Abstract
Background
NIBP/TRAPPC9 is expressed in brain neurons, and human NIBP mutations are associated with neurodevelopmental disorders. The cellular distribution and function of NIBP in the enteric nervous system (ENS) remain unknown.
Methods
Western blot and RT-PCR analysis were used respectively to identify the protein and mRNA expression of NIBP and other neuronal markers. Multilabeled immunofluorescent microscopy and confocal image analysis were used to examine the cellular distribution of NIBP-like immunoreactivity (IR) in whole mount intestine. Enteric neuronal cell line (ENC) was infected with lentivirus carrying NIBP or its shRNA expression vectors and treated with vehicle or TNFα.
Key Results
NIBP is expressed at both mRNA and protein levels in different regions and layers of the mouse intestine. NIBP-like-IR was co-localized with various neuronal markers, but not with glial, smooth muscular, or ICC markers. A small population of NIBP-expressing cells and fibers in extra-ganglionic and intra-ganglionic area were negative for pan-neuronal markers HuD or Peripherin. Relatively high NIBP-like-IR was found in 35-44% of myenteric neurons and 9-10% of submucosal neurons. Approximately 98%, 87% and 43% of these relatively high NIBP-expressing neurons were positive for ChAT, nNOS and Calretinin, respectively. NIBP shRNA knockdown in ENC inhibited TNFα-induced NFkB activation and neuronal differentiation, whereas NIBP overexpression promoted it.
Conclusions & Inferences
NIBP is extensively expressed in the ENS with relatively high level in a subpopulation of enteric neurons. Various NIBP expression levels in different neurons may represent dynamic trafficking or posttranslational modification of NIBP in some functionally-active neurons and ultimately regulate ENS plasticity.
Keywords: Enteric nervous system, NFκB, TRAPPC9/NIBP, Cytokines, Immunohistochemistry, Confocal image, Enteric neuronal cell line
1. Introduction
Nuclear factor κB (NFκB) regulates a wide array of genes that are important in the processes of inflammation, immunity, cancer, and neural plasticity1-3. Canonical and non-canonical pathways for NFκB activation have been identified4. The canonical pathway is triggered by classical stimuli, such as tumor necrosis factor (TNF) α and interleukin (IL)-1β and depends on the signalsome of IκB kinase (IKK). IKK phosphorylates the inhibitory proteins of NFκB (IκBs) to induce their ubiquitination and degradation, resulting in nuclear translocation of NFκB dimers (mainly p65/p50) and activation of target genes. The non-canonical pathway relies on phosphorylation of IKK1 by NFκB-inducing kinase (NIK) to induce p100 processing and nuclear translocation of RelB/p52 dimers. In order to identify novel regulators for NFκB signaling, we previously performed yeast two-hybrid screening from a human brain cDNA library using NIK as bait and identified a novel protein called NIK- and IKK2-Binding Protein (NIBP)5. We demonstrated that NIBP enhances cytokine-induced NFκB activation through potentiating IKK2 kinase activity5, which was confirmed by two recent studies6, 7. In contrast to the canonical IKK1/2/γ complex, a novel subcomplex of NIK-NIBP-IKK2 lacking IKK1 and IKKγ exists in brain tissue and neurons5, and regulates neuronal differentiation in PC12 cells5. NIBP contains a highly conserved domain, TRS120, and may function as a key member of the transport protein particle (TRAPP) complex II, designated TRAPPC98-12. Mutation or deletion of NIBP/TRAPPC9 gene is closely correlated with autosomal recessive mental retardation, maternal autism, neonatal microcephaly, hearing loss, and stroke7, 13-18. These findings suggest that NIBP may play important roles in neurogenesis, mental development, and neural functional integrity.
The enteric nervous system (ENS), as “the second brain in the gut”, plays an important role in regulating gut motility19-21, secretion22-25, absorption23, and mucosal homeostasis25. Intestinal inflammation alters the function and plasticity of the ENS26-29. However, little is known about the role of NFκB signaling in regulating the ENS30. NIBP is extensively expressed in the central nervous system5, 7. Bioinformatics analyses, such as Unigene, ACEVIEW, and Gene Expression Omnibus, showed that NIBP transcripts are present in the tissues of human gastrointestinal tract. Thus, we hypothesized that NIBP is expressed in the ENS. The aim of this study was to characterize the expression pattern and cellular distribution of NIBP in mouse ENS. We also examined the role of NIBP/NFκB signaling in regulating enteric neuronal differentiation using an enteric neuronal cell line (ENC)31, 32. Some of the data were presented in abstract form in the 2010 Joint International Neurogastroenterology and Motility Meeting, Boston, USA.
2. Experimental procedures
2.1 Animal and tissue collections
This study focused on mouse tissues for the continued investigation of transgenic or knockout mice33, 34. Animal protocols were reviewed and approved by the Temple University Institutional Animal Care and Use Committee (IACUC). Adult C57BL/6 mice (2-4 months-old) were obtained from Jackson Lab. CO2 inhalation was used for euthanization. The gut tissues were removed for further analysis.
2.2 Mammalian expression vector cloning, cell cultures and transfection
The N-terminally Flag-epitope-tagged cytomegalovirus(CMV) promoter-based pCMV-Tag vector (Agilent Technology, Santa Clara, CA) constructs encoding mouse NIBP isoform I with 960 amino acids, designated as NIBP(960) was previously described5. Human NIBP(1148)-expressing pRK7-Flag vector was generated by PCR cloning. The N-terminal domain (ND, residue 1-210) mutant vector pRK-Flag-NIBP(ND) was generated from pRK-Flag-NIBP(1148) by double digestion with PpuMI and Sma I followed by blunting and ligation. All vectors were confirmed by sequencing, immunocytochemistry, and Western blot analysis with corresponding Tag antibodies. The human embryonic kidney (HEK) 293T cells were maintained in high-glucose DMEM containing 10% fetal bovine serum and 1% penicillin/streptomycin. Mouse immorto-post natalENC (IM-PEN) was cultured and differentiated as described previously31. For immunofluorescent microscopy, cells were seeded on eight-well plastic chamber slides. For Western blot and immunoprecipitation experiments, cells were seeded on 10 cm dishes. HEK293T cells were transfected 24 h after seeding by the standard calcium phosphate precipitation. ENCs were transfected with pn-Fect™ transfection reagent (Neuromics, Edina, MN).
Lentivirus-mediated NIBP knockdown or overexpression in ENCs
Lentiviral vectors encoding enhanced green fluorescent protein (EGFP) as an internal marker together with shRNA targeting the C-terminal region (CR, 5′-caaagttgaacttgtccac-3′) and 3′-untranslated region (UTR, 5′-gcaacataaatacacagac-3′) of mouse NIBP were generated using a modified PCR-based strategy as previously described5, 35. Lentiviral vector encoding mouse NIBP(960) was generated by cloning the NIBP(960) PCR fragment (coding sequence) into the pRRL-EGFP vector36, 37 through SalI/SpeI digestion sites.
Packaging, purification and titer determination of the lentivirus were performed as described previously38, 39. All recombinant lentiviruses were produced by calcium phosphate-mediated transient transfection of HEK293T cells according to standard protocols. Briefly, HEK293T cells were co-transfected with lentiviral transfer vector (10 μg) and the lentiviral packaging vectors pRSV-REV (2 μg), pMDLg/pRRE (5 μg), and the vesicular stomatitis virus G glycoprotein (VSVG) expression vector pMD2G (3 μg). The viruses were collected from the culture supernatants on days 2 and 3 post-transfection, concentrated by ultracentrifugation for 1.5 h at 25,000 rpm, and then resuspended in phosphate-buffered saline (PBS). Titers were determined by infecting HEK293T cells with serial dilutions of concentrated lentivirus and counting EGFP-expressing cells after 48 h under fluorescent microscopy. For a typical preparation, the titer was approximately 4–10×108 infectious units per ml. For lentiviral vector transduction in ENCs, the cells were cultured in a 6-well plate with culture media containing lentivirus at 50 multiplicity of infection (MOI) for 24 h. The transduction efficiency (∼95%) was determined by examining EGFP-expressing cells. To obtain a pure NIBP-shRNA stable cell line, fluorescence activated cell sorting was performed at a core facility.
2.3 Western blot and immunoprecipitation
The intestine tissues were placed in cold 0.1 M PBS (pH7.4) containing 1 μM nicardipine. The entire small intestine was divided into 3 parts of equal length as duodenum (DE), jejunum (JE) and ileum (IL), and the entire colon was cut into proximal (PC) and distal colon (DC) sections. The tissue segments were then opened along the mesentery border and pinned flat on an agar plate with mucosa face up. Under a stereomicroscope, the mucosa layers were scraped off, and sequential dissection of muscularis mucosae (for colon), submucosal plexuse (SMP), and circular muscle/myenteric plexuse/longitudinal muscle (MP) layers was performed. After quick centrifugation to remove PBS, the dissected tissues were snap-frozen instantly in liquid nitrogen, and stored at -80°C until further processing.
Snap-frozen tissues, or transfected cells were solubilized in Triton X-100-based lysis buffer containing 20 mM Tris-HCl (pH 7.4), 1% Triton X-100, 5 mM ethylenediaminetetraacetic acid, 5 mM dithiothreitol, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1× nuclear extraction proteinase inhibitor cocktail (Cayman Chemical, Ann Arbor, MI), 1 mM sodium orthovanadate and 30 mM NaF. Lysates were incubated through end-over-end mixing at 4°C for 30 min. Nuclear and cellular debris was cleared by centrifugation at 20,000 g for 20 min at 4°C. The protein concentrations of the supernatant were determined with Pierce BCA Protein Assay Kit (cat# 23225). Equal amount of protein lysates (20 μg) were denatured via boiling for 5 min in sodium dodecyl sulphate (SDS) sample buffer, fractionated by SDS-polyacrylamide gel electrophoresis in tris-glycine buffer, and transferred to nitrocellulose membrane (BioRad). The SeeBlue prestained standard (Invitrogen) was used as molecular weight reference. Blots were blocked in 5% nonfat dry milk/tris-buffered saline (pH 7.6) plus 0.1% Tween-20 (TBS-T) for 1 h and then incubated overnight at 4°C with affinity-purified rabbit anti-NIBP polyclonal antibody (1:3,000), designated anti-NIBP(417), against the peptide 417-VYNPMPFELRVENMGLLTSGVEF-439 of mouse NIBP(960) as described previously5. Other primary antibodies for the Western blot were obtained from commercial vendors as follows: p-p65 (1:2,000), p-IκBα (1:1,000) from Cell Signal Technology; IKK1/2 (1:2,000), GAPDH (1:2,000) from Santa Cruz Biotechnology; and NSE (1:500), Flag (1:2,000), β-actin (1:10,000) from Sigma. After incubation for 1 h with horseradish peroxidase-conjugated corresponding secondary antibody (1/2000; 10 μg/ml, Pierce) in TBS-T plus 1% milk, immunoreactive proteins were visualized using SuperSignal Femto maximum sensitivity substrate kit (Thermo Fisher Scientific, Rockford, IL). All washing steps were performed with TBS-T. In some cases, Odyssey CL× Infrared Fluorescent Western Blot system (LI-COR, Lincoln, NE) was used following the manufacture's instruction. Briefly, the nitrocellulose membrane was blocked with Odyssey blocking buffer containing 0.1% (v/v) Tween 20. Membranes were incubated overnight at 4°C with rabbit anti-NIBP(417) and mouse anti-GAPDH primary antibodies and then washed before incubation with fluorescently conjugated secondary antibodies (IRDye 680LT-conjugated anti-mouse and IRDye 800CW-conjugated anti-rabbit) for 1 h at room temperature. Membranes were scanned and analyzed using an Odyssey Infrared Imaging System. Relevant signal intensity was determined using LI-COR imaging software and exported to Microsoft Excel. Relative fold changes in NIBP protein expression were calculated after GAPDH normalization.
For immunoprecipitation, the lysates from ENCs transfected with Flag-NIBP(960) or empty control vector were incubated overnight at 4°C with 0.5 μg anti-NIBP antibody or rabbit control IgG. The immune complexes were precipitated with protein A/G plus agarose, washed four times with 0.6 M NaCl lysis buffer, and then eluted by boiling for 5 min in SDS sample buffer. The eluted proteins were fractionated on SDS-polyacrylamide gel electrophoresis and detected by Western blot with anti-NIBP polyclonal antibody (1: 3,000) or anti-Flag monoclonal antibody (1:2,000).
2.4 Conventional and quantitative reverse transcription (RT)-PCR
For total RNA extraction, the ENCs, muscle layers containing plexuses (SMP, MP), mucosa, and brain tissues were processed with the RNeasy Mini kit (Qiagen) following the manufacturer's instructions. The potentially-contaminated genomic DNA was removed by on-column DNase digestion with RNase-Free DNase Set (Qiagen). One μg of RNA for each sample was used to synthesize cDNA using the High Capacity cDNA Reverse Transcription Kit (Invitrogen, Grand Island, NY) with random hexanucleotide primer. Conventional PCR was performed on the cDNA using HotMasterTM Taq DNA polymerase kit (5-Prime Inc, Gaithersburg, MD). The primer sequences for mouse NIBP (NM_001164641) were 5′-actacatgcagtgcgcagaggatcac-3′(forward) and 5′-ctgggtcaatgagaacttcttgcgccc-3′(reverse), generating a fragment of 872 bp covering nucleotides 267-1139. The primer sequences for mouse β-actin were obtained from RealTimePrimers (Elkins Park, PA). The PCR product was purified and cloned into T-A vector for confirmation by sequencing.
Quantitative PCR (qPCR) analysis was carried out on the LightCycler480 (Roche) using SYBR® Green PCR Master Mixe Kit (Applied Biosystems) as described previously40. The RT reaction was diluted to 5 ng/μl total RNA and 2 μl was used in a total 20 μl PCR reaction. The sequences for three pairs of mouse NIBP primers targeting the N-terminal (exon 5-7), middle (exon 13-14) and C-terminal (exon 27-28) regions of mouse NIBP (NM_029640.2) are as follows: 5′- atttatcattatcccggaggaact-3′ (forward) and 5′-ctcagtgctggtgtcaggatttat-3′ (reverse) for generating 170 bp PCR product covering nucleotides 244-411; 5′-tgtgaagttcagctgatggtgtac-3′ (forward) and 5′-gctgcaggaagagactcaaact-3′ (reverse) for 103 bp covering 1276-1379; and 5′-agaacggtgtgcacaactatgac-3′ (forward) and 5′-gatgttgaggaagaagtcacctgt-3′ (reverse) for 150 bp covering 2687-2835, respectively. The primers for the housekeeping genes GAPDH, β-actin, β2-microglobulin (B2M), β-glucuronidase (Gusb), and phosphoglycerate kinase 1(PGK1) were obtained from RealTimePrimers (Elkins Park, PA). Each reaction was performed in triplicate. Cycle threshold (Ct) values were obtained graphically. Absolute quantification of NIBP was performed using mouse NIBP cDNA plasmid as the standard (0.1 pg to 100 ng). The copy number for NIBP in the tested samples was calculated from the standard cure (R=0.99) and expressed as copies per ng of total RNA. The generalized average Ct value (ΔCt) of each gene in all experimental samples was subtracted from individual Ct value to obtain ΔΔCt values and relative fold change in gene expression was calculated as 2-ΔΔCt. The ΔCt value of each housekeep gene was also used to normalize the Ct value of NIBP in each experimental sample.
2.5 Adenovirus-mediated NFκB-luciferase reporter assay
ENCs were cultured at 33°C in a 96-well plate and infected with adenovirus carrying the NFκB-firefly-luciferase vector (Vector Biolabs) at 100 MOI for 24 h. Cells were cultured at 39°C and treated with or without TNFα (10 ng/ml) for 24h, and the cell lysate was used for measurement of firefly luciferase activity with the ONE-Glo luciferase assay system (Promega). Luminescence was measured on a 2104 EnVision® Multilabel Reader (PerkinElmer). Three separate experiments were conducted and, in each experiment, data were calculated as the average of 4-6 samples.
2.6 Immunohistochemistry and confocal imaging analysis
The whole mount preparation of mouse intestine was performed as described previously with minor modification41, 42. Briefly, the intestinal tissue segments were placed in cold 0.1 M phosphate buffered saline (PBS, pH7.4) containing 1 μM nicardipine, opened along mesentery border, mounted on a strip of whatman filter paper and fixed for 1-2 h with 4% paraformaldehyde/PBS. After washing, the samples were cryopreserved with buffered 30% sucrose/PBS at 4°C for 2-3 d and stored at -35°C before use. Under a stereomicroscope, the mucosa, SMP layers and MP layers described above were dissected. In some cases (for confocal scanning), only the superficial mucosa was removed, leaving the whole colon mounts consisting of the deep mucosa/muscularis mucosae, SMP, circular muscle, MP, longitudinal muscle and serosa.
The whole mounts were permeated for 30 min with 0.5% Triton X-100/0.01M PBS and blocked with 10% normal donkey serum for 30 min and then incubated with various primary antibodies (Table 1) in 0.1% Triton X-100/0.01M PBS for 1-2 d at 4°C. Tissues were then washed in PBS for 3 × 10 min and incubated in corresponding Alexa Fluor® conjugated donkey secondary antibody (1:400; Invitrogen, Grand Island, NY) for 1 h at room temperature. For multi-labeled immunofluorescent staining, the samples were subsequently or simultaneously incubated with another primary antibody from a different host species. The results of single staining were evaluated for all antibodies and compared with those from double labeling. The specificity of all the primary antibodies was tested by including controls without primary antibody or with irrelevant secondary antibody. Hoechst 33258 was used for counterstaining of nuclei. After 3×10 min washing, the whole mounts were mounted on 1% gelatinized slides and dried for 30 min. Slides bearing whole mounts or transverse sections were coverslipped with anti-fading aqueous mounting media. The fluorescences were captured using sequential acquisition under the fluorescent inverted microscope (Nikon, Japan) equipped with a cooled CCD camera using Slidebook 5.0 digital imaging software. In some cases, the three-dimensional reconstructions were made using nearest-neighbor deconvolution of fluorescent images taken in a Z-stack of focal planes (0.5 μm).
Table 1. Primary antibodies used for immunohistochemical staining.
| Primary antibody | Immunogens | Sources | Species and clonality | Catalog (lot) Number | Dilution |
|---|---|---|---|---|---|
| NIBP(417) | VYNPMPFELRVENMGLLTSGVEF | Self-made | Rabbit polyclonal | 1:500 | |
| Peripherin | Full-length rat protein | Neuromics | Chicken polyclonal | CH23016 (400644) | 1:500 |
| HuD(N-15) | aa 1-15 of human origin (accession # 123734) | Santa Cruz | Goat polyclonal | Sc-5977 (I0104) | 1:400 |
| ChAT | Human placental enzyme | Millipore | Mouse monoclonal | AB144P | 1:40 |
| Calretinin (N-18) | aa 1-18 of human origin (accession #NP_001731.1). | Santa Cruz | Goat polyclonal | Sc-11644 (E0306) | 1:1,000 |
| nNOS (A-11) | aa 2-300 of human NOS1 | Santa Cruz | Mouse monoclonal | Sc-5302 (K1009) | 1:400 |
| GFAP (C-19) | aa 411-432 of human origin (accession # P14136) | Santa Cruz | Goat polyclonal | Sc-6170 (D0109) | 1:500 |
| S100β (EP1576Y) | aa 78-92 of human S100-β (accession #p04271) | Millipore | Rabbit monoclonal | 04-1054 (jbc1771181) | 1:400 |
| c-Kit (D13A2) | DHAEAALyKNLLHSK | CellSignal | Rabbit monoclonal | 3074 (2) | 1:400 |
| Flag M2 | DYKDDDDK | Sigma | Mouse monoclonal | F7425 | 1:500 |
| α-Actin (1A4) | aa 1-12 of human origin (accession # NP_001135417) | Sigma | Mouse monoclonal | A5228 | 1:2,000 |
For confocal imaging analysis, samples were viewed on a Leica SP5 confocal microscope system. Differential visualization of three or four fluorophores Alexa-488, Alexa-594, Alexa-657 and Hoechst 33258 was obtained via specific filter combinations. Samples were scanned sequentially to avoid any potential for fluorophore bleedthrough. The Z-stack scanning images (1,024 × 1,024 pixels) through 0.5 or 1.0 μm optical sections were obtained under identical exposure conditions and processed through the build-in processing tools. In some cases, the entire thickness of intestine tissues covering serosa, longitudinal muscle, MP, circular muscle, SMP, muscularis mucosae and deep crypts was scanned through 1 μm optical sections to show the dimensional correlations between various layers. Tiff images and 3-D movies were explored for analysis.
2.7 Quantitative analysis and statistics
The MPs and SMPs in the whole mount preparation were identified under a fluorescent inverted microscope at 40× oil objective by adjusting the focus on different layers. The proportions of double-labeled cells in MPs or SMPs were assessed in at least three animals. Around 2 cm of colonic muscle strip per animal was analyzed. For each whole mount sample, three sites separated by at least 0.5 cm were positioned for five sequential micrographs (fields) containing 2-3 MPs or SMPs per micrograph. The number of positive cells per micrograph was counted under the microscope and further validated using Image J software. The nuclear staining was used to validate the cell body and distinguish SMCs (small or spindle nucleus) from non-SMCs. The percentage of coexpressed cells for individual fluorescent color (representing one type of cells) was calculated for each animal.
Differentiation of ENCs was assessed by determining the percentage of neurite-bearing cells and measuring the neurite length using Image J software43. Cells bearing neurites that were double the length of the diameter of the cell body were considered as neurite-bearing differentiated cells. Four to five micrographs were randomly selected for each condition. For each micrograph, 50 cells were measured for the neurite length and 10 cells for the neuronal size. The evaluation was conducted in a blinded fashion.
For statistical analysis, the data represented mean ± standard deviation from 3-5 independent experiments, and were evaluated by Student's t-test or ANOVA and Newman-Keuls multiple comparison test. A p< 0.05 or 0.01 was considered as statistically significant.
3. Results
3.1 Expression of NIBP mRNA and protein in mouse small intestine and colon
In our previous study, Northern blot analysis of multiple human tissues showed very low expression of NIBP mRNA in the small intestine and undetectable expression in the colon5. However, bioinformatics analyses, such as Unigene, ACEVIEW, and Gene Expression Omnibus, showed that NIBP transcripts were present in the tissues of human gastrointestinal tract. To address whether NIBP is expressed in mouse intestine, we performed RT-PCR and Western blot analysis. Conventional RT-PCR analysis showed the existence of NIBP mRNA in the whole tissues of mouse colon and small intestine (Fig. 1A). Further quantitative analysis of the different layers (mucosa, SMP and MP) by real-time RT-qPCR validated the presence of 14-4,600 copies of NIBP mRNA per ng total RNA, with lower expression in SMP and higher expression in mucosa and MP (Fig. 1B). To determine if there was any regional difference in NIBP mRNA expression, an equal length of small intestine (duodenum, ileum, and jejunum) or colon (half proximal and half distal) were harvested for total RNA extraction and equal amount of total RNA (1 μg) was used for RT-PCR. The copy number of NIBP mRNA in MP layers was higher in the colon and ileum while the mucosal NIBP mRNA copies were higher in distal colon and duodenum (Fig. 1B). Using the average expression level of all the tested samples as a generalized reference, the SMP layers showed a 2.5∼6.2-fold decrease and the MP layers showed a 1.1∼5.3-fold increase (Fig. 1C). After normalization with GAPDH, which is a widely used housekeeping gene, the mucosal layers exhibited higher copies of NIBP mRNA in all regions except for duodenum region, but the MP layers showed lower copies in distal colon (Fig. 1D, upper panel). High variability of GAPDH mRNA expression was observed among different layers and different regions (Fig. 1D, lower panel). Several other housekeeping genes (β-actin, B2M, Gusb and PGK1) also showed high but similar variability under these conditions (data not shown). In particular, the NIBP mRNA expression in SMP layers presented higher levels in almost all regions after normalization with these housekeeping genes compared to non-normalized data. Since normalization with different housekeeping genes demonstrated an inconsistent pattern of NIBP mRNA expression among the layers and regions of the intestine, we decided to use the non-normalized data for further interpretation, which was supported by the following studies using Western blot and immunohistochemistry approaches.
Figure 1. NIBP mRNA expression in mucosal (Mucosa), submucosal plexus (SMP) and myenteric plexus (MP) layers of mouse duodenum (DU), jejunum (JE), ileum (IL), proximal colon (PC) and distal colon (DC).
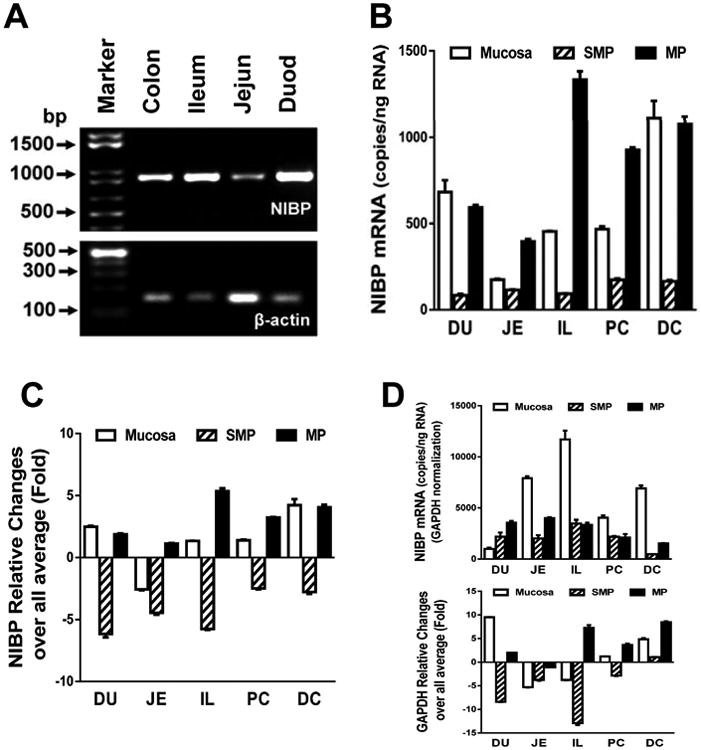
A. Conventional RT-PCR analysis showed the expression of NIBP mRNA in various regions of mouse intestine. B-D. Quantitative RT-PCR analysis identified higher NIBP mRNA levels in MP/Mucosa than SMP as well as DC, PC and IL. An equal amount of reverse transcribed cDNA (10 ng/reaction) was used for real time PCR. The copies of NIBP mRNA were calculated using a cDNA standard curve (B). Relative fold changes in NIBP (C) and the housekeeping gene (D) were calculated using 2-ΔΔCt method.
Western blot analysis was performed using the affinity-purified anti-NIBP(417) antibody. The specificity of this antibody has been previously validated by antigen-preabsorption, immunoprecipitation and NIBP-shRNA knockdown5. Here, the specificity was further validated in mouse ENC overexpressing Flag-tagged mouse NIBP(960) (Fig. 2A) and intestinal tissues (Fig. 2B-D). A higher concentration of EDTA in the lysis buffer increased the stability of NIBP protein at the predicted size (106 kDa) (Fig. 2B). The endogenous NIBP in mouse colonic muscle layers and mucosa was identified as a specific band at the predicted size (Fig. 2C), which matched the band from an overexpressed Flag-tagged NIBP(960) and was abolished by antigen preabsorption (1:100) (Fig. 2C). The MP and SMP layers showed higher level of NIBP protein expression (for the predicted size) than the mucosal layers in the colon (Fig. 2C, D) and small intestine (Fig. 2D). Among the muscle layers, the MP layers exhibited the highest level of NIBP protein expression in all of the examined regions with or without GAPDH normalization (Fig. 2D). The expression of β-actin was also highly variable among the layers and regions (data not shown). The mucosal layers showed additional bands at around 40, 55, 130 and 170 kDa (Fig. 2C, D), which were all abolished by antigen preabsorption (Fig. 2C). The MP layers in all regions presented additional bands at around 140, 280 and 350 kDa while the SMP layers in colon had an additional band at 280 kDa (Fig. 2D). The molecular weight of NIBP-immunoreactive band in the cerebral cortex (as positive control) was slightly smaller than that in the gut tissues (Fig. 2D), which may indicate tissue-specific isoforms or posttranslational modifications. Taken together, these results suggest that NIBP is expressed at both mRNA and protein levels in the mouse intestine.
Figure 2. NIBP protein expression in mucosal (Muc), submucosal plexus (SMP) and myenteric plexus (MP) layers of mouse duodenum (DU), jejunum (JE), ileum (IL), proximal colon (PC) and distal colon (DC).
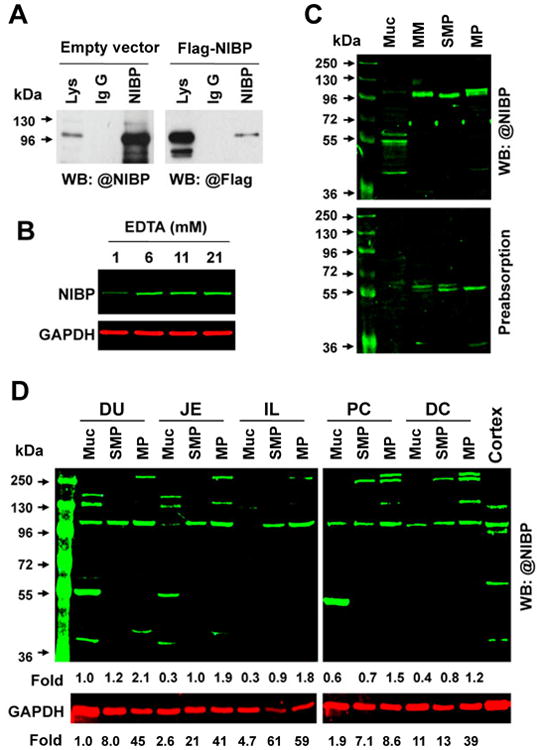
A. Endogenous NIBP and exogenous Flag-tagged NIBP(960) in the lysate (Lys) of a mouse enteric neuronal cell line were specifically immunoprecipitated with anti-NIBP antibody and determined by Western blot with an anti-NIBP or anti-Flag antibody. B. Effect of EDTA on NIBP protein stability. C. Infrared fluorescent Western blot analysis shows a specific band of predicted size for endogenous NIBP in four layers of Muc, muscularis mucosae (MM), SMP and MP from mouse colon (upper), which was abolished by antigen preabsorption (lower). D. Regional changes of NIBP protein in different layers. The number below the blot indicates the relative fold compared with the DU-Muc with or without GAPDH normalization. Cerebral cortex lysate was used as the positive control.
3.2 NIBP-like immunoreactivity is abundant in mouse small intestine and colon
To examine the cellular distribution of NIBP protein expression, we performed immunohistochemistry with an anti-NIBP(417) antibody using whole mount preparation from mouse small intestine and colon. The binding specificity of this anti-NIBP(417) antibody for immunohistochemistry was tested previously5. To further validate the specificity, mammalian expression vectors expressing Flag-tagged mouse NIBP(960) and the C-terminal or N-terminal fragment flanking the region recognized by anti-NIBP(417) antibody NIBP were transfected into HEK293T cells followed by immunocytochemistry. As expected, this anti-NIBP(417) antibody recognized the full-length Flag-NIBP (Fig. 3A) but not C-terminal NIBP mutant (Fig. 3B). As shown in Figure 3C-F, NIBP-like immunoreactivity (IR) was abundant in the MPs, the interganglionic fiber strands and the branches. A similar expression pattern was observed among different regions of the small intestine and colon (Fig. 3C-F). The positive NIBP-like-IR staining was blocked in a dose-dependent manner (1:1-1:100) by the preabsorption of anti-NIBP(417) antibody (2 μg/ml) with a corresponding antigenic peptide (2-200 μg/ml).
Figure 3. Specificity of affinity-purified rabbit anti-NIBP(417) polyclonal antibody for immunoreactive staining (A-D) and NIBP-like-immunoreactivity in mouse duodenum (DU), jejunum (JE), ileum (IL) and distal colon (DC).
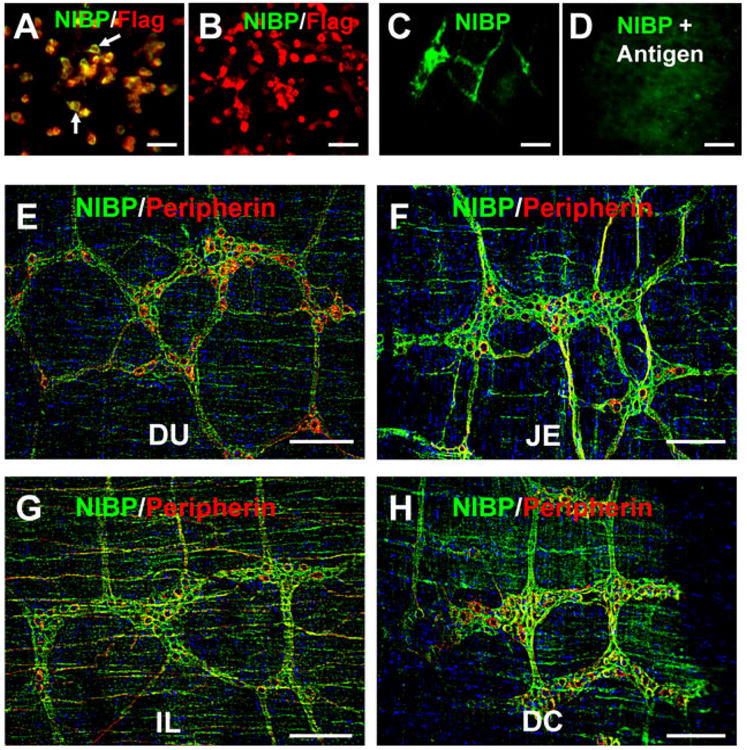
A, B. Immunocytochemistry of HEK293T cells transfected with pRK-Flag-NIBP(960) (A) or pRK-Flag-NIBP (ND, residue 1-210) vectors (B) showing the specific binding of NIBP antibody to Flag-NIBP(960) as indicated by the white arrows. C, D. Peptide preabsorption (1:10) completely blocked the NIBP-like-immunoreactive staining in myenteric plexuses. E-H. Representative deconvolution microscopic images of NIBP-positive and Peripherin-positive cells as well as fibers in myenteric plexuses of the indicated intestine regions. The nuclei (blue) were counterstained with Hoechst 33258. Scale bars = 20 μm.
3.3 NIBP-like-IR is absent in intestinal smooth muscle cells, ICC, and glial cells
To identify the cell types expressing NIBP-like-IR within colon tissue, double-labeled fluorescent immunohistochemical staining was performed using whole mount preparation. The location and morphology of NIBP-like-IR-positive cells within the ENS ruled out the possibility of its expression in smooth muscle cells and interstitial cells of Cajal (ICC). This conclusion was verified by double-labeled immunohistochemistry using the anti-NIBP(417) antibody and an antibody against smooth muscle specific α-actin (Fig. 4A) or ICC marker c-Kit (Fig. 4B). Since the ENS is mainly composed of glial cells and neurons, and enteric glial cells share the properties of GFAP-positive astrocytes and S100β-positive Schwann cells44, we examined whether NIBP is expressed in enteric glial cells using GFAP and S100β as markers. We found that NIBP-like-IR was not present in GFAP or S100β-positive glial cells (Fig. 4C, 4D).
Figure 4. Absence of NIBP-like-immunoreactivity in colonic smooth muscle cells, ICC, and glial cells.
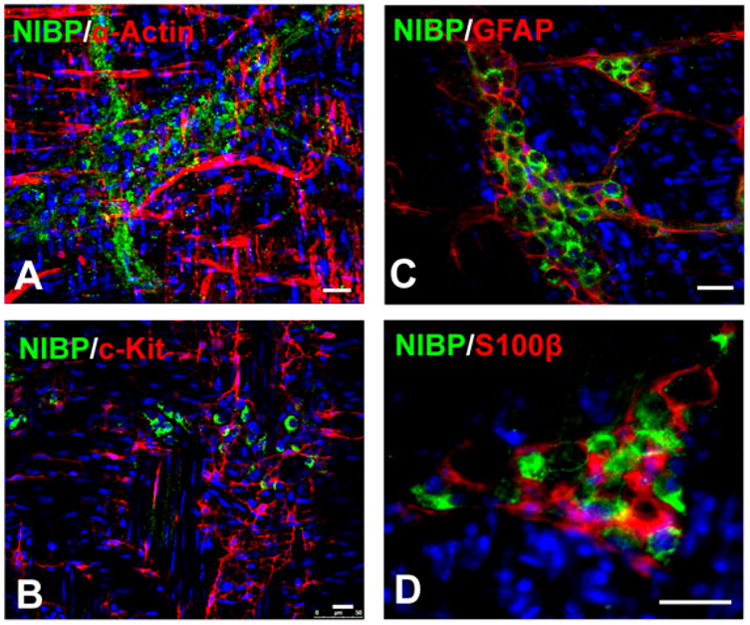
Whole mount myenteric plexus strips of mouse colons were immunostained first with anti-NIBP antibody and then with primary antibodies against α-actin (A), c-Kit (B), GFAP (C), and S100β (D). The nuclei (blue) were counterstained with Hoechst 33258. Scale bars = 20 μm
3.4 NIBP-like-IR is expressed in enteric neurons to various extents
To determine whether NIBP is expressed in enteric neurons, we examined the co-localization of NIBP-like-IR with the pan-neuronal marker HuD. As previously reported45-47, HuD-like-IR was predominantly detected in the nucleus and weakly in the cell body of enteric neurons, but not in the neurites (Fig. 5). Confocal image analysis of series z-stack optic sections from the full-thickness intestine preparation (containing longitudinal muscle layers, MPs, circular muscle layers, SMPs, muscularis mucosae and deep crypts) under the same imaging conditions showed that NIBP-like-IR was strongest in the cells and fibers at the MP layers (Fig. 5B-D), followed by SMPs (Fig. 5H-J) and intramuscular fibers (Fig.5A, 5E-G). The staining of NIBP-like-IR appeared around the HuD-positive nucleus with various intensities among different size of HuD-positive cells. The strongest expression of NIBP was found in HuD neurons with round nucleus, which also presented punctate HuD staining in the cytoplasm and may reflect a unique population of enteric neurons, but weaker expression or no expression was found in HuD neurons with irregular nucleus (Fig. 5M-R). The majority of HuD neurons in MP (Fig. 5M-O) and few cells in SMP (Fig. 5P-R) were stained highly positive for NIBP. Interestingly, a small population of NIBP-expressing cells was negative for HuD mainly in extraganglionic area (Fig. 5C′, red arrow) and occasionally within the ganglionic plexuses(Fig. 5O, red arrow). A pattern of bright and larger particle (puncta) staining for NIBP-like-IR was consistently observed in a subpopulation of HuD-positive neurons (Fig. 5O, R), which may reflect the dynamic trafficking of NIBP in these neurons and their neurites. Quantitative assessment of these highly positive cells under 40× oil objective showed that 34.9 ± 4.0% of the total number of HuD-positive cells (mean ± SD, cohort size 625-881 cells for each mouse, n = 3) were brightly stained for NIBP-like-IR in the MPs and 9.7 ± 2.1% of HuD neurons (cohort size 224-336 cells for each mouse, n = 3) in the SMPs.
Figure 5. Co-localization of NIBP-like-immunoreactivity with pan-neuronal marker HuD in the myenteric plexuses (MP) and submucosal plexuses (SMP).
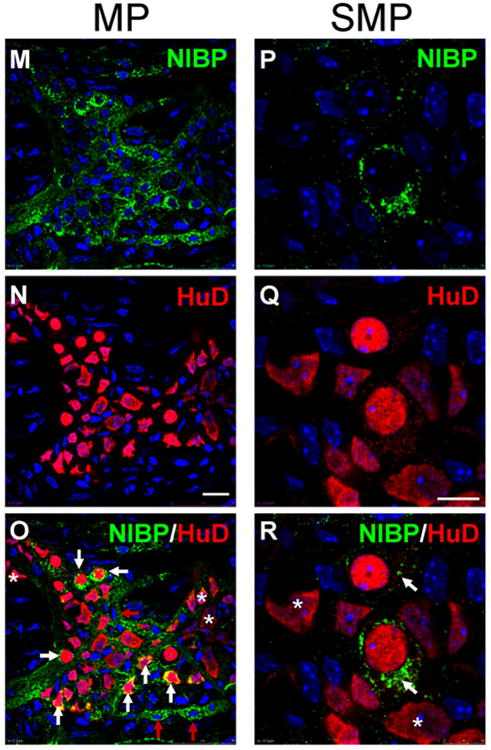
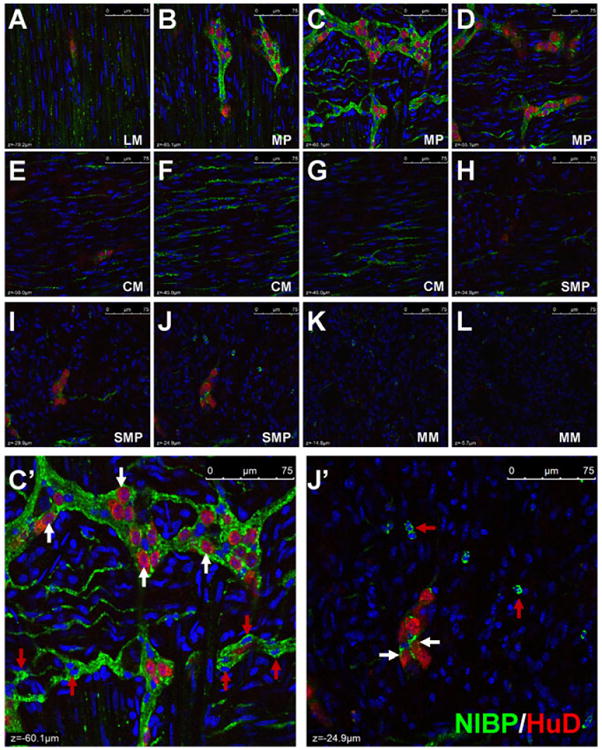
The whole mount preparations of mouse colon were immunostained sequentially with primary antibodies against NIBP(green) and HuD (red). The nuclei (blue) were counterstained with Hoechst 33258. A-L. Representative series z-stack (1 μm optic section) confocal images of full-thickness colon (75 μm) containing longitudinal muscle (LM), MP, circular muscle (CM), SMP and muscularis mucosae (MM) show the highest NIBP expression in MP and its adjacent intramuscular fibers (B-F) and weaker expression in SMP and MM (J-L) under the same image acquiring condition. A-J is 5 μm apart and J-L is 10 μm. C′ and J′ at higher amplification of C and J show NIBP/HuD colocalization (white arrow) mostly in MP but limited colocalization in SMP. M-R. Representative confocal images of MP and SMP at higher amplification show complete colocalization (yellow) of NIBP and HuD in round and small neurons and partial colocalization in irregular and larger neurons. The white arrows show co-localization and the red arrows indicate only NIBP and the stars indicate only HuD. Scale bars = 75 μm in A-L, 20 μm in M-O and 10 μm in P-R.
Another pan-neuronal marker Peripherin31, 48, 49 was used to confirm the expression of NIBP protein in enteric neurons. As described previously48, 50, the staining intensity and structural pattern of Peripherin-like-IR varied among enteric neurons (Fig. 3, 6). Various extents of NIBP-positive staining were found in all Peripherin-expressing neurons in the MPs, even at a very low dilution (1:10,000) of the primary anti-NIBP antibody, which was completely blocked by antigen preabsorption. Counting of the brightly-stained punctate cells in the MPs under 40× oil objective showed that 44.3 ± 3.7% of total Peripherin-expressing neurons (cohort size 464-499 cells for each mouse, n = 3) expressed relatively high level of NIBP-like-IR. In the SMPs, 8.9 ± 4.0% of Peripherin-positive neurons (cohort size 136-182 cells for each mouse, n = 3) were highly positive for NIBP-like-IR. In most plexuses, NIBP and Peripherin staining overlapped entirely within the cytoplasm of the positive neurons (Fig. 3, 6, white arrow). In some plexuses, NIBP-positive staining distributed around the penumbral regions of Peripherin-positive cell body, but was absent in the perinuclear region, where Peripherin-like-IR was intensively stained (Fig. 6D-F). In particular, mutually exclusive staining for NIBP and Peripherin was observed frequently in the MPs and SMPs, and occasionally the lamina propria between crypts (Fig. 6D-I). Such mutually exclusive distribution indicates a possibility that NIBP-like-IR was expressed in axon ending or neurites that originate from other plexuses or other neurons, which requires further investigation using neuronal tracing methods. Similar to HuD co-labeling, a small population of NIBP-expressing cells and fibers were negative for Peripherin in extra- and intra-ganglionic areas (Fig. 6, red arrow), suggesting the existence of a non-neuronal cell population that needs further identification.
Figure 6. Double-labeled immunofluorescent confocal images showing the correlation of NIBP-like-immunoreactivity with pan-neuronal marker Peripherin in representative myenteric plexuses (A-F), the submucosal plexuses (G, H) and lamina propria (I) of mouse colon.
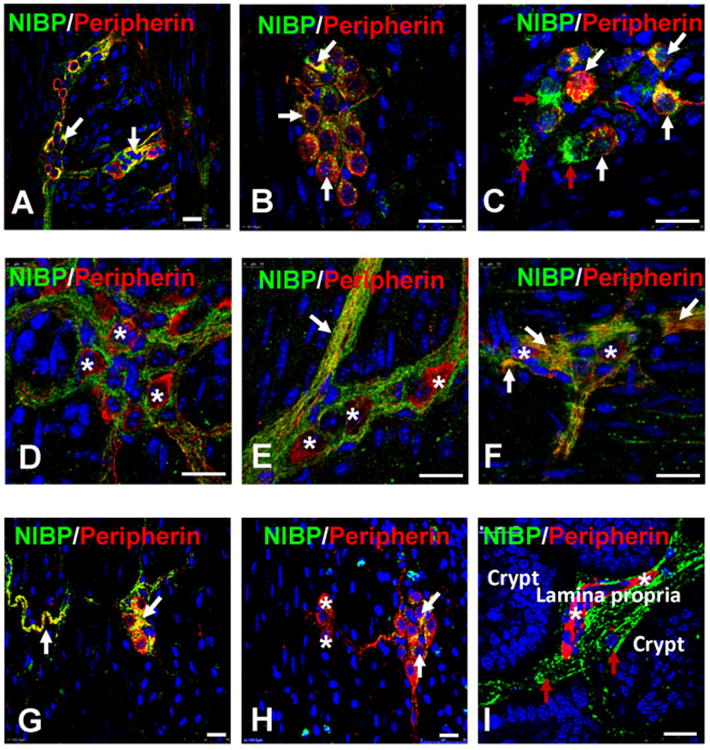
The white arrows show the co-localization, the red arrows indicate only NIBP and the stars indicate only Peripherin. Scale bars = 20 μm
Taken together, these data suggest that NIBP protein is expressed extensively in enteric neurons and neurites. The varying level of NIBP expression in different enteric plexuses as well as different neurons indicates that the protein most likely has different functions in different subgroups of enteric neurons.
3.5 NIBP-like-IR is present in a subpopulation of enteric neurons
In the mouse ENS, 11 subtypes of myenteric neurons and 5 subtypes of submucosal neurons have been identified51-53. To identify the chemical coding properties of NIBP-positive neurons, selected subtypes of neuronal markers choline acetyltransferase (ChAT), neuronal nitric oxide synthase (nNOS), and Calretinin were assessed by multi-labeled fluorescent immunohistochemistry.
Acetyl choline is a major excitatory neurotransmitter in the ENS, which is synthesized by ChAT. ChAT-like-IR has been widely used to identify cholinergic neurons in the ENS51, 54, 55. Multi-labeled confocal imaging analysis showed that ChAT-like-IR was extensively expressed and completely colocalized with NIBP-like-IR in the enteric neurons (Fig. 7A-F). Of the brightly stained NIBP cells, 97.7 ± 1.8% (cohort size 100-177 cells for each mouse, n = 4) were positive for ChAT. In the majority of intraganglionic fibers and intramuscular nerve fibers, ChAT staining completely overlapped with NIBP staining (Fig. 7G-I). In some locations, however, ChAT-positive fibers were NIBP-negative or vice visa (Fig. 7J-L, stars and red arrows). As described above, a small population of NIBP-expressing cells and fibers was negative for ChAT and Peripherin mainly in the extraganglionic area (Fig. 7, red arrows).
Figure 7. Triple-labeled immunofluorescent confocal images showing complete colocalization of NIBP with cholinergic neuronal marker ChAT and pan-neuronal marker Peripherin in myenteric plexuses(A-F) and some intramuscular nerve fibers (G-I). In some regions, NIBP and ChAT were not colocalized in the intramuscular fibers (J-L).
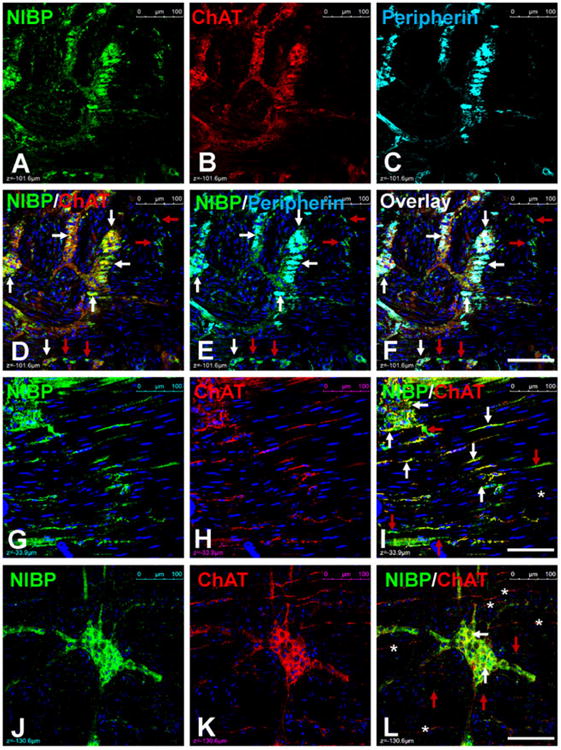
The white arrows show the co-localization, the red arrows indicate only NIBP and the stars indicate only Peripherin. Scale bars = 100 μm
In the ENS, nNOS synthesizes nitric oxide, which is a primary neurotransmitter of inhibitory muscle motor neurons in the ENS51, 56-58. Previous studies have shown that approximately 26-32% of all myenteric neurons were positive for NOS-like-IR using HuD51, neuron-specific enolase52, or PGP9.559 as pan-neuronal markers. As shown in Fig. 8A-E, NIBP-like-IR was detected in most of the nNOS-positive cells in the MPs. Of all NIBP-like-IR cells including punctate and non-punctate staining, 26.5 ± 1.4% (cohort size 194-285 cells for each mouse, n = 3) were positive for nNOS, whereas 97.2 ± 2.5% (cohort size 53-94 cells for each mouse, n = 7) of the total nNOS-positive cells presented with bright punctate staining for NIBP-like-IR in the MPs. Of the brightly stained NIBP cells, 86.9 ± 9.6% (cohort size 98-109 cells for each mouse, n = 4) were positive for nNOS. In the SMPs from adult mouse colon, NIBP-like-IR was weakly expressed in the majority of enteric neurons and partially co-localized with nNOS-like-IR (Fig. 8F), which has been shown to be rarely expressed in SMPs59.
Figure 8. Double-labeled immunohistochemistry with NIBP and subneuronal marker nNOS (A-F) or Calretinin (G-N).
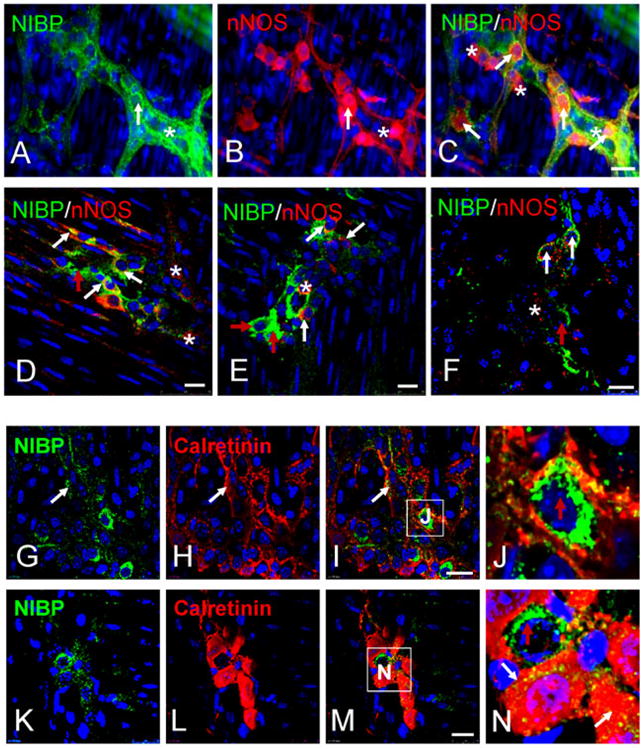
A-C. Representative micrographs of regular fluorescent microscopy. D-N. Representative confocal images. A-E and G-J show the myenteric plexuses. F and K-N show the submucosal plexuses. Higher magnified insets (J and N) show mutually exclusive staining between NIBP and Calretinin. The white arrows show co-localization, the red arrows indicate only NIBP and the stars indicate only nNOS or Calretinin. Scale bars = 20 μm.
Calretinin-like-IR has been observed in approximately 52% of enteric neurons in the MPs51 and in processes that innervate both longitudinal and circular muscles51. In the MPs, 43.4 ± 11.3% (cohort size 55-263 cells for each mouse, n = 3) of the brightly stained NIBP cells were positive for Calretinin, whereas 50.7 ± 30.3% (cohort size 124-163 cells for each mouse, n = 3) of the total Calretinin-positive cells were highly positive for NIBP-like-IR. Both Calretinin and NIBP staining frequently occurred in individual cell bodies and neurites of enteric neurons in MPs, but the staining labeled distinct subcellular structures (Fig. 8G-J). In SMPs, almost all enteric neurons express Calretinin, whereas only a fraction of that population also contains NIBP-like-IR (Fig. 8K-N).
3.6 NIBP/NFκB signaling regulates enteric neuronal differentiation
NIBP enhances cytokine-induced NFκB activation by binding to IKK2 and increasing its kinase activity5, 7. Recent clinical studies show that NIBP mutations are closely correlated with autosomal-recessive mental retardation and postnatal microcephaly, suggesting an essential role of NIBP in brain development7, 8, 17, 60. Since NIBP is widely expressed in the ENS, we hypothesized that NIBP/NFκB signaling may play a role in enteric neuronal differentiation. To test this, we first validated the existence of NIBP/NFκB signaling in a recently-established ENC31. The interaction of NIBP with endogenous IKK2 but not IKK15 was corroborated by co-immunoprecipitation (Fig. 9A). NIBP knockdown stable cell line was established by lentivirus-mediated NIBP shRNA expression5 and fluorescence activated cell sorting. The efficacy of NIBP shRNA knockdown was verified by RT-qPCR analysis using 3 sets of PCR primers that generated PCR fragments covering exon 5-7, 13-14 and 27-28 of mouse NIBP gene. Consistent with our previous report in PC12 cells5, NIBP-CR in ENCs effectively inhibited NIBP mRNA expression even after 5-10 passages (Fig. 9B). Our recent studies demonstrated that TNFα treatment of ENCs induced NFκB activation as determined by the dramatic increase in adenovirus-mediated NFκB-luciferase reporter activity and phosphorylation of IKK2/p65/IκBα32. As shown in Figure 9C and 9D, NIBP-CR inhibited TNFα-induced NFκB activation. NIBP-UTR was ineffective in knocking down NIBP expression and inhibiting NFκB activation, and thus served as an additional negative control in subsequent experiments.
Figure 9. NIBP enhances TNFα-induced NFκB activation by interacting with IKK2 in an enteric neuronal cell line (ENC).
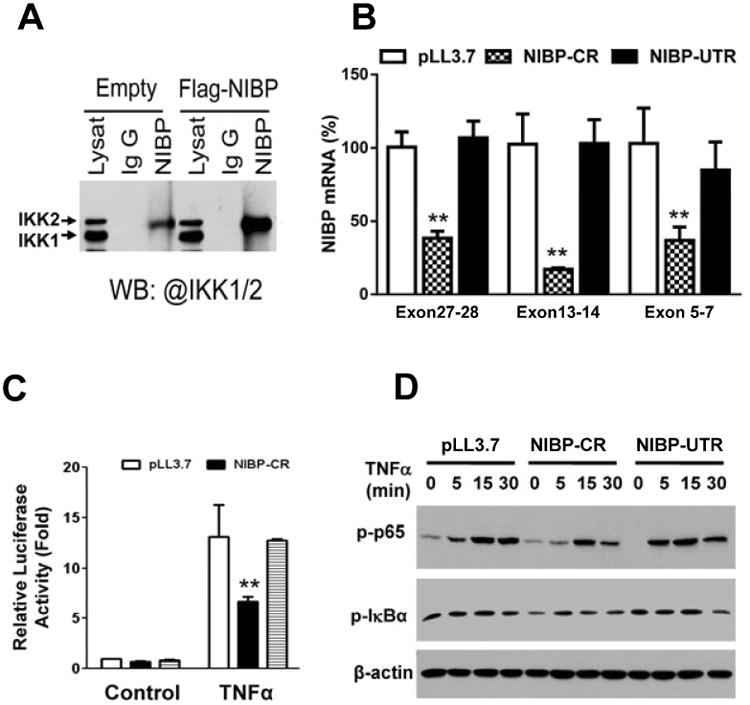
A. Co-immunoprecipitation and Western blot analysis (WB) validated the interaction of endogenous and exogenous NIBP with IKK2 but not IKK1. B. Realtime RT-qPCR analysis confirmed the effective knockdown of lentivirus-mediated NIBP shRNA targeting C-terminal region (CR) compared with empty vector pLL3.7 or the ineffective NIBP shRNA targeting the 3′-untranslated region (UTR). C. An adenovirus-mediated NFκB-luciferase reporter assay identified significant inhibition of NIBP-CR on TNFα (10 ng/ml)-induced activation of NFκB reporter. D. Western blot analysis showed the inhibition of NIBP-CR on TNFα-induced phosphorylation of p65 and IκBα. Data represent the mean ± SEM of 3 independent experiments. ** p<0.01 indicates a statistically significant decrease using a student's t test compared to the corresponding pLL3.7 control.
We next evaluated the effect of NIBP stable knockdown on neuronal differentiation of ENCs by Western blot and RT-qPCR analysis. Temperature switch from 33 to 39°C induced neuronal differentiation as assessed by measurement of neurite length and determining the percentage of neurite-bearing cells (Fig. 10A, B)31, 43. TNFα treatment further induced a significant increase in neuronal differentiation (Fig. 10A, B). Such differentiation was attenuated by stable knockdown of endogenous NIBP with the effective NIBP-CR shRNA but was not affected by the ineffective NIBP-UTR shRNA or control empty pLL3.7 vector (Fig. 10A, B). Knockdown of endogenous NIBP also led to a reduction in baseline and TNF-α-induced neuronal protein (NSE) expression (Fig. 10C). Furthermore, NIBP-CR inhibited the mRNA expression of most pan-neuronal markers (PGP9.5, NSE, Tuj1) and well-differentiated neuronal markers (NEFM)(Fig. 10D), suggesting that NIBP influences neuronal differentiation. To further determine the role of NIBP in regulating ENC neuronal differentiation, we overexpressed NIBP in ENCs via lentivirus-mediated gene delivery. As shown in Figure 10E, the NIBP-expressing stable cell line exhibited higher mRNA levels of the neuronal marker PGP9.5 under constitutive and TNFα-inducible conditions. Taken together, these results showed that lentivirus-mediated NIBP shRNA efficiently knocked down NIBP expression and inhibited cytokine-induced NFκB activation and neuronal differentiation in ENC, whereas NIBP overexpression promoted constitutive and inducible expression of PGP9.5.
Figure 10. NIBP shRNA stable knockdown inhibits neuronal differentiation in an enteric neuronal cell line (ENC) and NIBP overexpression promotes it.
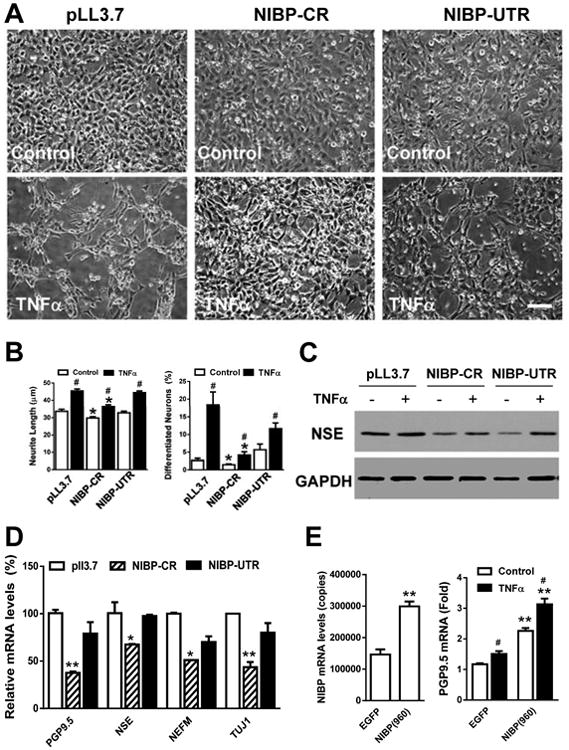
A. Representative phase contrast micrographs showing the inhibitory effect of NIBP-CR on neuronal differentiation of ENCs cultured at 39°C for 3 d in the presence or absence of TNFα (10 ng/ml). Scale bar = 50 μm. B. Quantitative analysis shows that NIBP-CR significantly decreases the neurite length and number of differentiated neurons in both the control and TNFα treatments. C. Western blot analysis identified the inhibition of NIBP-CR on constitutive and TNFα-induced expression of neuronal markers NSE. D. RT-qPCR analysis shows that NIBP-CR inhibits the expression of PGP9.5, NSE, NEFM and Tuj1. E. NIBP overexpression significantly increases constitutive and TNFα-induced expression of PGP9.5. Data represent the mean ± SEM of 3-5 independent experiments. * p<0.05 and ** p<0.01 indicate statistically significant changes using a student's t test compared to the corresponding pLL3.7 or LV-EGFP empty vector controls. # p<0.05 indicates significant changes compared to the control treatment.
4. Discussion
NIBP is a novel protein that regulates NFκB signaling5, 7 and may also play a role in the protein trafficking8, 9, 12. NIBP mRNA and protein are widely expressed in neurons of the brain5, 7, and NIBP is required for nerve growth factor-induced neuronal differentiation of PC12 cells5. Recent work has identified a correlation between specific NIBP mutations and mental retardation7, 13, 15-18, 60, 61. In the present study, we demonstrated for the first time that NIBP is extensively expressed in enteric neurons and is implicated in enteric neuronal differentiation. The ENS has been called “the second brain in the gut”, and it plays a critical role in regulating gut motility, secretion, absorption, and mucosal homeostasis. Functional disorders of the adult ENS are implicated in inflammatory bowel disease and irritable bowel syndrome62-64, which are two major gastrointestinal diseases. Therefore, further characterization of NIBP cellular distribution and functional regulation may advance our understanding of the molecular mechanisms underlying ENS functional disorders.
The muscle whole mount preparations from non-inflamed colon are predominantly composed of smooth muscle cells, ICC, and ENS. The high expression of NIBP mRNA and protein in the muscle whole mount preparations detected by RT-PCR and Western blot analysis likely reflects expression in the ENS only, because no evidence of NIBP-like-IR was observed in smooth muscle cells and ICC. The mammalian ENS consists of neurons and glial cells that are distributed throughout the gut wall and mainly form myenteric and submucosal plexuses. Double-labeled fluorescent microscopy and confocal imaging analysis showed that NIBP-like-IR-positive cells are not co-localized with GFAP and S100β-positive enteric glial cells. However, the possibility of NIBP expression in GFAP and S100β-negative enteric glial cells cannot be ruled out. A small population of non-neuronal cells (HuD/Peripherin-negative) was observed in extra-ganglionic and intra-ganglionic area. Given recent evidence showing the extra-and intra-ganglionic existence of enteric neural stem cells both in vitro65-67 and in vivo34, 68, we speculate that such non-neuronal/non-glial NIBP-expressing cells may be the long-sought and un-defined enteric neural stem cells. This hypothesis is supported by the facts that 1) NIBP is highly expressed in the developing mouse and human brain7, 2) NIBP mutation leads to mental retardation7, 13, 15-18, 60, 61, 3) NIBP is required for neuronal differentiation in PC12 cells5 and ENC, and 4) NIBP is highly expressed in cultured enteric neural stem cells69. Therefore, an NIBP-reporter transgenic animal model is warranted to determine if NIBP is a novel marker for enteric neural stem cells.
In this study, we focused on the neuronal and subneuronal expression of NIBP in the mouse colon. NIBP-like-IR is correlated extensively with the expression of pan-neuronal markers HuD and Peripherin in the MPs and weakly expressed in the SMPs in the normal mice. In approximately 35-44% of myenteric neurons and 9-10% of submucosal enteric neurons, NIBP expression presented as a bright punctate structure. These data suggest that the dynamic trafficking or shuttling of NIBP from the cytoplasm to punctate structures and/or from the soma to dendrites/axons occurs in some neurons that likely exhibit different functions. Such punctate accumulation may also indicate a higher level of NIBP protein expression or posttranslational modification, which may reflect selective regulation of NFκB signaling in some neurons. Further characterization of NIBP co-expression with subcellular structures, in particular the Golgi network, and protein trafficking8-12 is important for identifying novel functions of NIBP in the ENS.
Confocal image analysis of NIBP co-localization with various subpopulations of enteric neurons demonstrated that a portion of NIBP positive enteric neurons and nerve fibers encode several yet independent sub-neuronal markers, such as ChAT, nNOS and Calretinin. The co-localization of these proteins mainly occurs in the ganglia and the first branches of the nerve fibers (primary plexuses) between ganglia70. Approximately 87% and 43% of the brightly-stained NIBP neurons were positive for nNOS and Calretinin, respectively. In contrast, almost all ChAT- and nNOS-expressing neurons were highly immunoreactive for NIBP, but half of Calretinin-expressing neurons were positive for NIBP. Therefore, active expression of NIBP in the enteric neurons is likely involved in the functional regulation of cholinergic excitatory neurons, nitrergic inhibitory neurons or Calretinin-containing neurons in the ENS. Mutually exclusive staining was observed between NIBP and these sub-neuronal markers in a few neurons within the ganglia. In particular, NIBP-like-IR in the second to fourth branches do not markedly co-localize with the ChAT, nNOS and Calretinin positive nerve fibers. Therefore, NIBP-IR cells may represent a unique subpopulation of enteric neurons and may regulate the function of various interneurons in the gut. NIBP-positive nerve fibers may originate from neurons that are either intrinsic or extrinsic to the intestine. Neuronal tracing studies are needed to identify the sources and directional projections of NIBP-positive fibers and neurons in the gut71.
NFκB has been widely shown to play a crucial role in neuronal plasticity, learning and memory, neuroinflammation, neurogenesis, and neurodegeneration1, 32, 72, 73. However, the role of NFκB signaling in ENS functional regulation remains unknown. ENS abnormalities in various inflammatory diseases of the gut have been reported74. The direct effects of inflammatory mediators on the ENS could be the activation of enteric neurons, suppression of neurotransmitter release, or the alteration of receptor expression. For instance, interleukine-1β activates specific populations of enteric neurons (NOS and VIP)75. The ENCs derived from mouse enteric neural stem/progenitor cells were maintained at the permissive temperature (33°C) and differentiated into neurons at 39°C31, 43. We previously demonstrated that TNFα and IL-1β induced NFκB activation and neuronal differentiation in cultured ENCs32. Here we show that NIBP is required for cytokine-induced NFκB activation in the ENCs. NIBP knockdown inhibited ENC neuronal differentiation while NIBP overexpression promoted it. We predict that NIBP/NFκB signaling may regulate neural plasticity of the ENS during intestinal inflammation76. The expression pattern and functional regulation of NIBP/NFκB signaling in the ENS after intestinal inflammation are currently under investigation.
NIBP is also expressed in the mucosal layers as shown by RT-PCR and Western blot analysis. The type of NIBP-positive cells within mucosa and their roles in regulating intestinal secretion/absorption need further investigation.
NIBP has been implicated in three biological processes: NFκB signaling5, 7, trans-Golgi network8, 9, 12 and antiviral defense6. NIBP is a large protein with 5 isoforms reported in Genebank. The transcriptional regulation and posttranscriptional modification for these isoforms remain elusive. Our Western blot analysis identified a consistent band of 106 kDa in various layers of both small intestine and colon, which matches the predicted molecular weight of isoform I (960 aa) of mouse NIBP (NP_083916). However, several additional specific bands (abolished by antigen preabsorption) were identified and varied with different layers and regional segments of mouse intestine. These bands may represent various post-translational modifications and/or cleavage, although their specificity and biological relevance warrant further investigation. In the mucosal layers, NIBP mRNA levels were the highest among the layers but NIBP protein levels at the predicted size were the lowest, leaving several additional smaller bands. This suggests that NIBP cleavage preferentially occurs in the mucosa layers.
Selection of housekeeping genes for RT-qPCR normalization remains a challenge. Many housekeeping genes commonly used exhibit a large variability with treatment or physiological state77-79. Taking liver tissue as an example, physiological state, food intake, or dietary treatment influenced the mRNA expression of commonly used housekeeping genes80. In this study, we observed high variability of several housekeeping genes among different regions and layers of the intestine, which undergoes dynamic roles similar to liver80. Although a stable housekeeping gene for intestine tissues remains to be identified, we used NIBP mRNA changes without housekeeping gene normalization for data interpretation as they correlate with the changes in protein expression and immunostaining. The relative differences of NIBP expression in regions or layers may have implications in the role of NIBP in different parts or layers of the intestine, however, future experiments will address this.
In conclusion, we have found that NIBP is extensively expressed in the enteric neurons and nerve fibers and influences enteric neuronal differentiation. Highly immunoreactive NIBP expression may represent a dynamic functional regulation of NIBP in the demanding enteric neurons. NIBP-positive cells encode various types of interneuronal peptides or neurotransmitters and may regulate their functions. Further studies on the functional significance of NIBP-expressing enteric neurons may provide insight into the mechanisms of neurogastrointestinal disorders.
Acknowledgments
This work was supported by Grant DK075964 (WH) and Grant DK080684 (SS) from the National Institutes of Diabetes and Digestive and Kidney Diseases.
Abbreviations
- ChAT
choline acetyltransferase
- CMV
cytomegalovirus
- ENC
enteric neuronal cell line
- ENS
enteric nervous system
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- HEK
human embryonic kidney
- ICC
interstitial cells of Cajal
- IκB
inhibitor of NFκB
- IKK
inhibitor of NFκB kinase
- IR
immunoreactivity
- kDa
kilodalton
- MP
myenteric plexus
- NFκB
nuclear factor κB
- NIK
NFκB-induced kinase
- nNOS
neuronal nitric oxide synthase
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- RT
reverse transcription
- SDS
sodium dodecyl sulphate
- SMP
submucosal plexus
- TBS-T
tris-buffered saline plus Tween-20
- TRAPP
transport protein particle
Footnotes
Disclosure statement: None of the authors have any actual or potential conflict of interests.
YGZ critically co-designed and performed the experiments, analyzed the data, and co-wrote the manuscript with WH. DB, AAPF and FL performed immunohistochemistry, Western blot and quantification. SL, HW, FY and SA performed RT-PCR, Western blot, cloning and imaging analysis. JG and SS provided materials, concept and design. WH co-designed the experiments and overall project, supervised and performed the experiments, analyzed the data, and wrote the manuscript.
References
- 1.Mattson MP, Meffert MK. Roles for NF-kappaB in nerve cell survival, plasticity, and disease. Cell Death Differ. 2006;13:852–60. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan NC, Croydon L, McGettigan PA, et al. Hippocampal region-specific regulation of NF-kappaB may contribute to learning-associated synaptic reorganisation. Brain Res Bull. 2010;81:385–90. doi: 10.1016/j.brainresbull.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 4.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–8. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Hu WH, Pendergast JS, Mo XM, et al. NIBP, a novel NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation. J Biol Chem. 2005;280:29233–41. doi: 10.1074/jbc.M501670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahoor MA, Yamane D, Mohamed YM, et al. Bovine viral diarrhea virus non-structural protein 5A interacts with NIK- and IKKbeta-binding protein. J Gen Virol. 2010;91:1939–48. doi: 10.1099/vir.0.020990-0. [DOI] [PubMed] [Google Scholar]
- 7.Mochida GH, Mahajnah M, Hill AD, et al. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85:897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong M, Satoh A, Yu MK, et al. TRAPPC9 mediates the interaction between p150 and COPII vesicles at the target membrane. PLoS One. 2012;7:e29995. doi: 10.1371/journal.pone.0029995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox R, Chen SH, Yoo E, et al. Conservation of the TRAPPII-specific subunits of a Ypt/Rab exchanger complex. BMC Evol Biol. 2007;7:12. doi: 10.1186/1471-2148-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westlake CJ, Baye LM, Nachury MV, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci U S A. 2011;108:2759–64. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zong M, Wu XG, Chan CW, et al. The adaptor function of TRAPPC2 in mammalian TRAPPs explains TRAPPC2-associated SEDT and TRAPPC9-associated congenital intellectual disability. PLoS One. 2011;6:e23350. doi: 10.1371/journal.pone.0023350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kummel D, Oeckinghaus A, Wang C, et al. Distinct isocomplexes of the TRAPP trafficking factor coexist inside human cells. FEBS Lett. 2008;582:3729–33. doi: 10.1016/j.febslet.2008.09.056. [DOI] [PubMed] [Google Scholar]
- 13.Abou Jamra R, Wohlfart S, Zweier M, et al. Homozygosity mapping in 64 Syrian consanguineous families with non-specific intellectual disability reveals 11 novel loci and high heterogeneity. Eur J Hum Genet. 2011;19:1161–6. doi: 10.1038/ejhg.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riendeau N. Graduate Thesis. The University Of British Columbia; 2009. Autism Spectrum Disorders: Identification of Novel Microdeletions and Microduplications and their Associated Phenotypes; pp. 56–59. [Google Scholar]
- 15.Philippe O, Rio M, Carioux A, et al. Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:903–8. doi: 10.1016/j.ajhg.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koifman A, Feigenbaum A, Bi W, et al. A homozygous deletion of 8q24.3 including the NIBP gene associated with severe developmental delay, dysgenesis of the corpus callosum, and dysmorphic facial features. Am J Med Genet A. 2010;152A:1268–72. doi: 10.1002/ajmg.a.33319. [DOI] [PubMed] [Google Scholar]
- 17.Marangi G, Leuzzi V, Manti F, et al. TRAPPC9-related autosomal recessive intellectual disability: report of a new mutation and clinical phenotype. Eur J Hum Genet. 2013;21:229–32. doi: 10.1038/ejhg.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakar N, Goebel I, Daud S, et al. A homozygous splice site mutation in TRAPPC9 causes intellectual disability and microcephaly. Eur J Med Genet. 2012;55:727–31. doi: 10.1016/j.ejmg.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Burzynski G, Shepherd IT, Enomoto H. Genetic model system studies of the development of the enteric nervous system, gut motility and Hirschsprung's disease. Neurogastroenterol Motil. 2009;21:113–27. doi: 10.1111/j.1365-2982.2008.01256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molenaar JC, Tibboel D, van der Kamp AW, et al. Diagnosis of innervation-related motility disorders of the gut and basic aspects of enteric nervous system development. Prog Pediatr Surg. 1989;24:173–85. doi: 10.1007/978-3-642-74493-8_19. [DOI] [PubMed] [Google Scholar]
- 21.Schemann M. Control of gastrointestinal motility by the “gut brain”--the enteric nervous system. J Pediatr Gastroenterol Nutr. 2005;41(1):S4–6. doi: 10.1097/01.scs.0000180285.51365.55. [DOI] [PubMed] [Google Scholar]
- 22.Bywater RA, Taylor GS, Furukawa K. The enteric nervous system in the control of motility and secretion. Dig Dis. 1987;5:193–211. doi: 10.1159/000171174. [DOI] [PubMed] [Google Scholar]
- 23.Guandalini S. Enteric nervous system: intestinal absorption and secretion. J Pediatr Gastroenterol Nutr. 1997;25(1):S5–6. doi: 10.1097/00005176-199700002-00003. [DOI] [PubMed] [Google Scholar]
- 24.Hogan DL, Yao B, Steinbach JH, et al. The enteric nervous system modulates mammalian duodenal mucosal bicarbonate secretion. Gastroenterology. 1993;105:410–7. doi: 10.1016/0016-5085(93)90714-n. [DOI] [PubMed] [Google Scholar]
- 25.Tixier E, Lalanne F, Just I, et al. Human mucosa/submucosa interactions during intestinal inflammation: involvement of the enteric nervous system in interleukin-8 secretion. Cell Microbiol. 2005;7:1798–810. doi: 10.1111/j.1462-5822.2005.00596.x. [DOI] [PubMed] [Google Scholar]
- 26.Margolis KG, Stevanovic K, Karamooz N, et al. Enteric neuronal density contributes to the severity of intestinal inflammation. Gastroenterology. 2011;141:588–98. 598 e1–2. doi: 10.1053/j.gastro.2011.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakhan SE, Kirchgessner A. Neuroinflammation in inflammatory bowel disease. J Neuroinflammation. 2010;7:37. doi: 10.1186/1742-2094-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasina V, Barbara G, Talamonti L, et al. Enteric neuroplasticity evoked by inflammation. Auton Neurosci. 2006:126–127. 264–72. doi: 10.1016/j.autneu.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 29.Mawe GM, Strong DS, Sharkey KA. Plasticity of enteric nerve functions in the inflamed and postinflamed gut. Neurogastroenterol Motil. 2009;21:481–91. doi: 10.1111/j.1365-2982.2009.01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Malley D, Liston M, Hyland NP, et al. Colonic soluble mediators from the maternal separation model of irritable bowel syndrome activate submucosal neurons via an interleukin-6-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. 2011;300:G241–52. doi: 10.1152/ajpgi.00385.2010. [DOI] [PubMed] [Google Scholar]
- 31.Anitha M, Joseph I, Ding X, et al. Characterization of fetal and postnatal enteric neuronal cell lines with improvement in intestinal neural function. Gastroenterology. 2008;134:1424–35. doi: 10.1053/j.gastro.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Liu J, Yao S, et al. Nuclear factor kappa B signaling initiates early differentiation of neural stem cells. Stem Cells. 2012;30:510–24. doi: 10.1002/stem.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furness JB, Robbins HL, Xiao J, et al. Projections and chemistry of Dogiel type II neurons in the mouse colon. Cell Tissue Res. 2004;317:1–12. doi: 10.1007/s00441-004-0895-5. [DOI] [PubMed] [Google Scholar]
- 34.Liu MT, Kuan YH, Wang J, et al. 5-HT4 receptor-mediated neuroprotection and neurogenesis in the enteric nervous system of adult mice. J Neurosci. 2009;29:9683–99. doi: 10.1523/JNEUROSCI.1145-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu W, Huang J, Mahavadi S, et al. Lentiviral siRNA silencing of sphingosine-1-phosphate receptors S1P1 and S1P2 in smooth muscle. Biochem Biophys Res Commun. 2006;343:1038–44. doi: 10.1016/j.bbrc.2006.03.079. [DOI] [PubMed] [Google Scholar]
- 36.Follenzi A, Naldini L. HIV-based vectors. Preparation and use. Methods Mol Med. 2002;69:259–74. [PubMed] [Google Scholar]
- 37.Blits B, Kitay BM, Farahvar A, et al. Lentiviral vector-mediated transduction of neural progenitor cells before implantation into injured spinal cord and brain to detect their migration, deliver neurotrophic factors and repair tissue. Restor Neurol Neurosci. 2005;23:313–24. [PubMed] [Google Scholar]
- 38.Follenzi A, Naldini L. Generation of HIV-1 derived lentiviral vectors. Methods Enzymol. 2002;346:454–65. doi: 10.1016/s0076-6879(02)46071-5. [DOI] [PubMed] [Google Scholar]
- 39.Rubinson DA, Dillon CP, Kwiatkowski AV, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 40.Hu W, Mahavadi S, Li F, et al. Upregulation of RGS4 and downregulation of CPI-17 mediate inhibition of colonic muscle contraction by interleukin-1beta. Am J Physiol Cell Physiol. 2007;293:C1991–2000. doi: 10.1152/ajpcell.00300.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li ZS, Pham TD, Tamir H, et al. Enteric dopaminergic neurons: definition, developmental lineage, and effects of extrinsic denervation. J Neurosci. 2004;24:1330–9. doi: 10.1523/JNEUROSCI.3982-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sang Q, Williamson S, Young HM. Projections of chemically identified myenteric neurons of the small and large intestine of the mouse. J Anat. 1997;190(Pt 2):209–22. doi: 10.1046/j.1469-7580.1997.19020209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anitha M, Shahnavaz N, Qayed E, et al. BMP2 promotes differentiation of nitrergic and catecholaminergic enteric neurons through a Smad1-dependent pathway. Am J Physiol Gastrointest Liver Physiol. 2010;298:G375–83. doi: 10.1152/ajpgi.00343.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Boyen GB, Steinkamp M, Reinshagen M, et al. Proinflammatory cytokines increase glial fibrillary acidic protein expression in enteric glia. Gut. 2004;53:222–8. doi: 10.1136/gut.2003.012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izbeki F, Wittman T, Rosztoczy A, et al. Immediate insulin treatment prevents gut motility alterations and loss of nitrergic neurons in the ileum and colon of rats with streptozotocin-induced diabetes. Diabetes Res Clin Pract. 2008;80:192–8. doi: 10.1016/j.diabres.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 46.Phillips RJ, Hargrave SL, Rhodes BS, et al. Quantification of neurons in the myenteric plexus: an evaluation of putative pan-neuronal markers. J Neurosci Methods. 2004;133:99–107. doi: 10.1016/j.jneumeth.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Lin Z, Gao N, Hu HZ, et al. Immunoreactivity of Hu proteins facilitates identification of myenteric neurones in guinea-pig small intestine. Neurogastroenterol Motil. 2002;14:197–204. doi: 10.1046/j.1365-2982.2002.00317.x. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda H, Hirato J, Kuroiwa M, et al. Histopathological and immunohistochemical study of the enteric innervations among various types of aganglionoses including isolated and syndromic Hirschsprung disease. Neuropathology. 2006;26:8–23. doi: 10.1111/j.1440-1789.2006.00649.x. [DOI] [PubMed] [Google Scholar]
- 49.Szabolcs MJ, Visser J, Shelanski ML, et al. Peripherin: a novel marker for the immunohistochemical study of malformations of the enteric nervous system. Pediatr Pathol Lab Med. 1996;16:51–70. [PubMed] [Google Scholar]
- 50.Sibaev A, Yuce B, Kemmer M, et al. Cannabinoid-1 (CB1) receptors regulate colonic propulsion by acting at motor neurons within the ascending motor pathways in mouse colon. Am J Physiol Gastrointest Liver Physiol. 2009;296:G119–28. doi: 10.1152/ajpgi.90274.2008. [DOI] [PubMed] [Google Scholar]
- 51.Qu ZD, Thacker M, Castelucci P, et al. Immunohistochemical analysis of neuron types in the mouse small intestine. Cell Tissue Res. 2008;334:147–61. doi: 10.1007/s00441-008-0684-7. [DOI] [PubMed] [Google Scholar]
- 52.Sang Q, Young HM. Chemical coding of neurons in the myenteric plexus and external muscle of the small and large intestine of the mouse. Cell Tissue Res. 1996;284:39–53. doi: 10.1007/s004410050565. [DOI] [PubMed] [Google Scholar]
- 53.Young HM, Bergner AJ, Muller T. Acquisition of neuronal and glial markers by neural crest-derived cells in the mouse intestine. J Comp Neurol. 2003;456:1–11. doi: 10.1002/cne.10448. [DOI] [PubMed] [Google Scholar]
- 54.Sang Q, Young HM. The identification and chemical coding of cholinergic neurons in the small and large intestine of the mouse. Anat Rec. 1998;251:185–99. doi: 10.1002/(SICI)1097-0185(199806)251:2<185::AID-AR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 55.Hao MM, Bornstein JC, Young HM. Development of myenteric cholinergic neurons in ChAT-Cre;R26R-YFP mice. J Comp Neurol. 2013 doi: 10.1002/cne.23354. [DOI] [PubMed] [Google Scholar]
- 56.Ward SM, Dalziel HH, Khoyi MA, et al. Hyperpolarization and inhibition of contraction mediated by nitric oxide released from enteric inhibitory neurones in guinea-pig taenia coli. Br J Pharmacol. 1996;118:49–56. doi: 10.1111/j.1476-5381.1996.tb15365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyster DJ, Bywater RA, Taylor GS. Neurogenic control of myoelectric complexes in the mouse isolated colon. Gastroenterology. 1995;108:1371–8. doi: 10.1016/0016-5085(95)90684-3. [DOI] [PubMed] [Google Scholar]
- 58.Rivera LR, Poole DP, Thacker M, et al. The involvement of nitric oxide synthase neurons in enteric neuropathies. Neurogastroenterol Motil. 2011;23:980–8. doi: 10.1111/j.1365-2982.2011.01780.x. [DOI] [PubMed] [Google Scholar]
- 59.Young HM, Ciampoli D. Transient expression of neuronal nitric oxide synthase by neurons of the submucous plexus of the mouse small intestine. Cell Tissue Res. 1998;291:395–401. doi: 10.1007/s004410051009. [DOI] [PubMed] [Google Scholar]
- 60.Mir A, Kaufman L, Noor A, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85:909–15. doi: 10.1016/j.ajhg.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshida T, Kato K, Yokoi K, et al. Association of genetic variants with hemorrhagic stroke in Japanese individuals. Int J Mol Med. 2010;25:649–56. doi: 10.3892/ijmm_00000388. [DOI] [PubMed] [Google Scholar]
- 62.Gross KJ, Pothoulakis C. Role of neuropeptides in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:918–32. doi: 10.1002/ibd.20129. [DOI] [PubMed] [Google Scholar]
- 63.Neunlist M, Van Landeghem L, Bourreille A, et al. Neuro-glial crosstalk in inflammatory bowel disease. J Intern Med. 2008;263:577–83. doi: 10.1111/j.1365-2796.2008.01963.x. [DOI] [PubMed] [Google Scholar]
- 64.Villanacci V, Bassotti G, Nascimbeni R, et al. Enteric nervous system abnormalities in inflammatory bowel diseases. Neurogastroenterol Motil. 2008;20:1009–16. doi: 10.1111/j.1365-2982.2008.01146.x. [DOI] [PubMed] [Google Scholar]
- 65.Becker L, Kulkarni S, Tiwari G, et al. Divergent fate and origin of neurosphere-like bodies from different layers of the gut. Am J Physiol Gastrointest Liver Physiol. 2012;302:G958–65. doi: 10.1152/ajpgi.00511.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joseph NM, He S, Quintana E, et al. Enteric glia are multipotent in culture but primarily form glia in the adult rodent gut. J Clin Invest. 2011;121:3398–411. doi: 10.1172/JCI58186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laranjeira C, Sandgren K, Kessaris N, et al. Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J Clin Invest. 2011;121:3412–24. doi: 10.1172/JCI58200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corpening JC, Deal KK, Cantrell VA, et al. Isolation and live imaging of enteric progenitors based on Sox10-Histone2BVenus transgene expression. Genesis. 2011;49:599–618. doi: 10.1002/dvg.20748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y, Li F, Hu W. NIBP/NfκB Signaling Regulates Neuronal Differentiation of Enteric Neural Stem Cells. Gastroenterology. 2011;140:S3–4. [Google Scholar]
- 70.Furness JB, Clerc N, Lomax AE, et al. Shapes and projections of tertiary plexus neurons of the guinea-pig small intestine. Cell Tissue Res. 2000;300:383–7. doi: 10.1007/s004410000210. [DOI] [PubMed] [Google Scholar]
- 71.Zagorodnyuk VP, Brookes SJ, Spencer NJ. Structure-function relationship of sensory endings in the gut and bladder. Auton Neurosci. 2010;153:3–11. doi: 10.1016/j.autneu.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 72.Li J, Tang Y, Cai D. IKKbeta/NF-kappaB disrupts adult hypothalamic neural stem cells to mediate a neurodegenerative mechanism of dietary obesity and pre-diabetes. Nat Cell Biol. 2012;14:999–1012. doi: 10.1038/ncb2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Camandola S, Mattson MP. NF-kappa B as a therapeutic target in neurodegenerative diseases. Expert Opin Ther Targets. 2007;11:123–32. doi: 10.1517/14728222.11.2.123. [DOI] [PubMed] [Google Scholar]
- 74.De Giorgio R, Guerrini S, Barbara G, et al. Inflammatory neuropathies of the enteric nervous system. Gastroenterology. 2004;126:1872–83. doi: 10.1053/j.gastro.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 75.Tjwa ET, Bradley JM, Keenan CM, et al. Interleukin-1beta activates specific populations of enteric neurons and enteric glia in the guinea pig ileum and colon. Am J Physiol Gastrointest Liver Physiol. 2003;285:G1268–76. doi: 10.1152/ajpgi.00073.2003. [DOI] [PubMed] [Google Scholar]
- 76.Lomax AE, Fernandez E, Sharkey KA. Plasticity of the enteric nervous system during intestinal inflammation. Neurogastroenterol Motil. 2005;17:4–15. doi: 10.1111/j.1365-2982.2004.00607.x. [DOI] [PubMed] [Google Scholar]
- 77.Waxman S, Wurmbach E. De-regulation of common housekeeping genes in hepatocellular carcinoma. BMC Genomics. 2007;8:243. doi: 10.1186/1471-2164-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huggett J, Dheda K, Bustin S, et al. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–84. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 79.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 80.Janovick-Guretzky NA, Dann HM, Carlson DB, et al. Housekeeping gene expression in bovine liver is affected by physiological state, feed intake, and dietary treatment. J Dairy Sci. 2007;90:2246–52. doi: 10.3168/jds.2006-640. [DOI] [PubMed] [Google Scholar]


