Abstract
Hepatocellular carcinoma (HCC), the primary form of human adult liver malignancy, is a highly aggressive tumor with average survival rates that are currently less than a year following diagnosis. Although bioinformatic analyses have indicated differentially expressed genes and cancer-related mutations in hepatocellular carcinoma (HCC), integrated genetic and metabolic pathway analyses remain to be investigated. Herein, gene (i.e. mRNA) enrichment analysis was performed to delineate significant alterations of metabolic pathways in HCC. The objective of this study was to investigate the pathway of aspartate metabolism in HCC of humans. Coupled with transcriptomic (i.e. mRNA) and NMR-based metabolomics of human tissue extracts, we utilized liquid chromatography mass spectrometry (LC-MS/MS)-based metabolomics analysis of stable [U-13C6]glucose metabolism or [U-13C5,15N2]glutamine metabolism of HCC cell culture. Our results indicated that aspartate metabolism is a significant and differentiable metabolic pathway of HCC compare to non-tumor liver (p-value < 0.0001). In addition, branched-chain amino acid metabolism (p-value< 0.0001) and tricarboxylic acid metabolism (p-value = < 0.0001) are significant and differentiable. Statistical analysis of measurable NMR metabolites indicated that at least two of the group means were significantly different for the metabolites alanine (p-value = 0.0013), succinate (p-value = 0.0001), lactate (p-value = 0.0114), glycerophosphoethanolamine (p-value = 0.015), and inorganic phosphate (p-value = 0.0001). However, 13C isotopic enrichment analysis of these metabolites revealed less than 50% isotopic enrichment with either stable [U-13C6]glucose metabolism or [U-13C5,15N2]glutamine. This may indicate the differential account of total metabolite pool versus de novo metabolites from a 13C labeled substrate. The ultimate translation of these findings will be to determine putative enzyme activity via 13C labeling, to investigate targeted therapeutics against these enzymes, and to optimize the in vivo performance of 13C magnetic resonance imaging techniques.
Keywords: hepatocellular carcinoma, cancer, aspartate, transcriptomics, metabolomics, branched-chain amino acids, genetics
INTRODUCTION
In recent years, detailed bioinformatic analyses have indicated differentially expressed genes and cancer-related mutations in various cancer models (1). These differentially expressed genes and mutations may affect signaling pathways and cell processes that ultimately have a profound effect on the metabolism of cancer in order to support their proliferation. To meet these needs, cancer cells acquire alterations in their metabolic phenotype of all four major classes of macromolecules: carbohydrates, proteins, lipids and nucleic acids (1). As technological improvements increase the feasibility of studying cancer metabolism, a growing number of studies have investigated the molecular basis of malignant transformation and cellular metabolism.
The vast majority (91%) of liver cancer is hepatocellular carcinoma (HCC), and it is the sixth most common cancer worldwide and the third most lethal cancer (2). Most patients with HCC are diagnosed when the disease is already at an advanced stage, thereby limiting therapeutic options and leading to a dismal one-year cause-specific survival rate (3). Although hepatitis B virus (HBV) and hepatitis C virus (HCV) are known risk factors for HCC, aspartate transaminase (AST) or alanine transaminase (ALT) concentrations of 25 IU/L or higher have been shown as risk factors to HCC even in subjects without HBV or HCV (4). This may warrant the need to investigate the metabolic pathways associated with AST or ALT in order to elucidate metabolic phenotypes of HCC.
Imaging modalities commonly considered diagnostic for liver cancers include contrast-enhanced ultrasound (CEUS), contrast-enhanced computerized tomography (CT), positron emission tomography (PET), and gadolinium-enhanced magnetic resonance imaging (MRI) (5). The techniques utilized in these modalities provide little to no biochemical information of metabolism that is evident in pathological tissues. In vivo magnetic resonance spectroscopy (MRS) is capable of discriminating benign from malignant tissues based on metabolic signatures, however this capability has yet to be fully implemented in clinical practice. Defining the metabolic signatures associated with malignancy is a critical step to developing MRS techniques for HCC. Currently, metabolites of aspartate metabolism pathway have not been quantified in HCC primarily due to the inability of any single methodology to provide a comprehensive measurement of these intermediate metabolites. Combining metabolomic approaches including liquid chromatography mass spectrometry (LC-MS) (6,7), gas chromatography mass spectrometry (GC-MS) (8), and MRS (9) may overcome this limitation. The metabolism of HCC has being investigated with high-resolution magic-angle spinning (1H HRMAS) nuclear magnetic resonance spectroscopy technique to examined the metabolic characteristics of human HCC tissues (10,11), human serum from liver cirrhosis (12), human liver transplant tissues (13), chronic hepatitis pathological tissues (14), and human urine samples of HCC patients (15). Mass spectrometry (16,17) have shown higher sensitivity and identified more metabolites than 1H HRMAS, but the extraction methods for metabolites in mass spectrometry are not trivial, are destructive, and may require derivatization of the metabolites, all of which could potentially lead to a loss of intrinsic metabolic signatures. Combining both approaches have been used to distinguish tumor metabolism of colorectal cancer from their matched normal mucosae (18) and to compare breast cancer to normal human mammary epithelial cell lines (19).
We present here an integrated system of genetic (i.e. mRNA) and metabolic profiling of human HCC as compared to non-tumor liver and cirrhotic liver. We performed gene enrichment analysis to determine significant alterations of metabolic pathways in HCC. We then perform high-resolution magnetic resonance spectroscopic analysis of HCC, cirrhotic liver and non-tumor tissues to quantify the metabolites associated with the relevant metabolic pathway(s). The objective of this study is to investigate the pathway of aspartate metabolism in hepatocellular carcinoma from human tissues and to further characterize putative metabolites in human HCC and HCC cell lines. Coupled with transcriptomic (i.e. mRNA) and metabolomic analyses of human tissues, we utilized LC-MS/MS-based metabolomics analysis of stable [U-13C6]glucose metabolism or [U-13C5,15N2]glutamine metabolism in HCC cell culture.
EXPERIMENTAL
Genomics Samples Characteristics
De-identified human tissues (i.e. hepatocellular carcinoma, cirrhosis, and non-tumor liver) were approved for use by the University of Pennsylvania Institutional Review Board (IRB) prior to study initiation. The tissues characterized as non-tumor are defined liver tissues that are not carcinoma and are not cirrhosis. All 24 genomics data were archived into the National Center for Biotechnology Information’s Gene Expression Omnibus repository (accession number GSE45050) supporting a compliance with Minimum Information About a Microarray Experiment (MIAME). The verified pathological reports of these patients indicated tumor grades G1 to G3 of type IIIA minimum stage grouping in accordance with the American Joint Committee on Cancer (AJCC) staging. All 24 patients were males with ages ranging from 26 years to 86 years. The RNA integrity numbers (RIN) ranged from 8.1 to 9.6. The RNA quality/purity determined by UV absorbance (A260/A280 ratio) ranged from 2.07 to 2.20. The specific details of each sample can be found in the supplementary material.
RNA Isolation, Microarray Hybridization and Scanning
Homogenized lysate of human tissues were processed to obtain about 5 µg of high-quality total RNA using RNeasy Midi Kit (QIAGEN, Valencia, CA, USA). The tissue samples, diagnosed by certified pathologists, were obtained from tissue biorepository (OriGene Technologies, Inc., Rockville, MD, USA). The RNA integrity and quality control of 2 µL sample was performed with NanoDrop® ND-1000 UV spectrophotometer and Agilent® 2100 Bioanalyzer at the Molecular Profiling Facility at our institution. Degraded samples were eliminated from the study based on their measured OD A260/A280 ratios, electrophoresis results, 28S/18S rRNA ratios, and RNA Integrity Number (RIN). The Ambion® WT Expression kit was used as suggested by the manufacturer to generate sense strand cDNA from 250 ng total RNA followed by fragmentation and labeling of 5.5 µg of sense strand cDNA with the GeneChip® WT Terminal labeling kit. Target hybridization was performed on Affymetrix Human GeneChip® Gene 1.0 ST Arrays (Affymetrix, Inc., Santa Clara, CA, USA) according to the manufacturer’s procedures in the GeneChip® Hybridization Oven 645, followed by washing and staining in the GeneChip® Fluidics Station 450. Data were acquired with GeneChip® Scanner 3000 7G (Affymetrix, Inc., Santa Clara, CA, USA).
Statistical Analysis of the Gene Expression Data
The CEL files, containing probe-level data from all arrays, were imported into Partek® Genomics SuiteTM software (Partek Incorporated, St. Louis, MO) where Robust Multichip Analysis (RMA) normalization was applied yielding log2-transformed expression intensities. One-way ANOVA was performed on the genes across the 3 groups (i.e. hepatocellular carcinoma, cirrhosis, and non-tumor liver) to provide p-values, and false positive rates (step-up p-values) were computed using the Benjamini-Hochberg step-up algorithm as implemented in Partek. Pairwise comparisons were made between tissues types providing p-values, log2 ratios, and fold-change values.
Metabolic Pathways Analyses
We performed functional pathway analyses using Ariadne Pathway Studio® version 9.0 (Elsevier, Rockville, MD, USA). The Elsevier’s Pathway Studio® was suited with MedScan® Reader 5.0 and the ResNet 9 Mammalian database. The experimental results, gene expression data, were imported into the ResNet 9 database to implement the Gene Set Enrichment Analysis (GSEA) algorithm for interpreting microarray data into biological mechanism. The gene expression results from two comparisons (HCC vs. cirrhosis, HCC vs. non-tumor) were ranked (Mann-Whitney U-Test) by absolute fold change with step-up p-value cut-off at 0.05, and the statistical enrichment of curated pathways was determined from known gene sets. The results of the analysis are a list of the most significantly enriched pathways ranked by p-value.
Human Tissue Procurement for MR Spectroscopy
After local IRB approval, tissue samples of human liver were provided by the Cooperative Human Tissue Network (CHTN), which is funded by the National Cancer Institute. The CHTN provides prospective investigator-defined procurement of malignant, benign, diseased and uninvolved (normal adjacent) tissues. These tissues were obtained at surgical resection of human liver, which were frozen in liquid nitrogen and stored at −80°C. These human tissues were characterized as non-tumor or normal liver (N=5), hepatocellular carcinoma (N=11), and cirrhosis (N=11) tissues.
Perchloric Acid Extraction of Metabolites for MR Spectroscopy
The snap-frozen samples were mixed with six times the weight in volume (mL) of 12% perchloric acid into a 15 mL polypropylene conical tube (BD Bioscience, Franklin Lakes, NJ). The mixture was homogenized while on ice with a Polytron® PT 2500E (Kinematica AG, Luzern, Switzerland). The homogenate was centrifuged at 3,452 ×g for 30 min at 4°C using a Sorvall® Legend® RT centrifuge (Harlow Scientific, Arlington, MA). The supernatant was removed from the pellet and titrated to a pH of 7±0.34 using 3 M KOH (potassium hydroxide) and a 0.6% perchloric acid, which resulted to the precipitation of potassium perchlorate. The buffered solution was centrifuged at 3,452 ×g for 30 min at 4°C, and the supernatant was extracted and lyophilized to a powder form.
MR Spectroscopy Data Acquisition and Quantification
MR spectroscopy was performed at 400 MHz on a Bruker Avance III™ wide-bore spectrometer using the Topspin Bruker software (Bruker BioSpin MRI GmbH, Ettlingen, Germany). Each lyophilized tissue extract was reconstituted in 0.45 mL of D2O, the pH adjusted to 7.0 and the solution introduced in a 5 mm MR tube. An external chemical shift and concentration standard made in a sealed capillary tube containing a 0.3 mM 3-(trimethylsilyl) propionic 2,2,3,3-d4 acid, sodium salt (TSP) (Cambridge Isotope Laboratories, Inc., Andover, MA) in D2O was introduced in the NMR tube. Fully relaxed proton spectra were acquired with a 5 mm BBO probe. Standard acquisition conditions were as follows: pulse width (PW) 45°, repetition time (TR) 8 s, water saturation during the relaxation delay, sweep width (SW) 6.7 kHz, data point (TD) 64 k, 4 dummy scans and 64 scans. Following the 1H NMR 0.150 µL of 50 mM EDTA was added to the sample, the pH adjusted to 8.0 and a sealed calibrated capillary of 30 mM methylene diphosphonate (MDP) (Sigma Chemical Co., St. Louis, MO) was introduced in the MR tube. Phosphorus spectra were acquired with the inversegate pulse program and the following conditions: SW 13 kHz, TD 64 k, AQ 2.5 s, D1 1.5 s, PW 45°, TR 4 s, 128 dummy scans, 2000 scans. Under these acquisition conditions the phosphorus signals are fully relaxed except for the inorganic phosphate (Pi). The post-processing and quantification of spectra were executed with 1 Hz line broadening followed by Fourier transformation. Assignments of metabolites resonance peaks were made by comparing chemical shifts to those of standard compounds measured in PBS/D2O. Cellular metabolites were quantified by computing the integral of assigned resonances using Lorentzian line-shapes to fit the 1D 1H spectra. The concentration of metabolites were computed with respect to the concentrations of the standards and expressed in units of µmoles per gram of wet weight in tissue.
Extraction of Cellular Metabolites for LC-MS/MS
Morris hepatoma McA-RH7777 cells were cultured in 75 cm2 flasks for 48–72 h to 70% confluence in formulated Dulbecco's Modified Eagle's Medium (DMEM) supplemented with fetal bovine serum to a final concentration of 10% (v/v). The cells and culture media were obtained from American Type Culture Collection (ATCC), Manassas, VA. DMEM contained 4 mM L-glutamine, 24.98 mM glucose, 1 mM pyruvate and 17.86 mM sodium bicarbonate. A 24.98 mM [U-13C6]glucose (Sigma Chemical Co., St. Louis, MO) stock solution was prepared in glucose free DMEM. Cells were split and cultured with labeled or unlabeled glucose for 6 hours in an incubator at 37°C and 5% carbon dioxide in air. In a separate experiment, 2.48 mM [U-13C5,15N2]glutamine (Sigma Chemical Co., St. Louis, MO) stock solution was prepared in glucose and glutamine free DMEM. Similarly in the separate experiment, cells were split and cultured with labeled or unlabeled glutamine and for 6 hours in an incubator at 37°C and 5% carbon dioxide in air. The cells were harvested with 25% Trypsin-0.53 mM EDTA, counted with a hemacytometer, and 13.3×106 cells were analyzed in triplicate. Cell suspension was transferred to centrifuge tubes and spun at 125 ×g for 10 minutes. Cell pellets were resuspended in 500 µL of 10% trichloroacetic acid for the extraction of acyl-CoA species or resuspended in 500 µL of 80% methanol for the extraction of organic acids.
The cell pellet was pulse-sonicated for 30 sec in 500 µL while on ice using a sonic dismembranator (Fisher Scientific, Pittsburg, PA). Following sonication, the lysate was centrifuged at 15000 ×g for 5 min. The supernatant was transferred to a fresh tube and the pellet was discarded. Short chain acyl-CoA species were purified from the sonicated lysate using Oasis HLB 1cc (30 mg) solid phase extraction columns (Waters Corporation, Milford, MA). The eluted solution was dried down under a vapor stream of nitrogen. The acyl-CoA samples were resuspended with 50 µL 5% sulfo-salicylic acid into LC-MS certified tubes with PTFE/silicone septa (Waters Corporation, Milford, MA). The organic acids samples were resuspended into 150 µL methoxylamine in pyridine (20 mg/L) and incubated for 1 hour in a water bath at 40°C. Subsequently, 50 µL of N-tert-butyldimethylsilyl-N-methyltrifluoro acetamide (MTBSTFA) was added and incubated for 1 hour in a water bath at 60°C. Then 20 µL of the final solution was mixed with 980 µL of acetonitrile into a LC-MS certified tube with PTFE/silicone septa (Waters Corporation, Milford, MA).
Intermediate metabolites were analyzed by modified methodologies of LC-MS/MS previously described for acyl-CoA species (6) and for organic acids (20). Samples were maintained at 4°C in a Leap CTC autosampler (CTC Analytics, Switzerland) during sample batch runs. 10 μL injections were used for LC-MS analysis. High-performance liquid chromatography (HPLC) was performed using a reversed phase Waters X-Bridge C18 column (2.1×150 mm, pore size 3 μm) with 10 μL injections at a constant flow rate of 0.2 mL/min into an Agilent 1100 HPLC system using solvents mixtures of ammonium acetate, water, acetonitrile, and formic acid. All solvents were Optima® grade for HPLC and spectrometry analysis (Fisher Scientific, Pittsburg, PA). Mass analyses were performed using an API 4000 triple quadrupole mass spectrometer (Applied Biosystems, Foster City, CA) in the positive electrospray ionization (ESI) mode and analytes were quantified using Analyst software. A matrix-based isotopomer analysis (21,22) was performed for quantification of [U-13C6]glucose isotopic enrichment into metabolites using custom script written in Matlab (The MathWorks, Inc., Natick, MA, USA). The isotopic mass distribution (M) of each metabolite was performed for unlabeled isotopomer (M0), one 13C isotopomer (M1), two 13C isotopomer (M2), three 13C isotopomer (M3) and four 13C isotopomer (M4).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software version 5.0 for Mac OS X (GraphPad, San Diego, California USA) and STATA® software version 12.0 (StataCorp LP, College Station, TX, USA). We tested the null hypothesis against the alternative hypothesis with a two-sided type I error rate of 5% at a 90% power.
RESULTS
Aspartate metabolism is a significant and differentiable metabolic pathway of HCC
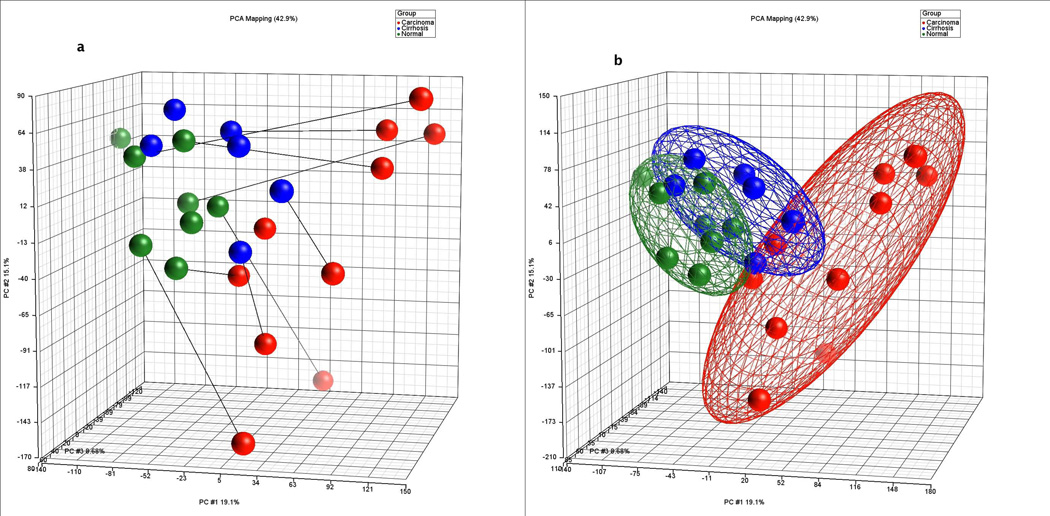
As graphically displayed in principal component analysis by clustering the samples at two standard deviations from the mean of each group (Figure 1), marked differences in gene expression were observed between HCC and cirrhosis as well as HCC and non-tumor tissues whereas modest differences were observed between cirrhotic and non-tumor tissues (see GEO database for detailed gene list and fold change). To identify metabolic pathways specifically altered in HCC, the GSEA algorithm was performed on the gene expression data at a step-up p-value threshold of 0.05. This pathway analysis revealed significant metabolic pathways of which the 13 most significant are outlined in Table 1.
Figure 1.
Principal Components Analysis (PCA) in three-dimensional plot of all hybridizations (a) showing individual samples with connecting lines to matched pairs from the same patient and (b) showing ellipsoids drawn at two standard deviations from the group/cohort centroid. Carcinoma = red, Cirrhosis = blue, and non-tumor = green.
Table 1. Enriched Metabolic Pathways in Hepatocellular Carcinoma.
Gene set enrichment analysis significantly expressed genes of hepatocellular carcinoma to identify metabolic pathways. The results are threshold (---) at top 100 pathways and an enrichment p-value cut-off 0.05.
| Metabolic Pathways Names | Total Entities |
Measured Entities |
Carcinoma vs Non-tumor p-value < |
Carcinoma vs Cirrhosis p-value < |
|---|---|---|---|---|
| Branched chain amino acids metabolism | 99 | 48 | 0.0001 | 0.0001 |
| Tryptophan metabolism | 112 | 138 | 0.0001 | 0.0001 |
| Malonate, propanoate and beta-alanine metabolism | 99 | 50 | 0.0001 | 0.0001 |
| Fatty acid oxidation | 73 | 40 | 0.0001 | 0.001 |
| Aspartate metabolism | 66 | 29 | 0.0001 | 0.0001 |
| Glycogen metabolism | 37 | 32 | 0.0001 | 0.001 |
| Arachidonic acid metabolism | 129 | 100 | 0.001 | 0.0001 |
| Glucose metabolism | 68 | 64 | 0.001 | --- |
| Glut/Gln/Pro metabolism | 80 | 39 | 0.001 | 0.01 |
| Pentose-phosphate shunt | 46 | 20 | 0.01 | --- |
| Vitamin K metabolism | 36 | 18 | 0.01 | 0.001 |
| Ethanol metabolism | 15 | 18 | 0.01 | 0.05 |
| Tricarboxylic acid cycle | 55 | 30 | 0.05 | --- |
The most significantly expressed pathway between HCC and non-tumor liver involved branched chain amino acid (BCAA) metabolism, specifically expression of BCAA transaminase, an enzyme critical to the reversible transamination of branched-chain alpha-keto acids to branched-chain L-amino acids such as leucine, isoleucine, and valine. Their respective tRNA ligases were also significantly expressed. In addition, methylmalonyl-CoA epimerase and 3-hydroxybutarate dehydrogenase were over expressed in the branched-chain amino acid pathway. Changes in the branched-chain amino acid pathway were similarly significant comparing HCC with cirrhotic and comparing HCC with non-tumor liver. Thus, BCAA pathway exhibits highly expressed genes differentiating HCC from cirrhotic and non-tumor liver.
Another pathway with significantly expressed genes differentiating HCC from cirrhotic and non-tumor liver was the tricarboxylic acid cycle (TCA) pathway. We identified most differentially expressed genes in the tricarboxylic acids circle pathway. The enzymes significantly expressed in HCC were pyruvate kinase, dihydrolipoyllysine-residue acetyltransferase, ATP citrate synthase, citrate (Si)-synthase, isocitrate dehydrogenase (NAD+), dihydrolipoyllysine-residue succinyltransferase, dihydrolipoyl dehydrogenase, and malate dehydrogenase.
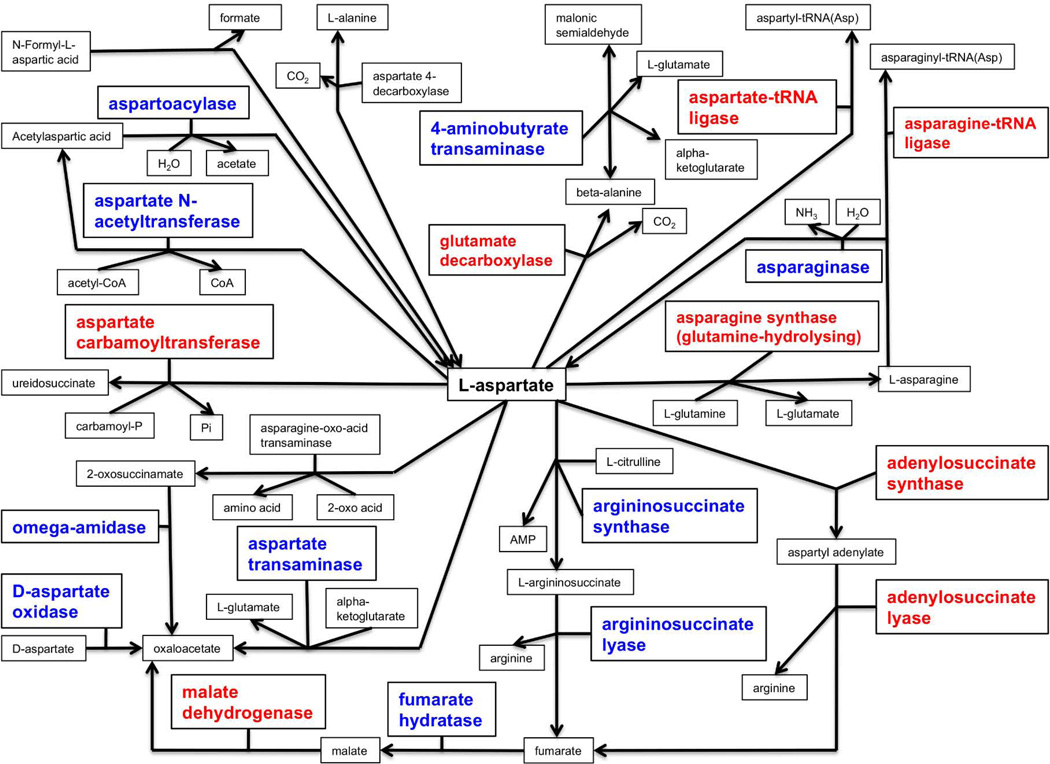
We were particularly interested in asparate metabolism, which we found to be significantly enriched in HCC as compared to non-tumor tissue (p < 0.0001). This aspartate metabolism pathway is shown in Figure 2. The enzymes significantly over-expressed (in red) were aspartate-tRNA ligase, asparagine-tRNA ligase, glutamate decarboxylase, aspartate carbamoyltransferase, asparagine synthase, malate dehydrogenase, adenylosuccinate synthase, and adenylosuccinate lyase. The significant expressions of adenylosuccinate lyase and malate dehydrogenase are critical to the production of fumarate, malate, and oxaloacetate in this aspartate metabolism pathway.
Figure 2.
Aspartate metabolism pathway composed of entities (i.e. protein, functional class, small molecules, etc) significantly up-regulated (in red) or significantly down-regulated (in blue) for the differential expression in hepatocellular carcinoma versus non-tumor.
Measurable NMR metabolites of significant interest are alanine, succinate, lactate, glycerophosphoethanolamine, and inorganic phosphate
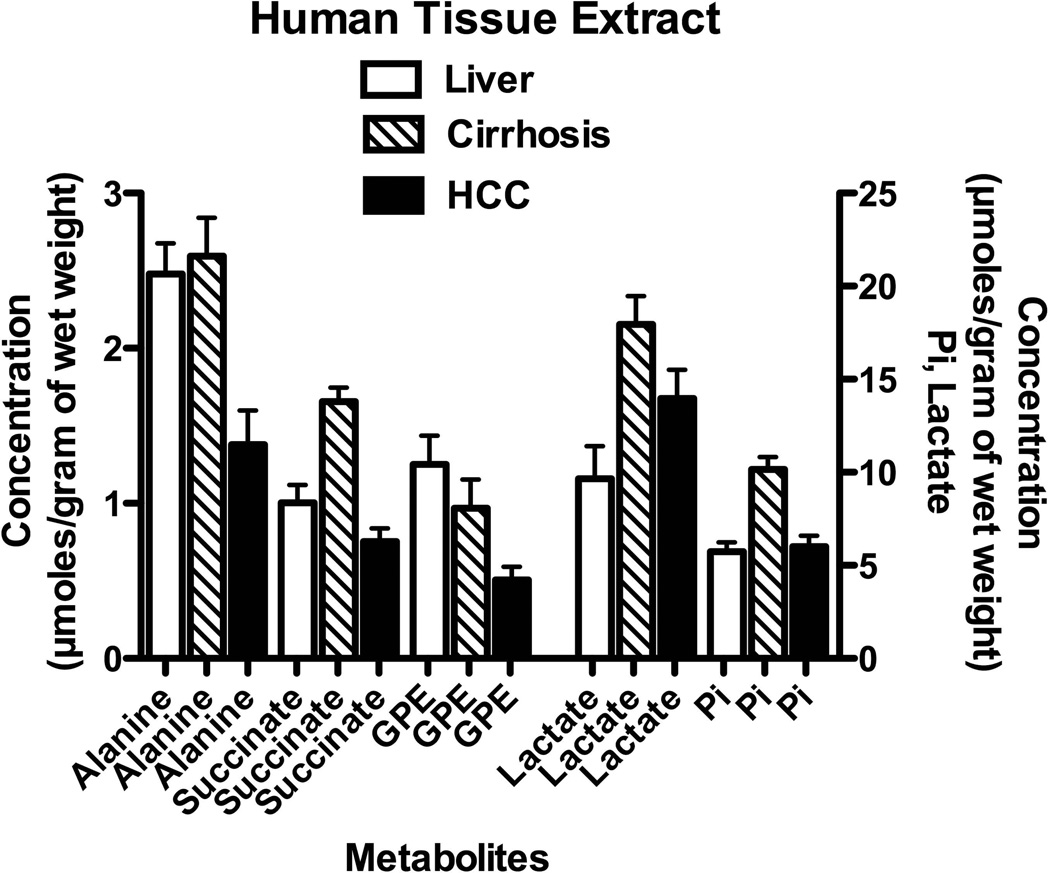
To verify the effect of these genetic expressions on the metabolism of these tissue types, we examined MR spectroscopic results of HCC, cirrhotic and non-tumor liver as shown in Figure 3. Representative spectra are shown and described in the supplementary material. Although these results were obtained from different patient cohort and not from the gene study cohort, putative metabolites in these identified metabolic pathways were measured. The leucine resonance peak at a chemical shift of 0.98 ppm and the isoleucine resonance peak at a chemical shift of 0.94 ppm were not quantified due to the distorted baseline in that region of the spectra (see Supplementary Material). A one-way analysis-of-variance (ANOVA) was used to compare the mean value of metabolites in various human tissues (i.e. non-tumor liver, cirrhosis, malignant-HCC) as shown in Figure 3. The overall (global) F test for the one-way ANOVA at the 95% confidence level indicated that at least two of the group means were significantly different for the metabolites alanine (p = 0.0013), succinate (p = 0.0001), lactate (p = 0.0114), glycerophosphoethanolamine (GPE) (p = 0.015), and inorganic phosphate (p = 0.0001). The Bonferroni’s method of multiple comparisons indicated that the concentration of alanine in cirrhotic liver was significantly elevated relative to the amount of alanine in HCC tissues (p = 0.002), and interestingly alanine levels in HCC were significantly lower compared to the alanine levels observed in non-tumor liver (p = 0.021). The concentration of succinate in cirrhotic tissues was significantly greater compared to succinate in HCC (p = 0.0001) and non-tumor liver (p = 0.001). The lactate pool in cirrhotic liver was significantly elevated than that of non-tumor liver (p = 0.010). We found that the amount of GPE in HCC was significantly lower than GPE found in non-tumor liver (p = 0.015). Also, the concentration of inorganic phosphate in cirrhosis was significantly greater compared to inorganic phosphate in HCC (p = 0.0001) and compare to inorganic phosphate in non-tumor liver (p = 0.0001). Although the test for normality of each metabolite’s distribution did not show a Gaussian distribution for all metabolites, we also performed a nonparametric test. In addition, the Kruskal-Wallis nonparametric test resulted to a significant difference of at least two of the group means for the creatine pools (p = 0.0373).
Figure 3.
Selected metabolite concentrations from human liver tissues (i.e. HCC, cirrhosis, non-tumor) after perchloric acid extraction: alanine, succinate, glycerophosphoethanolamine (GPE), lactate, and inorganic phosphate (Pi). All metabolites are referenced against MDP or TSP. All bar graphs are displayed as mean±SEM.
Isotopic 13C enrichment of metabolites may delineate the de novo metabolite from its steady-state pool
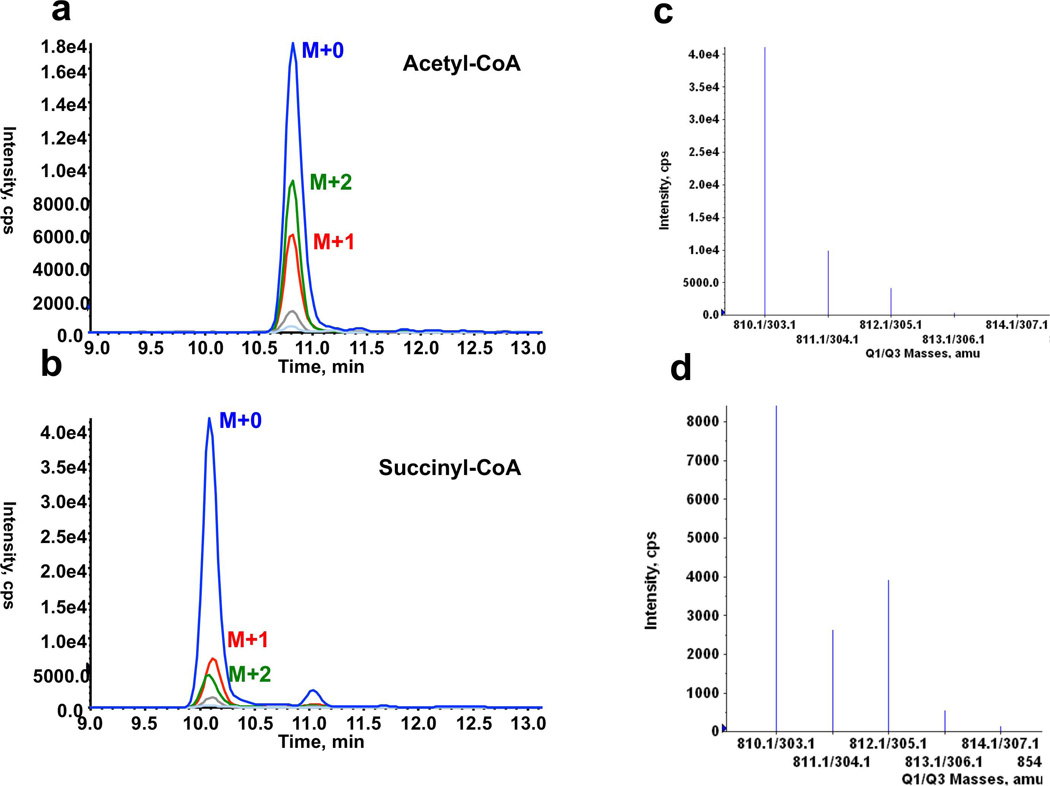
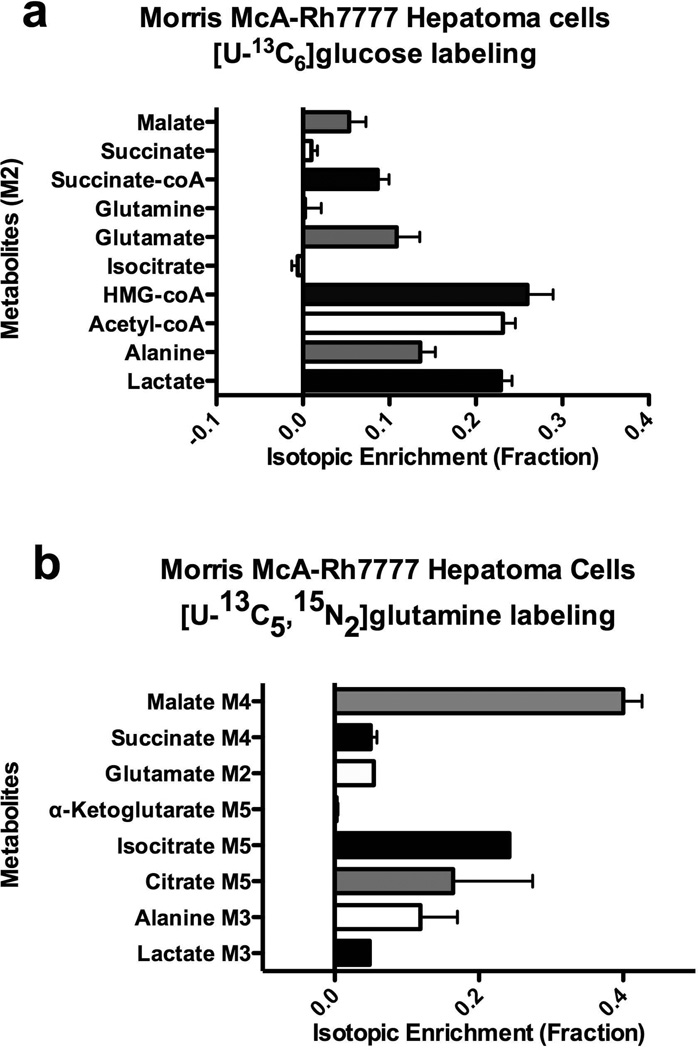
Representative chromatograms and spectra quality achievable with these LC-MS/MS methods are illustrated in Figure 4. The 13C enrichment in acetyl-CoA (Figure 4d) was quite notable when the cells were labeled with [U-13C6]glucose compare to the isotope distribution in acetyl-CoA (Figure 4c) where the cells were labeled with [U-12C6]glucose. Isotopic enrichments of acyl-CoA species and organic acids in HCC cells are shown in Figure 5 where bar graphs are displayed as mean±SEM. Under these conditions, it appears that the cells incorporated up to 50% of 13C carbon in acyl-CoA species and organic metabolites. Measurements for the two 13C isotopomer (M2), three 13C isotopomer (M3) and four 13C isotopomer (M4) are illustrated.
Figure 4.
LC-MS/MS chromatograms of (a) acetyl-CoA and of (b) succinyl-CoA, (c) spectrum indicating isotope distribution in acetyl-CoA with [U-12C6]glucose, and corresponding (d) spectrum indicating isotope distribution in acetyl-CoA with [U-13C6]glucose.
Figure 5.
Isotopic enrichment analysis of LC-MS/MS data for HCC cell culture (a) uniformly labeled [U-13C6]glucose followed by quantified 13C isotope enrichment in M+3 for alanine and lactate, and M+2 of acetyl-CoA, citrate, glutamate, glutamine, succinate-CoA, succinate and malate. Unlabeled [U-12C6]glucose data was used to compute the basis set of the isotopomer matrix, (b) uniformly labeled [U-13C5,15N2]glutamine followed by quantified 13C isotope enrichment in lactate, succinate, glutamate, α-ketoglutarate, isocitrate, citrate, alanine and lactate.
DISCUSSION
This study of transcriptomics and metabolomics from human tissues provides an integrated biological analysis of human hepatocellular carcinoma. Our results showed that alterations of aspartate metabolism are a signature of hepatocellular carcinoma. It appears that in HCC, asparagine synthase hydrolyzes glutamine to glutamate thereby depleting the steady-state glutamine pool, while glutamate is converted to α-ketoglutarate by glutamate decarboxylase. In addition, malate dehydrogenase depletes the malate pool by converting malate to oxaloacetate. Sookoian et al (23) investigated the role of AST and ALT in metabolic pathways and observed that ALT is a major contributor to the steady-state glutamate levels. While this may hold true for its role in the intermediary metabolism of glucose and amino acids, our observations suggest a more elaborate aspartate metabolism pathway. Perhaps, the steady-state pools of metabolites may not entirely elucidate the metabolic activities in HCC. Over expressed enzymes in the aspartate metabolism pathway may be contributing to the steady-state pool of metabolites. However, it is important to note that the isotopic enrichment with 13C label substrates revealed only a fractional enrichment into the steady-state pool.
It is known that the oxidation to acetyl-CoA to CO2 by the TCA pathway is key to the continuous function of oxidative metabolism in mammalian cells. However, the intermediate metabolites of the TCA pathway can be converted to glucose, fatty acids and amino acids for use into biosynthetic pathways. Then these intermediate metabolites are replenished via routes different from acetyl-CoA by a process term anaplerosis. Conversely, cataplerosis is the process whereby 4- and 5-carbon intermediates not fully oxidized are purged out of the TCA pathway to prevent the accumulation of anions in the mitochondrial matrix. Based on our study, we underscore the existence of both processes in the TCA pathway of HCC cells. HCC cells labeled with [U-13C6]glucose converted the glucose to 3-carbon molecules as pyruvate. Subsequent decarboxylation of pyruvate resulted to a 2-carbon molecule, acetyl, which was condensed with CoA and detected as M2 isotopic enrichment. This M2 isotopic enrichment into the intermediate metabolites is evidence of oxidative metabolism.
Another possible way to obtain these 13C labeled metabolites may be explained by cataplerosis and anaplerosis of [U-13C5,15N2]glutamine. In this scenario, the glutamine is oxidized to α-ketoglutarate, which is subsequently converted to malate in the mitochodria. Based on the transcriptomic analysis, malate dehydrogenase is over expressed in aspartate metabolism pathway. The expelled malate from the mitochondria is oxidized in the cytosol to oxaloacetate by malate dehydrogenase. The oxaloacetate is further decarboxylated to phosphoenolpyruvate (PEP), and pyruvate kinase converts PEP to pyruvate. In addition, pyruvate is converted to lactate detected as M3, is transaminated to alanine detected as M3, or is decarboxylated for entry into the TCA pathway as actyl-CoA. Also, we observed M2 isotopic enrichment into lactate and alanine when HCC cells were incubated with [U-13C6]glucose. On the other hand, other studies have shown reductive carboxylation of glutamine metabolism under hypoxic conditions within mammalian cell lines (24–29). These studies were performed mostly with melanoma or lymphoma cell lines carrying c-myc mutation or mutations for isocitrate dehydrogenase 1 (IDH1) or IDH2. Metallo et al (26) suggested that the reductive glutamine metabolism contributes to fatty acid synthesis. These findings may suggest the relevance of cataplerosis and its supporting role in biosynthetic pathways.
Another fascinating aspect of these observations is the isotopic 13C labeling from glucose into the fatty acid synthesis pathways. In our study with Morris hepatoma cell lines using [U-12C6]glucose, labeling into acetyl-CoA then HMG-CoA could suggest a path to fatty acid synthesis. It was previously shown from rat liver perfusates that the mass isotope enrichments of free fatty acids were comparative to acyl-CoAs when perfused with labeled acetic and propionic acid generated from [3,4-13C2]-4-hydroxynonanoate and [5,6,7-13C3]heptanoate (22), respectively. In an anaplerotic path, β-oxidation of heptanoic acid could produce propionyl-CoA that can convert to succinyl-CoA and thereby contribute to TCA intermediates. Detecting 13C-enriched fatty acids in hepatoma cells would verify the utilization of [U-12C6]glucose into fatty acid synthesis under normoxic conditions. Such experiment will require much longer incubation periods than the six-hour incubation periods in our study in order to obtain high 13C enrichment into fatty acids.
Another observation in this study indicated alanine level in HCC was significantly lower compared to the alanine level in non-tumor liver. Conversely, our previous study with hyperpolarized [1-13C]pyruvate MRS imaging of rat livers showed an increased level of alanine in HCC tumors relative to normal liver, which was attributed to the increased level and activity of alanine transaminase in HCC tumors (30). Perhaps, this discordant finding in alanine levels supports the need for isotopic 13C labeling experiments in HCC, which will delineate the de novo alanine pool from the steady-state pool of alanine.
The interpretations of our findings may be limited to hepatocellular carcinomas developed in white males who were HCV negative or HBV negative at the time of diagnosis. Nonetheless, our study was a systematic investigation designed to develop or contribute to generalizable knowledge of HCC. Further investigations are also needed in sub-populations of various etiology of HCC to integrate genetic and metabolic signatures.
Perhaps, this approach can be use to predict the sensitivity of 13C labeling in potential metabolites for the development of MR spectroscopic imaging.
Conclusions
We found that the integrated transcriptomics and metabolomics analyses of HCC permitted quantification of significant metabolic pathways such as aspartate metabolism. The results herein suggest that gene enrichment analysis determined significant alterations of metabolic pathways in HCC, especially for the aspartate metabolism pathway, as important characteristics of HCC. Changes in the overall metabolic profile of HCC indicated that aspartate metabolism is a relevant metabolic signature of HCC, and may be crucial to the progression of HCC. Such molecular signature of HCC could provide an impetus to developing novel enzyme inhibitors as therapeutic agents. Hyperpolarized 13C 3D MRS imaging is a potential diagnostic tool for detection of HCC and may become an important new imaging tool to measure surrogate markers and endpoints for drug treatment.
Supplementary Material
ACKNOWLEDGEMENTS
We very much appreciate the scholarship, support, or scientific contribution of Nathaniel Synder and Kavindra Nath. We very much appreciate the critical review by James Eberwine, Janet Sawicki, James Delikatny, Andrew Cucchiara, and Bruce Sachais. Funding sources were provided by the following NIH grants; KL2RR024132, KL2TR000139 and CA101871. The content of this project is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
List of Abbreviations
- 1D
one-dimensional
- BCAT-1 or -2
branched-chain aminotransferase-1 or -2
- cDNA
complementary deoxyribonucleic acid
- CoA
coenzyme A
- EDTA
ethylenediaminetetraacetic acid
- GSEA
gene set enrichment analysis
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HMG-CoA
3-hydroxyl-3-methylglutaryl-CoA
- Hz
hertz
- LC-MS/MS
liquid chromatography mass spectrometry
- mRNA
messenger ribonucleic acid
- MRSI
magnetic resonance spectroscopic imaging
- PBS
phosphate buffered saline
- rRNA
ribosomal ribonucleic acid
- TCA
tricarboxylic acid
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
REFERENCES
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature Reviews. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global Cancer Statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular Carcinoma Incidence, Mortality, and Survival Trends in the United States From 1975 to 2005. Journal of Clinical Oncology. 2009;27(9):1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen CP, Lin J, Yang YC, Tsai MK, Tsao CK, Etzel C, Huang M, Hsu CY, Ye Y, Mishra L, Hawk E, Wu X. Hepatocellular carcinoma risk prediction model for the general population: the predictive power of transaminases. J Natl Cancer Inst. 2012;104(20):1599–1611. doi: 10.1093/jnci/djs372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franca AV, Elias Junior J, Lima BL, Martinelli AL, Carrilho FJ. Diagnosis, staging and treatment of hepatocellular carcinoma. Braz J Med Biol Res. 2004;37(11):1689–1705. doi: 10.1590/s0100-879x2004001100015. [DOI] [PubMed] [Google Scholar]
- 6.Basu SS, Blair IA. Rotenone-mediated changes in intracellular coenzyme A thioester levels: Implications for mitochondrial dysfunction. Chemical Research in Toxicology. 2011;24:1630–1632. doi: 10.1021/tx200366j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plumb R, Granger J, Stumpf C, Wilson ID, Evans JA, Lenz EM. Metabonomic analysis of mouse urine by liquid-chromatography-time of flight mass spectrometry (LC-TOFMS): detection of strain, diurnal and gender differences. Analyst. 2003;128:819–823. doi: 10.1039/b304296k. [DOI] [PubMed] [Google Scholar]
- 8.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative Metabolome Profiling of Colon and Stomach Cancer Microenvironment by Capillary Electrophoresis Time-of-Flight Mass Spectrometry. Cancer Research. 2009;69(11):4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 9.Opstad KS, Bell BA, Griffiths JR, Howe FA. An assessment of the effects of sample ischaemia and spinning time on the metabolic profile of brain tumour biopsy specimens as determined by high-resolution magic angle spinning 1H NMR. NMR in Biomedicine. 2008;21(10):1138–1147. doi: 10.1002/nbm.1296. [DOI] [PubMed] [Google Scholar]
- 10.Gao H, Lu Q, Liu X, Cong H, Zhao L, Wang H, Lin D. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100(4):782–785. doi: 10.1111/j.1349-7006.2009.01086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Li C, Nie X, Feng X, Chen W, Yue Y, Tang H, Deng F. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6(7):2605–2614. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- 12.Amathieu R, Nahon P, Triba M, Bouchemal N, Trinchet JC, Beaugrand M, Dhonneur G, Le Moyec L. Metabolomic approach by 1H NMR spectroscopy of serum for the assessment of chronic liver failure in patients with cirrhosis. J Proteome Res. 2011;10(7):3239–3245. doi: 10.1021/pr200265z. [DOI] [PubMed] [Google Scholar]
- 13.Duarte IF, Stanley EG, Holmes E, Lindon JC, Gil AM, Tang H, Ferdinand R, McKee CG, Nicholson JK, Vilca-Melendez H, Heaton N, Murphy GM. Metabolic assessment of human liver transplants from biopsy samples at the donor and recipient stages using high-resolution magic angle spinning 1H NMR spectroscopy. Anal Chem. 2005;77(17):5570–5578. doi: 10.1021/ac050455c. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Granados B, Monleon D, Martinez-Bisbal MC, Rodrigo JM, del Olmo J, Lluch P, Ferrandez A, Marti-Bonmati L, Celda B. Metabolite identification in human liver needle biopsies by high-resolution magic angle spinning 1H NMR spectroscopy. NMR Biomed. 2006;19(1):90–100. doi: 10.1002/nbm.1005. [DOI] [PubMed] [Google Scholar]
- 15.Shariff MI, Gomaa AI, Cox IJ, Patel M, Williams HR, Crossey MM, Thillainayagam AV, Thomas HC, Waked I, Khan SA, Taylor-Robinson SD. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation study. J Proteome Res. 2011;10(4):1828–1836. doi: 10.1021/pr101096f. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Wang W, Lv S, Yin P, Zhao X, Lu X, Zhang F, Xu G. Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal Chim Acta. 2009;650(1):3–9. doi: 10.1016/j.aca.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Xue R, Dong L, Liu T, Deng C, Zeng H, Shen X. Metabolomic profiling of human urine in hepatocellular carcinoma patients using gas chromatography/mass spectrometry. Anal Chim Acta. 2009;648(1):98–104. doi: 10.1016/j.aca.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 18.Chan ECY, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance. Journal of Proteome Research. 2009;8:352–361. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- 19.Yang C, Richardson AD, Smith JW, Osterman A. Comparative metabolomics of breast cancer. Pacific Symposium on Biocomputing. 2007;12:181–192. [PubMed] [Google Scholar]
- 20.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nature Protocols. 2008;3(8):1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez CA, Rosiers CD, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. Journal of Mass Spectrometry. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Tomcik K, Ibarra RA, Sadhukhan S, Han Y, Tochtrop GP, Zhang G-F. Isotopomer enrichment assay for very short chain fatty acids and its metabolic applications. Analytical Biochemistry. 2011;410:110–117. doi: 10.1016/j.ab.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sookoian S, Pirola CJ. Alanine and aspartate aminotransferase and glutamine-cycling pathway: their roles in pathogenesis of metabolic syndrome. World J Gastroenterol. 2012;18(29):3775–3781. doi: 10.3748/wjg.v18.i29.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filipp FV, Scott DA, Ronai ZA, Osterman AL, Smith JW. Reverse TCA cycle flux through isocitrate dehydrogenases 1 and 2 is required for lipogenesis in hypoxic melanoma cells. Pigment Cell Melanoma Res. 2012;25(3):375–383. doi: 10.1111/j.1755-148X.2012.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, Zimmerman LJ, Liebler DC, Siebos RJC, Lorkiewicz PK, Higashi RM, Fan TWM, Dang CV. Glucose-Independent Glutamine Metabolism via TCA Cycling for Proliferation and Survival in B Cells. Cell Metabolism. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, Osterman AL, Smith JW. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286(49):42626–42634. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darpolor MM, Yen YF, Chua MS, Xing L, Clarke-Katzenberg RH, Shi W, Mayer D, Josan S, Hurd RE, Pfefferbaum A, Senadheera L, So S, Hofmann LV, Glazer GM, Spielman DM. In vivo MRSI of hyperpolarized [1-(13)C]pyruvate metabolism in rat hepatocellular carcinoma. NMR in Biomedicine. 2011;24(5):506–513. doi: 10.1002/nbm.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.