abundance of full-length calpastatin increased in both genotypes up until 10 wk, before significantly decreasing at 20 and 30 wk (Table 4; P < 0.001). This decrease corresponded with the appearance of an isoform at ~27 kDa. Although this pattern of expression was observed in both genotypes, it was greater in the KO mice (P = 0.004), resulting in significant genotype × age effect at 5 wk (P = 0.04) and a trend at 10 wk (P = 0.09).
Calpastatin activity did not exhibit the same pattern as protein expression (Table 4). In general, calpastatin activity decreased with age (P < 0.001), with activity at 3 and 5 wk greater than all other age groups. In the KO mice, there was a tendency for decreased calpastatin activity (P = 0.06) compared with WT mice, with KO mice at 3 wk having significantly less activity (genotype × age; P = 0.02).
Caspase 3/7 activity decreased with age (P < 0.001) in both genotypes, with activity at 3 wk significantly greater than all other age groups. A significant increase in caspase 3/7 activity was detected in KO mice in comparison with WT mice (P < 0.01; Table 4), and a genotype × age interaction observed at 5 wk (P = 0.05). The differences in caspase 3/7 activity between genotypes diminished with age.
The proteasome assay used in this study measured the 3 proteolytic activities associated with the proteasome separately. Each component of the proteasome followed the same pattern and therefore activity was combined and total proteasome activity is reported. In contrast to the other proteolytic systems analyzed, there was a trend for less proteasome activity in KO mice (P = 0.079; Table 4). At 5 wk, proteasome activity was significantly decreased in KO (genotype × age interaction; P = 0.013), corresponding with genotype differences in m-calpain and caspase 3/7.
Muscle Histology
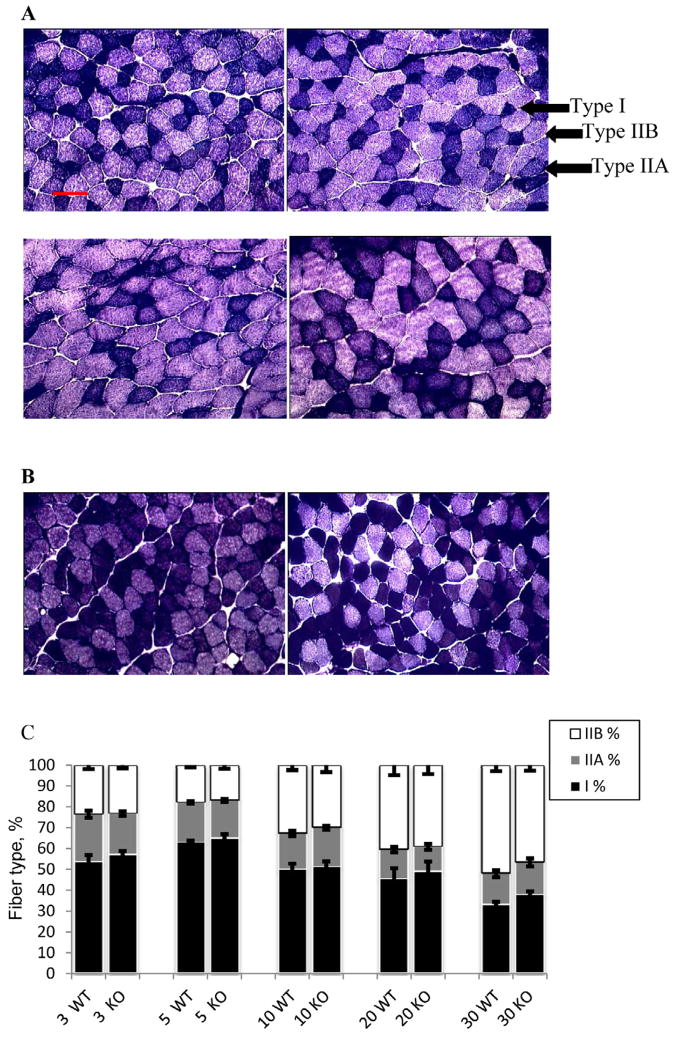
Silencing the Capn1 gene significantly affected the histology profile in both the slow-twitch red oxidative soleus and fast-twitch white glycolytic TA (Fig. 2, Table 5; P < 0.05). In TA, there was a trend for KO mice to have more Type IIA and less Type I fibers than WT mice at 5 wk (P = 0.06; data not shown). Additionally, the KO mice had more Type IIB fibers (P = 0.05; Table 5), with a genotype × age interaction observed at 30 wk (P = 0.03; data not shown), which appeared to be at the expense of Type IIA fibers (P = 0.02; Fig. 2A). Furthermore, at 30 wk, the Type IIB fibers in KO mice were significantly larger than those in the WT animals (genotype × age × fiber type interaction, P = 0.03).
Figure 2.
Muscle histology. A) Representative cross-sectional fibers of tibialis anterior muscle from wild-type (WT) and knockout (KO) mice at 5 and 30 wk, simultaneously stained for NADH and myofibrillar ATPase activities. Clockwise from top left: 5 wk WT, 5 wk KO, 30 wk WT, 30 wk KO. B) Representative cross-sectional fibers of soleus muscle from WT and KO mice at 10 wk simultaneously stained for NADH and myofibrillar ATPase activities. Left to right: 10 wk WT, 10 wk KO. C) Changes in percentage fiber type distribution in soleus muscle in KO and WT mice, with age. Age and sex were significant sources of variation in fiber size (P < 0.001) and genotype had significant effect on fiber type distribution in tibialis anterior (P < 0.05), and a trend effect in the soleus (P < 0.1). Bar equals 50 μm for all panels.
Table 5.
Least squares means for muscle fiber type distribution, fiber area, and fiber diameter for soleus and tibialis anterior
| Item | % distribution | Area, mm2 | Diam., mm | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Type IIB | Type IIA | Type I | Type IIB | Type IIA | Type I | Type IIB | Type IIA | Type I | |
| Soleus muscle | |||||||||
| Genotype | |||||||||
| WT | 33.34 | 17.74 | 48.92 | 402.6 | 318.91 | 316.55 | 25.82 | 23.03 | 23.07 |
| KO | 31.20 | 16.99 | 51.81 | 395.43 | 308.03 | 266.31 | 25.59 | 22.18 | 21.36 |
| SEM | 0.64 | 0.30 | 0.71 | 6.54 | 11.22 | 9.34 | 0.25 | 0.33 | 0.31 |
| P-value | 0.24 | 0.39 | 0.049 | 0.71 | 0.63 | 0.015 | 0.74 | 0.21 | 0.018 |
| Gender | |||||||||
| Female | 34.13 | 17.3 | 48.56 | 396.01 | 293.66 | 265.7 | 25.54 | 22.01 | 21.03 |
| Male | 30.4 | 17.43 | 52.16 | 402.02 | 333.27 | 317.15 | 25.87 | 23.19 | 23.39 |
| SEM | 0.43 | 0.3 | 0.64 | 9.53 | 7.63 | 12.51 | 0.35 | 0.43 | 0.22 |
| P-value | 0.04 | 0.88 | 0.01 | 0.76 | 0.08 | 0.008 | 0.64 | 0.08 | 0.01 |
| Age | |||||||||
| 3 wk | 23.39 | 21.40 | 55.21 | 255.86 | 206 | 226.61 | 19.62 | 17.81 | 18.24 |
| 5 wk | 17.38 | 18.80 | 63.82 | 316.62 | 268.95 | 230.32 | 23.11 | 21.03 | 19.90 |
| 10 wk | 31.37 | 18.17 | 50.47 | 434.13 | 322.1 | 311.76 | 28.01 | 23.88 | 23.76 |
| 20 wk | 39.84 | 13.08 | 47.08 | 458.6 | 324.23 | 324.96 | 27.65 | 23.04 | 23.72 |
| 30 wk | 49.38 | 15.38 | 35.24 | 529.87 | 446.06 | 363.48 | 30.16 | 27.27 | 25.45 |
| SEM | 1.01 | 0.47 | 0.75 | 11.01 | 12.51 | 14.52 | 0.55 | 0.37 | 0.35 |
| P-value | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 |
| Tibialis anterior muscle | |||||||||
| Genotype | |||||||||
| WT | 50.12 | 29.91 | 19.32 | 570.11 | 350.93 | 252.47 | 31.7 | 24.59 | 20.84 |
| KO | 52.26 | 28.92 | 19.95 | 558.665 | 347.63 | 237.7 | 31.11 | 24.34 | 20.18 |
| SEM | 0.53 | 0.54 | 0.59 | 16.91 | 5.15 | 8.57 | 0.44 | 0.31 | 0.29 |
| P-value | 0.05 | 0.35 | 0.27 | 0.65 | 0.82 | 0.23 | 0.51 | 0.67 | 0.27 |
| Gender | |||||||||
| Female | 50.17 | 30.18 | 19.64 | 555.6 | 336.09 | 232.66 | 31.23 | 24.04 | 20.08 |
| Male | 51.13 | 29.54 | 19.33 | 573.16 | 362.46 | 257.51 | 31.57 | 24.89 | 20.94 |
| SEM | 0.98 | 0.61 | 0.59 | 8.91 | 7.25 | 4.25 | 0.31 | 0.22 | 0.24 |
| P-value | 0.49 | 0.45 | 0.77 | 0.49 | 0.07 | 0.047 | 0.71 | 0.16 | 0.15 |
| Age | |||||||||
| 3 wk | 49.87 | 30.55 | 19.57 | 307.04 | 178.84 | 120.47 | 22.45 | 17.19 | 14.18 |
| 5 wk | 48.95 | 32.76 | 18.28 | 452.24 | 276.36 | 203.54 | 28.55 | 21.66 | 18.93 |
| 10 wk | 51.07 | 27.28 | 21.58 | 593.05 | 366.93 | 247.91 | 32.57 | 24.02 | 21.05 |
| 20 wk | 52.92 | 29.57 | 17.49 | 782.07 | 479.43 | 348.87 | 38.08 | 28.48 | 25.04 |
| 30 wk | 53.14 | 26.91 | 20.49 | 689.93 | 444.82 | 304.62 | 35.37 | 28.07 | 23.36 |
| SEM | 0.75 | 0.48 | 0.47 | 14.04 | 8.15 | 6.55 | 0.49 | 0.47 | 0.33 |
| P-value | 0.46 | 0.05 | 0.024 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 |
WT = wild-type; KO = knockout.
In the soleus, KO mice had smaller Type I fibers in both area (P = 0.02) and diameter (P = 0.018), but were more abundant in numbers (P < 0.05; Table 5). A shift in fiber type distribution was observed in the soleus muscle, with an increase in Type IIB fibers as the animals aged at the expense of both types of oxidative fibers (P < 0.001; Fig. 2C). Although sex influenced muscle fiber size (P < 0.001), with males having larger fibers than females (Table 5), the effects of genotype and age were independent of gender (P > 0.09). Age had the largest effect on muscles, with both fiber area and diameter increasing with age (P < 0.001).
Indicators of Muscle Development and Atrophy
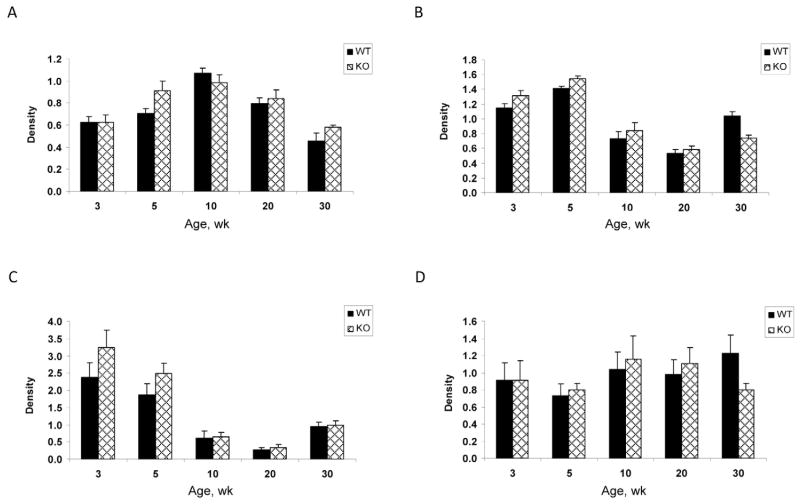
Myogenin protein abundance (Fig. 3A) increased until 10 wk before, decreasing as the animals aged, in both genders and genotypes (P < 0.001), which was consistent with the fastest growth rates. Overall, myogenin protein abundance tended to be greater in KO mice (P = 0.078) and a significant age × genotype interaction was detected at 5 wk (P = 0.035).
Figure 3.
Protein abundance of: A) myogenin, B) MyoD, C) protein tyrosine phosphatase 1B (PTP1B), and D) neural cell adhesion molecule (NCAM) in skeletal muscle of wild-type (WT) and μ-calpain knockout mice (KO), at 3, 5, 10, 20, and 30 wk of age. Protein abundance is expressed as mean arbitrary units of densitometry ± SEM (n = 6 per genotype). The effect of age was significant for myogenin, MyoD, and PTP1B (P < 0.001). A genotype effect on protein expression was present for myogenin at 5 wk (genotype × age; *P ≤ 0.05), MyoD at 30 wk (genotype × age, **P ≤ 0.01), PTP1B at 3 wk (genotype × age; *P ≤ 0.05), and NCAM at 30 wk (genotype × age; *P ≤ 0.05).
MyoD protein expression was significantly affected by age (P < 0.001), with abundance at 3 and 5 wk greater than at 10 and 20 wk (Fig. 3B). Between 20 and 30 wk, there was a significant increase in myoD abundance in WT mice (P < 0.001), whereas in KO mice, abundance was not significantly altered. Subsequently, at 30 wk, myoD abundance in KO was lower than WT (genotype × age; P = 0.003).
Protein tyrosine phosphatase 1B (PTP1B) was greater in the KO mice in comparison with WT (P = 0.06), with a genotype × age interaction observed at 3 wk (P = 0.029; Fig. 3C). The expression of PTP1B decreased as animals aged (P < 0.001); however, protein abundance significantly increased between the 20- and 30-wk ages (P < 0.02), irrespective of genotype.
Protein abundance of NCAM was unaffected by the main effects of sex, age, and genotype (Fig. 3D). However, at 30 wk, KO mice showed significantly less NCAM in comparison with WT animals (genotype × age; P = 0.0231).
DISCUSSION
This study investigated the role of the calpain proteolytic system in muscle growth, development, and atrophy, using μ-calpain null mice, and evaluated the subsequent effects of silencing this gene on other proteolytic systems. Knocking out the μ-calpain catalytic subunit had no effect on muscle mass of the heart, nor any of the skeletal muscles quantitatively dissected in this study. Body weights of the mature mice at 30 wk were greater in WT in comparison with KO, and this difference was attributed to increased liver weights. Although no differences in muscle mass were detected between the 2 genotypes, both m-calpain protein expression and activity were significantly greater in the KO mice, indicating that m-calpain was up regulated to compensate for the lack of μ-calpain expression. This up regulation was particularly apparent in the younger mice when animals were rapidly growing, as indicated by the genotype × age interactions observed. These differences in m-calpain abundance between genotypes became minimal as the animals matured, which coincided with differences in BW.
Calpastatin is the specific inhibitor of μ- and mcalpains (Parr et al., 2004). Protein expression of fulllength calpastatin was significantly greater in the μ-calpain KO mice in comparison with WT. Calpastatin expression peaked at 10 wk, whereas m-calpain was greatest at 5 wk, suggesting that the increase in m-calpain abundance observed preceded that of calpastatin. This, in turn, indicates that calpastatin is up regulated in the KO mice in response to the increased m-calpain abundance detected, as twice as much calpastatin is required to control m-calpain as μ-calpain (Goll et al., 2003). There were no differences between the genotypes in protein abundance of any of the cleaved calpastatin isoforms detected, suggesting that increased expression of full-length calpastatin occurred via an up regulation in the KO mice, rather than an increase in degradation in WT. In contrast to protein expression, calpastatin activity tended to be less in the KO mice and decreased with age, which could be a reflection of the complex of calpastatin. For example, multiple isoforms of calpastatin exist, which are controlled at the transcriptional, translational, and post-translational levels, and each has discrete functionality in controlling calpain activity (Parr et al., 2004; Raynaud et al., 2005). Consequently, the observed up regulation at the translational level was primarily to control m-calpain, but the actual inhibitory effect of calpastatin was unaffected by genotype.
Given the numerous interactions reported between the calpain and caspases systems (Wang, 2000; Barnoy and Kosower, 2003; Neumar et al., 2003; Harwood et al., 2005), it was imperative that activity of effector caspases 3 and 7 were assessed. Caspases 3/7 are the end point of the apoptotic pathway, responsible for targeting and cleaving specific substrates (Chua et al., 2000). Like m-calpain, caspase 3/7 activity was significantly greater in KO mice, suggesting that caspases were also up regulated to compensate for the lack of μ-calpain and that m-calpain and caspase 3/7 were working in conjunction with each other. This difference coincided with when the largest differences in m-calpain and calpastatin were detected. Although m-calpain and caspases were both up regulated in KO mice during the early growth period (3 to 10 wk), patterns of expression and activity were different; although m-calpain increased during this period before declining, caspase activity continuously decreased. The observed decrease in caspase 3/7 activity as the animals aged is consistent with previous observations (Ruest et al., 2002), indicating that, in skeletal muscle, effector caspases are regulated at the post-transcriptional level. Thus, it is likely that although these 2 proteolytic systems are working in conjunction with each other, the signaling pathways involved and the roles of caspases and calpains in muscle growth and development are, in fact, very distinct.
In contrast to m-calpain and caspases 3/7, μ-calpain KO mice had lower proteasome activity and, again, this difference in activity was most apparent at 5 wk of age. However, as the mice aged, the genotype differences in proteasome activity disappeared, corresponding with the ages when no differences in either m-calpain or caspase 3/7 were observed. These results agree with calpain 3 KO mice studies, where mRNA abundance of 20S, the proteolytic unit of the proteasome, was decreased in the KO mice in comparison with WT (Combaret et al., 2003). In muscle-wasting conditions, proteasome-mediated degradation is often secondary to calpain-mediated proteolysis, indicative that its pathway is downstream to that of calpains (Briguet et al., 2008). Thus, perhaps the decrease in proteasome activity observed is a protective mechanism ensuring that mass protein degradation and atrophy does not occur, and is necessary to maintain a homeostasis of proteolytic activity, protein turnover, and muscle mass.
To gain further understanding of how the genotype affected muscle growth and development, DNA, RNA, and protein contents and concentrations were measured, reflecting the activities of protein accumulation. The temporal abundance of total DNA and RNA content reflected the differences observed in growth rates and muscle mass among age groups and between genders, rather than between genotypes. However, differences in ratios of nucleic acids and protein were detected between genotypes. At 30 wk, KO mice had greater protein:DNA ratio, an indicator of myonuclear domain size and RNA:DNA ratio, an indirect measure of ribosomal abundance (Fiorotto et al., 2003). This observation suggests that at 30 wk, the μ-calpain KO mice have an increased potential for protein synthesis and muscle hypertrophy, in comparison with their age-matched counterparts. However, this potential was not realized in the current experiment.
Differences in growth and development between genotypes were further highlighted in body composition analysis. In WT mice, protein content steadily decreased after 10 wk, whereas in the KO mice, a reduction in protein content was not observed until 20 wk. Together with the increased ratios of protein:DNA and RNA:DNA, these data suggest that KO mice have an increased ability to maintain protein content. Furthermore, this apparent ability to maintain protein content is accompanied by a reduction in lipid accumulation. Although genetic variation in the gene encoding for calpain 10 has been associated with obesity and type II diabetes in humans (Horikawa et al., 2000), there is no direct evidence to suggest a role for μ-calpain in lipid metabolism. Nonetheless, accretion rates of lipid varied significantly between genotypes, with KO mice having greater lipid accretion rates between the 3- to 5-wk period than WT, which in turn is reversed between the 5- to 10-wk growth period. Although we do not have a mechanistic explanation for this observation, it is important to highlight that these differences coincide with when the largest variations in protease expression and activity occurred between the genotypes.
Histology analysis also identified differences in muscle growth between genotypes. In TA, KO mice had an increase in the proportion of Type IIB fibers. At 30 wk, these fibers were significantly larger than those in WT mice. This coincided with the increased protein:DNA and RNA:DNA ratios, signifying an increased ability to accumulate and/or maintain protein mass, as seen in the percentage of protein composition at 20 and 30 wk in the KO mice. This increase in Type IIB fibers corresponds with muscle analysis in callipyge sheep, a phenotype caused by massive up regulation of calpastatin. Muscles affected by the callipyge phenotype undergo substantial postnatal muscle hypertrophy, characterized by an increase in the proportion and size of Type IIB fibers (Carpenter et al., 1996), and an increase in DNA, RNA, and protein content, and RNA:DNA ratio (Koohmaraie et al., 1995). Additionally, in agreement with our body composition data, callipyge sheep have increased lean muscle mass and decreased fat content (Jackson et al., 1997), further implicating the calpain system in lipid metabolism.
Silencing the Capn1 gene also affected soleus muscle histology. In KO mice, soleus muscle, Type I fibers, the predominant fiber type, was smaller in both diameter and area, but more numerous than in WT mice, thus neither the muscle mass nor the metabolic capacity differed between genotypes. In addition, transition from Type I and IIA to Type IIB fibers in the soleus was observed. Fiber type conversion is not uncommon, yet the extent and the mechanisms behind these transformations vary between species and muscles, and can be affected by external factors including nutrition and physical stimulation (Picard et al., 2002). Thus, the decrease in Type IIB fibers observed at 5 wk was probably in response to weaning at 3 wk and reflects this change in nutrition. Changes in fiber type also occur in atrophying muscle, a condition in which calpains are known to be involved (Pette and Staron, 2001). Thus, the observed fiber plasticity suggests that down-regulating calpains could protect against muscle degeneration.
Regulation of muscle mass is influenced by numerous transcription factors, including myogenic regulatory factors (MRF), myoD, and myogenin, which have a major role in myogenesis (Favier et al., 2008). Myogenin protein abundance tended to be greater in the KO mice in comparison with WT, with significantly greater abundance detected at 5 wk. These results are consistent with those of Moyen et al. (2004), who showed overexpression of μ-calpain decreased myogenin expression. Moreover, this difference in myogenin expression coincided with when the major differences in protease activities were observed. However, what impact this increase in myogenin expression has on growth and development is unclear as no differences were observed between genotypes at 5 wk for any of the growth parameters measured. Based on these observations, we can only speculate that this increase in myogenin in the KO mice is a reflection of ensuring correct muscle development, rather than enhanced growth. In contrast to myogenin, no differences in myoD protein abundance were detected during the early growth period between the genotypes. Moreover, the KO mice showed reduced abundance of this MRF in comparison with WT at 30 wk. This difference in myoD protein expression between genotypes could be indicative of another function of myoD, promoting muscle regeneration in response to muscle damage. In myoD−/− mice, muscle regeneration is completely impaired, such that myogenic stem cells undergo several rounds of division but return to their quiescent state, rather than engaging in the developmental program (Sabourin and Rudnicki, 2000). Interestingly, in myoD−/− myogenic cells, desmin, a key structural protein in the myofibrillar lattice and a known calpain substrate (Goll et al., 2008), is not expressed (Sabourin and Rudnicki, 2000). The decreased protein abundance of myoD at 30 wk indicates that muscle from KO mice had been exposed to less muscle damage and was undergoing less regeneration. Less muscle regeneration in KO mice is further substantiated by reduced NCAM abundance at 30 wk. Neural cell adhesion molecule is a good marker for regeneration, revealing newly regenerating myofibers, which in turn indicates damage previously experienced by the muscle (Murphy et al., 2008).
Protein tyrosine phosphatase 1B (PTP1B) has been identified as a physiological target of μ-calpain in mouse platelets (Kuchay et al., 2007). Similarly, in the current study, PTP1B protein expression in skeletal muscle was lower in WT mice than KO mice at 3 wk of age. The PTP1B can act as a negative regulator of insulin and IGF-I signaling, dephosphorylating the phosphotyrosine residues of the type 1 IGF receptor and insulin receptor subtrate-1 (Klaman et al., 2000). Insulin-like growth factor-I is a known anabolic agent of skeletal muscle activating the Akt/TSC2/mTOR pathway; which are crucial regulators of skeletal muscle hypertrophy and promote protein synthesis (Favier et al., 2008). Like PTP1B, IGF abundance in skeletal muscle is high at birth and decreases as muscles mature. Therefore, the increased PTP1B abundance detected at 3 to 5 wk could indicate that increased expression is required to control insulin and IGF-I during this rapid growth phase. Furthermore, Trümpler et al. (2009) demonstrated that insulin signaling was diminished in μ-calpain−/− cells and this may explain the differences in lipid deposition observed between genotypes. These observations suggest a potential role for calpains in mediating insulin and IGF-I signaling through its actions on PTP1B.
In conclusion, silencing the μ-calpain gene induces up regulation of m-calpain and caspases 3/7 in a compensatory manner. Because no differences in muscle mass were observed between genotypes, it is likely that this compensation is predominantly to maintain muscle protein homeostasis and ensure normal growth and development, particularly during the early growth phase. This is further substantiated with the lower proteasome activity observed in the KO mice and indicates that these protease systems are functionally associated within skeletal muscle. Furthermore, we demonstrated that μ-calpain KO mice exhibit greater protein:DNA and RNA:DNA ratios, larger and more numerous Type IIB fibers, and increased protein body composition, suggesting an increased capacity to accrue and maintain muscle protein content. Moreover, the decreased expression of muscle regeneration factors myoD and NCAM indicates less muscle degeneration and lower levels of fiber type plasticity, which are indicative of muscle atrophy. The potential changes in protein synthesis and degradation and/or atrophy likely limited changes in muscle mass in the current study. Collectively, these data suggest that there are opportunities to target the calpain system to promote the growth and/or restoration of skeletal muscle mass.
Footnotes
Mention of trade names, proprietary products, or specified equipment does not constitute a guarantee or warranty by USDA and does not imply approval to the exclusion of other products that may be suitable. The USDA is an equal opportunity provider and employer.
LITERATURE CITED
- AOAC. Official methods of analysis. 15. Assoc. Off. Anal. Chem; Gaithersburg, MD: 1997. [Google Scholar]
- Ashmore CR, Doerr L. Comparative aspects of muscle fiber types in different species. Exp Neurol. 1971;31:408–418. doi: 10.1016/0014-4886(71)90243-3. [DOI] [PubMed] [Google Scholar]
- Azam M, Andrabi SS, Sahr KE, Kamath L, Kuliopulos A, Chishti AH. Disruption of the mouse mu-calpain gene reveals an essential role in platelet function. Mol Cell Biol. 2001;21:2213–2220. doi: 10.1128/MCB.21.6.2213-2220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badalamente MA, Stracher A. Delay of muscle degeneration and necrosis in mdx mice by calpain inhibition. Muscle Nerve. 2000;23:106–111. doi: 10.1002/(sici)1097-4598(200001)23:1<106::aid-mus14>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Barnoy S, Kosower NS. Caspase-1-induced calpastatin degradation in myoblast differentiation and fusion: Cross-talk between the caspase and calpain systems. FEBS Lett. 2003;546:213–217. doi: 10.1016/s0014-5793(03)00573-8. [DOI] [PubMed] [Google Scholar]
- Briguet A, Erb M, Courdier-Fruh I, Barzaghi P, Santos G, Herzner H, Lescop C, Siendt H, Henneboehle M, Weyermann P, Magyar JP, Dubach-Powell J, Metz G, Meier T. Effect of calpain and proteasome inhibition on Ca2+-dependent proteolysis and muscle histopathology in the mdx mouse. FASEB J. 2008;22:4190–4200. doi: 10.1096/fj.07-099036. [DOI] [PubMed] [Google Scholar]
- Brule C, Dargelos E, Diallo R, Listrat A, Bechet D, Cottin P, Poussard S. Proteomic study of calpain interacting proteins during skeletal muscle aging. Biochimie. 2010;92:1923–1933. doi: 10.1016/j.biochi.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Carpenter CE, Rice OD, Cockett NE, Snowder GD. Histology and composition of muscles from normal and callipyge lambs. J Anim Sci. 1996;74:388–393. doi: 10.2527/1996.742388x. [DOI] [PubMed] [Google Scholar]
- Chua BT, Guo K, Li P. Direct cleavage by the calcium-activated protease calpain can lead to inactivation of caspases. J Biol Chem. 2000;275:5131–5135. doi: 10.1074/jbc.275.7.5131. [DOI] [PubMed] [Google Scholar]
- Combaret L, Bechet D, Claustre A, Taillandier D, Richard I, Attaix D. Down-regulation of genes in the lysosomal and ubiquitin-proteasome proteolytic pathways in calpain-3-deficient muscle. Int J Biochem Cell Biol. 2003;35:676–684. doi: 10.1016/s1357-2725(02)00357-6. [DOI] [PubMed] [Google Scholar]
- Dargelos E, Poussard S, Brule C, Daury L, Cottin P. Calcium-dependent proteolytic system and muscle dysfunctions: A possible role of calpains in sarcopenia. Biochimie. 2008;90:359–368. doi: 10.1016/j.biochi.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Dayton WR, Goll DE, Zeece MG, Robson RM, Reville WJ. A Ca2+-activated protease possibly involved in myofibrillar protein turnover. Purification from porcine muscle. Biochemistry. 1976;15:2150–2158. doi: 10.1021/bi00655a019. [DOI] [PubMed] [Google Scholar]
- Dedieu S, Poussard S, Mazeres G, Grise F, Dargelos E, Cottin P, Brustis JJ. Myoblast migration is regulated by calpain through its involvement in cell attachment and cytoskeletal organization. Exp Cell Res. 2004;292:187–200. doi: 10.1016/j.yexcr.2003.08.014. [DOI] [PubMed] [Google Scholar]
- Favier FB, Benoit H, Freyssenet D. Cellular and molecular events controlling skeletal muscle mass in response to altered use. Pflugers Arch. 2008;456:587–600. doi: 10.1007/s00424-007-0423-z. [DOI] [PubMed] [Google Scholar]
- Fiorotto ML, Schwartz RJ, Delaughter MC. Persistent IGF-I overexpression in skeletal muscle transiently enhances DNA accretion and growth. FASEB J. 2003;17:59–60. doi: 10.1096/fj.02-0289fje. [DOI] [PubMed] [Google Scholar]
- Geesink GH, Kuchay S, Chishti AH, Koohmaraie M. Micro-calpain is essential for postmortem proteolysis of muscle proteins. J Anim Sci. 2006;84:2834–2840. doi: 10.2527/jas.2006-122. [DOI] [PubMed] [Google Scholar]
- Goll DE, Neti G, Mares SW, Thompson VF. Myofibrillar protein turnover: The proteasome and the calpains. J Anim Sci. 2008;86:E19–E35. doi: 10.2527/jas.2007-0395. [DOI] [PubMed] [Google Scholar]
- Goll DE, V, Thompson F, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–780. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- Harwood SM, Yaqoob MM, Allen DA. Caspase and calpain function in cell death: Bridging the gap between apoptosis and necrosis. Ann Clin Biochem. 2005;42:415–431. doi: 10.1258/000456305774538238. [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, del Bosque-Plata L, Horikawa Y, Oda Y, Yoshiuchi I, Colilla S, Polonsky KS, Wei S, Concannon P, Iwasaki N, Schulze J, Baier LJ, Bogardus C, Groop L, Boerwinkle E, Hanis CL, Bell GI. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Miller MF, Green RD. Phenotypic characterization of Rambouillet sheep expressing the callipyge gene: II. Carcass characteristics and retail yield. J Anim Sci. 1997;75:125–132. doi: 10.2527/1997.751125x. [DOI] [PubMed] [Google Scholar]
- Klaman LD, Boss O, Peroni OD, Kim JK, Martino JL, Zabolotny JM, Moghal N, Lubkin M, Kim YB, Sharpe AH, Stricker-Krongrad A, Shulman GI, Neel BG, Kahn BB. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol Cell Biol. 2000;20:5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koohmaraie M, Shackelford SD, Wheeler TL, Lonergan SM, Doumit ME. A muscle hypertrophy condition in lamb callipyge: Characterization of effects on muscle growth and meat quality traits. J Anim Sci. 1995;73:3596–3607. doi: 10.2527/1995.73123596x. [DOI] [PubMed] [Google Scholar]
- Kuchay SM, Kim N, Grunz EA, Fay WP, Chishti AH. Double knockouts reveal that protein tyrosine phosphatase 1B is a physiological target of calpain-1 in platelets. Mol Cell Biol. 2007;27:6038–6052. doi: 10.1128/MCB.00522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyen C, Goudenege S, Poussard S, Sassi AH, Brustis JJ, Cottin P. Involvement of micro-calpain (CAPN 1) in muscle cell differentiation. Int J Biochem Cell Biol. 2004;36:728–743. doi: 10.1016/S1357-2725(03)00265-6. [DOI] [PubMed] [Google Scholar]
- Murphy JL, Blakely EL, Schaefer AM, He L, Wyrick P, Haller RG, Taylor RW, Turnbull DM, Taivassalo T. Resistance training in patients with single, large-scale deletions of mitochondrial DNA. Brain. 2008;131:2832–2840. doi: 10.1093/brain/awn252. [DOI] [PubMed] [Google Scholar]
- Neumar RW, Xu YA, Gada H, Guttmann RP, Siman R. Cross-talk between calpain and caspase proteolytic systems during neuronal apoptosis. J Biol Chem. 2003;278:14162–14167. doi: 10.1074/jbc.M212255200. [DOI] [PubMed] [Google Scholar]
- Parr T, Jewell KK, Sensky PL, Brameld JM, Bardsley RG, Buttery PJ. Expression of calpastatin isoforms in muscle and functionality of multiple calpastatin promoters. Arch Biochem Biophys. 2004;427:8–15. doi: 10.1016/j.abb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Transitions of muscle fiber phenotypic profiles. Histochem. Cell Biol. 2001;115:359–372. doi: 10.1007/s004180100268. [DOI] [PubMed] [Google Scholar]
- Picard B, Lefaucheur L, Berri C, Duclos MJ. Muscle fiber ontogenesis in farm animal species. Reprod Nutr Dev. 2002;42:415–431. doi: 10.1051/rnd:2002035. [DOI] [PubMed] [Google Scholar]
- Raynaud P, Gillard M, Parr T, Bardsley R, Amarger V, Leveziel H. Correlation between bovine calpastatin mRNA transcripts and protein isoforms. Arch Biochem Biophys. 2005;440:46–53. doi: 10.1016/j.abb.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Ruest LB, Khalyfa A, Wang E. Development-dependent disappearance of caspase-3 in skeletal muscle is post-transcriptionally regulated. J Cell Biochem. 2002;86:21–28. doi: 10.1002/jcb.10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- Shackelford SD, Koohmaraie M, Cundiff LV, Gregory KE, Rohrer G, Savell JW. Heritabilities and phenotypic and genetic correlations for bovine postrigor calpastatin activity, intramuscular fat content, Warner-Bratzler shear force, retail product yield, and growth rate. J Anim Sci. 1994;72:857–863. doi: 10.2527/1994.724857x. [DOI] [PubMed] [Google Scholar]
- Solomon MB, Dunn MC. Simultaneous histochemical determination of three fiber types in single sections of ovine, bovine and porcine skeletal muscle. J Anim Sci. 1988;66:255–264. doi: 10.2527/jas1988.661255x. [DOI] [PubMed] [Google Scholar]
- Sorimachi H, Imajoh-Ohmi S, Emori Y, Kawasaki H, Ohno S, Minami Y, Suzuki K. Molecular cloning of a novel mammalian calcium-dependent protease distinct from both m- and mu-types. Specific expression of the mRNA in skeletal muscle. J Biol Chem. 1989;264:20106–20111. [PubMed] [Google Scholar]
- Strucksberg KH, Tangavelou K, Schroder R, Clemen CS. Proteasomal activity in skeletal muscle: A matter of assay design, muscle type, and age. Anal Biochem. 2010;399:225–229. doi: 10.1016/j.ab.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Thompson VF, Saldana S, Cong J, Goll DE. A BODIPY fluorescent microplate assay for measuring activity of calpains and other proteases. Anal Biochem. 2000;279:170–178. doi: 10.1006/abio.1999.4475. [DOI] [PubMed] [Google Scholar]
- Trümpler A, Schlott B, Herrlich P, Greer PA, Böhmer FD. Calpain-mediated degradation of reversibly oxidized protein- tyrosine phosphatase 1B. FEBS J. 2009;276:5622–5633. doi: 10.1111/j.1742-4658.2009.07255.x. [DOI] [PubMed] [Google Scholar]
- Veiseth E, Shackelford SD, Wheeler TL, Koohmaraie M. Effect of postmortem storage on mu-calpain and m-calpain in ovine skeletal muscle. J Anim Sci. 2001;79:1502–1508. doi: 10.2527/2001.7961502x. [DOI] [PubMed] [Google Scholar]
- Wagner KD, Wagner N, Schley G, Theres H, Scholz H. The Wilms’ tumor suppressor Wt1 encodes a transcriptional activator of the class IV POU-domain factor Pou4f2 (Brn-3b) Gene. 2003;305:217–223. doi: 10.1016/s0378-1119(02)01231-3. [DOI] [PubMed] [Google Scholar]
- Wang KK. Calpain and caspase: Can you tell the difference? Trends Neurosci. 2000;23:20–26. doi: 10.1016/s0166-2236(99)01536-2. [DOI] [PubMed] [Google Scholar]
- Wheeler TL, Koohmaraie M. The extent of proteolysis is independent of sarcomere length in lamb longissimus and psoas major. J Anim Sci. 1999;77:2444–2451. doi: 10.2527/1999.7792444x. [DOI] [PubMed] [Google Scholar]