SUMMARY
Embryonic stem cell (ESC) self-renewal and differentiation are governed by a broad-ranging regulatory network. Although the transcriptional regulatory mechanisms involved have been investigated extensively, post-transcriptional regulation is still poorly understood. Here we describe a critical role of the THO complex in ESC self-renewal and differentiation. We show that THO preferentially interacts with pluripotency gene transcripts through Thoc5, and is required for self-renewal at least in part by regulating their export and expression. During differentiation, THO loses its interaction with those transcripts due to reduced Thoc5 expression, leading to decreased expression of pluripotency proteins that facilitates exit from self-renewal. THO is also important for the establishment of pluripotency, as its depletion inhibits somatic cell reprogramming and blastocyst development. Together, our data indicate that THO regulates pluripotency gene mRNA export to control ESC self-renewal and differentiation, and therefore uncover a role for this aspect of post-transcriptional regulation in stem cell fate specification.
INTRODUCTION
ESCs are derived from the inner cell mass (ICM) of blastocysts. They are pluripotent and can differentiate into all cell types from the three germ layers, and they can also maintain the pluripotent state through the process of self-renewal (Evans, 2011; Irion et al., 2008). ESC pluripotency and self-renewal is governed by a complex transcription network that integrates input from extracellular signal transduction pathways, transcription factors, and epigenetic regulators (Dejosez and Zwaka, 2012; Jaenisch and Young, 2008; Ng and Surani, 2011; Young). Master transcription factors Oct4, Nanog, and Sox2 form the core of this network. They positively regulate their own expression through an auto-regulatory loop. At the same time, they activate other self-renewal genes while repressing lineage specification genes (Boyer et al., 2005; Loh et al., 2006). Other pluripotency transcription factors, as well as signal transduction pathway components and epigenetic regulators, further modulate and refine the transcription circuitry established by Oct4, Nanog, and Sox2 to maintain proper gene expression in ESCs (Chen et al., 2008; Kim et al., 2008).
In addition to the transcriptional network, recent studies have begun to reveal an important role of post-transcriptional regulation in the maintenance of ESC pluripotency and self-renewal. For example, alternative splicing and post-translational modification of key pluripotency factors have been shown to modulate their function in ESCs (Buckley et al., 2012; Han et al., 2013; Lu et al., 2009). In addition, noncoding RNAs control ESC fate by regulating pluripotency gene mRNA decay and translation (Guttman et al., 2011; Melton et al., 2010; Tay et al., 2008; Wang et al., 2008). However, compared to the extensive knowledge on the transcriptional control, post-transcriptional regulation of pluripotency and self-renewal remains poorly understood.
To systematically study self-renewal, we and others have previously carried out genome-wide RNAi screens in ESCs and identified a number of novel candidate regulators (Chia et al.; Ding et al., 2009; Fazzio et al., 2008; Ivanova et al., 2006; Kagey et al., 2010; Lu et al., 2009; Zheng et al., 2012). Among those, two components of the THO complex, Thoc2 and Thoc5, were listed as potential self-renewal factors (Ding et al., 2009; Lu et al., 2009). THO is a nuclear protein complex present from yeast to mammals and functions at the interface between mRNA transcription and export. Together with other proteins, such as Alyref and Uap56, THO forms the transcription/export (TREX) complex (Chang et al., 2013; Katahira, 2012) that facilitates the proper formation and export of messenger ribonucleoprotein complexes (mRNP) by helping to recruit other factors involved in mRNA processing and transport (Luna et al., 2012; Rondon et al., 2010). Although the general role of THO in mRNA export is thought to be conserved, THO has different compositions and may also have distinct functions in different species (Jimeno and Aguilera, 2010; Luna et al., 2012; Rondon et al., 2010). In yeast, THO consists of Tho2, Hpr1, Mft1 and Thp2, and is involved in mRNA export, processing, transcription elongation, and prevention of transcription-mediated replication obstacles (Piruat and Aguilera, 1998). In mammals, THO is composed of Thoc1 (homologue of yeast Hpr1), 2 (homologue of yeast Tho2), 3, 5, 6 and 7, among which Thoc2 functions as a scaffold and Thoc5 acts as an adaptor for mRNA export (Chi et al., 2013; Griaud et al., 2013; Guria et al., 2011; Katahira et al., 2009; Pena et al., 2012; Ramachandran et al., 2011; Viphakone et al., 2012). THO plays a pivotal role in normal development and cellular differentiation, as its disruption leads to early embryonic lethality, as well as defects in hematopoietic progenitor survival and testis development (Mancini et al., 2010; Wang et al., 2006; Wang et al., 2009).
In this study, we show that two subunits of the THO complex, Thoc2 and Thoc5, are required for mouse ESC self-renewal. Thoc2 preferentially interacts with and facilitates the export and expression of the pluripotency gene transcripts in a Thoc5-dependent manner. Furthermore, reduced expression of Thoc5 is an important priming step for ESC differentiation. Finally, Thoc2 and Thoc5 are required for somatic cell reprogramming and blastocyst formation ex vivo, and thereby play critical roles in the establishment of pluripotency. Our study reveals a novel regulatory mechanism for the delicate regulation of ESC self-renewal and differentiation at the post-transcriptional level.
RESULTS
Thoc2 and Thoc5 Expression Correlates with the Pluripotent State
Thoc2 and Thoc5, two components of the THO complex, have recently been identified in genome-wide RNAi screens as potential self-renewal factors in mouse ESCs (Ding et al., 2009; Lu et al., 2009). Thoc2 is the largest subunit in the complex and has been suggested to act as a scaffold (Pena et al., 2012), while Thoc5 is thought to be an adaptor for mRNA export (Chi et al., 2013; Griaud et al., 2013; Guria et al., 2011; Katahira et al., 2009; Viphakone et al., 2012). To examine their roles in ESCs, we first determined their expression pattern during differentiation. Reverse transcription followed by quantitative polymerase chain reaction (RT-qPCR) revealed a modest decrease in the Thoc2 mRNA and a more pronounced decrease in the Thoc5 mRNA during differentiation, in parallel with a known ESC marker Oct4 (Figure 1A). We next compared their expression between ESCs and other types of stem cells, epiblast stem cells (EpiSCs) and trophoblast stem cells (TSCs). Thoc2 and Thoc5 were expressed at comparable levels in ESCs and EpiSCs, but were significantly down-regulated in TSCs (Figure S1A, S1B). Consistently, immunofluorescence staining confirmed that they are highly expressed in ESCs, and they largely localize in the nucleus (Figure S1C). Because ESCs and EpiSCs are both pluripotent stem cells while TSCs are not, these results suggest that the expression of Thoc2 and Thoc5 correlates with the pluripotent state, and they may thus be important for the maintenance of pluripotency.
Figure 1. Thoc2 and Thoc5 expression correlates with the pluripotent state of ESCs.

(A) Expression of Thoc2 and Thoc5 during ESC differentiation. Thoc2, Thoc5, and Oct4 expression was determined by RT-qPCR at the indicated time points during ESC differentiation. Differentiation was induced by LIF-withdrawal, retinoic acid (RA) treatment, or embryoid body (EB) formation. Expression was normalized to day-0 by β-actin and plotted as mean ± SEM. (B) Oct4GiP reporter assay: the Oct4GiP reporter ESCs were transfected with non-targeting (NT), luciferase (Luc), Thoc2, or Thoc5 siRNAs, and the reporter activity was determined by fluorescence-activated cell sorting (FACS) analysis 96 hours after transfection. The percentage of differentiation was determined by the percentage of GFP-negative cells and plotted as mean ± SEM. (C) ESC morphology after Thoc2 and Thoc5 silencing. ESCs were transfected with the indicated siRNAs, and cellular morphology was examined 96 hours after transfection. See also Figure S1, S2.
Thoc2 and Thoc5 are Required for ESC Self-renewal
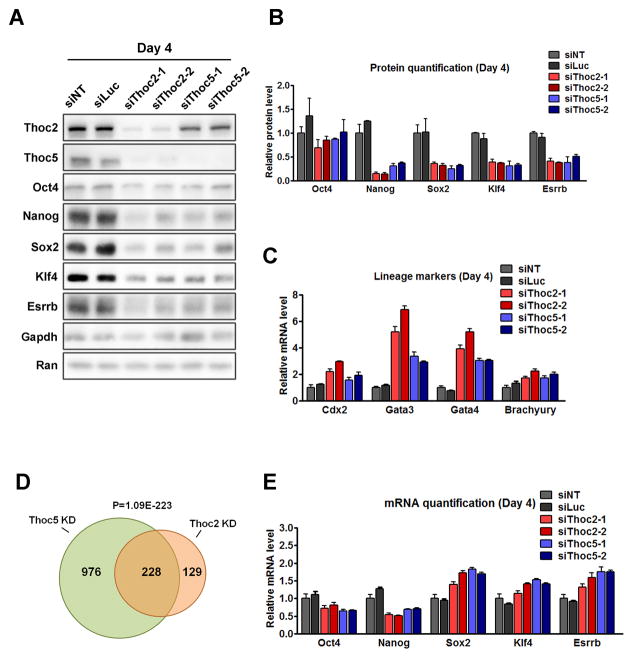
To test the role of Thoc2 and Thoc5 in ESCs, we silenced them with siRNAs. We used two different siRNAs for each gene to reduce the likelihood of off-target effects, and we confirmed that all the siRNAs effectively silenced their targets by RT-qPCR (Figure S1D). Using the Oct4GiP reporter ESCs (Zheng and Hu, 2012), in which the expression of the enhanced green fluorescent protein (EGFP) is driven by the ESC-specific Oct4 promoter (Ying and Smith, 2003), we found that either Thoc2 or Thoc5 silencing led to loss of self-renewal, as determined by the loss of EGFP expression in a significant fraction of the cells (Figure 1B). This result validated previous findings from the RNAi screens and indicated a critical role for THO in self-renewal. Consistently, Thoc2 or Thoc5 depletion by siRNAs, either individually or simultaneously, resulted in loss of typical ESC morphology and alkaline phosphatase (AP) staining (Figure 1C, S1E–G), and similar results were obtained using Thoc2 or Thoc5 shRNAs. (Figure S1H, S1I). In addition, their depletion also led to decreased expression of pluripotency genes such as Oct4, Nanog, Sox2, Esrrb, and Klf4 as determined by western blot (Figure 2A, 2B), as well as increased expression of differentiation markers such as Cdx2, Gata3, Gata4 and Brachyury based on RT-qPCR (Figure 2C). The expression of the differentiation genes was further elevated 6 days after siRNA transfection (Figure S1J).
Figure 2. Thoc2 or Thoc5 depletion results in loss of ESC self-renewal.
(A) Expression of the pluripotency genes after Thoc2 and Thoc5 silencing. Expression was determined by western blot 96 hours after siRNA transfections. GAPDH and RAN were used as loading controls. (B) Quantitation of the pluripotency protein expression based on western blot. Expression was normalized by GAPDH and compared to the NT-siRNA transfected cells, and plotted as mean ± SEM from four independent experiments. (C) Lineage marker expression after Thoc2 and Thoc5 silencing. Expression was determined by RT-qPCR 96 hours after siRNA transfection. Values are plotted as mean ± SEM from three independent experiments. (D) Venn diagram of genes differentially expressed 96 hours after Thoc2 or Thoc5 siRNA transfection based on microarray. (E) Pluripotency gene mRNA levels were determined by RT-qPCR 96 hours after siRNA transfection. Values are plotted as mean ± SEM from three independent experiments. See also Figure S3, Table S1, S2.
We next sought to determine the gene expression changes caused by Thoc2 or Thoc5 knockdown (KD). Microarray analysis revealed that Thoc2 or Thoc5 KD altered the expression of 357 and 1204 genes (Table S1), respectively. This result is consistent with previous reports that deletion of THO components only affect the steady-state level of a fraction of genes in the genome at the transcript level (Gomez-Gonzalez et al., 2011; Guria et al.; Rehwinkel et al., 2004). There was a significant overlap between genes differentially expressed in Thoc2 or Thoc5 KD (228 in common, p = 1.09E-223, Figure 2D), suggesting that they may function as a complex in ESCs. Indeed, reciprocal immunoprecipitation of Thoc2 or Thoc5 showed that they interact with each other in ESCs (Figure S1K). Moreover, immunoprecipitation followed by mass spectrometry (IP-MS) using the HA-tagged Thoc5 as a bait showed that Thoc5 interacted with all the other known components of the THO complex in ESCs (Table S2). Together, these results strongly support the idea that Thoc2 and Thoc5 regulates ESC self-renewal as a complex.
THO forms the TREX complex with several other proteins, such as Alyref and Uap56, to carry out its function in mRNA export (Chang et al., 2013; Katahira, 2012). Therefore, we tested whether Alyref and Uap56 are also involved in self-renewal. We found that they were down-regulated during ESC differentiation (Figure S2A). However, in contrast to Thoc2 and Thoc5, Alyref and Uap56 silencing did not induce significant changes in cell morphology or the expression of pluripotency genes (Figure S2B–D), suggesting that they are not rate-limiting factors in ESC maintenance. Thus, we focused on the THO complex for the rest of our studies in ESCs.
Thoc2 and Thoc5 Depletion Inhibits the Export and Expression of Pluripotency Gene Transcripts
Surprisingly, the microarray analysis (Table S1) showed that the vast majority of pluripotency genes were not significantly down-regulated at the mRNA level and was inconsistent with the western blot result (Figure 2A, 2B). We therefore carried out RT-qPCR for several pluripotency genes. Indeed, we found a clear discrepancy between the mRNA and the protein level upon Thoc2 and Thoc5 depletion for Nanog, Sox2, Esrrb, and Klf4. Despite a large decrease in their protein levels, their mRNA levels did not show changes to the same extent and in some cases were increased (Figure 2A, 2B, 2E). To determine whether the reduction of the pluripotency proteins is likely a direct consequence of Thoc2 and Thoc5 KD, we examined their expression as early as 48 hours after siRNA transfection. Again, we found that Thoc2 and Thoc5 depletion markedly reduced Nanog, Sox2, Esrrb, and Klf4 protein expression, without perturbing their mRNA levels even at this early time point (Figure S3A–G). These results indicate that the THO complex may regulate pluripotency gene expression at the post-transcriptional level.
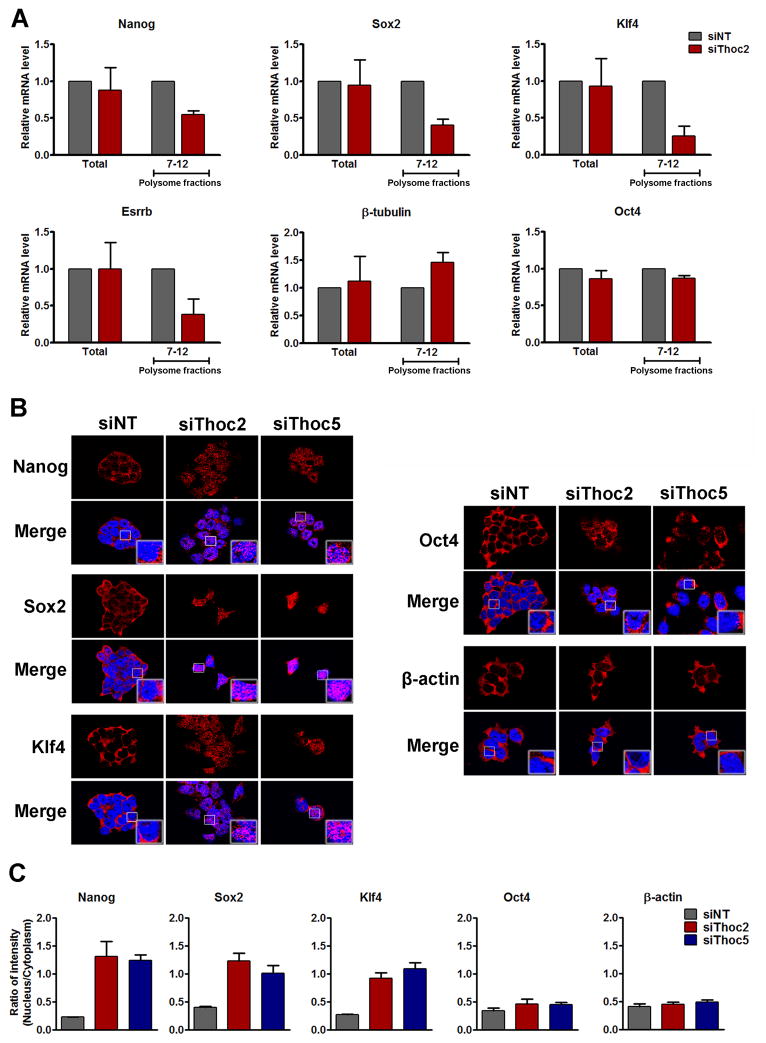
The reduction of the pluripotency gene proteins can be caused either by a reduction in their production or an increase in their degradation. We found that Thoc2 or Thoc5 KD resulted in decreased pluripotency gene protein expression even when protein degradation was inhibited with the proteasome inhibitor MG132 (Figure S3H, S3I), suggesting that THO silencing may lead to lower production of the pluripotency gene products. To further test this idea, we performed polysome fractionation, as polysome association generally correlates with the rate of protein synthesis for a given transcript. Consistent with the above results (Figure 2A, 2B), marked reduction of Nanog, Sox2, Klf4 and Esrrb mRNAs in the polysome fractions was observed upon Thoc2 depletion, whereas Oct4 and β-tubulin mRNAs were not affected (Figure 3A). Thus, THO depletion inhibits the production of those pluripotency genes post-transcriptionally.
Figure 3. Thoc2 or Thoc5 depletion inhibits the export of pluripotency gene mRNAs.
(A) Effect of Thoc2 silencing on polysome-associated RNAs. ESCs were transfected with NT or Thoc2 siRNAs, and cells were harvested 96 hours after transfection. Polysome-associated RNAs were purified by sucrose-gradient ultracentrifugation. Relative abundance of mRNAs in total (Total) or polysome-associated (Fraction 7–12) RNAs was determined by RT-qPCRs, and plotted as mean ± SEM from three independent experiments. (B) Effect of Thoc2 or Thoc5 silencing on mRNA export. ESCs were transfected with NT, Thoc2, or Thoc5 siRNAs. RNA Fluorescence in situ hybridization (RNA-FISH) was carried out for Nanog, Sox2, Klf4, Oct4 and β-actin 96 hours after transfection. Red: fluorescence signals from the FISH probes. Blue: DAPI staining of the nuclei. (C) Ratio of the RNA FISH signals in the nucleus versus cytoplasm was quantified by Metamorph from at least 10 images from three independent experiments. Values are plotted as mean ± SEM. See also Figure S4.
It has been reported that THO complex is responsible for coupling mRNPs biogenesis to export. Therefore, we asked whether depletion of THO impaired pluripotency genes expression by impeding mRNA export. We carried out RNA flurorescence in situ hybridization (RNA-FISH) to examine the subcellular localization of several pluripotency gene transcripts. In agreement with the above results, while Nanog, Sox2, and Klf4 transcripts displayed predominant cytoplasmic localization in the control, they appeared abnormally accumulated in the nucleus following Thoc2 or Thoc5 depletion, with a concomitant reduction in the cytoplasm (Figure 3B, S4). Quantification of the fluorescence intensity ratios between the nucleus and cytoplasm revealed a 2.5- to 6-fold increase after Thoc2 or Thoc5 depletion for these transcripts (Figure 3C). In contrast, transcripts of Oct4 and β-actin did not show nuclear accumulation (Figure 3B, 3C, S4). Collectively, these observations suggest that THO may play an important role in the export and expression of pluripotency gene transcripts to maintain ESC self-renewal.
Thoc2 Interacts with Pluripotency Gene Transcripts in a Thoc5-Dependent Manner
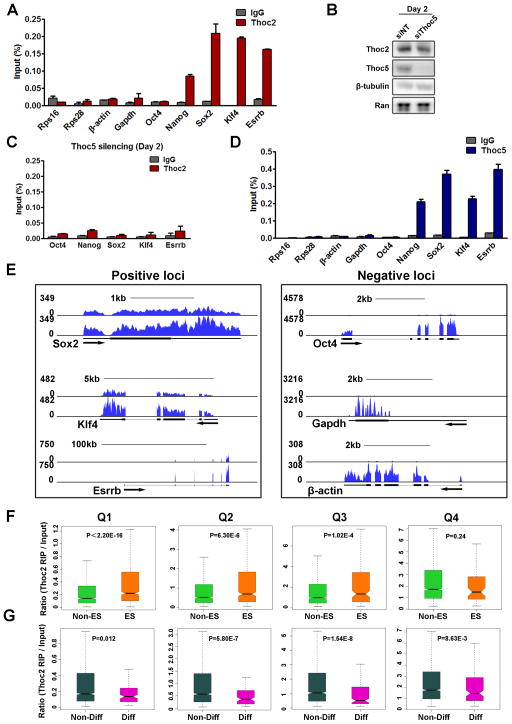
To test if THO regulates mRNA export in ESCs via direct binding, we carried out RNA immunoprecipitation (RIP) (Keene et al., 2006) using an antibody against Thoc2. In line with our previous results, considerable amounts of Nanog, Sox2, Klf4, and Esrrb mRNAs co-immunoprecipitated with Thoc2 (Figure 4A). In comparison, Oct4 transcripts were not pulled down efficiently, consistent with our previous results that Thoc2 and Thoc5 silencing did not dramatically affect its protein expression (Figure 4A). Furthermore, housekeeping gene transcripts, such as β-actin, Gapdh, Rps16, and Rps28, were also not pulled down efficiently (Figure 4A), suggesting that Thoc2 preferentially interacts with a subset of pluripotency gene mRNAs.
Figure 4. Thoc2 preferentially binds to pluripotency gene mRNAs in a Thoc5-dependent manner.
(A) Thoc2 interaction with mRNAs. RIP was performed in ESCs with either IgG or Thoc2 antibody. Relative abundance of the immunoprecipitated mRNAs was determined by RT-qPCRs and normalized to Input, and plotted as mean ± SEM from three independent experiments. (B) Thoc2 protein level 48 hours after Thoc5 silencing. (C) Effect of Thoc5 silencing on Thoc2 RIP. ESCs were transfected with Thoc5 siRNAs, and RIP was carried out 48 hours after transfection. Values are plotted as mean ± SEM. (D) Thoc5 interaction with mRNAs. RIP was performed in ESCs. Relative abundance of the immunoprecipitated mRNAs was determined by RT-qPCRs and normalized to Input, and plotted as mean ± SEM from three independent experiments. (E) Thoc2 RIP-sequencing. Thoc2 RIP was performed in ESCs, and the total and Thoc2-interacting RNAs were subjected to high-throughput sequencing. Representative images from the genome browser were shown for the indicated genes. (F–G) Statistical analysis of Thoc2 RIP-sequencing. Genes detected in the sequencing were ranked by their expression level in ESCs and divided into 4 equal groups (Q1–Q4). Thoc2 binding was determined by the ratio of the reads per kilo base per million (RPKM) in Thoc2 RIP/Input, and was compared between non-ES cell genes (Non-ES) and ES cell genes (ES) or non-differentiation (Non-Diff) and differentiation (Diff) genes in each quarter. ES genes were defined as those down-regulated during ESC differentiation, while differentiation genes were defined as those up-regulated (see methods for details). p-values were calculated by the Wilcoxon rank sum test.
Among all the subunits of THO, Thoc5 has been suggested as an adaptor for mRNA export (Chi et al., 2013; Griaud et al., 2013; Guria et al., 2011; Katahira et al., 2009; Ramachandran et al., 2011; Viphakone et al., 2012). Interestingly, Thoc5 was significantly down-regulated during ESC differentiation (Figure 1A). Therefore, we hypothesized that Thoc5 may be a key factor regulating the interaction between THO and the pluripotency mRNAs. To test this idea, we performed RIP against Thoc2 following Thoc5 depletion. Because Thoc5 silencing can lead to reduced Thoc2 protein level (Figure 2C), we performed Thoc2 RIP-qPCR 48 hours after Thoc5 siRNA transfection. Even at this earlier time point when Thoc2 level did not change significantly (Figure 4B), Thoc5 depletion led to much reduced binding of Thoc2 with the pluripotency mRNAs (Figure 4C). In addition, we performed Thoc5 RIP-qPCR and found that, similar to Thoc2, Thoc5 also interacted with Nanog, Sox2, Esrrb, and Klf4 mRNAs (Figure 4D). Thus, Thoc5 plays a critical role in mediating the interaction between THO and the pluripotency gene transcripts in ESCs. To further test the idea that THO preferentially interacts with a subset of pluripotency gene mRNAs at the global level, we carried out RIP followed by high-throughput sequencing (RIP-seq) for Thoc2 in ESCs.
Consistent with the RT-qPCR results (Figure 4A), significant amounts of the pluripotency gene mRNAs such as Sox2, Esrrb, and Klf4 were recovered in the Thoc2 RIP sample compared to the input, whereas mRNAs including Oct4, Gapdh, and β-Actin were not efficiently co-purified (Figure 4E). More importantly, there was a significant increase in Thoc2 binding with transcripts that are associated with the ESC state in general, as the ratio of reads per kilobase per million (RPKM) of Thoc2-RIP/Input was higher for ESC vs. non-ESC mRNAs (Figure 4F). On the contrary, there was a decrease in Thoc2 binding with transcripts associated with differentiation (Figure 4G). Together, our data suggest that THO may preferentially interact with pluripotency gene mRNAs via Thoc5, and regulates their export and expression to maintain ESC self-renewal.
Thoc5 Regulates the Exit from Self-renewal
Given the importance of THO in self-renewal, we next asked whether it also plays a role during differentiation. Intriguingly, we found that during ESC differentiation induced by LIF withdrawal, RA, or N2B27, pluripotency gene proteins were down-regulated more significantly than their mRNAs (Figure 5A, 5B, S5A–F), suggesting that they may indeed be subjected to post-transcriptional regulation. In addition, Thoc5 protein level decreased promptly at the early stages of differentiation, coinciding with the decrease of the pluripotency proteins (Figure 5A, S5A, S5B). Thoc2 protein level, on the other hand, did not decrease significantly (Figure 5A).
Figure 5. Thoc5 regulates the exit from the pluripotent state.
(A–B) Expression of Thoc5 and pluripotency genes during ESC differentiation induced by LIF withdrawal. Protein expression was determined by western blot (A). mRNA expression was determined by RT-qPCRs (B). Values were plotted as mean ± SEM from three independent experiments. (C) Thoc2 RIP after LIF withdrawal. RIP was carried out 24 hours after LIF withdrawal. Immunoprecipitated mRNAs were quantitated by RT-qPCRs and normalized to Input, and plotted as mean ± SEM from three independent experiments. (D) Effect of Thoc5 overexpression on Thoc2 interaction with the pluripotency gene mRNAs. RIP was carried out in the GFP (GFPOE) or Thoc5 overexpression (Thoc5OE) ESCs 24 hours after LIF withdrawal. Immunoprecipitated mRNAs were quantitated by RT-qPCRs and normalized to the input, and plotted as mean ± SEM from three independent experiments. *: p < 0.05. (E) Effect of Thoc5 overexpression on pluripotency gene expression. Expression of pluripotency gene proteins in GFP (Ind-GFP) or Thoc5 overexpression ESCs (two different lines Ind-Thoc5-1 and Ind-Thoc5-2) at the indicated time points during LIF withdrawal was determined by western blot. (F–H) Effect of Thoc5 overexpression on ESC self-renewal. (F) GFP (Ind-GFP) or Thoc5 (Ind-Thoc5-1, Ind-Thoc5-2) overexpression ESCs were cultured without LIF for the indicated amount of time, and the different types of colonies formed were counted and plotted as percentage of total. Values are plotted as mean from three independent experiments. (G) GFP (GFPOE) or Thoc5 overexpression (Thoc5OE) ESCs were cultured without LIF for the indicated amount of time. The expression of the differentiation markers was determined by RT-qPCRs and plotted as mean ± SEM from three independent experiments. (H) GFP (GFPOE) or Thoc5 (Thoc5OE) overexpression ESCs were cultured without LIF for 48 hours, and then re-plated at clonal density in ESC culture conditions. The number of ESC colonies formed 8 days later was counted and plotted as mean ± SEM from three independent experiments. Representative images of differentiated and undifferentiated colonies are shown at the top. See also Figure S5, S6.
Therefore, we hypothesized that reduced Thoc5 may lead to reduced interaction between THO and the pluripotency gene transcripts, and thus the reduced expression of those genes. To test this hypothesis, we assessed the binding of Thoc2 to pluripotency gene transcripts in ESCs differentiated by LIF withdrawal. We found that the interactions between Thoc2 and Nanog, Sox2, Esrrb, and Klf4 mRNAs were greatly diminished in ESCs cultured without LIF (Figure 5C). Moreover, the interaction between Thoc2 and the pluripotency gene transcripts could be partially restored by Thoc5 overexpression (Figure 5D), supporting our hypothesis.
We further tested whether Thoc5 overexpression could sustain pluripotency gene expression during differentiation. Using inducible ESC lines that express Thoc5 in the presence of doxycycline, we found that Thoc5 overexpression significantly delayed the down-regulation of Nanog, Sox2, and Esrrb proteins, without affecting their mRNAs and without affecting Thoc2 protein levels, during LIF-withdrawal (Figure 5E, S5G). Consistent with the molecular changes, Thoc5-overexpressing ESCs showed less differentiation compared to the control cells based on cellular morphology (Figure 5F, S5H) and lineage marker expression (Figure 5G). When cells were cultured without LIF for two days and then re-plated into ESC medium, Thoc5 overexpressing cells also produced more ESC colonies compared to the control cells (Figure 5H). Further, Thoc5 overexpression was not able to sustain ESC self-renewal in the absence of LIF in prolonged culture (Figure S6A, S6B), suggesting that its function may be limited to the early stage of differentiation. Taken together, these results strongly suggest that decreased expression of Thoc5 is an important step for the quick down-regulation of pluripotency proteins during ESC differentiation. Thus, in addition to its function in ESC maintenance, THO also plays a critical role in controlling the exit from self-renewal.
As EpiSCs are derived from post-implantation embryos and represent a developmental stage immediately after ESCs (Tesar et al., 2007), we examined whether Thoc2 or Thoc5 silencing leads to differentiation into EpiSCs. Based on RT-qPCRs, Thoc2 and Thoc5 silencing did not elicit marker expressions that are consistent with the EpiSC state (Figure S6C) (Tesar et al., 2007). In addition, several pluripotency regulators that distinguish between ESCs and EpiSCs, such as Nanog (Osorno and Chambers, 2011), are expressed heterogeneously in ESCs. On the contrary, Thoc2 and Thoc5 expression were homogeneous in ESCs based on immunofluorescence staining and FACS analylsis (Figure S6D, S6E). Therefore, although THO regulates the early stages of ESC differentiation, it does not seem to regulate the transition from ESCs to EpiSCs under our culture conditions.
THO is Required for Efficient Somatic Cell Reprogramming and Blastocyst Development
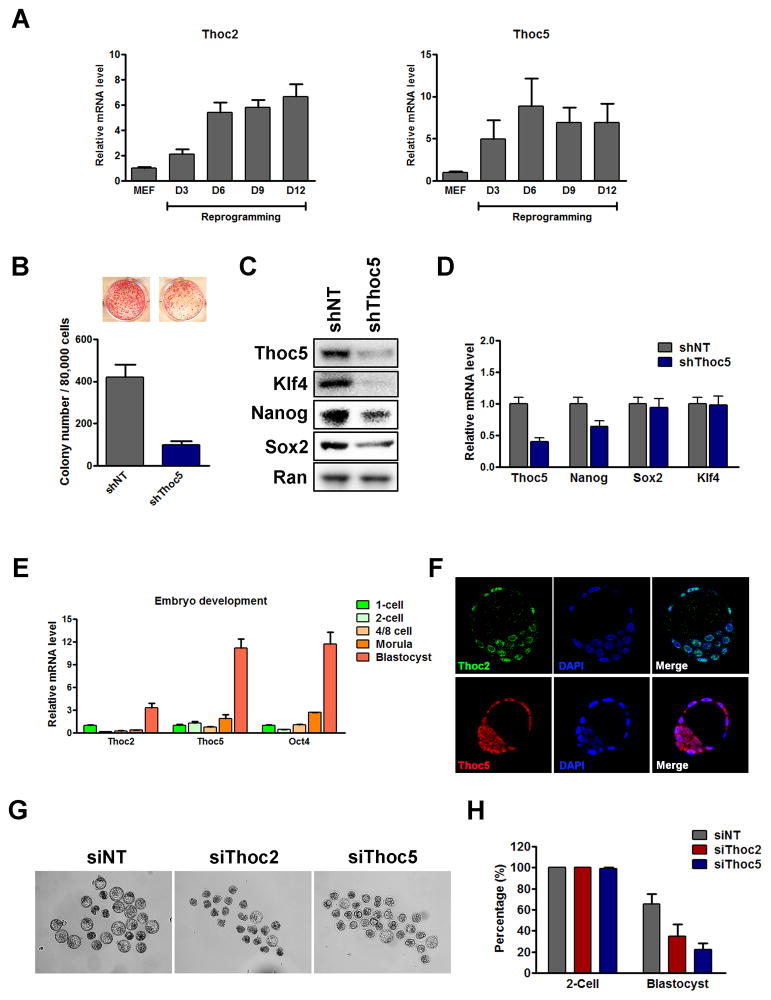
As THO is important in the maintenance of the pluripotent state in ESCs, we asked whether it also functions in the establishment of the pluripotent state. We first tested its role in somatic cell reprogramming. Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) that possess similar characteristics to ESCs (Takahashi and Yamanaka, 2006). We found that Thoc2 and Thoc5 expression increased quickly during the course of reprogramming. Their expression peaked at 6 days after the transduction of the reprogramming factors (Figure 6A), coinciding with the time when the endogenous pluripotency genes started to become activated (Polo et al., 2012). Because Thoc5 is a key determinant in the regulation of pluripotency gene mRNAs by THO, we examined its role in reprogramming by gene silencing (Figure S7A). As complete deletion of Thoc5 impairs the viability of MEFs (Guria et al., 2011), we used a lentivirus-based shRNA to silence Thoc5 so that we could modulate the degree of knockdown. When transduced at lower multiplicity of infection (MOI), Thoc5 silencing by shRNA did not dramatically affect the growth and survival of MEFs (Figure S7A, S7B). However, its silencing significantly reduced the number of alkaline phosphatase-positive iPSC colonies formed during reprogramming (Figure 6B). To unveil possible mechanisms underlying this phenotype, we measured pluripotency gene expression at both the mRNA and protein levels. Nanog, Sox2, and Klf4 proteins were dramatically reduced in Thoc5 KD cells, but their mRNAs showed only modest or no reduction (Figure 6C, 6D). This result is consistent with our findings in ESCs, and suggests that post-transcriptional regulation of pluripotency gene expression by THO may also be important for the establishment of pluripotency in vitro.
Figure 6. THO depletion inhibits reprogramming and blastocyst development.
(A) Thoc2 and Thoc5 expression during reprogramming as determined by RT-qPCRs. Values are plotted as mean ± SEM from three independent experiments. (B) Effect of Thoc5 KD on reprogramming. MEFs were transduced with NT or Thoc5 shRNA viruses, and then transduced with the OKSM viruses to initiate reprogramming. Number of AP-positive colonies was counted 9 days after the start of the reprogramming, and values are plotted as mean ± SEM from four independent experiments. (C-D) The expression of Thoc5 and pluripotency genes was determined at the end of reprogramming. The protein expression was determined by western blot (C). The mRNA expression was determined by RT-qPCRs (D). Values are plotted as mean ± SEM from three independent experiments. (E) Thoc2 and Thoc5 expression during early embryonic development in vivo. mRNA expression was determined by RT-qPCRs and normalized to the 1-cell stage. Values are plotted as mean ± SEM. (F) Immunofluorescence staining of Thoc2 (green) and Thoc5 (red) in E3.5 blastocysts. DAPI (Blue) was used to stain the cell nuclei. (G–H) Effect of Thoc2 or Thoc5 KD on blastocyst development. (G) 1-cell embryos were injected with NT, Thoc2, or Thoc5 siRNAs and cultured for 4 days ex vivo. Images of the embryos were taken on day-4. (H) The number of normal blastocyst formed on day-4 was counted and normalized to the number of normal 2-cell embryos formed on day-2, and plotted as mean ± SEM from three independent experiments. See also Figure S7.
In contrast to Thoc2 and Thoc5, the expression of the TREX components Alyref and Uap56 was only modestly elevated during reprogramming (Figure S7C, S7D). Furthermore, their silencing, even at moderate level, led to significant growth defect in MEFs (Figure S7E–H). Although Alyref and Uap56 silencing resulted in reduced reprogramming efficiency (Figure S7I, S7J), the concomitant defects in MEF growth suggest that they are required for reprogramming because of their roles in general cell proliferation and viability.
During blastocyst development in vivo, ICM cell specification requires the establishment of pluripotency. Therefore, we next asked whether Thoc2 and Thoc5 are involved in the normal blastocyst development. We first examined their expression in pre-implantation embryos. Based on RT-qPCR, both Thoc2 and Thoc5 expression increased at the blastocyst stage compared to 1-cell embryos (3.3- and 11.1-fold, respectively) (Figure 6E). Furthermore, they were both expressed in the ICM as determined by immunofluorescence staining, consistent with their roles in ESCs (Figure 6F). To directly test if they are important for the formation of the blastocyst, we injected siRNAs against Thoc2 or Thoc5 into one-cell embryos, and cultured the embryos ex vivo to allow development to proceed. RT-qPCR showed that the siRNA injection effectively silenced Thoc2 and Thoc5 (Figure S7K). Visual inspection of the embryos indicated that Thoc2 and Thoc5 silencing significantly inhibited blastocyst and ICM formation (Figure 6G, 6H). Interestingly, the effect of Thoc5 silencing was more pronounced, in agreement with our previous finding that Thoc5 is the key component mediating the binding of THO to the pluripotency gene transcripts (Figure 4C). These results suggest that THO is required for blastocyst development ex vivo. Together with our results in ESCs and iPSCs, we propose that THO may be important for ICM specification and the establishment of pluripotency in vivo.
DISCUSSION
In this study, we reveal a novel post-transcriptional regulatory mechanism that controls ESC self-renewal and differentiation. We show that in ESCs the pluripotency gene mRNAs are bound by the THO complex for proper export and expression. During differentiation, these genes are quickly down-regulated at the protein level due to reduced interaction with THO. We propose that by regulating the export and expression of pluripotency genes at the post-transcription level, THO provides an extra layer of control to fine tune the balance between self-renewal and differentiation (Figure 7). Our data further suggest that this regulatory mechanism may also contribute to the establishment of pluripotency during somatic cell reprogramming and ICM specification. However, we cannot exclude the possibility that THO may regulate self-renewal in other ways, such as regulating the export of other mRNAs in addition to the pluripotency gene mRNAs.
Figure 7. THO controls pluripotency gene mRNA export to regulate ESC self-renewal and differentiation.
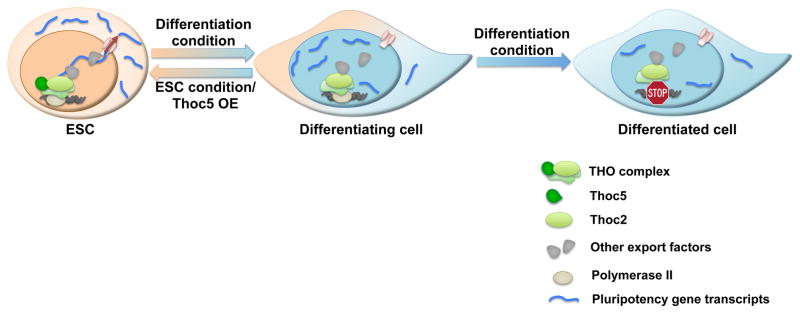
In ESCs, THO is necessary for the efficient export of pluripotency gene mRNAs and the maintenance of self-renewal. Upon differentiation or Thoc5 down-regulation, decreased Thoc5 level results in impaired pluripotency gene mRNA export and expression, leading to differentiation. By post-transcriptionally regulating pluripotency gene expression, THO provides an important layer of control to regulate ESC self-renewal and differentiation.
THO and TREX in pluripotency mRNA export
The THO complex interacts physically and functionally with other export factors, such as Uap56, Alyref, Ddx39, Uif, Tex1, and Chtop, to form the TREX complex (Chang et al., 2013; Luna et al., 2012). These factors play important roles in mediating the interactions between THO and the general export receptor Nxf1 that binds mRNAs and transports them through the nuclear pore complex (Viphakone et al., 2012). Our data show that Alyref and Uap56 may not be the limiting factors in ESC self-renewal. However, TREX components are known to have functional redundancy. For example, Alyref and Uif can bind to the same mRNAs to facilitate their export (Hautbergue et al., 2009). Uap56 and Ddx39 are structurally similar and exert similar functions in yeast (Pryor et al., 2004). Thus, it is possible that THO still functions as part of the TREX complex to regulate the export of pluripotency gene mRNAs, and it will be interesting to determine which TREX component(s) is involved in future studies.
Post-transcriptional Regulation of ESC Self-renewal and Differentiation
ESCs have a sophisticated gene expression program that controls the delicate balance between self-renewal and differentiation. While transcription may function as on-off switches in gene regulation, we show that THO modulates pluripotency gene expression through mRNA export without significantly impacting their mRNA level in ESCs. Indeed, recent studies have begun to reveal the importance of post-transcriptional mechanisms, such as non-coding RNAs, alternative splicing, polyadenylation, and RNA stability, in controlling the ESC state (Buckley et al.; Guttman et al., 2011; Han et al.; Tay et al., 2008; Wang et al., 2008). Thus, we propose that in addition to the transcriptional networks, there exist post-transcriptional modules that act in concert with transcription to control the gene expression network in ESCs.
Our results indicate that THO regulates many pluripotency mRNAs such as Nanog, Sox2, Esrrb, and Klf4. However, Oct4 mRNAs do not show similar dependence on THO. Interestingly, Oct4 is an essential gene for ESC maintenance and changes in its expression lead to irreversible changes in ESC fate (Niwa et al., 2000). Nanog (Chambers et al., 2007), Esrrb (Martello et al., 2012), and Klf4 (Jiang et al., 2008), on the other hand, are important but not required for ESC maintenance. Sox2 is required in mouse ESCs, but its function can be substituted in part by Oct4 (Masui et al., 2007), and low levels of Sox2 are permissive in human ESCs (Wang et al., 2012). Thus, we speculate that by regulating pluripotency genes that are important but maybe not essential for ESC self-renewal, such as Nanog, Sox2, Esrrb, and Klf4, THO may provide a mechanism to regulate ESC fate in a fast and reversible manner.
Compared to the known self-renewal regulatory network, genes involved in the exit from the ESC state are poorly understood (Hu and Wade, 2012). In addition to its role in self-renewal, our data suggest that THO also functions to regulate ESC differentiation and the level of Thoc5 may be a rate-limiting step in differentiation. Intriguingly, we observed that the reduction of some of pluriptency gene proteins occurs with a faster kinetics than that of their mRNAs, suggesting that post-transcriptional regulation may play an important role in ESC fate specification. Together, we propose that by regulating pluripotency gene expression at the level of mRNA export, THO provides a quick and non-committal mechanism to control differentiation and lineage specification.
THO and mRNA Export in the Establishment of Pluripotency
ESCs and ICM cells have been suggested to exist in a similar pluripotent state (Nichols and Smith, 2009). iPSCs are somatic cells reprogrammed into an ESC-like pluripotent state by forced expression of pluripotent factors (Takahashi and Yamanaka, 2006). Many factors that regulate ESC self-renewal and pluripotency play important roles in the establishment of both the embryonic and induced pluripotency. Consistent with that notion, we found that Thoc2 and Thoc5 are required for blastocyst formation and efficient reprogramming of somatic cells. Intriguingly, although Thoc2 and Thoc5 expression are much reduced in TSCs compared to ESCs, they are expressed in the trophectoderm cells in E3.5 blastocysts. It will thus be interesting to determine their expression at later blastocyst stages that are more closely mimicked by ESCs (Wray et al., 2010). Nonetheless, the siRNA injection results are in agreement with the per-implantation lethality phenotypes of Thoc1 and Thoc5 knockout mice (Mancini et al.; Wang et al., 2006). In addition, THO seems to regulate the pluripotency gene expression post-transcriptionally in reprogramming as well. Therefore, THO and mRNA export may be important for both the initiation and maintenance of pluripotency.
THO and mRNA Export in Development
Our data suggest that THO may preferentially interact with pluripotency gene mRNAs and regulate their export and expression in ESCs through Thoc5. It is consistent with previous reports that Thoc5 can act as an adaptor for the export of specific mRNAs (Chi et al., 2013; Griaud et al., 2013; Guria et al., 2011; Katahira et al., 2009; Viphakone et al., 2012), and supports the model that THO may regulate the export of different groups of mRNAs through the recruitment of different adaptors. Indeed, although THO is required for bulk mRNA export in yeast (Strasser et al., 2002), it has been shown to regulate subsets of mRNAs of the heat shock and developmental genes in flies and mice (Guria et al., 2011; Katahira et al., 2009; Rehwinkel et al., 2004). In addition, different subunits of the complex also play different roles in the regulation of bulk or specific subsets of mRNAs (Chi et al., 2013; Katahira et al., 2009; Viphakone et al., 2012). In human diseases, dysregulation of mRNA export and aberrant expression of export factors can alter the expression of specific genes involved in proliferation, survival, and oncogenesis (Culjkovic-Kraljacic and Borden, 2013). However, it is not clear how THO recognizes mRNAs at the molecular level. In yeast THO may be loaded during transcription (Gomez-Gonzalez et al., 2011), while in mammals THO may interact with the 5′-cap (Cheng et al., 2006) or splicing junctions (Masuda et al., 2005) of the transcripts. It will be interesting to continue to search for the mechanisms for the loading of THO and the specificity of the interaction between THO and different mRNAs.
Importantly, our study provides experimental support to the “RNA regulon” hypothesis (Keene, 2007), which suggests that mRNAs encoded by functionally related genes may be coordinately regulated as post-transcriptional RNA regulons by specific mRNP processing machineries. This coordinated regulation can happen at various steps during RNA processing. At the step of the mRNA export, our data support previous findings in yeast where mRNAs in different functional groups were found to associate with different export proteins (Hieronymus and Silver, 2003). We propose that by coordinating multiple mRNAs, THO ensures the efficient yet flexible expression of the pluripotency genes to allow ESCs to quickly respond to developmental cues. Interestingly, THO plays important roles in many aspects of development in mammals, ranging from embryogenesis, hematopoiesis, to testis development. Thus, mRNA export and post-transcriptional gene regulation by THO may be an important mechanism controlling development and cell fate specification.
EXPERIMENTAL PROCEDURES
Mouse ESC culture, differentiation, and transfection
J1 ESCs were obtained from the American Type Culture Collection. Oct4GiP cells were kindly provided by Dr. Austin Smith. ESCs culture and transfections were carried out as described before (Zheng et al., 2012).
Oct4GiP reporter assay
Oct4GiP reporter assay was carried out as described previously (Zheng et al., 2012; Zheng and Hu, 2012). Briefly, cells were transfected with lipids-only, non-targeting, Thoc2, or Thoc5 siRNAs in 96-well plates. Four days after transfection, cells were analyzed by FACS to determine the percentage of GFP-negative cells.
TSC and EpiSC culture
Mouse TSCs and EpiSCs were kindly provided by Dr. Janet Rossant and Dr. Konrad Hochedlinger. They were cultured based on the published protocol without feeders (Rossant, 2006; Tesar et al., 2007).
Antibodies
Primary antibodies used in this study were: Oct4 (Santa Cruz), Nanog (Millipore), Sox2 (Cell signaling), Klf4 (R&D Biosystems), Esrrb (Perseus Proteomics), Thoc2 (Abcam), Thoc5 (Bethyl), β-Tubulin (Santa Cruz), GAPDH (Santa Cruz), Ran (BD Biosciences), Hemagglutinin (HA) (Covance). RAN, GAPDH, Tubulin, were used for loading controls.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 0.5% BSA in PBS. They were then incubated with the primary antibodies: Thoc2 (1:100, Abcam), Thoc5 (1:100, Bethyl), Cdx2 (1:100, Cell Signaling) or Oct4 (1:100, Santa Cruz) at 4°C overnight. The next day, cells were incubated with the appropriate secondary antibodies (Life Technologies). Nuclei were counterstained with DAPI (Invitrogen), and confocal images were taken on the Zeiss LSM 710 microscope.
For embryo staining, mouse blastocyst stage embryos were collected at E3.5 dpc. They were fixed in 4% paraformaldehyde, permeabilized with 0.4% Triton X-100 in PBS, and blocked in 5% goat serum in PBS. They were then incubated with antibodies against Thoc2 (1:300) or Thoc5 (1:300) overnight at 4°C. The next day, embryos were washed and incubated with the appropriate secondary antibodies (Alexa Fluor® 488 or 594, goat anti-rabbit IgG, 1:1000, Life Technologies). The embryos were washed and stained with DAPI to identify cell nuclei. Confocal images were taken on the Zeiss LSM 710 microscope.
Microarray analysis
J1 ESCs were transfected with siRNAs as described above in duplicates, and were collected 96 hours after transfection. Gene expression analysis was carried out on the Agilent Whole Mouse Genome 4 × 44k arrays following the Agilent 1-color microarray-based gene expression analysis protocol.
Thoc5 overexpression ESCs
The mouse Thoc5 coding region was PCR cloned into the pHAGE-EF1a or pHAGE-Inducible destination expression vectors (see attached maps in supplemental information) using the Gateway Technology (Life Technologies). The resulting Thoc5 expression vectors were packaged into viruses. J1 ESCs were transduced with the packaged virues and drug-selected, and clonal lines were established.
Immunoprecipitation (IP) and mass spectrometry (MS)
For Thoc2 or Thoc5 immunoprecipitation (IP), J1 cells were lysed in lysis buffer (1% NP-40, 50mM Tris-Hcl pH 8.0, 150 mM NaCl, 10mM NaF, 1mM Na3VO4, phenylmethanesulfonylfluoride, and Roche EDTA-free Protease inhibitors). Lysates were sonicated and centrifuged to remove insoluble materials. Lysates were then incubated at 4°C with protein-A agarose beads for pre-clearing. The cleared lysates were divided into identical portions and incubated with IgG (Santa Cruz), Thoc2 (Abcam) or Thoc5 (Bethyl) antibodies at 4°C overnight. The next day, beads were washed and bound proteins were eluted with 2 × SDS-PAGE sample buffer.
For IP-MS, Thoc5-HA overexpression J1 cells were lysed and prepared as described above, and the exogenous Thoc5-HA was immunoprecipitated with the HA-antibody (Roche). The precipitated proteins were separted by SDS-PAGE and subjected to MS analysis.
Polysome fractionation and RNA fluorescence in situ hybridization (RNA-FISH)
Polysome fractionation was conducted as described in the published protocol (Sampath et al., 2011). RNA-FISH was performed following the instructions provided in the FISH Tag™ RNA Multicolor Kit (Life Technologies).
RNA immunoprecipitation, sequencing and analysis
RNA immunoprecipitation (RIP) was carried out using the Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to manufacturer’s instructions. The eluted RNAs were precipitated, quantified, and subjected to RT-qPCR or library generation for sequencing. RNA sequencing libraries were prepared with the TruSeq™ RNA Sample Preparation Kit (Illumina). The resulting libraries were sequenced on the MiSeq (Illumina) and the sequencing results were submitted to the GEO database (GSE48436).
Reads from both the Thoc2-RIP and Input samples were aligned using Bowtie 0.12.8 to an index of RefSeq transcripts. Up to ten alignments per read were allowed, with up to two mismatches. FPKM values were then calculated for each RefSeq transcript. For Figure 4D, 10,082 genes with sufficient sequencing coverage (total mapped reads > 5, RPKM > 1) were used for the analysis. They were grouped into 4 equal quartiles based on their input expression level: Q1 (High expression), Q2 (High-medium expression), Q3 (Medium-low expression), Q4 (Low expression). In each quartile, the RPKM ratio of Thoc2 RIP/Input for each ESC and non-ESC gene was calculated, and Wilcoxon rank-sum test was applied to assess the binding preference of Thoc2 between ESC and Non-ESC genes. Similarly, the RPKM ratio of Thoc2 RIP/Input for each differentiation and non-differentiation genes was calculated as well. ESC genes were defined as those showing at least 1.5 fold down-regulation during ESC differentiation into EB (at any time point up to day-9), based on the microarray from Stembase experiment 201 (GSE3749). Differentiation genes were defined as those showing at least 1.5 fold up-regulation during the same differentiation time course.
Mouse embryo microinjection
Microinjections were performed using a Leica DMI 6000B inverted microscope equipped with a XenoWorks Micromanipulator system and a PrimeTech PMM-150FU Piezo drill (Sutter Instruments). Thoc2, or Thoc5 siRNA was injected into the cytoplasm of 1-cell embryos, and the embryos were cultured in KSOM (Millipore) and inspected at indicated time points to determine developmental progress.
Reprogramming
MEFs were plated in 12-well plates at 2 × 105/well (day-0), and transduced first with the non-targeting, Thoc5, Alyref, or Uap56 shRNA viruses, and then with viruses encoding the Yamanaka factors. iPSC colony number was counted by AP staining on day-14.
Supplementary Material
HIGHLIGHTS.
THO is required for embryonic stem cell (ESC) self-renewal.
THO binds to pluripotency gene transcripts and controls their export through Thoc5.
Thoc5 level is a key determinant of ESC differentiation kinetics.
THO is required for somatic cell reprogramming and mouse blastocyst development.
Acknowledgments
We thank Drs. Karen Adelman, Perry Blackshear, Traci Hall, Raja Jothi, and Paul Wade for critically reading the manuscript. This study was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences 1ZIAES102745 (to L. W., X. Z., B. L., G. H.) and 1ZIAES102985 (to Y. M., C. W.), and NIH R01GM090056 and American Cancer Society Grant RSG-12-186 (to C. Y., Y. S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell stem cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chang CT, Hautbergue GM, Walsh MJ, Viphakone N, van Dijk TB, Philipsen S, Wilson SA. Chtop is a component of the dynamic TREX mRNA export complex. The EMBO journal. 2013;32:473–486. doi: 10.1038/emboj.2012.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Chi B, Wang Q, Wu G, Tan M, Wang L, Shi M, Chang X, Cheng H. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic acids research. 2013;41:1294–1306. doi: 10.1093/nar/gks1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Culjkovic-Kraljacic B, Borden KL. Aiding and abetting cancer: mRNA export and the nuclear pore. Trends in cell biology. 2013;23:328–335. doi: 10.1016/j.tcb.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M, Zwaka TP. Pluripotency and nuclear reprogramming. Annu Rev Biochem. 2012;81:737–765. doi: 10.1146/annurev-biochem-052709-104948. [DOI] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell stem cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Evans M. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol. 2011;12:680–686. doi: 10.1038/nrm3190. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Garcia-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marin A, Foiani M, Aguilera A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. The EMBO journal. 2011;30:3106–3119. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griaud F, Pierce A, Gonzalez Sanchez MB, Scott M, Abraham SA, Holyoake TL, Tran DD, Tamura T, Whetton AD. A pathway from leukemogenic oncogenes and stem cell chemokines to RNA processing via THOC5. Leukemia. 2013;27:932–940. doi: 10.1038/leu.2012.283. [DOI] [PubMed] [Google Scholar]
- Guria A, Tran DD, Ramachandran S, Koch A, El Bounkari O, Dutta P, Hauser H, Tamura T. Identification of mRNAs that are spliced but not exported to the cytoplasm in the absence of THOC5 in mouse embryo fibroblasts. RNA. 2011;17:1048–1056. doi: 10.1261/rna.2607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Walsh MJ, Snijders AP, Chang CT, Jones R, Ponting CP, Dickman MJ, Wilson SA. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Current biology: CB. 2009;19:1918–1924. doi: 10.1016/j.cub.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nature genetics. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- Hu G, Wade PA. NuRD and pluripotency: a complex balancing act. Cell stem cell. 2012;10:497–503. doi: 10.1016/j.stem.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Nostro MC, Kattman SJ, Keller GM. Directed differentiation of pluripotent stem cells: from developmental biology to therapeutic applications. Cold Spring Harbor symposia on quantitative biology. 2008;73:101–110. doi: 10.1101/sqb.2008.73.065. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nature cell biology. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Jimeno S, Aguilera A. The THO complex as a key mRNP biogenesis factor in development and cell differentiation. J Biol. 2010;9:6. doi: 10.1186/jbiol217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J. mRNA export and the TREX complex. Biochimica et biophysica acta. 2012;1819:507–513. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Katahira J, Inoue H, Hurt E, Yoneda Y. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. The EMBO journal. 2009;28:556–567. doi: 10.1038/emboj.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nature reviews Genetics. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nature protocols. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R, Rondon AG, Aguilera A. New clues to understand the role of THO and other functionally related factors in mRNP biogenesis. Biochimica et biophysica acta. 2012;1819:514–520. doi: 10.1016/j.bbagrm.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Mancini A, Niemann-Seyde SC, Pankow R, El Bounkari O, Klebba-Farber S, Koch A, Jaworska E, Spooncer E, Gruber AD, Whetton AD, et al. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol. 2010;8:1. doi: 10.1186/1741-7007-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Gottgens B, Niwa H, Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell stem cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes & development. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nature cell biology. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nature cell biology. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell stem cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature genetics. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Osorno R, Chambers I. Transcription factor heterogeneity and epiblast pluripotency. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2011;366:2230–2237. doi: 10.1098/rstb.2011.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena A, Gewartowski K, Mroczek S, Cuellar J, Szykowska A, Prokop A, Czarnocki-Cieciura M, Piwowarski J, Tous C, Aguilera A, et al. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. The EMBO journal. 2012;31:1605–1616. doi: 10.1038/emboj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. The EMBO journal. 1998;17:4859–4872. doi: 10.1093/emboj/17.16.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor A, Tung L, Yang Z, Kapadia F, Chang TH, Johnson LF. Growth-regulated expression and G0-specific turnover of the mRNA that encodes URH49, a mammalian DExH/D box protein that is highly related to the mRNA export protein UAP56. Nucleic acids research. 2004;32:1857–1865. doi: 10.1093/nar/gkh347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Tran DD, Klebba-Faerber S, Kardinal C, Whetton AD, Tamura T. An ataxia-telangiectasia-mutated (ATM) kinase mediated response to DNA damage down-regulates the mRNA-binding potential of THOC5. RNA. 2011;17:1957–1966. doi: 10.1261/rna.2820911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- Rondon AG, Jimeno S, Aguilera A. The interface between transcription and mRNP export: from THO to THSC/TREX-2. Biochimica et biophysica acta. 2010;1799:533–538. doi: 10.1016/j.bbagrm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Rossant J. Culturing Trophoblast Stem (TS) Cell Lines. CSH protocols. 2006;2006 doi: 10.1101/pdb.prot4406. [DOI] [PubMed] [Google Scholar]
- Sampath P, Lee QY, Tanavde V. Identifying translationally regulated genes during stem cell differentiation. Current protocols in stem cell biology. 2011;Chapter 1(Unit1B):8. doi: 10.1002/9780470151808.sc01b08s18. [DOI] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, Wilson SA. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun. 2012;3:1006. doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chang Y, Li Y, Zhang X, Goodrich DW. Thoc1/Hpr1/p84 is essential for early embryonic development in the mouse. Mol Cell Biol. 2006;26:4362–4367. doi: 10.1128/MCB.02163-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chinnam M, Wang J, Wang Y, Zhang X, Marcon E, Moens P, Goodrich DW. Thoc1 deficiency compromises gene expression necessary for normal testis development in the mouse. Mol Cell Biol. 2009;29:2794–2803. doi: 10.1128/MCB.01633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nature genetics. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell stem cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Wray J, Kalkan T, Smith AG. The ground state of pluripotency. Biochem Soc Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- Ying QL, Smith AG. Defined conditions for neural commitment and differentiation. Methods in enzymology. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Dumitru R, Lackford BL, Freudenberg JM, Singh AP, Archer TK, Jothi R, Hu G. Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem cells. 2012;30:910–922. doi: 10.1002/stem.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hu G. Oct4GiP reporter assay to study genes that regulate mouse embryonic stem cell maintenance and self-renewal. Journal of visualized experiments: JoVE. 2012 doi: 10.3791/3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.