Abstract
Purpose
We present here the first report of PID1 (Phosphotyrosine Interaction Domain containing 1; NYGGF4) in cancer. PID1 was identified in 2006 as a gene that modulates insulin signaling and mitochondrial function in adipocytes and muscle cells.
Experimental Design and Results
Using four independent medulloblastoma datasets, we show that mean PID1 mRNA levels were lower in unfavorable medulloblastomas (Groups 3 and 4, and anaplastic histology) compared with favorable medulloblastomas (SHH and WNT groups, and desmoplastic/nodular histology) and with fetal cerebellum. In two large independent glioma datasets PID1 mRNA was lower in glioblastomas (GBMs), the most malignant gliomas, compared to other astrocytomas, oligodendrogliomas and non-tumor brains. Neural and proneural GBM subtypes had higher PID1 mRNA compared to classical and mesenchymal GBM. Importantly, overall survival and radiation-free progression-free survival were longer in medulloblastoma patients with higher PID1 mRNA (univariate and multivariate analyses). Higher PID1 mRNA also correlated with longer overall survival in glioma and GBM patients. In cell culture, overexpression of PID1 inhibited colony formation in medulloblastoma, atypical teratoid rhabdoid tumor (ATRT) and GBM cell lines. Increasing PID1 also increased cell death and apoptosis, inhibited proliferation, induced mitochondrial depolarization, and decreased serum-mediated phosphorylation of AKT and ERK in medulloblastoma, ATRT and/or GBM cell lines, whereas siRNA to PID1 diminished mitochondrial depolarization.
Conclusions
These data are the first to link PID1 to cancer and suggest that PID1 may have a tumor inhibitory function in these pediatric and adult brain tumors.
Keywords: ATRT, apoptosis, brain cancer, glioma, glioblastoma, medulloblastoma, PID1 (NYGGF4), proliferation
INTRODUCTION
Here, we report on PID1 (Phosphotyrosine interaction domain containing 1) in two important groups of brain tumors: embryonal brain tumors (medulloblastomas and atypical teratoid rhabdoid tumors (ATRT)) and gliomas. Medulloblastomas are the most common malignant brain tumors in children, where in high-risk disease prognosis remains poor. Moreover, in younger children, therapies that include whole brain irradiation are associated with serious long-term sequelae that hamper quality of life of survivors (1, 2). ATRT is another poor-prognosis, highly malignant embryonal brain cancer of young children. Most ATRTs have lost expression and/or function of the INI1 (SMARCB1) tumor suppressor gene, which is part of the SWI/SNF chromatin-remodeling complex (3–5). Malignant gliomas, including glioblastoma multiforme (GBM), comprise the most common primary malignant brain tumors in adults and also carry poor prognosis (6, 7). Here, we provide novel clinical correlations of PID1 in medulloblastomas and gliomas, and demonstrate tumor-inhibitory effects of PID1 in cell lines of these three brain tumors.
The PID1 gene (also called NYGGF4 and PTB-containing, cubilin and LRP1-interacting protein; PCLI1; rhymes with “BID-1”) was identified in 2006 based on its differential expression in adipose tissue of obese compared to non-obese subjects (8). To date, PID1 has not been reported in the context of cancer. While PID1 is known to contain a phosphotyrosine binding (PTB) domain/phosphotyrosine interaction domain (PID) (8), the molecular mechanism(s) underlying its activities are poorly understood. PID1 mRNA increases during differentiation of 3T3-L1 pre-adipocytes to adipocytes (8), and is lower in brains of Alzheimer patients compared to controls (9). PID1 overexpression increases proliferation of 3T3-L1 pre-adipocytes, but does not alter their adipogenic differentiation (8). In NIH-3T3 cells, however, overexpression of PID1 amino acids 84–230, which includes its PTB domain, causes cell cycle arrest (10), suggesting that PID1 function may differ depending on cellular context.
Pilot microarray expression analysis identifying PID1 mRNA level as highly correlated with outcome in medulloblastomas. A similar correlation of PID1 mRNA levels in gliomas using publicly-available microarray datasets prompted us to study PID1 in more detail. Analysis revealed highly significant correlations between PID1 mRNA and survival and between PID1 mRNA and subgroup and/or grade in medulloblastomas and gliomas across six independent datasets. Moreover, ectopic expression of PID1 in embryonal brain tumor cell lines (medulloblastomas and ATRT) and GBM cell lines showed consistent growth-inhibitory effects. These data are the first to link PID1 to cancer in general and to brain tumors in particular, and to suggest that PID1 may have a growth-inhibitory function in medulloblastomas, glioblastomas and ATRT.
METHODS
Details for additional methods can be found in Supplemental Materials.
Patients, samples and mRNA expression data
Medulloblastoma specimens and clinical records from 81 children diagnosed at Children’s Hospital Los Angeles (CHLA) between 1989 and 2008 were obtained according to a protocol approved by the local institutional Review Board. Patient and sample characteristics are described in Supplemental Table 1. The Heidelberg microarray dataset includes 446 medulloblastomas, profiled on Affymetrix U133 plus2.0 arrays, and is a combination of published data on 230 patients obtained from the Gene Expression Omnibus (GSE10327, GSE12992, and GSE37418) (11–13) and unpublished data on 216 additional patients from Heidelberg (Kool and Pfister, unpublished data). The published Toronto microarray dataset includes 103 medulloblastomas profiled on Affymetrix Human Exon Array data (GSE21140), and the published Boston microarray dataset includes 194 medulloblastomas profiled on Affymetrix U133A (14, 15). Molecular subgroups of medulloblastoma were identified either by using available published data, or by cluster analyses to assign molecular subgroup for the unpublished datasets (11, 12, 14), or for the CHLA analysis, by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and a medulloblastoma gene signature derived from prior microarray studies (15, 16) and a CHLA study (manuscript in preparation). Glioma PID1 mRNA microarray data in REMBRANDT (REpository for Molecular BRAin Neoplasia DaTa) were analyzed using Affymetrix U133 plus2.0 arrays (17). Glioblastoma PID1 mRNA data in TCGA (The Cancer Genome Atlas; The Cancer Genome Atlas Research Network, National Cancer Institute and National Human Genome Research Institute, Bethesda, MD, USA) were analyzed using AgilentG4502A_07) (18). For GBM clinical and PID1 correlations, publicly available data for 196 patients was obtained from TCGA and Verhaak et al (19), and was accessed at https://tcga-data.nci.nih.gov/docs/publications/gbm_exp/ (files: unifiedScaled.txt and TCGA_unified_CORE_ClaNC840.txt) and https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm?mode=ApplyFilter&diseaseType=GBM (files: clinical_patient_gbm.txt and clinical_follow_up_v1.0_gbm.txt) on February 18, 2013. RNA from CHLA medulloblastoma samples was extracted and processed using a previously published method (20). PID1 mRNA levels were obtained by qRT-PCR (PID1: Forward Primer: 5′-GATTGCTGGCAACCACCTGATGT-3′, reverse primer: 5′-AAATGTAAAGGTTGGCAGGGCCAC-3′, probe: 5′-TCGAGGACATGTTCTTCTCCAGACGT-3′) and after normalization to three housekeeping genes (GAPDH/SDHA/HPRT1).
AnnexinV, cell proliferation and mitochondrial depolarization
Assays were performed on green fluorescent protein (GFP)-positive cells 24 hr after transfections. AnnexinV staining was done by flow cytometry using the APC AnnexinV kit (Cat. No. 550474, BD Pharmingen) according to manufacturer’s instructions. Cell proliferation and viability were assessed by flow cytometry using the APC BrdU Flow Kit (Cat. No. 552598, BD Pharmingen). Mitochondrial depolarization was measured by flow cytometry using the MitoProbe™ DilC1(5) Assay Kit for Flow Cytometry (Cat. No. M34151, Invitrogen).
Cell Culture
The cell lines used were: GBM (U87, U251, LN18, LN229, CHLA-07-BSGBM), medulloblastoma (D283MED, D425MED, UW-228-2, CHLA-259, CHLA-01-MED, CHLA-01R-MED) and ATRT (BT-12, CHLA-05-ATRT, CHLA-06-ATRT). Details for their culture are in Supplemental Materials.
Plasmids
PID1 variant 1 (NM_017933.4; PID1-1) and variant 2 (NM_001100818.1; PID1-2) human cDNA ORFs with a turbo-GFP (tGFP) C-terminal tag in pCMV6-AC-tGFP (pCMV6-AC-PID1-tGFP) were from OriGene (Cat# PS100010, RG212451, RG212505). PID1 in the pCLS-2A-eGFP plasmid (21) (from Dr. Lawlor, U. Michigan) was expressed 5′ to the foot-and mouth-disease virus-derived 2A self-cleaving peptide sequence. In pCIENS (a pcDNA3.1-based CMV promoter-driven expression vector that also expresses eGFP via an EMCV IRES), PID1 (variant 1) was expressed from the CMV promoter.
Statistical Analysis
Details of the statistical analysis of patient-related information can be found in the Supplemental Materials.
In vitro experiments were analyzed using GraphPad Prism version 5.0 for MAC (GraphPad Software, San Diego California USA, www.graphpad.com). Results are depicted as mean±SEM from at least three independent experiments unless stated otherwise. P-values represent unpaired two-sided Student’s t-test unless stated differently.
RESULTS
PID1 mRNA is higher in favorable medulloblastomas and correlates with longer rf-PFS and OS in medulloblastoma patients
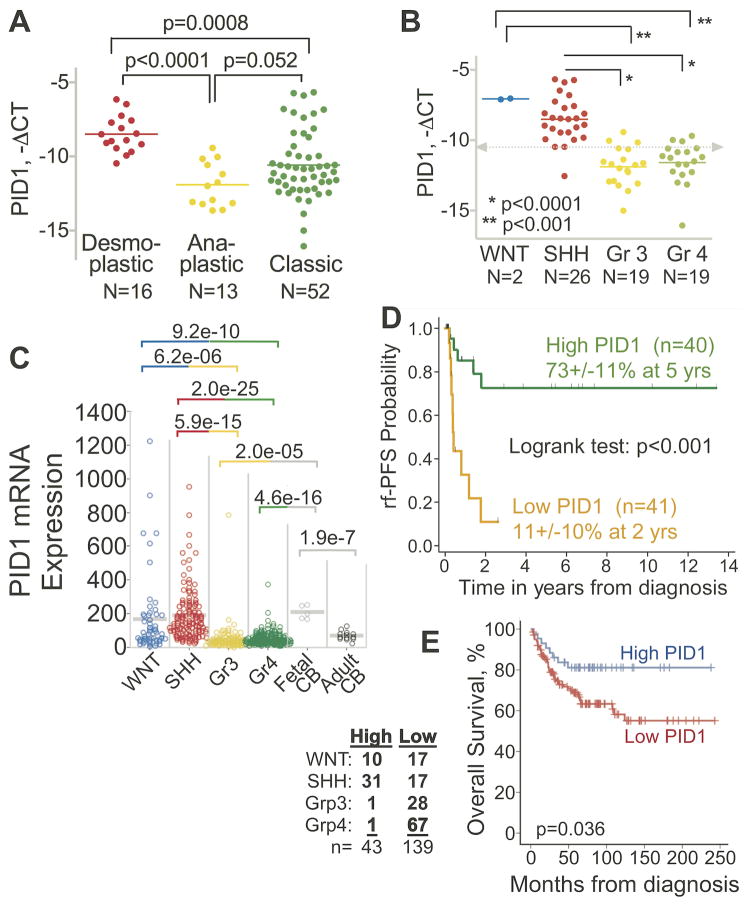
Quantitative RT-PCR of 81 pediatric medulloblastoma tumors showed that mean PID1 mRNA levels were significantly higher in medulloblastomas with desmoplastic/nodular histology compared to those with anaplastic histology (p<0.001; Fig 1A; patient characteristics in Supplemental Table 1). Supporting this, analysis of PID1 in the four medulloblastoma core molecular subgroups (11, 22, 23) showed that SHH group medulloblastomas had higher mean PID1 mRNA compared with the less favorable subgroup, Group 3 and Group 4 (Fig 1B). Microarray data from three other independent medulloblastoma datasets similarly revealed higher mean PID1 mRNA in the more favorable subgroups (SHH and WNT) compared to medulloblastomas in Groups 3 and 4 (Fig 1C and Supplemental Fig 1A) (11–13). Mean expression of PID1 mRNA was higher in fetal cerebellum compared to Groups 3 and 4 medulloblastomas, and was similar to PID1 mRNA in the WNT and SHH groups (Fig 1C).
Figure 1. Group 3 and Group 4 medulloblastomas have lower PID1 mRNA than WNT and SHH groups; higher PID1 mRNA correlates with longer rf-PFS and OS in medulloblastomas.
A) Relative mRNA expression (real time qRT-PCR, -ΔCT) of PID1 according to medulloblastoma histology (desmoplastic/nodular, classic, anaplastic) in 81 CHLA patients. B) Relative PID1 mRNA (-ΔCT) of 66 of the tumors described in A for which molecular subgroup classification (WNT, SHH, Group 3, and Group 4) by qRT-PCR was available. Lines depict mean mRNA for each group; grey dashed line represents median PID1 mRNA of all samples. C) PID1 mRNA in medulloblastomas from the Heidelberg series, analyzed on Affymetrix U133 plus2.0, according to molecular classification. This series (n=446) is a combination of published data (11–13) on 230 patients and unpublished data on 216 additional patients from Heidelberg (Kool and Pfister, unpublished data). The medulloblastoma subgroup distribution in this cohort is: 54 WNT, 123 SHH, 98 Group 3, 171 Group 4. These were compared with 5 fetal cerebella and 13 adult cerebella. p-values comparing the groups are listed above the panels and are color-coded according to the groups compared. mRNA values are expressed in arbitrary units, using the MAS5.0 normalized Affymetrix values; D) Radiation-free progression free survival (rf-PFS; Kaplan Meier curve) according to PID1 mRNA (variant 1, NM_017933.4; real time qRT-PCR) in 81 CHLA medulloblastomas. Green curve - patients with above-the-median PID1 mRNA; Orange curve - patients with below-the-median PID1 mRNA. E) Kaplan Meier curve of OS for 182 patients with medulloblastoma from the series shown in panel C for whom OS data were available, plotted according to PID1 mRNA above 100 (High PID1; blue) or below 100 (Low PID1; red). Molecular subgrouping for the tumors of patients in the High PID1 and Low PID1 curves are detailed in the table left of panel.
In univariate analysis of the CHLA medulloblastoma patients (n=81, Supplemental Table 1), rf-PFS was significantly longer in children whose tumor PID1 mRNA was higher than the median of the cohort, compared to those with tumor PID1 mRNA lower than median (rf-PFS 73%±11% vs. 11%±10%, respectively; p<0.001, Fig 1D). Difference in rf-PFS also remained significant when dividing the group into equal tertiles according to PID1 mRNA (Suppl Fig 1B). In the non-CHLA medulloblastoma patients from Fig 1C for whom survival data were available, OS was significantly longer in patients with higher tumor PID1 mRNA compared to those with lower PID1 mRNA (Fig 1E), supporting the findings in the CHLA cohort (Fig 1D). In multivariate analysis of the CHLA cohort, risk of radiation-free disease progression was significantly higher in patients with lower-than-median PID1 mRNA compared to those with higher-than-median PID1 mRNA (Table 1).
Table 1.
Cox Proportional Hazards Model for Progression Free Survival (Patients who received irradiation as part of the primary treatment were censored at time of irradiation)
| Variables | Univariate Analyses | Multivariate Analyses | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| HR1 | 95% CI1 | p1 | HR2 | 95% CI2 | p2 | HR3 | 95% CI3 | p3 | |
| Age at Diagnosis | |||||||||
| Relative risk increase per 1 year | 1.2 | (1.05, 1.4) | 0.017 | 1.0 | (0.84, 1.3) | 0.78 | Not included in the model | ||
| >=3 year vs. <3 year | 3.5 | (1.3, 9.1) | 1.2 | (0.36, 3.8) | |||||
|
| |||||||||
| Stage4 | |||||||||
| M0 | 1.0 | -- | 0.006 | 1.0 | -- | 0.004 | 1.0 | -- | 0.002 |
| M+ | 4.7 | (1.4, 15) | 16 | (2.0, 128) | 7.9 | (1.8, 34) | |||
|
| |||||||||
| Histology4 | |||||||||
| Classic | 1.0 | -- | 0.21 | 1.0 | -- | 0.27 | |||
| Desmoplastic/nodular | 0.42 | (0.09, 2.0) | 3.8 | (0.29, 49) | Not included in the model | ||||
| Anaplastic/Large Cell | 2.0 | (0.54, 7.6) | 0.35 | (0.06, 1.9) | |||||
|
| |||||||||
| Molecular Subgroup4 | |||||||||
| SHH/WNT | 1.0 | -- | 0.048 | 1.0 | -- | 0.75 | |||
| Group 3 | 6.2 | (1.5, 26) | 2.7 | (0.17, 44) | Not included in the model | ||||
| Group 4 | 3.2 | (0.58, 18) | 2.1 | (0.12, 37) | |||||
|
| |||||||||
| PID1 | |||||||||
| High | 1.0 | -- | 1.0 | -- | 1.0 | -- | |||
| Low | 6.8 | (2.2, 21) | <0.001 | 21 | (1.9, 221) | 0.005 | 12.1 | (2.8, 52) | <0.001 |
Abbreviations: HR, hazard ratio; CI, confidence interval.
HR and CI from univariate Cox models.
HR, CI and p-values from the Cox model that included age, stage, histology, molecular subgroup and PID1 expression.
HR, CI and p-values from the Cox model that included stage and PID1 expression.
Missing data for these variables were included in the models as a “missing” category.
These analyses demonstrate that the clinically favorable medulloblastomas (desmoplastic/nodular histology, or WNT and SHH molecular subgroups) had higher mean PID1 mRNA compared to clinically unfavorable medulloblastomas (anaplastic histology or Group 3 and Group 4), and that higher medulloblastoma PID1 mRNA correlated with longer rf-PFS and OS.
GBM have lower PID1 mRNA compared to other gliomas and non-tumor brains; Higher PID1 mRNA correlates with longer overall survival in glioma patients
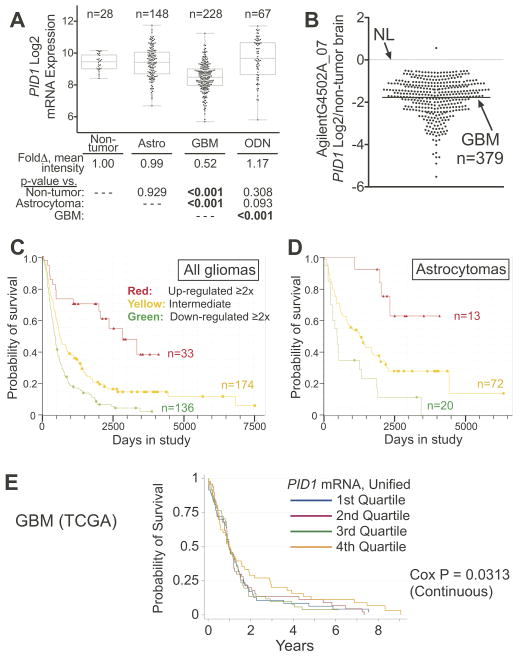
Analysis of PID1 mRNA data using the REMBRANDT glioma clinical genomics database (17) revealed that PID1 mRNA levels were lower in the highest grade gliomas, GBMs, compared to non-tumor brains, astrocytomas and oligodendrogliomas (Fig 2A). The TCGA glioma dataset (18) similarly showed that mean PID1 mRNA in GBM was significantly lower than its level in non-tumor brains (CI95% 0.34–0.38, p<0.0001; Fig 2B). For GBM patients for whom tumor molecular subtype (classical, mesenchymal, neural, proneural) and clinical data were available and unified PID1 mRNA expression values from three platforms in TCGA were analyzed (see Methods), there were significant differences in mean PID1 mRNA among the molecular subtypes (p<0.0001; Suppl Fig 2A, Supplemental Table 2). Neural and proneural GBM subtypes showed higher PID1 mRNA compared to classical and mesenchymal subtypes. All paired comparisons between GBM with known subtypes were also significantly different (after Bonferroni adjustment for multiple comparisons) with the exception of the Neural vs. Proneural pairing (p=0.88). Differences in PID1 mRNA between GBM subtypes remained significant even after adjusting for age at diagnosis (p<0.0001).
Figure 2. GBM tumors have lower PID1 mRNA compared to non-tumor brain and lower grade gliomas; higher tumor PID1 mRNA correlates with longer OS in patients with gliomas.
A) Relative PID1 mRNA in gliomas (GBM, non-GBM astrocytomas, oligodendrogliomas) compared to non-tumor brain and to each other; data from REMBRANDT (Affymetrix U133 plus2.0 arrays). Linear regression was utilized to analyze the PID1 mRNA Log2 expression by glioma type. The number of samples analyzed are depicted above each box/whiskers set. The table below the panel displays the mean fold-change of PID1 mRNA compared to non-tumor brain and the p-values for comparisons between the groups. Astro: astrocytomas, ODN: oligodendrogliomas. B) Log2 PID1 mRNA (219093_at probeset profiled on AgilentG4502A_07) of 379 GBM samples from TCGA compared to 10 non-cancer brain samples (downloaded March 2, 2013). The lines represent means. C–D) Kaplan Meier analysis (REMBRANDT) of OS according to PID1 mRNA in all glioma patients (≥2x: red; ≤2X: green; 0.5x to 2x: orange, compared to non-tumor brain) (C) or only in astrocytoma patients (D) using probeset 219093_at. The log rank p-values between the curves in panels C-D were significant among each of the comparisons and are detailed in Suppl Fig 2B. E) Survival of 196 patients with GBM according to PID1 unified mRNA level from the three platforms of publicly available data in TCGA (see methods). In Cox regression analysis (continuous), PID1 mRNA expression was univariately significantly associated with survival (p=0.031). Quartiles: 1st is lowest, 4th is highest.
Survival analysis using the REMBRANDT dataset revealed significant differences in OS among patients who were stratified according to PID1 mRNA. PID1 mRNA was directly correlated to OS in patients with any diagnosis of glioma, or within the subset of astrocytomas that does not include GBMs (Fig 2C–D, Suppl Fig 2B–E; using two available PID1 probesets). For GBMs in the TCGA dataset PID1 mRNA was univariately significantly associated with survival (Cox regression analysis, p=0.031; Fig 2E), but the REMBRANDT dataset did not reveal such correlation (not shown). While the PID1 mRNA association with OS in GBM (TCGA) was no longer significant after adjusting for molecular subtype (p=0.15), the relative failure rate decrease per unit increase in PID1 mRNA was only slightly attenuated in our analysis (0.83 ± 0.071 univariate, 0.85±0.094 multivariate). In Cox multivariate analysis, with age at diagnosis, and PID1 expression and subtype as variables (TCGA), PID1 was marginally significant (p=0.056). The relative failure rate decrease per unit increase in PID1 mRNA was 0.81 ± 0.091.
Thus, similar to our findings in medulloblastomas, the most malignant gliomas (i.e., GBMs) had lower PID1 mRNA compared to other gliomas or to non-tumor brains. Additionally, higher tumor PID1 mRNA correlated with longer OS in glioma patients.
PID1 confers growth disadvantage in brain tumor cell lines
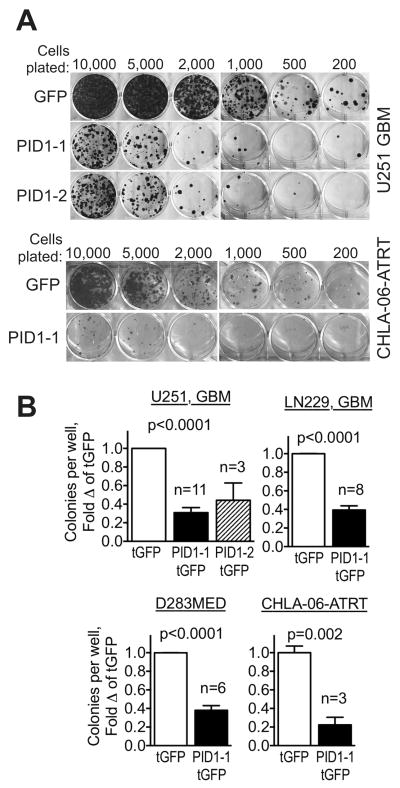
We next asked if the clinical correlations we found for PID1 mRNA pointed to a possible biological role for PID1 in brain tumors. To evaluate a role in growth, we assessed the effect of PID1 on colony formation in U251 GBM, LN229 GBM, D283MED medulloblastoma, and CHLA-06-ATRT brain tumor cell lines as follows: cells were transfected with PID1-tGFP or tGFP control for 24hr, tGFP-expressing cells were sorted by flow cytometry and plated at equal numbers, and colonies were allowed to form over two weeks without further selection. Each of the cell lines formed significantly fewer colonies when transfected with PID1-tGFP than did cells transfected with the control tGFP vector, despite an equal number of sorted tGFP+ cells plated for the tGFP control and PID1-tGFP transfected cells (Fig 3A–B). Both variants of PID1 (variant 1: NM_017933, variant 2: NM_001100818) showed similar effects in this assay. Similar inhibition of colony formation was also observed using the self-cleaving bicistronic pCLS-PID1-2A-eGFP compared with its control vector, pCLS-2A-eGFP (21) as well as with the bicistronic pCIENS-PID1 compared to pCIENS control vector (Suppl Fig 3A–B). Expression of PID1 at the time of flow-sorting and plating of the tGFP-expressing cells was verified by western blotting (24 hr after transfection, Suppl Fig 3C). These data indicate that PID1 confers growth and/or survival disadvantage upon brain tumor cell lines ectopically expressing PID1.
Figure 3. Overexpression of PID1 confers growth inhibition in medulloblastoma, GBM and ATRT cell lines.
A) Colony formation assays of flow-sorted tGFP-expressing U251 GBM and CHLA-06-ATRT cells transiently transfected with pCMV6-AC-tGFP or pCMV6-AC-PID1-tGFP (PID1 variants 1 or 2), seeded at the cell numbers indicated and stained with crystal violet stain after 2 weeks. B) Quantification of colony numbers in replicate experiments performed as in panel A. The number of independent experiments comprising the mean±SEM for each bar is depicted above the PID1 bars. Most experiments were performed in triplicates and the mean of each experiment was considered one value for calculation of the means of the experiments. P-values by Student’s t-test.
PID1 inhibits proliferation and promotes of cell death of cultured brain tumor cell lines
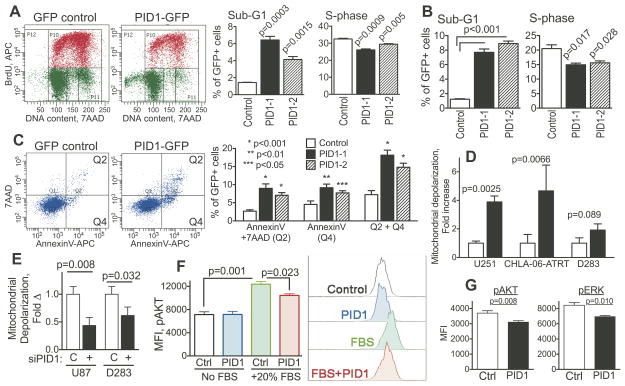
To characterize the PID1-induced growth disadvantage, we analyzed proliferation and cell death, measuring BrdU/7AAD uptake in tGFP-positive cells 24 hr after transient transfection with pCMV6-PID1-tGFP or pCMV6-tGFP control. GBM (U251) and medulloblastoma (UW-228-2) cell lines as well as primary medulloblastoma cells (CHLA-259) expressing PID1-tGFP had significantly more cells in the sub-G0/G1 phase compared to cells expressing tGFP+ control vector (Fig 4A–B and Suppl Fig 4), indicating that PID1 increased cell death. PID1-tGFP also decreased proliferation, as indicated by the decreased proportion of cells in S-phase (Figure 4A–B). Apoptosis accounted for at least some of the cell death, as annexinV binding was higher in PID1-tGFP-expressing cells compared to tGFP controls (Fig 4C). There were very few 7AAD-positive/annexinV-negative cells, indicating that necrosis was only minimal. As mitochondria are involved in metabolism and several forms of cell death and PID1 has been reported to induce mitochondrial dysfunction in adipocytes and myocytes (24–28), we examined the effect of PID1 on mitochondrial depolarization. U251 GBM, D283MED medulloblastoma and CHLA-06-ATRT cell lines transfected with PID1-tGFP showed increased depolarization of mitochondrial membrane potential compared with tGFP-transfected cells 24 hr after transfection (Fig 4D). Conversely, siRNA knockdown of PID1 in U87 GBM (U87 express PID1 protein; see Suppl Fig 5), diminished baseline depolarization of the mitochondrial membrane potential compared to non-silencing siRNA control (Fig 4E). siPID1 knockdown similarly diminished baseline mitochondrial depolarization in D283MED medulloblastoma cells (Fig 4E). These data demonstrate that the inhibitory effect of PID1 on brain tumor cell line growth is the result of both inhibition of proliferation and increased cell death, and is associated with depolarization of mitochondrial membrane potential.
Figure 4. Overexpression of PID1 diminishes proliferation, increases cell death and causes mitochondrial depolarization in brain tumor cell lines.
A) U251 GBM cells transiently transfected for 24 hr with pCMV6-AC-tGFP or pCMV6-AC-PID1-tGFP (PID1 variants 1 or 2) were analyzed by flow cytometry for BrdU uptake and 7AAD content in the tGFP-expressing cells. Upper panel: typical flow cytometry plot demonstrating decreased BrdU uptake and increased sub-G0/G1 in PID1-tGFP cells compared to controls. Lower panel: mean/SEM (n=3) from one of three similar experiments in U251 cells; B) BrdU and 7AAD uptake performed similarly to panel A using UW-228-2 medulloblastoma cells; n=3; C) AnnexinV and 7AAD staining in the tGFP-expressing U251 GBM cells 24 hr after transient transfection as in panel A. Top shows representative panels, bottom shows mean±SEM of three independent experiments; D) Mitochondrial depolarization measured by flow cytometry using MitoProbe assay in tGFP-expressing brain tumor cells 24 hr after transfection with tGFP control or PID1-tGFP; n=3–8 repeats. Plasmids were all pCMV6-AC-tGFP vs. pCMV6-AC-PID1-tGFP; E) Mitochondrial depolarization, measured as in D, in FAM-labeled U87 GBM and D283MED medulloblastoma cells treated for 48 hrs with non-silencing control siRNA (C) or siPID1; F) LN229 GBM cells transiently transfected with pCIENS (Control) or pCIENS-PID1 (PID1) and serum starved overnight were stimulated for 15 min with medium containing 20% FBS. Phosphorylation of AKT was measured by flow cytometry in the eGFP-expressing cells. Shown are means/SEM of triplicate measurements from one of two similar experiments and representative flow cytometry tracings; G) D283MED medulloblastoma cells grown in FBS-containing conditions were transiently transfected with pCIENS (Control) or pCIENS-PID1 overnight (PID1). Phosphorylation of AKT and ERK was measured in the eGFP-expressing cells by flow cytometry. Shown are means/SEM of one of three similar experiments performed in duplicate or triplicate. Supplemental Fig 6 shows a representative flow cytometry tracing of the phospho-AKT and phospho-ERK.
Last, to begin investigating the molecular mechanism of PID1, we examined its effect on serum-mediated phosphorylation of AKT and ERK, two effectors which are central to a number of proliferation and survival signaling pathways. LN229 GBM cells transiently transfected with PID1, serum starved overnight and acutely stimulated with 20% FBS showed decrease in the serum-induced phosphorylation of AKT compared with empty vector-transfected cells (Fig 4F). Additionally, transient transfection of PID1 into D283MED medulloblastoma cells grown in FBS-containing medium caused decrease in phosphorylation of both AKT and ERK (Fig 4G and Suppl Fig 6). This indicates that PID1 inhibits serum-mediated signaling pathways which involve AKT and ERK.
In summary, using six independent datasets, we have shown that PID1 mRNA is lower in the more aggressive medulloblastomas and gliomas compared to their relatively more favorable counterparts, and that higher PID1 mRNA correlates with longer rf-PFS and OS in medulloblastoma patients and longer OS in glioma and GBM patients. We also showed that PID1 conferred a growth-inhibitory effect on cell lines of three types of brain tumors (medulloblastoma, GBM and ATRT), which manifested as decreased proliferation and/or increased cell death. Last, PID1 induced mitochondrial depolarization and inhibition of serum-mediated phosphorylation of AKT and ERK. Taken together, these findings suggest that PID1 is a novel growth-inhibitor in gliomas and embryonal brain tumors.
DISCUSSION
The small number of articles reporting on PID1 to date have mostly been in the context of obesity and diabetes in adipocytes and muscle cells (8, 28–30) and in Alzheimer’s brains (9). Our work here is the first to report on PID1 in cancer.
Our clinical findings for PID1 using six independent datasets comprising two different types of brain cancers emphasize the validity of the association between lower PID1 mRNA and less favorable tumor subgroups. The in vitro experiments suggest that PID1 confers growth suppression due to combined inhibition of proliferation and increase in cell death. Regulation of PID1 expression in embryonal brain tumors and gliomas is currently unknown.
The molecular mechanism by which PID1 affects growth of brain tumor cells in culture is also unknown at this time. Our finding on depolarization of mitochondrial membrane potential by PID1 in brain tumor cell lines is consistent with reports that PID1 disrupts mitochondrial function in adipocytes and muscle cells (24–28). In adipocytes and muscle cells, PID1 also inhibits insulin-mediated phosphorylation of IRS-1 and AKT, and insulin-mediated translocation of the GLUT-4 glucose transporter to the membrane, resulting in decreased glucose uptake (28–30). Our experiments showing that PID1 inhibits phosphorylation of AKT and ERK suggest that PID1 may modulate signaling pathways involved in cell proliferation and survival. While insulin signaling is less likely to be a target of PID1 modulation in brain tumor cell lines due to their low and/or infrequent expression of the insulin receptor, it will be interesting to examine if PID1 modulates signaling through the insulin-like growth factor receptor (IGF1R), which is important in many brain tumors (31–40). The inhibition of insulin receptor signaling in adipocytes was hypothesized to be mediated via interaction of the PID1 PTB domain with the NPXY motif in the cytoplasmic tail of the insulin receptor (29); however, co-immunoprecipitation of the two proteins has not been reported yet. Using yeast two hybrid screens and pull-down experiments, several groups reported direct interaction between the PTB domain of overexpressed PID1 and the NPXY motif of LRP1 (Low density lipoprotein Receptor-Like-1) (9, 10). However, it is unlikely that LRP1 is a major player in the PID1 (aa84-230)-induced cell cycle arrest in NIH-3T3, because the arrest could not be overcome by overexpression of LRP1 (10).
Cellular effects of PID1 may differ between cell types. In 3T3-L1 pre-adipocytes, PID1 increased proliferation (8), but in NIH3T3 cells a PID1 fragment that includes the PTB domain (aa84-230) caused cell cycle arrest (10). Our findings that PID1 has a growth-suppressive effect in multiple brain tumor cell lines are consistent with the latter.
Examination of the Catalogue of Somatic Mutations in Cancer (COSMIC) (41) revealed that among 7055 tumors reported to date (July 18, 2013), there were no copy number variations of PID1 and only 27 mutations, of which 15 were non-synonymous. No mutations were found in the 510 brain tumors reported. The lack of PID1 copy number variations and PID1 mutations in brain tumors suggests that PID1 does not function as a classical tumor suppressor gene. The direct correlation of PID1 mRNA and survival in medulloblastoma and gliomas suggests that PID1 may render these brain tumors more susceptible to therapy, accounting for the better outcome in patients whose tumors expressed higher PID1 mRNA. This possibility is currently under investigation in our laboratory.
In summary, this is the first report to link PID1 to cancer and to brain tumors, to demonstrate correlation between PID1 mRNA level and survival in medulloblastomas and gliomas, and to show growth-inhibitory effect of PID1 in cultured medulloblastoma, GBM and ATRT cell lines. Our data therefore suggest that PID1 may have a growth-modulating function in brain tumors. It will now be important to further investigate the molecular mechanism(s) of PID1’s effects in pediatric and adult malignant brain tumors.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
We present here the first report of PID (Phosphotyrosine Interaction Domain containing 1; NYGGF4) in cancer. PID1 was identified in 2006, and its role and molecular mechanism are still poorly understood. Using six independent datasets in two common brain tumors, medulloblastomas and gliomas, we show that: 1) PID1 mRNA is lowest in least favorable subgroups of medulloblastomas and gliomas; 2) higher PID1 mRNA is directly correlated with longer patient survival; and 3) PID1 causes increased cell death and decreased proliferation in medulloblastoma, glioma, and atypical rhabdoid tumor cell lines. These data in two brain tumor types suggest that it may be possible to incorporate PID1 into personalized molecular prognostic signatures that predict patient response and outcome. Ongoing work on the molecular function of PID1 intends to utilize this knowledge in the design of improved approaches to therapy.
Acknowledgments
The authors are grateful to Mr. Jason Scott and Ms. Min Kang, and to Dr. Mike Sheard for their expert technical assistance and helpful discussions. We thank Dr. Martine Torres for critically reviewing the manuscript and Dr. Susan Lee for assistance in identification of PID1 by proteomic analysis. This work was supported by grant 1R21NS077007-01 from the National Institute of Neurological Disorders and Stroke, a Hyundai Hope on Wheels research grant, a ThinkCure research grant, and support from the Concern Foundation and Mrs. Mimi Feldman to AE-E. The work was also supported by grants from the American Cancer Society, St. Baldrick’s Foundation, and Alex’s Lemonade Stand Foundation to SA. MS was supported by a Rose Hills Foundation Science and Engineering Undergraduate Research Fellowship and a Provost’s Research Fellowship from the University of Southern California. NR was supported by a Fellowship award from the Hope Fund Foundation. This work was also supported in part by generous funding from The Nautica Malibu Triathlon Fund, Grayson’s Gift, The Rachel Ann Hage Neuro-Oncology Fund, The Brad Kaminsky Foundation and the Heroes of Hope Race Teams and The T.J. Martell Foundation for Leukemia, Cancer, and AIDS Research.
References
- 1.Packer RJ, Gajjar A, Vezina G, Rorke-Adams L, Burger PC, Robertson PL, et al. Phase III study of craniospinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin Oncol. 2006;24:4202–8. doi: 10.1200/JCO.2006.06.4980. [DOI] [PubMed] [Google Scholar]
- 2.Packer RJ, Vezina G. Management of and prognosis with medulloblastoma: therapy at a crossroads. Arch Neurol. 2008;65:1419–24. doi: 10.1001/archneur.65.11.1419. [DOI] [PubMed] [Google Scholar]
- 3.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–9. [PubMed] [Google Scholar]
- 4.Packer RJ, Biegel JA, Blaney S, Finlay J, Geyer JR, Heideman R, et al. Atypical teratoid/rhabdoid tumor of the central nervous system: report on workshop. J Pediatr Hematol Oncol. 2002;24:337–42. doi: 10.1097/00043426-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Biegel JA, Tan L, Zhang F, Wainwright L, Russo P, Rorke LB. Alterations of the hSNF5/INI1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res. 2002;8:3461–7. [PubMed] [Google Scholar]
- 6.Johnson DR, O’Neill BP. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107:359–64. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 7.Thumma SR, Fairbanks RK, Lamoreaux WT, Mackay AR, Demakas JJ, Cooke BS, et al. Effect of pretreatment clinical factors on overall survival in glioblastoma multiforme: a Surveillance Epidemiology and End Results (SEER) population analysis. World J Surg Oncol. 2012;10:75. doi: 10.1186/1477-7819-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang B, Zhang M, Ni YH, Liu F, Fan HQ, Fei L, et al. Identification and characterization of NYGGF4, a novel gene containing a phosphotyrosine-binding (PTB) domain that stimulates 3T3-L1 preadipocytes proliferation. Gene. 2006;379:132–40. doi: 10.1016/j.gene.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Kajiwara Y, Franciosi S, Takahashi N, Krug L, Schmeidler J, Taddei K, et al. Extensive proteomic screening identifies the obesity-related NYGGF4 protein as a novel LRP1-interactor, showing reduced expression in early Alzheimer’s disease. Mol Neurodegener. 2010;5:1–11. doi: 10.1186/1750-1326-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caratu G, Allegra D, Bimonte M, Schiattarella GG, D’Ambrosio C, Scaloni A, et al. Identification of the ligands of protein interaction domains through a functional approach. Mol Cell Proteomics. 2007;6:333–45. doi: 10.1074/mcp.M600289-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fattet S, Haberler C, Legoix P, Varlet P, Lellouch-Tubiana A, Lair S, et al. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol. 2009;218:86–94. doi: 10.1002/path.2514. [DOI] [PubMed] [Google Scholar]
- 13.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–8. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–14. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–30. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–31. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 17.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–67. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asgharzadeh S, Salo JA, Ji L, Oberthuer A, Fischer M, Berthold F, et al. Clinical significance of tumor-associated inflammatory cells in metastatic neuroblastoma. J Clin Oncol. 2012;30:3525–32. doi: 10.1200/JCO.2011.40.9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trichas G, Begbie J, Srinivas S. Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 2008;6:40. doi: 10.1186/1741-7007-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol (Berl) 2012;123:473–84. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol (Berl) 2012;123:465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang CM, Zeng XQ, Zhang R, Ji CB, Tong ML, Chi X, et al. Effects of NYGGF4 knockdown on insulin sensitivity and mitochondrial function in 3T3-L1 adipocytes. J Bioenerg Biomembr. 2010;42:433–9. doi: 10.1007/s10863-010-9313-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Zhang C, Chen X, Gao C, Ji C, Chen F, et al. Overexpression of NYGGF4 (PID1) induces mitochondrial impairment in 3T3-L1 adipocytes. Mol Cell Biochem. 2010;340:41–8. doi: 10.1007/s11010-010-0398-5. [DOI] [PubMed] [Google Scholar]
- 26.Shi CM, Wang YM, Zhang CM, Qiu J, Shen YH, Zhu JG, et al. Knockdown of NYGGF4 (PID1) rescues insulin resistance and mitochondrial dysfunction induced by FCCP in 3T3-L1 adipocytes. Mitochondrion. 2012;12:600–6. doi: 10.1016/j.mito.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Shi CM, Xu GF, Yang L, Fu ZY, Chen L, Fu HL, et al. Overexpression of TFAM Protects 3T3-L1 Adipocytes from NYGGF4 (PID1) Overexpression-Induced Insulin Resistance and Mitochondrial Dysfunction. Cell Biochem Biophys. 2012 doi: 10.1007/s12013-012-9496-1. [DOI] [PubMed] [Google Scholar]
- 28.Bonala S, McFarlane C, Ang J, Lim R, Lee M, Chua H, et al. Pid1 Induces Insulin Resistance in Both Human and Mouse Skeletal Muscle during Obesity. Mol Endocrinol. 2013;27:1518–35. doi: 10.1210/me.2013-1048. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Zhang CM, Chen XH, Wang B, Liu F, Chi X, Tong ML, et al. Over-expression of NYGGF4 inhibits glucose transport in 3T3-L1 adipocytes via attenuated phosphorylation of IRS-1 and Akt. Acta Pharmacol Sin. 2009;30:120–4. doi: 10.1038/aps.2008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu WL, Gan WH, Tong ML, Li XL, Dai JZ, Zhang CM, et al. Over-expression of NYGGF4 (PID1) inhibits glucose transport in skeletal myotubes by blocking the IRS1/PI3K/AKT insulin pathway. Mol Genet Metab. 2010 doi: 10.1016/j.ymgme.2010.11.165. [DOI] [PubMed] [Google Scholar]
- 31.Gammeltoft S, Ballotti R, Kowalski A, Westermark B, Van Obberghen E. Expression of two types of receptor for insulin-like growth factors in human malignant glioma. Cancer Res. 1988;48:1233–7. [PubMed] [Google Scholar]
- 32.Hahn H, Wojnowski L, Specht K, Kappler R, Calzada-Wack J, Potter D, et al. Patched target Igf2 is indispensable for the formation of medulloblastoma and rhabdomyosarcoma. J Biol Chem. 2000;275:28341–4. doi: 10.1074/jbc.C000352200. [DOI] [PubMed] [Google Scholar]
- 33.Wang JY, Del Valle L, Gordon J, Rubini M, Romano G, Croul S, et al. Activation of the IGF-IR system contributes to malignant growth of human and mouse medulloblastomas. Oncogene. 2001;20:3857–68. doi: 10.1038/sj.onc.1204532. [DOI] [PubMed] [Google Scholar]
- 34.Friend KE, Khandwala HM, Flyvbjerg A, Hill H, Li J, McCutcheon IE. Growth hormone and insulin-like growth factor-I: effects on the growth of glioma cell lines. Growth Horm IGF Res. 2001;11:84–91. doi: 10.1054/ghir.2000.0183. [DOI] [PubMed] [Google Scholar]
- 35.Del Valle L, Enam S, Lassak A, Wang JY, Croul S, Khalili K, et al. Insulin-like growth factor I receptor activity in human medulloblastomas. Clin Cancer Res. 2002;8:1822–30. [PubMed] [Google Scholar]
- 36.Rao G, Pedone CA, Del Valle L, Reiss K, Holland EC, Fults DW. Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene. 2004;23:6156–62. doi: 10.1038/sj.onc.1207818. [DOI] [PubMed] [Google Scholar]
- 37.Hartmann W, Koch A, Brune H, Waha A, Schuller U, Dani I, et al. Insulin-like growth factor II is involved in the proliferation control of medulloblastoma and its cerebellar precursor cells. Am J Pathol. 2005;166:1153–62. doi: 10.1016/S0002-9440(10)62335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu TJ, LaFortune T, Honda T, Ohmori O, Hatakeyama S, Meyer T, et al. Inhibition of both focal adhesion kinase and insulin-like growth factor-I receptor kinase suppresses glioma proliferation in vitro and in vivo. Mol Cancer Ther. 2007;6:1357–67. doi: 10.1158/1535-7163.MCT-06-0476. [DOI] [PubMed] [Google Scholar]
- 39.Corcoran RB, Bachar Raveh T, Barakat MT, Lee EY, Scott MP. Insulin-like growth factor 2 is required for progression to advanced medulloblastoma in patched1 heterozygous mice. Cancer Res. 2008;68:8788–95. doi: 10.1158/0008-5472.CAN-08-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh A, Ellsworth R, Hsieh D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J Cell Physiol. 2011;226:1118–27. doi: 10.1002/jcp.22433. [DOI] [PubMed] [Google Scholar]
- 41.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J, Erdreich-Epstein A, Gonzalez-Gomez I, Melendez EY, Smbatyan G, Moats RA, et al. Novel cell lines established from pediatric brain tumors. J Neurooncol. 2012;107:269–80. doi: 10.1007/s11060-011-0756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren X, Xu J, Cooper JP, Kang MH, Erdreich-Epstein A. c-Abl Is an upstream regulator of acid sphingomyelinase in apoptosis induced by inhibition of integrins αvβ3 and αvβ5. PLoS One. 2012;7:e42291. doi: 10.1371/journal.pone.0042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32:W130–4. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox D, Oakes D. Analysis of survival data. London; New York: Chapman and Hall; 1984. [Google Scholar]
- 46.von Levetzow C, Jiang X, Gwye Y, von Levetzow G, Hung L, Cooper A, et al. Modeling initiation of Ewing sarcoma in human neural crest cells. PLoS One. 2011;6:e19305. doi: 10.1371/journal.pone.0019305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.