Abstract
Nanomedicine is an emerging field that integrates nanotechnology, biomolecular engineering, life sciences and medicine; it is expected to produce major breakthroughs in medical diagnostics and therapeutics. Nano-scale structures and devices are compatible in size with proteins and nucleic acids in living cells. Therefore, the design, characterization and application of nano-scale probes, carriers and machines may provide unprecedented opportunities for achieving a better control of biological processes, and drastic improvements in disease detection, therapy, and prevention. Recent advances in nanomedicine include the development of nanoparticle-based probes for molecular imaging, nano-carriers for drug/gene delivery, multi-functional nanoparticles for theranostics, and molecular machines for biological and medical studies. This article provides an overview of the nanomedicine field, with an emphasis on nanoparticles for imaging and therapy, as well as engineered nucleases for genome editing. The challenges in translating nanomedicine approaches to clinical applications are discussed.
Key Terms: Nanomedicine, nanoparticles, molecular imaging probes, engineered nucleases, multifunctional nanoparticle, genome editing
Introduction
Recent advances in nanotechnology provide new abilities to measure, control and manipulate matter (including soft matter) at the nano-scale that is unthinkable with conventional tools. As an emerging field in biomolecular engineering and molecular medicine, nanomedicine focuses on the development and application of engineered nano-scale (1–100 nm) materials, structures and devices for better diagnostics of, and highly specific medical intervention in curing, human diseases1. Owing to the size-compatibility of nano-scale structures with proteins and nucleic acids in living cells, nanomedicine approaches have the potential to provide unprecedented opportunities for achieving a better control of biological processes, and drastic improvements in disease detection, therapy, and prevention, thus revolutionizing medicine.
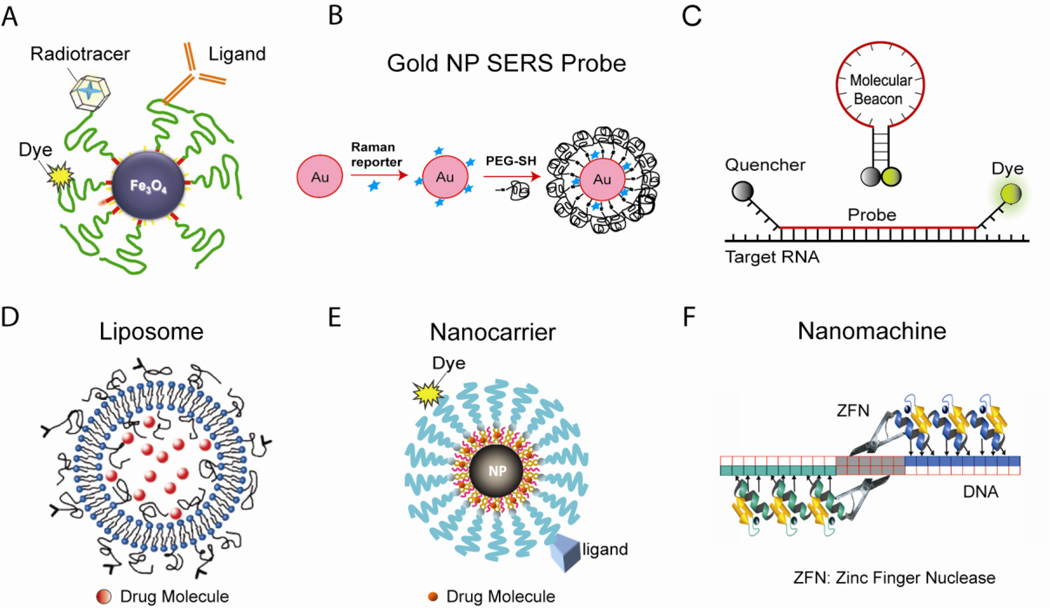
Over the last 15 years or so, significant efforts have been made to develop nanomedicine based approaches, and many potential applications of nanomedicine have been, or are being, explored, including nanoparticle-based molecular imaging probes for biological studies and disease detection (Figures 1a–1c); nanocarriers for targeted in vivo drug/gene delivery for more efficient therapy (Figures 1d–1e); nanoparticles as direct therapeutic agents; and nuclease-based biological nanomachines for genome editing (Figure 1f). For basic biological studies, the development of new nano-scale tools and devices have the potential to permit imaging of cellular structures at the nano-scale, rapid measurement of the dynamic behavior of protein complexes and molecular assemblies in living cells and animals, and a better control of intracellular machinery. It is expected that the multifunctional, targeted nanoparticles are capable of overcoming biological barriers to deliver therapeutic agents preferentially to diseased cells and tissues at high local concentrations, resulting in much enhanced efficacy and reduced toxicity. Nanomedicine approaches have the potential to allow clinicians to detect a disease in its earliest, most easily treatable, presymptomatic stage, and provide real-time assessments of therapeutic and surgical outcomes. Nano-scale tools may also be used to quickly identify new disease targets for drug development and predicting drug resistance.
Figure 1. A nanotechnology toolbox.
(a) Superparamagnetic iron oxide nanoparticle probes for multimodality molecular imaging. The iron oxide (Fe3O4) nanoparticle is a T2 contrast agent for MRI, and the addition of a radiotracer makes it a PET contrast agent. With a dye in/on the coating, the nanoparticle is also a fluorescent report. (b) Gold nanoparticle-based SERS probes for biomolecule detection. The gold nanoparticle is encoded with a Raman reporter, and stabilized with a layer of thiol-PEG. (c) Molecular beacons, dual-labeled hairpin oligonucleotide probes for RNA detection in living cells and clinical samples. (e) Targeted liposomes for drug/gene delivery. (f) Targeted magnetic nanocarriers for drug/gene delivery. The T2 contrast of the nanoparticle and the fluorescent report allow tracking of the nanocarrier in vivo. (D) Zinc finger nuclease-based nanomachines for genome editing. The zinc fingers bind to DNA with high specificity, and the nuclease domains of the pair of ZFNs dimerize and cleave DNA, inducing modification of the genome.
In this review, emphasis is placed on nanoparticle-based molecular imaging probes, multifunctional nanoparticles, and the design and validation of engineered nucleases for genome editing. The challenges in translating nanomedicine approaches to clinical applications are discussed. Due to space limitations, this is not intended to be a comprehensive review, but rather a review of selected research topics in nanomedicine. Other recent reviews of nanomedicine can be found in the literature 2–4.
Inorganic Nanoparticles
Many studies in nanomedicine involve the development and application of nanoparticles, including organic and inorganic nanoparticles. Attention here is placed primarily on inorganic nanoparticles; reviews of organic nanoparticles can be found elsewhere5–11. Inorganic nanoparticles are typically made of metal, metal oxide, semiconductor, or rare-earth element. They often possess unique electric, magnetic, optical and plasmonic properties due to the quantum mechanical effects at nanometer scales12. Due to significant progress in nanocrystal synthesis over the past decade or so, for most nanoparticle systems, their chemical composition, size, shape and other physical properties can be well controlled13–16.
Quantum Dots
Perhaps the most extensively studied nanoparticle system to date is quantum dots (QDs), especially fluorescence-emitting semiconductor quantum dots17, which usually have a core-shell structure such as CdSe core with ZnS shell (Figure 2a). Semiconductor QDs, typically 2–6 nm in diameter, have exceptionally bright fluorescence emission; their emission peaks are red-shifted as the size of QDs increases (Figure 2a). The optical properties of QDs are the results of quantum confinement of valence electrons at nanometer scales12; their fluorescence emission wavelengths rely on the energy band gap determined by the size and compositions of the QD18. In contrast to organic fluorophores, QDs are very photostable, their emission peaks are narrow, with absorption spectra range from UV to visible wavelength. Therefore, multiple QDs with different emission wavelengths can be excited simultaneously upon UV excitation, facilitating multicolor imaging. Further, QDs have Stokes shifts (separation between excitation and emission peaks) as large as 300–400 nm19. This reduces interference from tissue autofluorescence in biological specimens, which may bury signals from an organic dye. However, semiconductor QDs are often toxic; concerns over the in vivo toxicity have prompted search for biocompatible QDs, such as InP/ZnSe and InP/ZnS QDs20, 21.
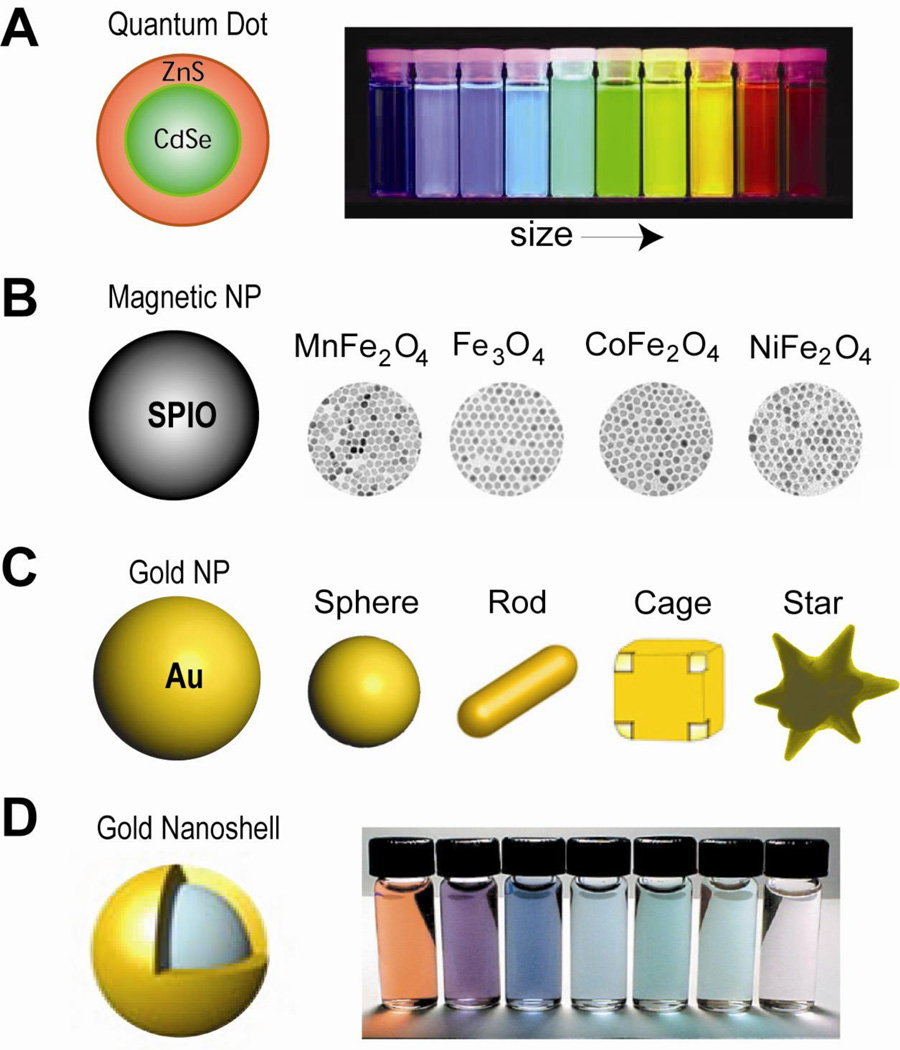
Figure 2. Inorganic nanoparticle systems.
(a) Quantum dots are core-shell nanocrystals. Under UV excitation, ZnS-capped CdSe quantum dots have tunable light emission. (b) Magnetic iron oxide nanoparticles. Magnetic nanoparticles (Fe3O4) and those doped with Mn, Co or Ni exhibit different mass magnetization and thus MRI T2 relaxivity. (c) Gold nanoparticles, including gold nanosphere, nanorod, gnanoshell, nanocage and nanostar, are excellent imaging contrast agents and photothermal therapeutic agents. (d) Gold nanoshells with various thicknesses have tunable light emission.
Magnetic Nanoparticles
Magnetic nanoparticles are mainly composed of iron oxides, including Fe3O4, Fe2O3, MnFe2O4, CoFe2O4 and NiFe2O4 (Figure 2b), and to a lesser extent of elementary iron and other magnetic elements 15, 22. With sizes < 20 nm, certain iron oxide nanoparticles become superparamagnetic at room temperature 23. Without an external magnetic field, neighboring superparamagnetic nanoparticles are free of inter-particle magnetic interactions, which is critical for the colloidal stability of the nanoparticle. However, when an external magnetic field is applied, the magnetic moment of the nanoparticles will align with the applied field and reach saturation at relatively low field strength. Magnetite (Fe3O4) and maghemite (Fe2O3), owing to their excellent biocompatibility, are the most common types of materials in generating superparamagnetic nanoparticles. Superparamagnetic nanoparticles of materials that have higher saturation magnetization have also been developed, including elementary iron, cobalt or iron oxide doped with manganese (Figure 2b) 24, 25.
Superparamagnetic iron oxide nanoparticles (SPIOs), mostly as a T2 contrast agent in magnetic resonance imaging (MRI), have the potential to significantly improve the sensitivity of disease detection26. The T2 relaxivity of SPIOs is determined by their magnetic moment and coating conformation, which are in turn controlled by the material and size of the core and coating layer, and the density of the coating molecules on nanoparticle surface24, 27. In general, SPIOs with higher magnetization, larger core size and thinner surface coating have higher T2 relaxivity. In particular, SPIOs with large size and high magnetization can have a T2 relaxivity about two orders of magnitude higher compared with the clinically used first-generation SPIOs on a per particle basis 27, therefore hold a great promise in MRI-based molecular imaging. Further, the T2 relaxivity of magnetic nanoparticles increases upon aggregation, a phenomenon referred to as “magnetic relaxation switch” 28. Accordingly, an approach to achieve high T2 relaxivity is to generate nanoparticles with multiple magnetite cores29.
Gold Nanoparticles
The most common type of gold nanoparticles is gold nanosphere (Figure 2c), which exhibits an intense ruby color in aqueous solutions. The intriguing optical properties of gold nanoparticles arise from localized surface plasmon resonance (LSPR), whereas valence electrons in gold nanoparticles oscillate coherently with incident light at specific frequency30. Part of the energy absorbed by gold nanoparticles is emitted in the form of scattered light, which is the basis of many gold nanoparticle-based optical imaging31. The rest of the energy decays in a non-radiative form, i.e., being converted into heat. Gold nanospheres with a wide range of sizes mainly absorb light at ~520 nm wavelength, at which light is rapidly attenuated by the tissue. For in vivo applications, the absorption peak should fall within the optical window of human tissues (650~1300 nm) so that the incident light could penetrate deeply32. The absorption spectrum of gold nanoparticles can be tuned by changing their geometry33. For example, in gold nanorods (Figure 2c), LSPR occurs in two directions along the short and the long axis. The frequency of oscillation along the long axis red-shifts from visible to the near infrared (NIR) region as the aspect ratio of nanorods increases34. Other gold nanostructures with tunable LSPR frequency are gold nanocages (Figure 2c) and gold nanoshells (Figure 2d), of which the absorption spectra change with the overall size, the thickness of the shell (nanoshell) or the wall (nanocage) 35, 36. Further, gold nanoshells and nanostars (Figure 2c) with the NIR photothermal property are ideal agents for photothermal therapy37–39. Aggregated gold nanospheres also exhibit significant NIR absorption as a result of coupled plasmon resonance. This has been used to create nanostructures with the NIR photothermal property for in vivo applications40.
Nanoparticle-based Probes for Molecular Imaging and Biomolecule Detection
One of the most fruitful areas of nanomedicine is molecular imaging using nanoparticle probes, including quantum dots, magnetic nanoparticles and gold nanoparticles. Nanoparticles possess unique characteristics that make them well suited as probes for molecular imaging 41. Nanoparticles, including metal, metal oxide and semi-conductor nanoparticles can be synthesized in a systematic fashion to have precise diameter with narrow size distributions. As nanoparticles become smaller, their surface area to volume ratio increases significantly. Engineering of nanoparticle surface chemistry allows the surface area to be decorated with therapeutic molecules, imaging agents, targeting ligands, or nucleic acids. To perform nanoparticle-based in vivo imaging of molecular markers associated with disease development, the nanoparticles must be functionalized with specific targeting ligands42. Distinct ligands and reporters can be attached to a single nanoparticle to allow multiplexing and multi-functionality. A single nanoparticle can be conjugated with a large number of targeting ligands, increasing the affinity of the nanoparticle to its biological target through multivalency. Further, a nanoparticle can be conjugated with a large number of reporter molecules (e.g. fluorophores, radiotracers), increasing signal-to-noise ratio in imaging applications. Described below are specific examples of nanoparticle-based molecular imaging probes, more comprehensive reviews in this area can be found in the literature.
Gold Nanoparticles for Imaging Applications
Gold nanoparticles, including nanospheres, nanoshells, nanorods and nanocages, can be used as contrast agents in photoacoustic imaging43–46. Gold nanoparticles scatter light strongly at their LSPR frequency and thus have broad applications in optical imaging47. Furthermore, gold nanoparticles increase local electromagnetic field due to LSPR. As a result, the signals of fluorophores and surface enhanced Raman scattering (SERS) reporters attached to gold surface can be drastically enhanced48. In in vivo applications, gold nanoparticles that scatter NIR light can provide good contrast for optical coherence tomography49. Gold nanospheres and gold nanorods have also been used as contrast agents in computed tomography (CT), taking advantage of the high material density and high atomic number of gold50, 51. Gold nanoshells and nanocages, even without targeting, can be used as blood pool contrast agents to enhance the visualization of vasculature in vivo (e.g., in the rat brain)43, 44.
Quantum Dots for Imaging Applications
Since QDs were first rendered water-soluble in 1998 thereby making them relevant for biological studies52, 53, they have been applied to cell tracking studies54, 55, cancer imaging 19, 56, flow-cytometry57, and labeling of membrane proteins58, 59. The applications of QDs have also been extend to fluorescence-based detection of enzymatic activities, particularly when QDs are utilized as either a donor or an acceptor for fluorescence (or Förster) resonance energy transfer (FRET)60–64.
For biological applications, it is necessary to coat QDs, making them water-soluble and functionalizable while avoiding deleterious effects on the optical properties65, 66. Strategies have been devised using direct adsorption of dihydrolipoic acid (DHLA) on the surface (leading to negative charges), and then electrostatic self-assembly to positively charged proteins67, 68. A novel approach is based on the adsorption of bifunctional ligands such as mercaptoacetic acid, mercaptosuccinic acid, dithiothreitol, glutathione or histidine directly to the QD surface69. Other strategies employ hydrophilic organic dendron ligands 70, and micellular encapsulation using phospholipids65. Importantly, amphiphilic polymer coated QDs conjugated to streptavidin have become commercially available66.
Quantum Dot – Fluorescent Protein FRET Probe for pH Sensing
QDs make excellent FRET donors because of their characteristic excitation and emission spectra, exceptional brightness and high quantum yields, and the ability to bind multiple acceptor molecules. A particular class of QD-based FRET probes is composed of multiple fluorescent proteins (FP) as FRET acceptors conjugated to a QD donor. Since the polypeptide sequence of a FP can be genetically modified to include structural and functional elements necessary for protein purification, signal transduction, and probe assembly, as well as intracellular delivery and localization, the use of fluorescent proteins as FRET acceptors offers clear advantages. For example, standard molecular biology techniques can easily be used to modify the fluorescent proteins to include a tag for effective conjugation, and a variety of amino acid sequences can be used as linkers between the protein bulk and the tag, adding to the functionality of the QD-FRET probe (such as a cleavage sequence for a protease). The variety of GFP-like fluorescent proteins now available provides a wide range of possible fluorescent protein acceptors; they can be readily expressed in E.coli in large quantities. QD-FP FRET pairs exhibit high energy transfer efficiencies and enable ratiometric measurements, resulting in heightened sensitivity by eliciting opposing changes in fluorescence emission at two wavelengths, while maintaining an internal control at an isosbestic point71.
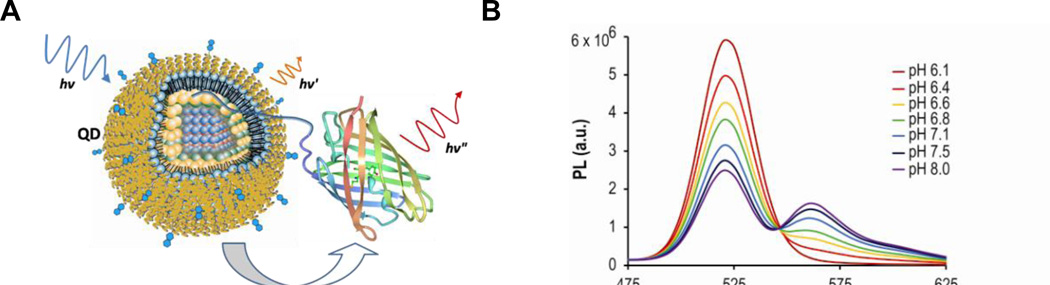
A specific application of the QD-FP FRET probes is the intracellular pH measurement in biological and disease studies. Intracellular pH (pHi) plays a critical role in the physiological and pathophysiological processes of cells72, and fluorescence imaging using pH-sensitive indicators can provide a powerful tool to assess the pHi of intact cells and sub-cellular compartments. As an example, shown in Figure 3a is the QD-based ratiometric pH sensor comprised of a carboxyl-functionalized QD as donor and pH-sensitive mOrange FPs as accepters73. Titration of the QD-FP probes showed that FRET between the QD and multiple FPs modulates the FP/QD emission ratio, exhibiting a >12-fold change between pH 6 and 8 (Figure 3b). The QD-FP probes can dramatically improve the sensitivity and photostability compared to BCECF, the most widely used fluorescent dye for pH imaging73. Further, the QD-FP probes facilitated the visualization of the acidification of endosomes in living cells following polyarginine-mediated uptake. Potential applications of the QD-FP FRET probes include tracking the endosomal release of nanocarriers for drug/gene delivery, and monitoring pH and/or metal ion concentration in both the intracellular and extracellular environment.
Figure 3. Quantum dot – fluorescent protein FRET probes for pH sensing.
(a) Schematic diagram of the FRET interaction between a quantum dot and a GFP-like fluorescent protein. A polyhistidine sequence inserted at the N-terminus of the fluorescent shown here coordinates to the ZnS capping layer of the QD, bringing the two into close proximity. Under excitation of the QD, energy is non-radiatively transferred to the fluorescent protein and sensitized emission is observed. (b) Titration of QD-FP probes containing the FP acceptors mOrange showing increased energy transfer at alkaline pHs with clear isosbestic points. Representative spectra of one of three independent titrations are shown.
Superparamagnetic Iron Oxide Nanoparticle based Imaging Contrast Agents
Nanoparticles are a very promising platform as MRI and PET contrast agent for in vivo molecular imaging. For example, compared to gadolinium-based MRI contrast agents, nanoparticle MRI contrast agents usually circulate in the blood for longer periods of time, and each nanoparticle may generate signal contrast several orders of magnitude higher than a gadolinium chelate, thus offering greater sensitivity with reduced side-effects 74. Perhaps one of the most promising nanoparticle systems is SPIOs 75, 76. SPIOs can induce an increased T2 relaxivity in MRI, which is determined by the translational diffusion of water molecules in the inhomogeneous magnetic field surrounding the SPIO77–79. In addition, iron oxide has little toxicity for in vivo applications. These features make SPIOs an appealing MRI contrast agent for the detection and diagnosis of human diseases 24, 80–82. For biomedical applications, the hydrophobic nanocrystal core of a SPIO is typically encapsulated with amphiphilic polymers such as Poloxamer®, Poloxamine® and lipid-PEG copolymer83–85. A variety of SPIOs with different cores and surface coatings are currently under clinical trials for imaging liver tumors and metastatic lymph nodes24, 85–89. SPIOs can accumulate in the tumor by enhanced permeability and retention effects (EPR), which facilitate tumor detection in tissues or organs that normally do not retain SPIOs.
Typically SPIOs can only provide a dark contrast in MRI, which might be difficult for quantitative determination of the contrast due to local nanoparticle accumulation. In contrast, nanoparticles conjugated with Mn2+ or Gd3+ chelates are capable of generating a bright signal in T1-weighted MRI images. Such nanoparticles include micelles, polymeric particles and liposomes. However, most T1 contrast agents have poor relaxivity compared with the T2 relaxivity of SPIOs. Recently, hollow MnO nanoparticles have been developed as a dual T1 and T2 contrast agent 90. Core-shell T1–T2 dual mode nanoparticles that combine both MnFe2O4 and gadolinium have also been synthesized91. It has shown that by separating gadolinium and MnFe2O4 with a silica oxide layer, the nanoparticles could achieve both high T1 and T2 relaxivity. Performing both T1- and T2-weighted imaging could help exclude faulty signals and enhance diagnostic accuracy.
The performance of nanoparticle probes as imaging contrast agents is determined by signal-to-noise ratio and detection sensitivity. It is therefore critical to synthesize SPIOs with substantial signal enhancement on a per-particle basis to improve sensitivity, and to tailor nanoparticle surface chemistry to optimize the specific accumulation (attachment) of SIPOs in/on diseased cells, tissues or organs. It has been established that T2 relaxivity of SPIOs increases with the magnetization and the size of iron oxide cores if the total amount of iron is constant77, 92. The magnetization of SPIOs can also be enhanced by using cores formed by elementary iron or by doping iron oxide with other magnetic elements such as nickel, cobalt and manganese24, 93, 94. Further, the core size of SPIOs can be increased by employing controllable crystallization through thermo-decomposition of iron complex in organic solvents87, 88, 94, 95.
Nanoparticles for Hyperthermia and Magnetic Targeting
In addition to in vivo imaging applications, over past few years, magnetic nanoparticles have received extensive studies for applications in hyperthermia and magnetic targeting. Magnetic nanoparticles can generate heat in oscillating magnetic field (100 kHz to 1 MHz), which enables hyperthermia in deep tissues96–98. Magnetic targeting has been widely used in various in vitro applications including cell separation, gene transfection and sample enrichment in detection assays. Recent studies have shown that in vivo magnetic targeting can be used for controlled delivery of therapeutic agents and cells99, 100, and stem cells and progenitor cells could be labeled with high amount of SPIOs without affecting cell viability and function101–103. Tracking immune cells labeled with SPIO has been an important approach in detecting atherosclerosis104. The magnetic mobility and heating capability of magnetic nanoparticles can also be employed to trigger cellular events in vivo105, 106.
Iron Oxide Nanoparticle–based Biomolecule Detection
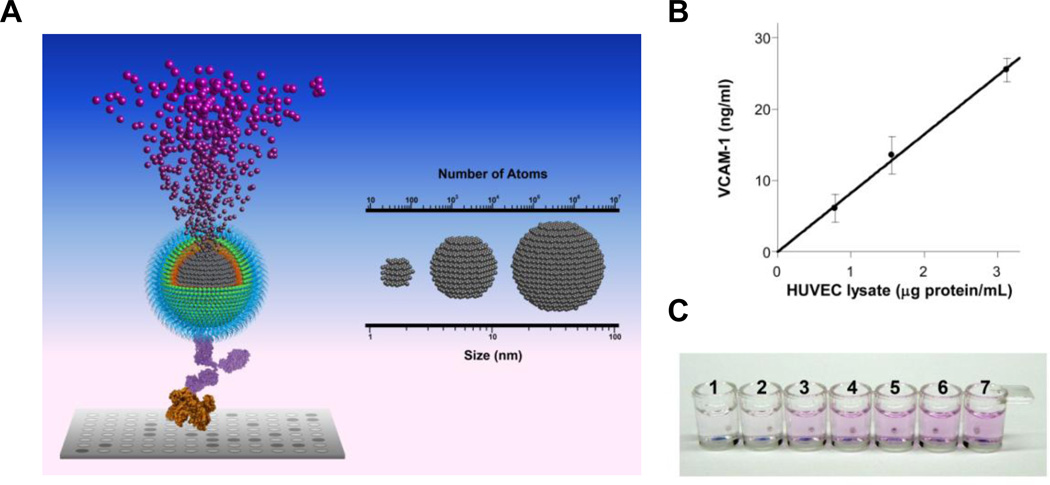
Many biological studies and clinical applications require sensitive detection of biomolecules, including lipids, proteins and nucleotides. Enzyme linked immunosorbent assay (ELISA) has been the industrial standard for quantification of protein and other macromolecule since its emergence in 1970s107, 108. However, enzyme-substrate interaction is subjected to a number of variables such as enzymatic activity and incubation conditions. Biomolecule quantification with enzyme-linked probes is hindered by the requirement of stringent control, costly calibration as well as the nonlinear nature of enzymatic catalysis. Recently, the iron oxide nanoparticle linked immunosorbent assay (ILISA) was developed by integrating the dissolution of nanocrystals and metal-induced stoichiometric chromogenesis, exploiting the dense atom packing in metallic nanocrystals.109 As shown in Figure 4a, antibody-functionalized wüstite (Fe1-xO) nanocrystals are bound to the target molecules (e.g., antigen) in a 96-well assay format. After unbound probes are removed, the nanocrystals are dissolved into individual metal ions that are then converted to chromophores. In ILISA, signal amplification is fully determined by the total number of Fe atoms in the nanocrystals bound to single target molecules. Since each metallic nanocrystal consists of thousands to millions of metal atoms, extremely high signal amplification can be achieved by stoichiometrically converting the nanocrystal to chromophores and quantifying them photometrically. Similar to ELISA, direct, indirect, competitive and sandwich ILISA can all be performed with high sensitivity. The signal amplification of ILISA can be further optimized by varying the size and number of nanocrystals bound to an individual analyte molecule109.
Figure 4. Iron oxide nanoparticle linked immunosorbent assay.
(a) Nanocrystal amplification. The nanocrystal conjugated with the probe is dissolved by acid into individual metal atoms which are converted to chromophores through a stoichiometric reaction. Signal amplification is fully determined by the total number of atoms in the nanocrystals bound to a single target molecule. (b) VCAM-1 in the lysate of LPS-stimulated HUVEC was quantified with a sandwich ILISA. Mean ± standard deviation of three measurements is plotted. (c) Instrument-free distinction of factor-X deficient vs. normal human plasma. Factor X in human plasma was detected with a sandwich ILISA.
Compared with enzyme-based signal amplification, ILISA provides a simple, highly sensitive and more reliable assay for biomolecule detection, facilitating the quantification of biomolecule concentration and binding kinetics in laboratory settings, and enabling instrument-free evaluation of disease markers for on-site diagnostics. To illustrate, sandwich ILISA was used to detect vascular cell adhesion molecule-1 (VCAM-1) in the lysate of lipopolysaccharide (LPS)-stimulated human umbilical vascular endothelial cells (HUVECs). It was found that VCAM-1 could be detected at low total protein concentrations (1 µg/mL) while achieving a good linearity (Figure 4b). Sandwich ILISA was also used to detect a blood coagulation factor factor-X in clinical plasma samples in an instrument-free fashion (Figure 4c). The chromogenic reactions can be either solution-based or surface-based and performed in aqueous or organic phase, supporting a variety of assay formats110. The nanocrystal-based amplification scheme can also be implemented with a rich selection of metal/metal oxide nanocrystals and metal-reactive chromogenic substrates, thus multiplexable. This nanoparticle-based signal amplification scheme adds a new dimension to current nanoparticle-based bioassays.
Multifunctional Nanoparticles for Combined Imaging and Therapy
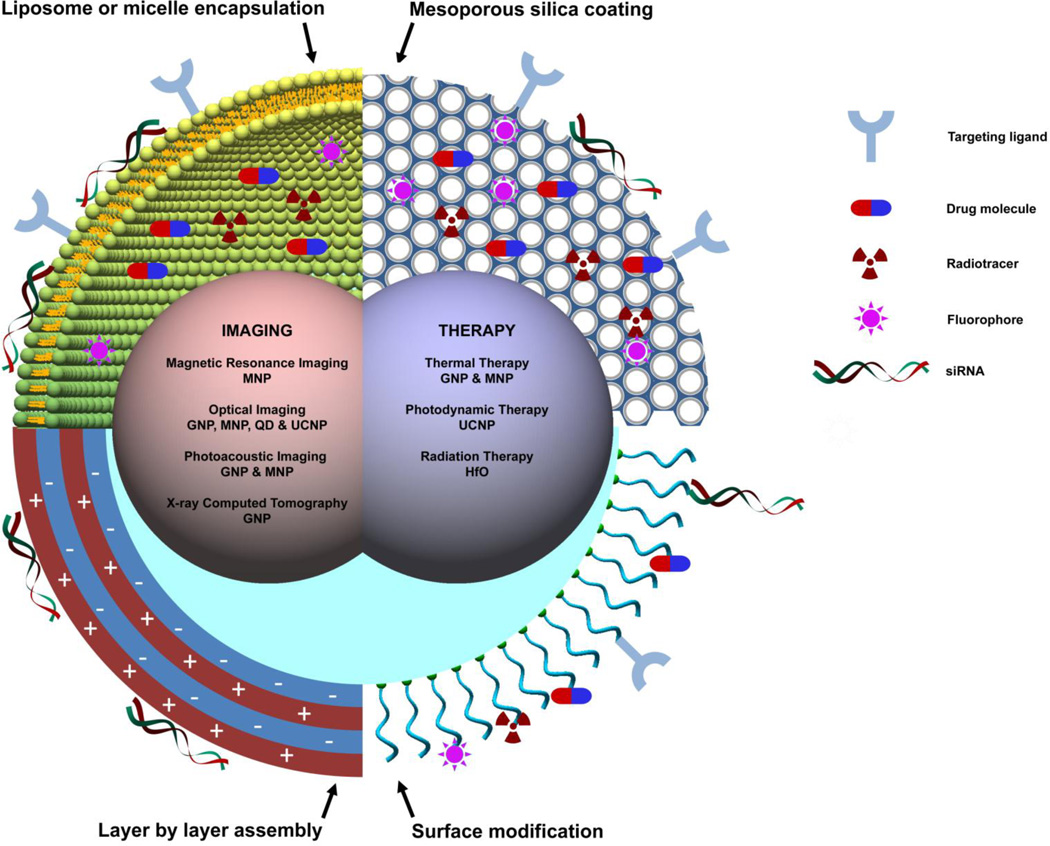
For medical applications, it is often desirable to integrate imaging probes and therapeutic agents into one, often referred to as a theranostic agent, to improve efficacy and reduce toxicity. With only a few exceptions, theranostic agents are developed using nanoparticle platforms. A typical theranostic nanoparticle may have part or all of the following components: (1) an imaging contrast agent, (2) chemical bonds or physical interactions for loading and releasing of drugs or genes, (3) targeting ligands, and (4) elements that respond to external triggers such as ultrasound, light and magnetic field (Figure 5). Recent advances in nanomaterials enable the synthesis of nanoparticles with rich physical and chemical properties that facilitate the incorporation of small molecular moieties. Multiple functionalities can be readily integrated using the same nanoparticle by modulating its structural and/or chemical compositions. For example, nanocrystals can be readily decorated with different coating strategies, including silica/mesoporous silica, micelle, liposome and layer-by-layer assembly. These coatings are useful for both physical/chemical adsorption of small molecules and encapsulation of multiple nanocrystals. Small molecules, including dyes, therapeutic agents and targeting ligands, can be conjugated to nanoparticles at substantial payload ratio using well controlled conjugation chemistry.
Figure 5. Schematic diagram of multifunctional nanoparticles.
Multifunctional nanoparticles can be generated by either combining nanocrystals with different functionalities or combining nanocrystals with functional small molecule cargos through different surface engineering strategies. Four typical coating developed for inorganic nanocrystals are: (1) liposome or micelle encapsulation, (2) mesoporous silica coating, (3) layer by layer assembly and (4) surface conjugation. Abbreviations used in this figure: GNP – gold nanoparticles, MNP – magnetic nanoparticles, QD – quantum dots, UCNP – upconversion nanoparticles, and HfO - hafnium oxide nanoparticles.
There have been a few classes of multifunctional, theranostic nanoparticles developed, including gold nanoparticles and magnetic iron oxide nanoparticles. In addition to be remarkable contrast agents for optical imaging, photoacoustic imaging and computed tomography (CT) 49, 50, gold nanoshells, nanorods, nanocages and nanostarts can be employed for photothermal therapies 34, 37. Recent studies have shown that gold nanoparticles can be heated by non-resonant shortwave radiofrequency fields, which allows targeted hyperthermia in deep tissue 111. Gold nanocages have also shown great potential as drug carriers for controlled release with near-infrared light112.
Superparamagnetic iron oxide nanoparticles (SPIOs) are the most well-studied theranostic nanoparticles owing to their excellent biocompatibility113, superior MRI T2 contrast27, magneto-mobility100 and capability to be heated with an alternating magnetic field97. In the past few years, several forms of SPIO-based drug carriers have been developed by modifying iron oxide nanocrystals with various surface coating. For example, it has been shown that mitoxantrone conjugated to starch-coated SPIOs could effectively treat VX-2 squamous cell carcinoma in a rabbit model114. In this study, the SPIOs administrated intra-arterially were retained at the tumor site by an electromagnet, thus significantly reducing systemic toxicity. However, drug loading via chemical conjugation is restricted to a small group of drug molecules with reactive moieties. In contrast, iron oxide nanocrystals with micellar coating have great promise as carriers for many small chemotherapeutic drug molecules115–118. In this approach, iron oxide nanocrystals are synthesized with nonpolar capping molecules. Water-dispersible nanoparticles are obtained by coating the nanocrystals with a monolayer of amphiphilic copolymers through hydrophobic interactions84, 119. Small hydrophobic molecules such as doxorubicin and paclitaxel can be stored in the hydrophobic layer of micellar coating. Nanoparticles with amphiphilic capping molecules covalently bound to iron oxide surface can also adsorb drug molecules by hydrophobic interactions120, 121. Another coating with broad drug loading capability is mesoporous silica122. Small drug molecules and fluorophores can be loaded into the porous silica shell through either chemical conjugation or electrostatic interactions. Drug loading by hydrophobic or electrostatic interactions often affords much higher loading capacity than that of chemical conjugation. However, drug molecules may be prematurely released from the nanoparticles during systemic circulation. Other major types of magnetic delivery vehicles include liposomes or cerasomes containing iron oxide nanocrystals123.
Magnetic iron oxide nanoparticles have also been used as the carriers for in vivo gene delivery. For example, multifunctional nanoparticles were constructed by simultaneously conjugating siRNA, near infrared fluorophores (Cy5.5) and cell membrane translocation peptides to dextran coated iron oxide nanoparticles124. Tumor accumulation of the nanoparticles was confirmed with both T2 contrast in MRI and fluorescence imaging, which was in good agreement with the knockdown effects of siRNAs observed. In a similar preparation, siRNA, Cy5 and RGD peptides were conjugated to the surface of MnFe2O4 nanoparticles that possess high T2 relaxivity 41. Magnetic liposomes that bound siRNAs by electrostatic interactions were also developed, which inhibited tumor angiogenesis and proliferation by suppressing the expression of EGF receptor in a mouse tumor model100.
To date, most of the studies in nanomedicine involve the development of nanoparticle-based approaches, especially multifunctional nanoparticle systems as imaging contrast agents and/or drug/gene delivery vehicles, utilizing a combination of targeting, delivery, reporting (contrast), and physical effector functions. Although still in its infancy, the development of engineered molecular machines has the potential to significantly advance nanomedicine. These molecular machines are typically multi-component and multi-functional, with target recognition, actuation, manipulation, self-assembly and disassembly functions, enabling the quantification, control and precise modification of biological processes in living cells. Discussed below is a class of molecular machines that perform highly specific modification of DNA sequences in living cells for genome editing applications.
Engineered Molecular Machines for Genome Editing
A major goal of nanomedicine is to enable highly specific medical intervention at the molecular scale for curing disease and/or repairing tissue. To achieve this goal, it is necessary to precisely control and manipulate biomolecules and supramolecular assemblies in living cells, exploiting nature’s design of biological machines, pathways and molecular structures.
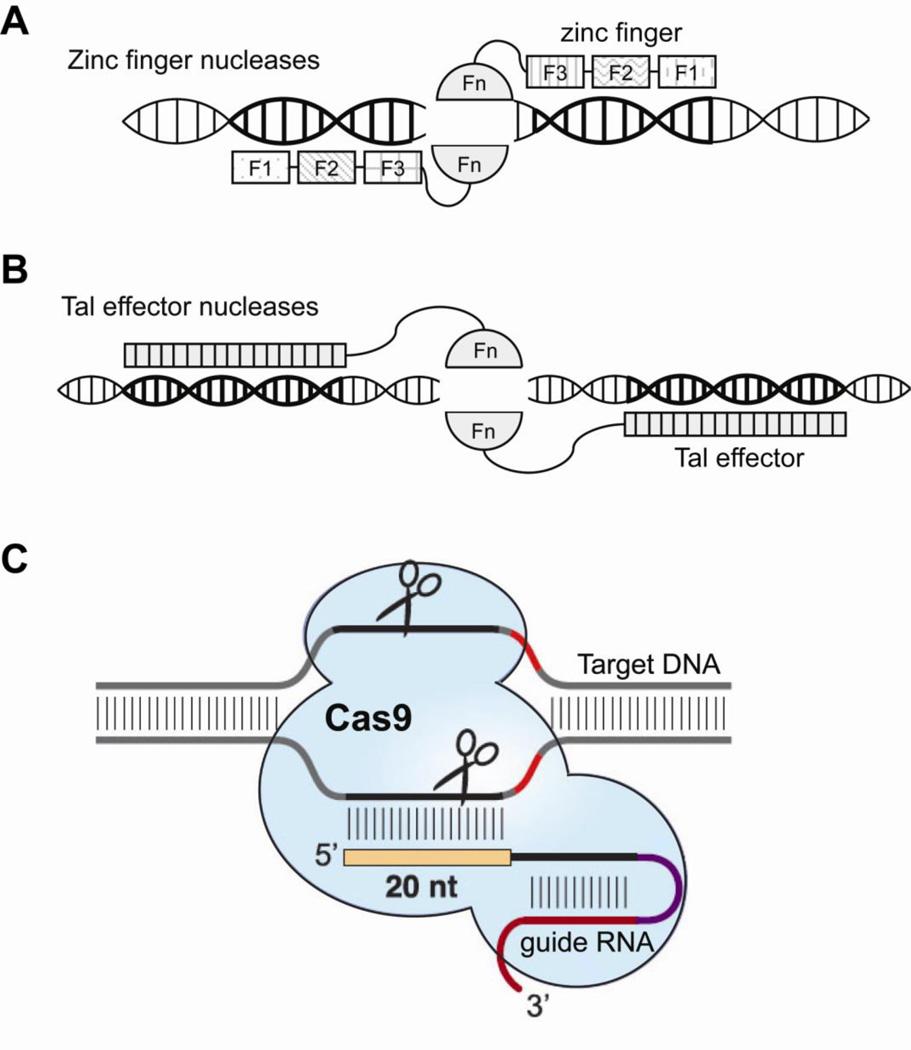
To date, the most promising molecular machines for nanomedicine applications are engineered nucleases, including Zinc Finger Nucleases (ZFNs), Tal Effector Nucleases (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR) systems as powerful tools for genome editing through specifically cleaving genomic sequences 125–127. As shown in Figure 6, the primary forms of targetable nucleases are zinc finger nucleases (ZFNs) 128 (Figure 6a) and transcription activator-like effector nucleases (TALENs) 128, 129 (Figure 6b), each containing a DNA binding domain (zinc finger or TAL effector, respectively) fused to the FokI non-specific DNA cleavage domain130. These nucleases can be designed to create a DNA double-strand break (DSB) in a specific (predetermined) target sequence, which can be processed by the cellular DNA repair machinery. The primary means of re-ligation of the DSB is error-prone non-homologous end joining (NHEJ), which often results in frame-shift mutations that can disrupt a coding sequence of the gene. If a donor DNA template is supplied along with the nucleases, the DSB can be repaired using the homologous recombination (HR) pathway. This process can lead to precise genome editing of the endogenous gene if the supplied donor sequence is designed for a specific purpose, such as gene correction (using wild-type template), gene disruption (inducing mutations and deletions), or adding new gene function (with insertion). In particular, nuclease-induced DSB near a disease-related mutation site can greatly increase the rate of HR that incorporates the supplied donor template131, thereby enabling gene correction and the isolation of gene-corrected cells. The high efficiency of genome editing has greatly enhanced the capability to edit genomic sequences for basic research, as well as medical applications.
Figure 6. Engineered nucleases for genome editing.
(a) Zinc finger nucleases (ZFNs) formed by coupling zinc finger proteins to FokI nuclease domains can bind to a targeted DNA sequence and cut the DNA. Each zinc finger binds to 3 DNA bases, so a pair of 3-finger ZFNs can recognize an 18 bp DNA sequence. (b) A transcription activator-like effector nucleases (TALEN) is formed by biding the FokI nuclease domain to a transcription activator-like effector (TALE) consisting an array of tandem repeats that mediate DNA recognition. Each repeat sequence contains a RVD (repeat variable di-residue) that determines base preference. A pair of nucleases (ZFNs or TALENs) is required to generate a double strand break or a nick on the DNA. (c)
The specificity of ZFNs and TALENs is greatly increased by the need for binding of a pair of nucleases for cleavage to occur. If a pair of nucleases binds to the two half-sites of a target sequence with the correct orientation and spacing, the FokI domains dimerize, resulting in cleavage of the intervening DNA132, 133 (Figure 6). A large amount of work in genome editing has been performed using ZFNs, which were developed over a decade ago. For example, ZFNs have been used to modify the genomes of multiple model organisms and human cells134. The DNA-binding domains of individual ZFNs typically contain between three and six individual zinc finger repeats, each finger can recognize 3 DNA bases. To engineer zinc fingers to bind desired sequences, both "modular assembly" and selection approaches have been used, and methods have been developed for identifying ZFN target sites135–137 or for rational design of ZFNs138, 139. However, the use of ZFNs has been significantly hindered by the difficulties in designing zinc fingers that can bind to a specific DNA sequence with high affinity and specificity.
In contrast to ZFNs, TALEN design is greatly simplified due to the straightforward relationship between the protein sequences of TALENs and their DNA targets129, 140. Specifically, TALENs contain a series of modular DNA binding elements (repeats); each ~34 amino acid repeat is well conserved and fairly constant except for the two amino acids termed the ‘repeat-variable di-residues’ (RVDs), which determine the nucleotide specificity of the repeat. RVDs are chosen to specify each nucleotide in the target sites: Asn-Ile (NI) target adenines, His-Asp (HD) target cytosine and Asn-Gly (NG) to target thymines. The most commonly used guanine-targeting RVD, Asn-Asn (NN), appears to be less specific compared to RVDs targeting other nucleotides, since it binds both guanine and adenine with similar affinity. Several alternative guanine-targeting RVDs have thus been studied to increase the target specificity141–143. Assembling TALENs was a challenge due to their repetitive nature and the large size of the genes, until a number of methods developed that allow high-throughput TALEN construction144–146.
In addition to being easier to design, TALENs appear to be superior to ZFNs in other aspects. Large-scale experiments confirmed a success rate significantly higher than that for ZFNs, with 88% of the TALENs constructed showing detectible cleavage activity144. TALENs can bind to a wide range of sequences with high affinity, whereas ZFNs only target G-rich sequences well. TALEN target sites are longer than ZFN target sites and therefore may result in lower levels of off-target activity (although this has not been well studied). Cells transfected with TALEN plasmids appear to have less cytotoxicity than those with ZFNs, possibly due to lower off-target cleavage144, 147, 148. One potential issue with TALENs is their large size, which may limit their delivery by viral-based vectors or nucleofection. It is likely that TALENs will enjoy a wide range of applications due to the ease of design, high success rate and activity, and low off-target effect and cytotoxicity compared with ZFNs134.
The successful application of custom designed, engineered nucleases for treating human diseases requires identification of the possible off-target cleavage events (cleavage at sites other than the intended target site), which can lead to point mutations, deletions, insertions or chromosomal rearrangements149, 150. To date only very limited studies on nuclease off-target cleavage have been performed, revealing that both ZFNs and TALENs have off-target cleavage, as identified by detecting NHEJ directed mis-repair at putative sites151, or through methods monitoring DNA breaks or cytotoxicity148, 152.
Several methods have been introduced to improve the specificity of pairs of custom nucleases. The first is the modification of the FokI domains to require that dimerization only occurs with one “left” and one “right” nuclease hetero-dimerizing127, 153, 154. This method can potentially eliminate cleavage at homo-dimeric off-target sites that allow either two left or two right nucleases to bind, dimerize and cut. The rate of NHEJ was also greatly reduced through the use of nuclease pairs where one of the two nucleases (a “nickase”) contains a mutation that renders it catalytically inactive, resulting in single-stranded breaks (nicks) as opposed to double strand breaks. The decrease in NHEJ could lead to an increase in the ratio of HR to NHEJ, which is advantageous in gene correction applications. However, the “nickases” typically have a decreased HR activity compared to wild-type nucleases155.
There are many potential medical applications of engineered nucleases, including the establishment of HIV-1 resistance in CD4+ T cells by genome editing156, and correcting genetic defects (including mutations, deletions or insertions) in treating single-gene disorders. An example is ZFN/TALEN based treatment of sickle cell disease (SCD). SCD is predominantly caused by a single (A-T) mutation in the beta-globin gene and is a painful and life shortening disease that afflicts primarily persons of African ancestry157. To use engineered nuclease to treat SCD, it will be necessary to design and optimize ZFNs or TALENs that bind specifically to the beta-globin gene, and deliver them, as well as donor templates containing the normal (healthy) beta-globin DNA sequence, into the nuclei of hematopoietic stem and progenitor cells (HSPCs). The nucleases introduce a DSB or a nick at the targeted site near the beta-globin locus, which will stimulate the homologous recombination (HR) pathway to incorporate the donor sequence for gene correction158–160. The autologous gene-corrected HSPCs can then be re-engrafted in the SCD patient to produce healthy red blood cells and replace sickle cells. HSPCs are the normal precursors of all blood cells, including the oxygen-carrying erythrocytes rendered dysfunctional in sickle cell patients. These cells are relatively rare in the body, but possess potent regenerative potential in that transplantation of a small amount of HSPCs is sufficient to rebuild the entire blood system of an organism. Thus, by isolating HSPCs that carry the sickle mutation, correcting this mutation ex vivo, and then transplanting the gene-corrected HSPCs back into affected recipients, it will be possible to provide enduring replacement of the blood-producing cells of SCD patients with unaffected precursors, thereby supplying healthy red blood cells and effectively curing the disease.
Although the gene correction approach for treating SCD rests on established scientific principles without any conceptual barrier to its implementation, there are many practical and technological challenges in translating the nuclease-based gene correction approach to clinical practice. These include shifting repair pathway choice from NHEJ toward HR160, increasing the spontaneous rate of gene correction by many orders of magnitude, identifying and minimizing unwanted off-targeting effects and gene rearrangements, and establishing a high-throughput, high efficiency delivery capability. These challenges can be overcome by optimizing nuclease and donor template designs161, exploring alternative delivery methods, and applying novel imaging probes and methods to observe and systematically optimize each step in the gene correction process. It is likely that the methods and technologies developed in nuclease-based SCD treatment can be applied to treating other diseases. It has been estimated that there are ~10,000 human single-gene disorders, which impose a significant burden on human health worldwide. Indeed, several of these diseases have already been corrected using nuclease technology in vitro; single point mutations causing epidermolysis bullosa162, α1-antitrypsin deficiency, and inactive versions of the p53 tumor suppressor protein163, as well as the three basepair deletion causing most forms of cystic fibrosis (Δ508)164 have all been repaired using ZFNs or TALENs. Therefore, the development of nanomedicine approaches for treating single-gene disorders based on engineered nucleases may have a significant impact to human health.
The recent discovery of a bacterial defense system that uses RNA-guided DNA cleaving enzymes and clustered, regularly interspaced, short palindromic repeats (CRISPR) 165–169 (Figure 6c) are an exciting alternative to ZFNs and TALENs. CRISPR systems provide a form of acquired immunity in bacteria against previously seen foreign DNA via RNA-guided DNA cleavage170. Type II CRISPR/Cas systems are comprised of short segments of foreign DNA (‘spacers’) integrated within the CRISPR genomic loci. When these sequences are transcribed and processed into short RNAs, they guide the cleavage of specific DNA sequences.
Unlike ZFNs and TALENs, the CRISPR-associated (Cas) protein remains the same for different gene targets; only the short sequence of the guide RNA needs be changed to redirect the site-specific cleavage of novel sequences171. With the matching guide RNA, a Type II CRISPR/Cas system is able to target a 22 nt genomic sequence immediately before a required protospacer adjacent motif (PAM). In stark contrast to ZFNs and TALENs, CRISPR guide strands are very simple to clone, and allow test within days. When cleavage is directed by the short RNA sequences of the guide strands, the target sites are limited to ~23 bp sequences including the PAM. Although CRISPR/Cas induced DNA cleavage was initially thought to be fairly specific, off-target cleavage was found to occur in endogenous gene sequences with up to five mismatches to the guide strands, though mismatches close to or inside the PAM were less tolerated149, 172, 173.
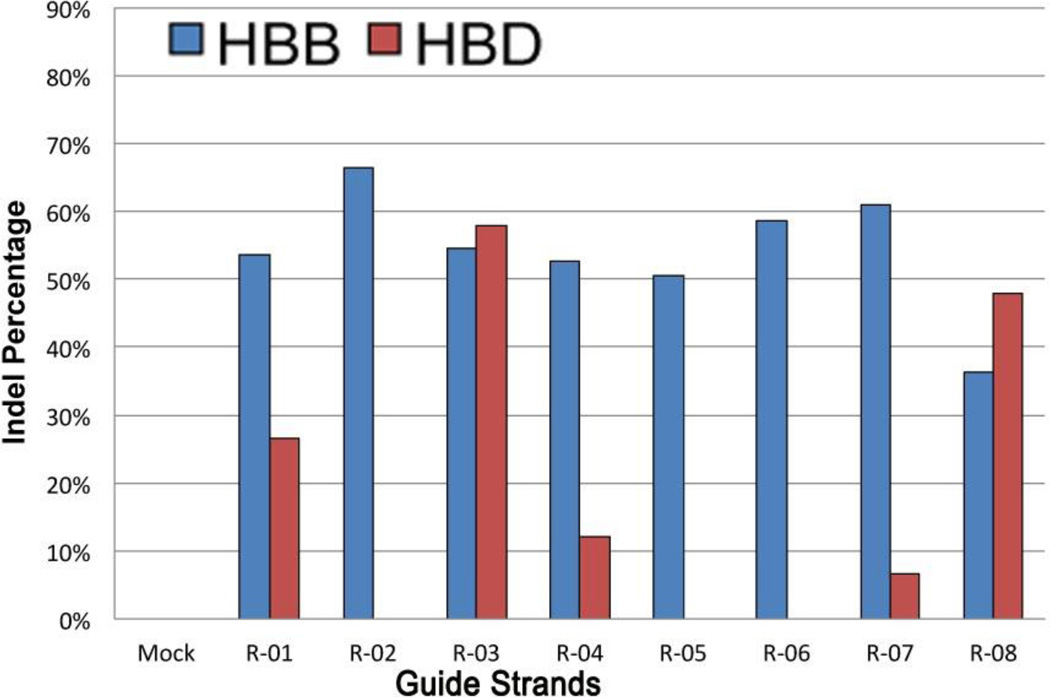
Recent experiments targeting HBB with CRISPR exemplify many of these features149. Specifically, eight pairs of oligonucleotides were cloned into the Cas9 expression vector, allowing efficient cleavage of HBB, as quantified by NHEJ mutagenesis rate (Figure 7). The average HBB mutation rate was 54%, but there was significant off-target cleavage, especially at similar sites in hemoglobin delta (HBD). Off-target cleavage results in mutagenesis and can lead to chromosomal rearrangements, as demonstrated by the R-03 guide strand with high on- and off-target cleavage rates. In cells transfected with this CRISPR system, the region between HBB and HBD was deleted in 13% of the chromosomes. In some cases, transfecting lower levels of the CRISPR plasmids resulted in decreased levels of off-target cleavage149, 172. The full extent of CRISPR off-target cleavage, however, has yet to be determined. Recent results suggest that it is difficult to identify sequences with no off-target cleavage in a mammalian genome due to the tolerance of up to 5 mismatches in a ~23-bp sequence 174. New methods to determine the possible off-target cleavage sites need be developed in hopes of monitoring and improving specificity. A new study using paired CRISPR/Cas9 nickases for generating DSBs may be a promising way to increase the specificity, though the off-target effects for this nickase system still need to be further investigated174.
Figure 7.
A comparison of CRISPR on- and off-target mutation rates at HBB and HBD, respectively (reproduced from 149). Cells were transfected with CRISPR plasmids with eight different HBB-directed guide strands: R-01 to R-08. After three days in culture, genomic DNA was amplified and the indel percentage determined using the T7E1 mutation detection assay.
Opportunities and Challenges in Nanomedicine
Nanomedicine research and development that utilize nano-scale (1–100 nm) features of materials and structures at atomic, molecular and macromolecular levels have the potential to provide fundamental understanding of biological processes in living cells that would be otherwise unthinkable, establish the ability to precisely measure, control and manipulate the functions of biomolecules in vivo, and create medical reagents, devices and systems that have novel properties and functions because of their nano-scale features. For example, the diagnosis and treatment of atherosclerosis represents an area where targeted nanoparticles have great potential for noninvasive diagnosis, targeted therapy, and plaque stabilization. Specifically, it is possible to have nano-scale multifunctional devices that could detect thrombotic events in vivo and deliver therapeutic agents such as anticoagulants as needed. Therapeutic nanoparticles have the potential for curing inflammatory lung diseases, including biodegradable nano- and microparticles releasing anti-oxidant and anti-inflammatory drug molecules, and nanoparticles capable of sensing alveolar function and releasing drugs only when needed, restricting drug delivery to affected areas in heterogeneous disease conditions. Nanomedicine approaches may enable early detection of cancer, and the targeted delivery of anti-cancer drugs into tumor tissue, dramatically increasing their efficacy and decreasing the side effects. Nanoparticle (NP) based imaging probes may enable us to detect cancer stem cells and reveal the interaction between normal and cancer cells during the earliest stages of the cancer development, thus having the potential to eliminate death and suffering from cancer.
To successfully develop nanomedicine as a new field, there are significant challenges in achieving high efficacy and safety, and in technology translation and commercialization. Some of the major challenges in these areas are summarized in Table 1. Clearly, for nanomedicine to generate a large clinical impact, it has to produce methods, devices, drugs, procedures, tools or reagents that are better than existing counterparts, such as simpler, faster, cheaper or safer; more sensitive, specific or robust; or having entirely new functionalities. Achieving these will require concerted efforts by researchers in nano-science, nano-engineering, biology, chemistry, medicine as well as experts in manufacturing, commercialization, regulation, safety, and environmental protection.
Table 1.
Major challenges in nanomedicine
| Efficacy | Safety | Translation | Commercialization |
|---|---|---|---|
| Quality of NP synthesis & functionalization Sensitivity & specificity Signal-to-noise Tunable blood circulation half-life Optimal biodistribution Efficiency & throughput Controllability & tunability Optimal size & surface chemistry |
Cytotoxicity Biocompatibility NP clearance NP degradation Off-target effects FDA approval Environmental effects |
Impact to medicine Imaging probes & contrast agents Targeted delivery vehicles Molecular devices NP based drugs Disease diagnosis, treatment & intervention Tissue repair/regeneration Theranostics Clinical trials |
Scale-up Barriers in regulation Robustness Cost Stability & storage life Quality control & BMP Standardization Packaging Venture capital |
Although nanoparticle-based contrast agents for MRI or combined MR/PET imaging have a great potential, there are also significant challenges in translating the nanotechnology into clinical use. Clearly, the nanoparticle probes must have good biocompatibility and minimal toxicity 175. The clearance of nanoparticles from the body, if they are not biodegradable, is a very important aspect. It is also essential to have a sufficient accumulation of nanoparticles inside (or on the surface) of diseased cells, tissues (such as tumor) or organs in order to generate a high enough signal-to-noise ratio in disease detection. Achieving a good balance among sensitivity, signal-to-noise ratio and safety, the size, functionalization (the type and amount of targeting ligand), surface chemistry (coating, reporters) and the total amount of contrast agent delivered need to be optimized.
To effectively develop nanomedicine, we must take a systems approach in addressing the major challenges. For example, for nanoparticles as an in vivo imaging contrast agent, the size and surface chemistry of nanoparticles need to be tailored to achieve optimal sensitivity, signal-to-noise, biodistribution, and the amount required in a well-balanced fashion. Similarly, for nanoparticle-based in vivo drug/gene delivery, the nano-carriers need to be well designed to achieve adequate cargo loading, controlled release, specific organ/cell internalization and accumulation, and quick clearance/degradation of the nanoparticles. For disease treatment using engineered nucleases, it is necessary to optimize the design of ZFNs, TALENs or CRISPR/Cas9 systems so that both high gene correction efficiency and minimal off-target effect can be realized. Most of the nanomedicine approaches involve cellular and/or in vivo delivery of nanoparticles, proteins, DNA/RNA or drug molecules, which is a common challenge, therefore requires a systematical study. To commercialize nanotechnologies for medical applications, new FDA policies, regulations and guidelines are being developed in the US and the rest of the world,
Despite the significant challenges and regulatory barriers in developing nanomedicine and its commercialization, a huge progress has been made in nanomedicine over the last ten years and many nanotechnology related clinical trials have been, or are being, conducted. Many commercial products based on nanomedicine approaches have emerged, and will continue to emerge, which may significantly impact on all areas of medicine. Nanomedicine will dramatically exceed what has occurred to date in the history of medicine, and will likely revolutionize medicine.
Acknowledgements
This work was supported by the National Heart Lung and Blood Institute of the National Institutes of Health (NIH) as a Program of Excellence in Nanotechnology Award (HHSN268201000043C to GB), by an NIH Nanomedicine Development Center Award (PN2 EY018244 to GB), and by the National Science Foundation as a Science and Technology Center Grant (CBET- 0939511).
References
- 1.Wagner V, Dullaart A, Bock AK, Zweck A. The emerging nanomedicine landscape. Nat Biotechnol. 2006;24:1211–1217. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- 2.Nie SM, Yun X, Gloria JK, Simmons JW. Nanotechnology Applications in Cancer. Annual Review of Biomedical Engineering. 2007;9:257–288. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 3.Kim BY, Rutka JT, Chan WC. Nanomedicine. N Engl J Med. 2010;363:2434–2443. doi: 10.1056/NEJMra0912273. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A. Multifunctional nanoparticles: cost versus benefit of adding targeting and imaging capabilities. Science. 2012;338:903–910. doi: 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pridgen EM, Langer R, Farokhzad OC. Biodegradable, polymeric nanoparticle delivery systems for cancer therapy. Nanomedicine (Lond) 2007;2:669–680. doi: 10.2217/17435889.2.5.669. [DOI] [PubMed] [Google Scholar]
- 6.Doshi N, Mitragotri S. Designer Biomaterials for Nanomedicine. Advanced Functional Materials. 2009;19:3843–3854. [Google Scholar]
- 7.Caldorera-Moore M, Peppas NA. Micro- and nanotechnologies for intelligent and responsive biomaterial-based medical systems. Adv Drug Deliv Rev. 2009;61:1391–1401. doi: 10.1016/j.addr.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chilkoti A, Dreher MR, Meyer DE, Raucher D. Targeted drug delivery by thermally responsive polymers. Adv Drug Deliv Rev. 2002;54:613–630. doi: 10.1016/s0169-409x(02)00041-8. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Szoka FC., Jr Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 10.Hook AL, et al. High throughput methods applied in biomaterial development and discovery. Biomaterials. 2010;31:187–198. doi: 10.1016/j.biomaterials.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder A, et al. Treating metastatic cancer with nanotechnology. Nature reviews. Cancer. 2012;12:39–50. doi: 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- 12.Alivisatos P. The use of nanocrystals in biological detection. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 13.Champion JA, Katare YK, Mitragotri S. Particle shape: a new design parameter for micro- and nanoscale drug delivery carriers. Journal of controlled release : official journal of the Controlled Release Society. 2007;121:3–9. doi: 10.1016/j.jconrel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Euliss LE, DuPont JA, Gratton S, DeSimone J. Imparting size, shape, and composition control of materials for nanomedicine. Chem Soc Rev. 2006;35:1095–1104. doi: 10.1039/b600913c. [DOI] [PubMed] [Google Scholar]
- 15.Jana NR, Chen YF, Peng XG. Size- and shape-controlled magnetic (Cr, Mn, Fe, Co, Ni) oxide nanocrystals via a simple and general approach. Chem Mater. 2004;16:3931–3935. [Google Scholar]
- 16.Xu ZC, Hou YL, Sun SH. Magnetic core/shell Fe3O4/Au and Fe3O4/Au/Ag nanoparticles with tunable plasmonic properties. Journal of the American Chemical Society. 2007;129:8698-+. doi: 10.1021/ja073057v. [DOI] [PubMed] [Google Scholar]
- 17.Alivisatos AP, Gu W, Larabell C. Quantum dots as cellular probes. Annu Rev Biomed Eng. 2005;7:55–76. doi: 10.1146/annurev.bioeng.7.060804.100432. [DOI] [PubMed] [Google Scholar]
- 18.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 19.Gao X, Cui Y, Levenson RM, Chung LW, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 20.Brunetti V, et al. InP/ZnS as a safer alternative to CdSe/ZnS core/shell quantum dots: in vitro and in vivo toxicity assessment. Nanoscale. 2013;5:307–317. doi: 10.1039/c2nr33024e. [DOI] [PubMed] [Google Scholar]
- 21.Hong G, et al. In vivo fluorescence imaging with Ag2S quantum dots in the second near-infrared region. Angew Chem Int Ed Engl. 2012;51:9818–9821. doi: 10.1002/anie.201206059. [DOI] [PubMed] [Google Scholar]
- 22.Sun S, et al. Monodisperse MFe2O4 (M = Fe, Co, Mn) nanoparticles. J Am Chem Soc. 2004;126:273–279. doi: 10.1021/ja0380852. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan KM, et al. Nanomagnetism and spin electronics: materials, microstructure and novel properties. J Mater Sci. 2006;41:793–815. [Google Scholar]
- 24.Lee JH, et al. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat Med. 2007;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 25.Jang JT, et al. Critical Enhancements of MRI Contrast and Hyperthermic Effects by Dopant-Controlled Magnetic Nanoparticles. Angewandte Chemie-International Edition. 2009;48:1234–1238. doi: 10.1002/anie.200805149. [DOI] [PubMed] [Google Scholar]
- 26.Clement O, Siauve N, Cuenod CA, Frija G. Liver imaging with ferumoxides (Feridex): fundamentals, controversies, and practical aspects. Topics in magnetic resonance imaging : TMRI. 1998;9:167–182. [PubMed] [Google Scholar]
- 27.Tong S, Hou SJ, Zheng ZL, Zhou J, Bao G. Coating Optimization of Superparamagnetic Iron Oxide Nanoparticles for High T-2 Relaxivity. Nano Letters. 2010;10:4607–4613. doi: 10.1021/nl102623x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez JM, Josephson L, O'Loughlin T, Hogemann D, Weissleder R. Magnetic relaxation switches capable of sensing molecular interactions. Nat Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 29.Park JH, et al. Magnetic Iron Oxide Nanoworms for Tumor Targeting and Imaging. Adv Mater. 2008;20:1630–1635. doi: 10.1002/adma.200800004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer D, Tamarat P, Maali A, Lounis B, Orrit M. Photothermal imaging of nanometer-sized metal particles among scatterers. Science. 2002;297:1160–1163. doi: 10.1126/science.1073765. [DOI] [PubMed] [Google Scholar]
- 31.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine. J Phys Chem B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 32.Tromberg BJ, et al. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia. 2000;2:26–40. doi: 10.1038/sj.neo.7900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prodan E, Radloff C, Halas NJ, Nordlander P. A hybridization model for the plasmon response of complex nanostructures. Science. 2003;302:419–422. doi: 10.1126/science.1089171. [DOI] [PubMed] [Google Scholar]
- 34.Huang XH, El-Sayed IH, Qian W, El-Sayed MA. Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. Journal of the American Chemical Society. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 35.Skrabalak SE, Au L, Li XD, Xia YN. Facile synthesis of Ag nanocubes and Au nanocages. Nat Protoc. 2007;2:2182–2190. doi: 10.1038/nprot.2007.326. [DOI] [PubMed] [Google Scholar]
- 36.Halas NJ. Playing with Plasmons. Tuning the optical resonant properties of metallic nanoshells. Mrs Bull. 2005;30:362–367. [Google Scholar]
- 37.Hirsch LR, et al. Nanoshell-mediated near-infrared thermal therapy of tumors under magnetic resonance guidance. Proc Natl Acad Sci U S A. 2003;100:13549–13554. doi: 10.1073/pnas.2232479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dam DH, Culver KS, Sisco PN, Odom TW. Shining light on nuclear-targeted therapy using gold nanostar constructs. Ther Deliv. 2012;3:1263–1267. doi: 10.4155/tde.12.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodríguez-Oliveros R, Sánchez-Gil JA. Gold nanostars as thermoplasmonic nanoparticles for optical heating. Optics Express. 2012;20:621–626. doi: 10.1364/OE.20.000621. [DOI] [PubMed] [Google Scholar]
- 40.Zharov VP, Galitovskaya EN, Johnson C, Kelly T. Synergistic enhancement of selective nanophotothermolysis with gold nanoclusters: potential for cancer therapy. Lasers in surgery and medicine. 2005;37:219–226. doi: 10.1002/lsm.20223. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, et al. All-in-one target-cell-specific magnetic nanoparticles for simultaneous molecular imaging and siRNA delivery. Angew Chem Int Ed Engl. 2009;48:4174–4179. doi: 10.1002/anie.200805998. [DOI] [PubMed] [Google Scholar]
- 42.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, et al. Photoacoustic Tomography of a Nanoshell Contrast Agent in the in Vivo Rat Brain. Nano Letters. 2004;4:1689–1692. [Google Scholar]
- 44.Yang X, Skrabalak SE, Li ZY, Xia Y, Wang LV. Photoacoustic tomography of a rat cerebral cortex in vivo with au nanocages as an optical contrast agent. Nano Lett. 2007;7:3798–3802. doi: 10.1021/nl072349r. [DOI] [PubMed] [Google Scholar]
- 45.Mallidi S, Larson T, Aaron J, Sokolov K, Emelianov S. Molecular specific optoacoustic imaging with plasmonic nanoparticles. Opt Express. 2007;15:6583–6588. doi: 10.1364/oe.15.006583. [DOI] [PubMed] [Google Scholar]
- 46.Yang X, Stein EW, Ashkenazi S, Wang LV. Nanoparticles for photoacoustic imaging. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1:360–368. doi: 10.1002/wnan.42. [DOI] [PubMed] [Google Scholar]
- 47.Jain PK, Huang XH, El-Sayed IH, El-Sayed MA. Noble Metals on the Nanoscale: Optical and Photothermal Properties and Some Applications in Imaging, Sensing, Biology, and Medicine. Accounts of Chemical Research. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 48.Qian X, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 49.Gobin AM, et al. Near-infrared resonant nanoshells for combined optical imaging and photothermal cancer therapy. Nano Lett. 2007;7:1929–1934. doi: 10.1021/nl070610y. [DOI] [PubMed] [Google Scholar]
- 50.Popovtzer R, et al. Targeted gold nanoparticles enable molecular CT imaging of cancer. Nano Lett. 2008;8:4593–4596. doi: 10.1021/nl8029114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Maltzahn G, et al. Computationally Guided Photothermal Tumor Therapy Using Long-Circulating Gold Nanorod Antennas. Cancer Research. 2009;69:3892–3900. doi: 10.1158/0008-5472.CAN-08-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 53.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 54.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat Med. 2004;10:993–998. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- 55.Garon EB, et al. Quantum dot labeling and tracking of human leukemic, bone marrow and cord blood cells. Leuk Res. 2007;31:643–651. doi: 10.1016/j.leukres.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hama Y, Koyama Y, Urano Y, Choyke PL, Kobayashi H. Simultaneous two-color spectral fluorescence lymphangiography with near infrared quantum dots to map two lymphatic flows from the breast and the upper extremity. Breast Cancer Res Treat. 2007;103:23–28. doi: 10.1007/s10549-006-9347-0. [DOI] [PubMed] [Google Scholar]
- 57.Chattopadhyay PK, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 58.Howarth M, Takao K, Hayashi Y, Ting AY. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc Natl Acad Sci U S A. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonasio R, et al. Specific and covalent labeling of a membrane protein with organic fluorochromes and quantum dots. Proc Natl Acad Sci U S A. 2007;104:14753–14758. doi: 10.1073/pnas.0705201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang E, et al. Protease-activated quantum dot probes. Biochem Biophys Res Commun. 2005;334:1317–1321. doi: 10.1016/j.bbrc.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 61.Xu C, Xing B, Rao J. A self-assembled quantum dot probe for detecting beta-lactamase activity. Biochem Biophys Res Commun. 2006;344:931–935. doi: 10.1016/j.bbrc.2006.03.225. [DOI] [PubMed] [Google Scholar]
- 62.Clapp AR, et al. Quantum Dot-Based Multiplexed Fluorescence Resonance Energy Transfer. J Am Chem Soc. 2005;127:18212–18221. doi: 10.1021/ja054630i. [DOI] [PubMed] [Google Scholar]
- 63.Yao H, Zhang Y, Xiao F, Xia Z, Rao J. Quantum dot/bioluminescence resonance energy transfer based highly sensitive detection of proteases. Angew Chem Int Ed. 2007;46:4346–4349. doi: 10.1002/anie.200700280. [DOI] [PubMed] [Google Scholar]
- 64.Medintz IL, et al. Proteolytic activity monitored by fluorescence resonance energy transfer through quantum-dot-peptide conjugates. Nat Mater. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 65.Dubertret B, et al. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 66.Wu X, et al. Immunofluorescent labeling of cancer marker Her2 and other cellular targets with semiconductor quantum dots. Nat Biotechnol. 2003;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 67.Goldman ER, et al. Conjugation of luminescent quantum dots with antibodies using an engineered adaptor protein to provide new reagents for fluoroimmunoassays. Anal Chem. 2002;74:841–847. doi: 10.1021/ac010662m. [DOI] [PubMed] [Google Scholar]
- 68.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 69.Chan WCW, Prendergast TL, Jain M, Nie S. One-Step Conjugation of Biomolecules to Luminescent Nanocrystals. Proc. SPIE. 2000;3924:2–9. [Google Scholar]
- 70.Wang YA, Li JJ, Chen H, Peng X. Stabilization of inorganic nanocrystals by organic dendrons. J Am Chem Soc. 2002;124:2293–2298. doi: 10.1021/ja016711u. [DOI] [PubMed] [Google Scholar]
- 71.Dennis AM, Bao G. Quantum dot-fluorescent protein pairs as novel fluorescence resonance energy transfer probes. Nano Lett. 2008;8:1439–1445. doi: 10.1021/nl080358+. [DOI] [PubMed] [Google Scholar]
- 72.Busa WBaNR. Metabolic regulation via intracellular pH. Am J Physiol. 1984;246:R409–R438. doi: 10.1152/ajpregu.1984.246.4.R409. [DOI] [PubMed] [Google Scholar]
- 73.Dennis AM, Rhee WJ, Sotto D, Dublin SN, Bao G. Quantum dot-fluorescent protein FRET probes for sensing intracellular pH. ACS nano. 2012;6:2917–2924. doi: 10.1021/nn2038077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie J, Huang J, Li X, Sun S, Chen X. Iron oxide nanoparticle platform for biomedical applications. Curr Med Chem. 2009;16:1278–1294. doi: 10.2174/092986709787846604. [DOI] [PubMed] [Google Scholar]
- 75.Wang YX, Hussain SM, Krestin GP. Superparamagnetic iron oxide contrast agents: physicochemical characteristics and applications in MR imaging. Eur Radiol. 2001;11:2319–2331. doi: 10.1007/s003300100908. [DOI] [PubMed] [Google Scholar]
- 76.Bjornerud A, et al. Use of intravascular contrast agents in MRI. Acad Radiol. 1998;1(Suppl 1):S223–S225. doi: 10.1016/s1076-6332(98)80111-6. [DOI] [PubMed] [Google Scholar]
- 77.Gillis P, Koenig SH. Transverse relaxation of solvent protons induced by magnetized spheres: application to ferritin, erythrocytes, and magnetite. Magn Reson Med. 1987;5:323–345. doi: 10.1002/mrm.1910050404. [DOI] [PubMed] [Google Scholar]
- 78.Gillis P, Moiny F, Brooks RA. On T(2)-shortening by strongly magnetized spheres: a partial refocusing model. Magn Reson Med. 2002;47:257–263. doi: 10.1002/mrm.10059. [DOI] [PubMed] [Google Scholar]
- 79.Brooks RA, Moiny F, Gillis P. On T2-shortening by weakly magnetized particles: the chemical exchange model. Magn Reson Med. 2001;45:1014–1020. doi: 10.1002/mrm.1135. [DOI] [PubMed] [Google Scholar]
- 80.Nahrendorf M, et al. Noninvasive vascular cell adhesion molecule-1 imaging identifies inflammatory activation of cells in atherosclerosis. Circulation. 2006;114:1504–1511. doi: 10.1161/CIRCULATIONAHA.106.646380. [DOI] [PubMed] [Google Scholar]
- 81.Montet X, Montet-Abou K, Reynolds F, Weissleder R, Josephson L. Nanoparticle imaging of integrins on tumor cells. Neoplasia. 2006;8:214–222. doi: 10.1593/neo.05769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winter PM, et al. Molecular imaging of angiogenesis in early-stage atherosclerosis with alpha(v)beta3-integrin-targeted nanoparticles. Circulation. 2003;108:2270–2274. doi: 10.1161/01.CIR.0000093185.16083.95. [DOI] [PubMed] [Google Scholar]
- 83.Torchilin VP, Trubetskoy VS. Which Polymers Can Make Nanoparticulate Drug Carriers Long-Circulating. Advanced Drug Delivery Reviews. 1995;16:141–155. [Google Scholar]
- 84.Pellegrino T, et al. Hydrophobic nanocrystals coated with an amphiphilic polymer shell: A general route to water soluble nanocrystals. Nano Letters. 2004;4:703–707. [Google Scholar]
- 85.Yu WW, Chang E, Sayes CM, Drezek R, Colvin VL. Aqueous dispersion of monodisperse magnetic iron oxide nanocrystals through phase transfer. Nanotechnology. 2006;17:4483–4487. [Google Scholar]
- 86.Weissleder R, Bogdanov A, Neuwelt EA, Papisov M. Long-Circulating Iron-Oxides for MR Imaging. Advanced Drug Delivery Reviews. 1995;16:321–334. [Google Scholar]
- 87.Jun YW, et al. Nanoscale size effect of magnetic nanocrystals and their utilization for cancer diagnosis via magnetic resonance imaging. Journal of the American Chemical Society. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 88.Xu CJ, Sun SH. Monodisperse magnetic nanoparticles for biomedical applications. Polymer International. 2007;56:821–826. [Google Scholar]
- 89.Nitin N, LaConte LE, Zurkiya O, Hu X, Bao G. Functionalization and peptide-based delivery of magnetic nanoparticles as an intracellular MRI contrast agent. J Biol Inorg Chem. 2004;9:706–712. doi: 10.1007/s00775-004-0560-1. [DOI] [PubMed] [Google Scholar]
- 90.Shin JM, et al. Hollow Manganese Oxide Nanoparticles as Multifunctional Agents for Magnetic Resonance Imaging and Drug Delivery. Angewandte Chemie-International Edition. 2009;48:321–324. doi: 10.1002/anie.200802323. [DOI] [PubMed] [Google Scholar]
- 91.Choi JS, et al. Self-confirming “AND” logic nanoparticles for fault-free MRI. J Am Chem Soc. 2010;132:11015–11017. doi: 10.1021/ja104503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koenig SH, Kellar KE. Theory of 1/T1 and 1/T2 NMRD profiles of solutions of magnetic nanoparticles. Magn Reson Med. 1995;34:227–233. doi: 10.1002/mrm.1910340214. [DOI] [PubMed] [Google Scholar]
- 93.Jun YW, Seo JW, Cheon A. Nanoscaling laws of magnetic nanoparticles and their applicabilities in biomedical sciences. Accounts of Chemical Research. 2008;41:179–189. doi: 10.1021/ar700121f. [DOI] [PubMed] [Google Scholar]
- 94.Tromsdorf UI, et al. Size and surface effects on the MRI relaxivity of manganese ferrite nanoparticle contrast agents. Nano Letters. 2007;7:2422–2427. doi: 10.1021/nl071099b. [DOI] [PubMed] [Google Scholar]
- 95.Yu WW, Falkner JC, Yavuz CT, Colvin VL. Synthesis of monodisperse iron oxide nanocrystals by thermal decomposition of iron carboxylate salts. Chemical Communications. 2004:2306–2307. doi: 10.1039/b409601k. [DOI] [PubMed] [Google Scholar]
- 96.Rosensweig RE. Heating magnetic fluid with alternating magnetic field. J Magn Magn Mater. 2002;252:370–374. [Google Scholar]
- 97.Fortin JP, et al. Size-sorted anionic iron oxide nanomagnets as colloidal mediators for magnetic hyperthermia. Journal of the American Chemical Society. 2007;129:2628–2635. doi: 10.1021/ja067457e. [DOI] [PubMed] [Google Scholar]
- 98.Lee JH, et al. Exchange-coupled magnetic nanoparticles for efficient heat induction. Nat Nanotechnol. 2011;6:418–422. doi: 10.1038/nnano.2011.95. [DOI] [PubMed] [Google Scholar]
- 99.Dames P, et al. Targeted delivery of magnetic aerosol droplets to the lung. Nat Nanotechnol. 2007;2:495–499. doi: 10.1038/nnano.2007.217. [DOI] [PubMed] [Google Scholar]
- 100.Namiki Y, et al. A novel magnetic crystal-lipid nanostructure for magnetically guided in vivo gene delivery. Nat Nanotechnol. 2009;4:598–606. doi: 10.1038/nnano.2009.202. [DOI] [PubMed] [Google Scholar]
- 101.Lewin M, et al. Tat peptide-derivatized magnetic nanoparticles allowin vivo tracking and recovery of progenitor cells. Nat Biotechnol. 2000;18:410–414. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]
- 102.Thu MS, et al. Self-assembling nanocomplexes by combining ferumoxytol, heparin and protamine for cell tracking by magnetic resonance imaging. Nat Med. 2012;18:463–467. doi: 10.1038/nm.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Landázuri N, et al. Magnetic Targeting of Human Mesenchymal Stem Cells with Internalized Superparamagnetic Iron Oxide Nanoparticles. [2013 Jun 13];Small. 2013 doi: 10.1002/smll.201300570. [DOI] [PubMed] [Google Scholar]
- 104.Tang T, et al. Assessment of inflammatory burden contralateral to the symptomatic carotid stenosis using high-resolution ultrasmall, superparamagnetic iron oxide-enhanced MRI. Stroke; a journal of cerebral circulation. 2006;37:2266–2270. doi: 10.1161/01.STR.0000236063.47539.99. [DOI] [PubMed] [Google Scholar]
- 105.Stanley SA, et al. Radio-Wave Heating of Iron Oxide Nanoparticles Can Regulate Plasma Glucose in Mice. Science. 2012;336:604–608. doi: 10.1126/science.1216753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cho MH, et al. A magnetic switch for the control of cell death signalling in in vitro and in vivo systems. Nat Mater. 2012;11:1038–1043. doi: 10.1038/nmat3430. [DOI] [PubMed] [Google Scholar]
- 107.Engvall E, Perlmann P. Enzyme-Linked Immunosorbent Assay (Elisa) Quantitative Assay of Immunoglobulin-G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 108.Vanweeme Bk, Schuurs AHW. Immunoassay Using Antigen-Enzyme Conjugates. Febs Lett. 1971;15:232–236. doi: 10.1016/0014-5793(71)80319-8. [DOI] [PubMed] [Google Scholar]
- 109.Tong S, Ren B, Zheng Z, Bao G. Tiny grains give huge gains: nanocrystal - based signal amplification for immunosorbent assays. ACS nano. 2013;7:5142–5150. doi: 10.1021/nn400733t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nishi H. Photometric determination of traces of metals. Edn. 4th. New York: Wiley; 1978. [Google Scholar]
- 111.Gannon CJ, Patra CR, Bhattacharya R, Mukherjee P, Curley SA. Intracellular gold nanoparticles enhance non-invasive radiofrequency thermal destruction of human gastrointestinal cancer cells. Journal of nanobiotechnology. 2008;6:2. doi: 10.1186/1477-3155-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yavuz MS, et al. Gold nanocages covered by smart polymers for controlled release with near-infrared light. Nat Mater. 2009;8:935–939. doi: 10.1038/nmat2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weissleder R, et al. Superparamagnetic iron oxide: pharmacokinetics and toxicity. AJR Am J Roentgenol. 1989;152:167–173. doi: 10.2214/ajr.152.1.167. [DOI] [PubMed] [Google Scholar]
- 114.Alexiou C, et al. Locoregional cancer treatment with magnetic drug targeting. Cancer Res. 2000;60:6641–6648. [PubMed] [Google Scholar]
- 115.Park JH, von Maltzahn G, Ruoslahti E, Bhatia SN, Sailor MJ. Micellar hybrid nanoparticles for simultaneous magnetofluorescent imaging and drug delivery. Angewandte Chemie-International Edition. 2008;47:7284–7288. doi: 10.1002/anie.200801810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jain TK, et al. Magnetic nanoparticles with dual functional properties: drug delivery and magnetic resonance imaging. Biomaterials. 2008;29:4012–4021. doi: 10.1016/j.biomaterials.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guthi JS, et al. MRI-visible micellar nanomedicine for targeted drug delivery to lung cancer cells. Molecular pharmaceutics. 2010;7:32–40. doi: 10.1021/mp9001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liong M, et al. Multifunctional inorganic nanoparticles for imaging, targeting, and drug delivery. ACS nano. 2008;2:889–896. doi: 10.1021/nn800072t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tong S, Hou S, Ren B, Zheng Z, Bao G. Self-assembly of phospholipid-PEG coating on nanoparticles through dual solvent exchange. Nano Lett. 2011;11:3720–3726. doi: 10.1021/nl201978c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dilnawaz F, Singh A, Mohanty C, Sahoo SK. Dual drug loaded superparamagnetic iron oxide nanoparticles for targeted cancer therapy. Biomaterials. 2010;31:3694–3706. doi: 10.1016/j.biomaterials.2010.01.057. [DOI] [PubMed] [Google Scholar]
- 121.Santra S, Kaittanis C, Grimm J, Perez JM. Drug/Dye-Loaded, Multifunctional Iron Oxide Nanoparticles for Combined Targeted Cancer Therapy and Dual Optical/Magnetic Resonance Imaging. Small. 2009;5:1862–1868. doi: 10.1002/smll.200900389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim J, et al. Multifunctional uniform nanoparticles composed of a magnetite nanocrystal core and a mesoporous silica shell for magnetic resonance and fluorescence imaging and for drug delivery. Angew Chem Int Ed Engl. 2008;47:8438–8441. doi: 10.1002/anie.200802469. [DOI] [PubMed] [Google Scholar]
- 123.Nobuto H, et al. Evaluation of systemic chemotherapy with magnetic liposomal doxorubicin and a dipole external electromagnet. Int J Cancer. 2004;109:627–635. doi: 10.1002/ijc.20035. [DOI] [PubMed] [Google Scholar]
- 124.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nature Medicine. 2007;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 125.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 126.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Szczepek M, et al. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 128.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 129.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 130.Kim YG, Chandrasegaran S. Chimeric restriction endonuclease. Proc Natl Acad Sci U S A. 1994;91:883–887. doi: 10.1073/pnas.91.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci U S A. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smith J, et al. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Segal DJ, Meckler JF. Genome Engineering at the Dawn of the Golden Age. Annu Rev Genomics Hum Genet. 2013 doi: 10.1146/annurev-genom-091212-153435. [DOI] [PubMed] [Google Scholar]
- 135.Jamieson AC, Kim SH, Wells JA. In vitro selection of zinc fingers with altered DNA-binding specificity. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 136.Rebar EJ, Pabo CO. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 137.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc Natl Acad Sci U S A. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dreier B, Segal DJ, Barbas CF., 3rd Insights into the molecular recognition of the 5'-GNN-3' family of DNA sequences by zinc finger domains. J Mol Biol. 2000;303:489–502. doi: 10.1006/jmbi.2000.4133. [DOI] [PubMed] [Google Scholar]
- 139.Kim JS, Pabo CO. Getting a handhold on DNA: design of poly-zinc finger proteins with femtomolar dissociation constants. Proc Natl Acad Sci U S A. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 141.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 142.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Christian ML, et al. Targeting G with TAL Effectors: A Comparison of Activities of TALENs Constructed with NN and NK Repeat Variable Di-Residues. Plos One. 2012;7:e45383. doi: 10.1371/journal.pone.0045383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of designer TAL effectors by Golden Gate cloning. Plos One. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schmid-Burgk JL, Schmidt T, Kaiser V, Höning K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xiao A, et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cradick TJ, Ambrosini G, Iseli C, Bucher P, McCaffrey AP. ZFN-Site searches genomes for zinc finger nuclease target sites and off-target sites. BMC Bioinformatics. 2011;12:152. doi: 10.1186/1471-2105-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cornu TI, et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Molecular therapy : the journal of the American Society of Gene Therapy. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 153.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 154.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2010;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 155.Ramirez CL, et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Research. 2012 doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, Klug PP. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 158.Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 159.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 160.Bertolini LR, et al. Transient depletion of Ku70 and Xrcc4 by RNAi as a means to manipulate the non-homologous end-joining pathway. J Biotechnol. 2007;128:246–257. doi: 10.1016/j.jbiotec.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 161.Cristea S, et al. In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng. 2013;110:871–880. doi: 10.1002/bit.24733. [DOI] [PubMed] [Google Scholar]
- 162.Osborn MJ, et al. TALEN-based Gene Correction for Epidermolysis Bullosa. Mol Ther. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Herrmann F, et al. p53 Gene Repair with Zinc Finger Nucleases Optimised by Yeast 1-Hybrid and Validated by Solexa Sequencing. Plos One. 2011;6 doi: 10.1371/journal.pone.0020913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lee CM, Flynn R, Hollywood JA, Scallan MF, Harrison PT. Correction of the ?F508 Mutation in the Cystic Fibrosis Transmembrane Conductance Regulator Gene by Zinc-Finger Nuclease Homology-Directed Repair. Biores Open Access. 2012;1:99–108. doi: 10.1089/biores.2012.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 166.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 167.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 169.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 171.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013 doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013 doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013 doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhu MT, Wang B, Wang Y, Yuan L, Wang HJ, Wang M, Ouyang H, Chai ZF, Feng WY, Zhao YL. Endothelial dysfunction and inflammation induced by iron oxide nanoparticle exposure: Risk factors for early atherosclerosis. Toxicol Lett. 2011;203:162–171. doi: 10.1016/j.toxlet.2011.03.021. [DOI] [PubMed] [Google Scholar]