Abstract
Molecular diagnostics is critical for prevention, identification, and treatment of disease. Traditional technologies for molecular diagnostics using blood are limited to laboratory use because they rely on sample purification and sophisticated instruments, are labor- and time-intensive and expensive, and require highly trained operators. This review discusses the frontiers of point-of-care diagnostic technologies using a drop of blood obtained from a finger-prick. These technologies, including emerging biotechnologies, nanotechnologies, and microfluidics, hold the potential for rapid, accurate, and inexpensive disease diagnostics.
Keywords: Point-of-care diagnostics, molecular diagnostics, proteins, nucleic acids, biomolecules, circulating tumor cells
Blood as a target for molecular diagnostics
Blood is a bodily fluid that contains abundant information about the health status of the individual. The average human adult has a blood volume of approximately five liters continuously circulating throughout the body to deliver necessary nutrients and transport metabolic waste [1]. Blood consists of 54.3% plasma, 45% red blood cells (RBCs), and 0.7% white blood cells (WBCs) by volume [2]. Plasma is composed of proteins, nucleic acids, and nutrients or waste products, and it maintains electrolyte balance and protects the body from infection and blood disorders [3–5]. Serum is produced by removal of blood-clotting factors from plasma [6] and is the main source of samples used in blood-based molecular diagnostics. Because levels of molecular constituents in blood are directly associated with the physiological state of the body, detection of these molecules in serum is often used for prevention, identification, and treatment selection for a variety of diseases.
Traditional technologies for molecular diagnostics in blood include enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR), and mass spectrometry (MS) [7]. However, these technologies are limited to laboratory use because they rely on sample purification and sophisticated instruments, are time- and labor-intensive and expensive, and require highly trained operators. In addition, the sensitivity of some of these technologies is unsatisfactory for detecting trace levels of biomarkers. Therefore, there is still a great challenge to develop simple, inexpensive, rapid, and easy-to-use technologies for point-of-care (POC) blood molecular diagnostics. A typical POC assay is affordable, specific, sensitive, portable, rapid, and user-friendly, which also makes it suitable for use in low-resource settings [8, 9]. The first true POC device was the urine dipstick, which was developed in 1957 to measure urinary protein [8]. Whereas glucose meters and lateral flow devices are currently the most widely used devices in POC blood molecular diagnostics, they are not applicable if highly quantitative, sensitive, and high-throughput measurements are required. Emerging technologies, including biotechnologies, nanotechnologies, and microfluidics, hold the promise to improve the capabilities for future POC disease diagnostics [10–13].
In this review, we discuss recent developments in new technologies for molecular diagnostics using a drop of blood obtained from a finger prick. Technological developments for low-volume blood diagnostics may facilitate rapid, accurate, and inexpensive diagnosis of disease in the hospital clinic or self-monitoring at home. Taking blood from a finger prick is relatively painless, and it is suitable for POC and pediatric disease diagnostics because of the small samples required. Here, we provide a survey of applicable new technologies for measuring proteins, nucleic acids, and other molecules (e.g., hormones, metabolites, and drugs) as well as downstream molecular analyses based on cancer cells isolated from the blood. We discuss the advantages and disadvantages of each method (table 1).
Table 1.
List of POC platforms for molecular diagnostics
| Platform | Affordable | Specific | Sensitive | Portable | Rapid | Multiplex | Quantitative | User-friendly | Refs. |
|---|---|---|---|---|---|---|---|---|---|
| Proteins | |||||||||
| ELISA-based methods | |||||||||
| LFA | +++ | +++ | +++ | +++ | +++ | [18] | |||
| mChip | ++ | +++ | + | ++ | +++ | + | +++ | [19] | |
| Digital ELISA | ++ | +++ | ++ | [20] | |||||
| Plasmonic ELISA | ++ | ++ | +++ | + | ++ | [21] | |||
| Silicon nanoribbon chip | + | +++ | +++ | + | ++ | ++ | + | [22] | |
| GMR sensor | +++ | +++ | ++ | +++ | ++ | [23] | |||
| IBBC | +++ | ++ | +++ | ++ | [24, 25] | ||||
| P-ELISA | +++ | +++ | + | +++ | + | +++ | ++ | ++ | [26–28] |
| V-Chip | +++ | +++ | ++ | +++ | +++ | +++ | ++ | [29] | |
| Non-ELISA–based methods | |||||||||
| MPS | ++ | +++ | ++ | + | [32] | ||||
| GFP-AuNPs | ++ | +++ | ++ | [31] | |||||
| AuNPs (colorimetric) | +++ | ++ | + | ++ | +++ | + | ++ | +++ | [33] |
| AuNPs (DLS) | ++ | ++ | +++ | +++ | ++ | ++ | [34, 35] | ||
| Nucleic acids | |||||||||
| DNA | |||||||||
| μCICS | ++ | +++ | ++ | ++ | + | [44] | |||
| TAm-Seq | ++ | ++ | ++ | + | [45] | ||||
| CCP | + | ++ | +++ | ++ | ++ | ++ | [46] | ||
| RNA | |||||||||
| HPD-SENS | + | +++ | ++ | + | ++ | ++ | [48] | ||
| EMRS | + | ++ | +++ | + | + | ++ | [50] | ||
| Nanopore sensor | ++ | +++ | ++ | ++ | [51] | ||||
| Other types of biomolecules | |||||||||
| Graphene glucose sensor | +++ | ++ | ++ | ++ | ++ | ++ | [52] | ||
| PGM-aptamer sensor | ++ | + | ++ | +++ | + | +++ | + | [56] | |
| DMF | + | +++ | + | [57] | |||||
| Amperometric sensor | + | ++ | ++ | + | + | ++ | ++ | [60] | |
| CTCs | |||||||||
| Immunomagnetic assay | ++ | ++ | + | + | ++ | + | ++ | [66] | |
| Immunoassay chip | + | ++ | ++ | + | + | + | [67–70] | ||
| Size-based microchip | + | + | + | [71] | |||||
| Dielectric separation | + | + | + | + | [73] | ||||
(+++), high; (++), intermediate; (+), low. Data for blank cells are not traceable through literature search.
Detection of proteins
Proteins are well known to be required for numerous biological functions and processes, ranging from enzymatic reactions to hormone synthesis, maintenance of metabolic equilibrium, and tissue repair.[14] For clinical applications, levels of certain protein biomarkers directly reflect disease stages and have been regarded as one of the most convenient clinical sources for disease diagnosis. Blood contains more than 20,000 different proteins, with concentrations ranging from <1 ng×L−1 (troponin) [15] to 50 g×L−1 (serum albumin) [16, 17]. Thus, there are abundant blood proteins available as candidate biomarkers for disease detection.
ELISA-based methods
Currently, most methods for blood protein analysis are based on ELISA, which serves as the clinical gold standard. In traditional ELISA methods, colorimetric or fluorescent readout signals are used to visualize the binding of a protein to a specific recognition molecule [12]. Despite the development of numerous new ELISA-based technologies for protein detection, many challenges remain to their application in POC diagnostics. Among these are improvements to increase sensitivity, multiplicity, quantification, portability, speed of operation, clarity of readout, and reduce cost.
The traditional ELISA requires repeated washing steps, which makes the method time-consuming and cumbersome. The lateral flow assay (LFA) or immunochromatographic assay, originally introduced in 1987, is considered the most successful commercial technology that overcomes these limitations [18]. This technology combines the principles of thin-layer paper chromatography and ELISA, allowing rapid separation of plasma components in a drop of blood in a few minutes. Recently, a new technology was developed that integrates fluid handling and silver reduction in a microfluidic chip (mChip) and can simplify ELISA. Diagnosis of human immunodeficiency virus (HIV) based on this device requires minimal equipment, analysis can be completed within 20 min, and it requires as little as 1 μL of blood [19]. Although these devices are simple to use, these technologies still exhibit many limitations, such as low sensitivity, results that are only semi-quantitative, and low-throughput.
The ability to measure low concentrations of disease biomarkers can improve the standard of care in resource-limited areas. Sensitivity can be improved by reducing the ELISA reaction volume to ensure a high concentration of fluorescent substrate [20]. By confining the fluorophore-generation reaction to 50 fL, a digital ELISA method was developed to detect proteins in serum at subfemtomolar concentrations. An alternative method to improve sensitivity is to introduce new signal amplification approaches into ELISA. Controlling the growth of gold nanoparticles (AuNPs) by catalase results in a color change, and its incorporation into an ELISA method (plasmonic ELISA) results in ultrasensitive detection of blood proteins [21]. The detection limit in serum for spiked prostate-specific antigen and HIV type 1 (HIV-1) capsid antigen (p24), which is observable by a color change, is as low as 10 18 g×mL−1. Other efficient methods for highly sensitive detection are based on reducing the background signal. The microfluidic purification chip is able to capture protein biomarkers from blood and then release them into buffer for sensing based on silicon nanoribbon detectors [22]. Magnetic signal-based ELISA platforms (GMR sensors) avoid detectable magnetic background signals, produce low noise signal [23], and can detect attomolar concentrations of proteins in serum over a range of more than six orders of magnitude. However, these technologies are time-consuming to operate and require sophisticated instruments to read the results.
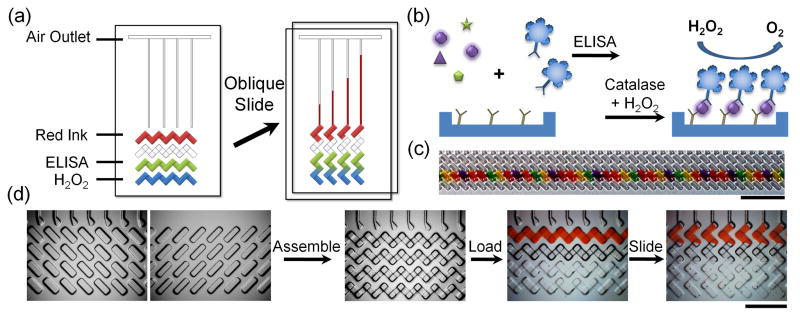
Clinical research has proven that multiprotein quantitative measurements provide more accurate diagnostic results. Microfluidic chips fabricated using photolithography show powerful applications in multiplex protein assays [12]. The recently introduced integrated blood barcode chip (IBBC) employs microscopic fluorescent barcodes as reporter signals for multiplex assays of blood [24, 25]. Paper-based ELISA (P-ELISA) has also been used to assay liver injury-related enzymes in human blood [26–28]. However, quantitative measurement still requires sophisticated instruments. The multiplexed volumetric bar-chart chip (V-Chip) integrates ELISA reactions with volumetric measurements of oxygen generated on a microfluidic chip (Figure 1), allowing instant and visual quantitation of biomarkers [29]. In addition, the V-Chip generates the visible bar chart without additional instrumentation, data processing, or graphic plotting. After a simple oblique sliding of the upper plate over the lower, the advance of ink in each individual channel indicates the amount of catalase that reacted in that well, which correlates with the concentration of the corresponding ELISA target protein. The V-Chip approach enables multiplex, quantitative measurement of target protein biomarkers. The sensitivity could be amplified by depositing multistage uniform platinum films in the device [30]. In the future, the technology may show wide application by integrating sample pretreatment in the device.
Figure 1.
V-Chip for detection of protein biomarkers. (a) Schematic design of the V-Chip. The left image shows assembled V-Chip, preloaded with ink and H2O2; ELISA is performed in the indicated lanes. Oblique sliding (arrow) of the upper plate across the lower plate interrupts the horizontal flow paths, forming isolated vertical channels in which catalase and H2O2 react to generate oxygen and propel ink through the channels. (b) Enzyme-linked immunosorbent assay (ELISA) reaction and mechanism of oxygen generation. (c) V-Chip can be loaded with 50 different antibodies using swab tips. Scale bar, 0.5 cm. (d) High-magnification microscopic images of typical operating steps in a 50-plex V-Chip. Scale bar, 2.5 mm. Reproduced with permission from Ref. [29].
Non-ELISA-based methods
The traditional clinical method for total serum analysis is electrophoresis combined with MS [31]. However, sensitivity and quantification remain issues for clinical applications. Effort is still needed to develop new optimal technologies for overall serum analysis. Thin films of mesoporous silica (MPS) were found to enhance enrichment of the circulating low molecular weight proteome (LMWP) from serum. The captured LMWP is then detected and analyzed by MS [32]. Nevertheless, MS is too expensive for use in POC diagnostics. In addition to MS, a new sensing platform relying on electrostatic interactions between green fluorescent protein (GFP) and AuNPs has recently been developed for sensing proteins in serum [31]. The GFP-AuNP conjugate mimics protein-protein surface interactions, and this is instrumental in reaching detection limits sufficiently low to measure biomedically relevant changes in protein concentration in undiluted human serum. Antibody-labeled AuNPs will aggregate in the presence of protein targets, which exhibits colorimetric or light scattering signals [33–35]. Colorimetric detection methods show promise for use in POC diagnostics because the signal is detectable by the naked eye, whereas dynamic light scattering (DLS) is able to detect very low concentrations of biomarker proteins. These assays are rapid and do not require any washing steps. However, direct analysis of proteins in blood requires further improvement and optimization.
Detection of nucleic acids
Nucleic acids are the intracellular carriers of genetic information [36–39] and are also found in small amounts in healthy and diseased human serum [40]. These circulating nucleic acid molecules are thought to be released from dying cells as they break down [41, 42], and their extraction and analysis from a drop of blood provides a relatively noninvasive, highly patient-compliant method for detection of genetic disease states. Current methods for detection of nucleic acids are mainly based on PCR, which limits use to the laboratory, and they are still relatively expensive and cumbersome. Although microfluidic devices provide the opportunity for rapid purification of nucleic acids from blood [43], a great challenge remains in integrating blood pretreatment with nucleic acid detection in an inexpensive, robust, and user-friendly format.
Measurement of DNA
Circulating DNA may arise from apoptotic or necrotic cells and is regarded as a noninvasive biomarker for a diverse array of human diseases. Analysis of fragment size shows promise for identification of DNA source in cancer and prenatal diagnostics. A microfluidic single-molecule spectroscopy technique (micro-cylindrical illumination confocal spectroscopy, μCICS) has been developed to directly analyze DNA biomarkers in serum [44]. This new technology allows for one-step analysis and quantification of circulating DNA in volumes smaller than 1 pL of serum with no additional steps required for DNA isolation or enzymatic amplification.
Identification of DNA mutations using serum samples is another important aspect for cancer therapy, as mutations can modulate the effectiveness of biological reagents on target-specific pathways [45]. Circulating DNA extracted from the serum of cancer patients always contains tumor-specific mutations. A new method to amplify and sequence large genomic regions from as little as a single copy of circulating DNA, tagged-amplicon deep sequencing (TAm-Seq) has been developed. It was found that the high sensitivity and specificity of TAm-Seq enables detection of mutation rates as low as 2% when 5,995 nucleotides were screened.
Methylated DNA is another promising biomarker for early detection of cancer and other genetic diseases. Combinations of changes in methylation show stronger associations with specific cancers than single alterations. Recently, a fluorescence resonance energy transfer method based on a water-soluble cationic conjugated polymer (CCP) was used to quantitate cellular DNA methylation levels in colon cancer patients [46]. This method exhibited high accuracy and sensitivity in discriminant analysis and cumulative detection. In the future, this technology may be extended to detect methylation of circulating DNA.
Measurement of RNA molecules
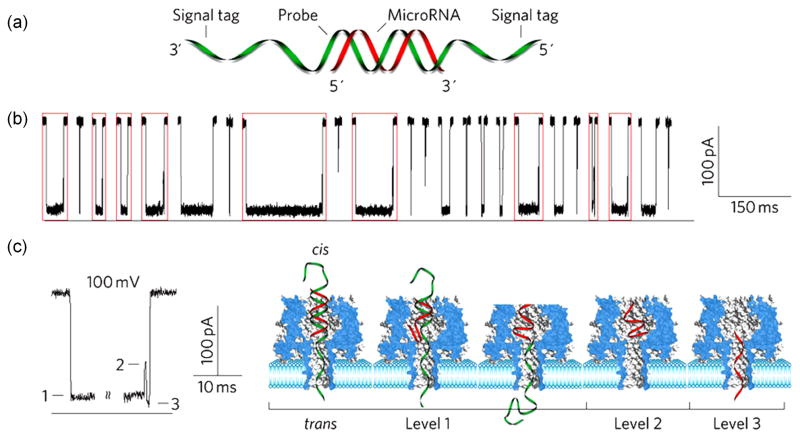
MicroRNAs (miRNAs) are small (approximately 22 nucleotides) regulatory RNA molecules that are frequently dysregulated in cancer and are promising candidates for biomarkers for cancer classification and prognosis [47]. Detection of miRNA in blood with high (at least femtomolar) sensitivity remains a challenge. Recently, a three-mode electrochemical sensor (HPD-SENS) with low detection limit and wide dynamic range was developed for quantitative detection of miRNA. The sensor can detect as little as 5 aM miRNA, with a dynamic range of 10 aM to 1 μM [48]. Nanotechnology-based methods have been employed to design highly sensitive sensors for miRNA detection [49]. DNA nanostructures can improve the sensitivity of an electrochemical miRNA sensor (EMRS), which is able to detect miRNAs at concentrations as low as attomolar [50]. In addition, a nanopore sensor based on the protein α-hemolysin was developed for detection of miRNAs in samples of plasma from lung cancer patients (Figure 2). This nanopore-based miRNA sensor can quantitate cancer-associated miRNAs at subpicomolar concentrations, as well as distinguish single-nucleotide differences [51].
Figure 2.
Nanopore-based sensor for miRNA analysis. (a) Schematic diagram of miRNA and probe used in nanopore sensor. (b) Signal for detection of miR-155. Red boxes show multilevel current pattern. (c) Signals (left) and corresponding molecular mechanism (right) of miR-155– probe hybrid dissociation and translocation in the nanopore cavity. Reproduced with permission from Ref. [51].
Detection of other types of biomolecules
Other types of biomolecules in blood come from metabolic processes or drug use. The levels of metabolites, including hormones and blood chemicals, are often indicators of disease, whereas the presence of addictive drugs, such as cocaine, in the blood, is often monitored to detect and prevent drug abuse and illicit trafficking [8]. Because of small size or similar chemical structure, commercially available antibodies recognizing these molecules always exhibit high crossreactivity in immunoassays. Therefore, existing methods for detection of small molecules often lack sufficient sensitivity or specificity.
Measurement of blood glucose is critical to maintaining the normal physiological range in controlling diabetes mellitus. The first glucose sensor for home use was marketed in 1981. Current glucose meters rely primarily on measuring the electrochemical signals [8]. Additional colorimetric glucose sensors based on the intrinsic peroxidase-like activity of graphene have also been developed [52]. Measurement of blood glucose using these devices is simple, inexpensive, and highly sensitive and selective. Other carbon nanomaterials, such as carbon nanotubes or graphene-hemin complexes, also exihibit peroxidase-like activity and could be used for detection of copper ions, DNA, lysozyme, and cancer cells [29, 53–55].
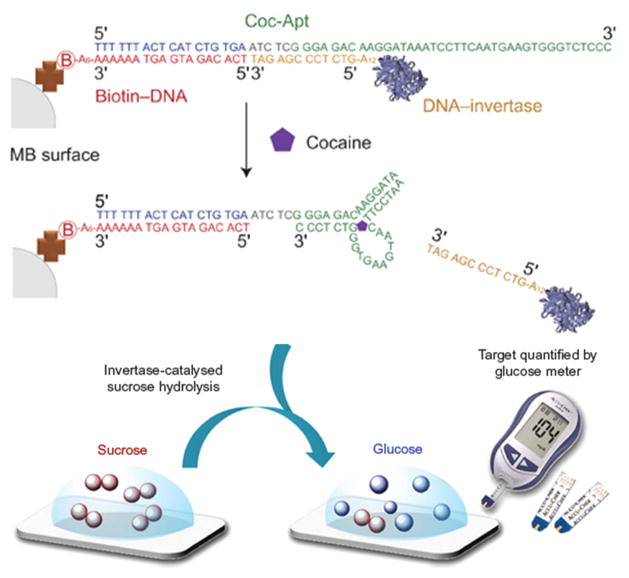
By combining the technology underlying personal glucose meters (PGMs) with newer functional DNA sensors, a new device has been developed that is able to quantitatively detect cocaine in blood and serum (Figure 3) [56]. This method shows high sensitivity, with detection limit of approximately 1.0 μg×mL−1. Other targets, including adenosine, interferon-γ, and toxic metal ions, can be detected by changing the sequence of the functional DNA.
Figure 3.
Combination of personal glucose meters (PGMs) and functional DNA sensors allows detection of cocaine or other addictive illicit substances in serum. Reproduced with permission from Ref. [56].
The steroid hormone estrogen is often used to identify women at risk for breast cancer or to monitor anti-estrogen therapy in breast cancer [57]. A compact, efficient, and intuitive method has recently been devised for estrogen analysis in blood samples as small as 1 μL. The underlying principle of digital microfluidics (DMF) involves movement of a sample through a set of electrodes by applying electric current. The method may be applied to most multistep sample-processing procedures and used for analysis of blood and serum samples [58, 59].
Nitric oxide (NO) is a metastable free radical synthesized by nitric oxide synthetases and involved in important physiological processes, such as wound healing, angiogenesis, and immune response [60]. A new microfluidic device based on a NO-selective xerogel polymer measures NO in small (250 μL) blood samples. This amperometric sensor shows excellent selectivity and high sensitivity, with detection limit of 840 pM in phosphate-buffered saline and 472 nM in blood.
Cell-based Molecular Analyses
Circulating tumor cells (CTCs) shed from the primary tumor site and present in the blood stream provide a rare model system for potential downstream molecular analyses to understand the mechanism of cancer development. Advanced technologies developed in genomics and proteomics can be further applied to determine the activity patterns of genes and proteins in different types of cancerous or precancerous cells. Such molecular signatures can significantly improve the clinician’s ability to diagnose and manage cancer.
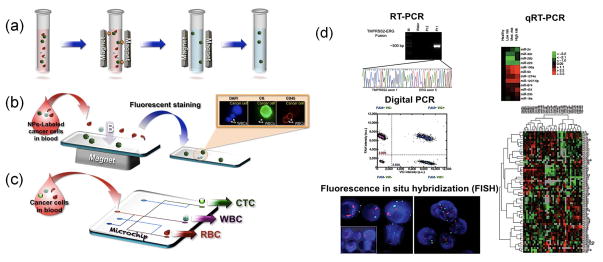
CTCs are rare (1 in 107 – 109 blood cells), which poses significant challenges for capture. Clinical applications would require novel platforms for such noninvasive “liquid biopsy” with high capture efficiency and sensitivity, and low cost. Among the recent developments in this area, microfluidic technology, immunomagnetic assay, and molecular analysis have been integrated to fulfill such a requirement. A commercial, FDA-approved screening system (CellSearch™) has been developed for the enumeration of CTCs in samples of whole blood. Integration of an immunomagnetic assay with a microchip is a widely used cell isolation method whereby magnetic force is applied to separate cancer cells (e.g., CTCs), labeled with magnetic particles, from unlabeled red and white blood cells for further molecular analysis. In this approach, magnetic particles/beads coated with antibodies specific for antigens expressed on cancer cells are used to tag target cells for magnetic capture (Figure 4) [61–66]. Dendrimers, polymer nanofibers, graphene, and other nanomaterials have also been introduced in microfluidic chips to increase capture efficiency [67–70]. Other than immunolabel-based isolation methods, label-free microfluidics-based methods have been proposed for application to cell separation. A size-based microchip has been developed to isolate cancer cells from other blood cells based on differences in physical properties, such as size, shape, density, and deformability of each cell type [71, 72]. Other label-free isolation technology, such as that exploiting dielectric properties of cancer cells, can be applied for separation purpose [73].
Figure 4.
Circulating tumor cells (CTC) isolation and downstream molecular analyses. (a) Anti-epithelial cell adhesion molecule (EpCAM)-conjugated ferrofluids were used to tag CTCs for magnetic separation. (b) A microchip-based immunomagnetic isolation system was proposed for separating magnetic-particle–labeled CTCs from other blood cells in whole blood samples for fluorescence analysis. (c) Other microchip-based approaches using nanomaterial-assisted, size-, mechanical property-, and electrophoresis-based microchips can also be used to isolate CTCs, white blood cell (WBCs), and red blood cells (RBCs) simultaneously for downstream analysis. (d) Downstream gene expression analysis of DNA or RNA extracted from captured CTCs using reverse transcription polymerase chain reaction (RT-PCR), quantitative reverse transcription polymerase chain reaction (qRT-PCR), digital PCR, or fluorescence in situ hybridization (FISH) [74–77]. Abbreviation: NP, nanoparticle. Reproduced with permission from Ref. [74–77].
To analyze the cells and acquire genomic information, such as cancer stage, epithelial to mesenchymal transition status of cancer cells, and molecular biomarkers for different cancer types, DNA and RNA are extracted from the collected cells. Several methods, such as fluorescence in situ hybridization (FISH), reverse transcription-polymerase chain reaction (RT-PCR), quantitative RT-PCR (qRT-PCR), and digital PCR have been used for the molecular analysis of cancer cells to provide diagnostic and prognostic information for cancer treatment strategy (Figure 4d) [74–77].
Concluding remarks and future perspectives
Molecular diagnostics provide the signatures of diseases throughout their development. As more and more blood biomarkers are discovered, diagnosis of disease by determining levels of proteins, nucleic acids, small molecules, or cell-derived molecules in blood remains an exciting area of modern medicine. In this review, we have presented an overview of recent developments in molecular diagnostic technologies that are applicable to a single drop of blood obtained by a simple finger prick. These new diagnostic technologies use optical, electronic, or other readable signals. Remaining challenges with some of these new molecular technologies include problems of consistency and reliability, and further optimization will be required before commercialization becomes feasible.
The future development of POC molecular diagnostics will be driven by the needs of resource-limited areas, such as developing countries or in the home, where insufficient health care facilities exist for diagnostic support. Therefore, low cost, delivery capability, and user-friendliness of the technologies are the main issues to be addressed in the future. In general, developments in nanotechnology and biotechnology have clearly improved the sensitivity and specificity of molecular diagnostics. However, these improvements were accompanied by sacrifices in cost and portability because they often require labor-intensive sample pretreatment and sophisticated instruments for reading results. Because microfluidic chips allow considerable throughput, portability, and the capacity for a high level of integration, they will fulfill the requirements of POC molecular diagnostics if sample preparation and advanced nanotechnologies/biotechnologies are introduced. Meanwhile, device costs may be reduced by use of inexpensive materials, small volumes of reagents, and massive levels of production.
Highlights.
We discuss the frontiers of point-of-care diagnostic technologies using a drop of blood obtained froma finger-prick.
A great challenge is still required to develop simple, inexpensive, rapid, and easy-to-use technologiesfor blood molecular diagnostics.
Proteins, nucleic acids, and other molecules as well as downstream molecular analyses based on cancer cells isolated from the blood are surveyed for molecular diagnostics in a drop of blood.
Various technologies, including emerging biotechnologies, nanotechnologies, and microfluidics, hold the potential for rapid, accurate, and non-expensive disease diagnostics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gutierrez G, et al. Clinical review: Hemorrhagic shock. Critical Care. 2004;8:373–381. doi: 10.1186/cc2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marieb EN. Essentials of Human Anatomy & Physiology. 9 Benjamin Cummings; 2008. [Google Scholar]
- 3.Huang SJ, et al. Clinical experience of hydroxyethyl starch (10% HES 200/0.5) in cerebral perfusion pressure protocol for severe head injury. Surg Neurol. 2006;66(Suppl 2):S26–31. doi: 10.1016/j.surneu.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Medoff BD, et al. Pathogenic T-cell recruitment into the airway in human disease. Ann N Y Acad Sci. 2005;1062:220–241. doi: 10.1196/annals.1358.026. [DOI] [PubMed] [Google Scholar]
- 5.Calder PC, et al. Early nutrition and immunity - progress and perspectives. Br J Nutr. 2006;96:774–790. [PubMed] [Google Scholar]
- 6.Maton AJH, McLaughlin Charles William, Johnson Susan, Warner Maryanna Quon, LaHart David, Wright Jill D. Human Biology and Health. Englewood Cliffs, New Jersey, USA: Prentice Hall; 1993. [Google Scholar]
- 7.Loonen AJ, et al. Highlights from the 7th European meeting on molecular diagnostics. Expert Rev Mol Diagn. 2012;12:17–19. doi: 10.1586/erm.11.89. [DOI] [PubMed] [Google Scholar]
- 8.Gubala V, et al. Point of care diagnostics: status and future. Anal Chem. 2011;84:487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
- 9.Yager P, et al. Point-of-care diagnostics for global health. Ann Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 10.Holland CA, Kiechle FL. Point-of-care molecular diagnostic systems--past, present and future. Curr Opin Microbiol. 2005;8:504–509. doi: 10.1016/j.mib.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Cheng MM, et al. Nanotechnologies for biomolecular detection and medical diagnostics. Curr Opin Chem Biol. 2006;10:11–19. doi: 10.1016/j.cbpa.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Gervais L, et al. Microfluidic Chips for Point-of-Care Immunodiagnostics. Adv Mater. 2011;23:H151–H176. doi: 10.1002/adma.201100464. [DOI] [PubMed] [Google Scholar]
- 13.Myers FB, Lee LP. Innovations in optical microfluidic technologies for point-of-care diagnostics. Lab chip. 2008;8:2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- 14.Yildiz F, editor. Advances in food biochemistry. CRC press; 2010. [Google Scholar]
- 15.Antman EM, et al. Cardiac-specific troponin I levels to predict the risk of mortality in patients with acute coronary syndromes. N Engl J Med. 1996;335:1342–1349. doi: 10.1056/NEJM199610313351802. [DOI] [PubMed] [Google Scholar]
- 16.Adkins JN, et al. Toward a human blood serum proteome - Analysis by multidimensional separation coupled with mass spectrometry. Mol Cell Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Pieper R, et al. The human serum proteome: Display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003;3:1345–1364. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 18.Warsinke A. Point-of-care testing of proteins. Anal Bioanal Chem. 2009;393:1393–1405. doi: 10.1007/s00216-008-2572-0. [DOI] [PubMed] [Google Scholar]
- 19.Chin CD, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011;17:1015–1138. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- 20.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de la Rica R, Stevens MM. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat Nanotechnol. 2012;7:821–824. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- 22.Stern E, et al. Label-free biomarker detection from whole blood. Nat Nanotechnol. 2010;5:138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaster RS, et al. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nat Med. 2009;15:1327–U1130. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan R, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin LD, et al. Self-powered microfluidic chips for multiplexed protein assays from whole blood. Lab chip. 2009;9:2016–2020. doi: 10.1039/b821247c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollock NR, et al. A Paper-Based Multiplexed Transaminase Test for Low-Cost, Point-of-Care Liver Function Testing. Sci Transl Med. 2012;4:152ra129. doi: 10.1126/scitranslmed.3003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vella SJ, et al. Measuring markers of liver function using a micropatterned paper device designed for blood from a fingerstick. Anal Chem. 2012;84:2883–2891. doi: 10.1021/ac203434x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng CM, et al. Paper-Based ELISA. Angew Chem In Ed. 2010;49:4771–4774. doi: 10.1002/anie.201001005. [DOI] [PubMed] [Google Scholar]
- 29.Song YJ, et al. Multiplexed volumetric bar-chart chip for point-of-care diagnostics. Nat Commun. 2012;3 doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y, et al. A multistage volumetric bar chart chip for visualized quantification of DNA. J Am Chem Soc. 2013;35:16785–16788. doi: 10.1021/ja4085397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De M, et al. Sensing of proteins in human serum using conjugates of nanoparticles and green fluorescent protein. Nat Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, et al. Tailoring of the Nanotexture of Mesoporous Silica Films and Their Functionalized Derivatives for Selectively Harvesting Low Molecular Weight Protein. Acs Nano. 2010;4:439–451. doi: 10.1021/nn901322d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, et al. A one-step homogeneous immunoassay for cancer biomarker detection using gold nanoparticle probes coupled with dynamic light scattering. J Am Chem Soc. 2008;130:2780–2782. doi: 10.1021/ja711298b. [DOI] [PubMed] [Google Scholar]
- 35.Jans H, Huo Q. Gold nanoparticle-enabled biological and chemical detection and analysis. Chem Soc Rev. 2011;41:2849–2866. doi: 10.1039/c1cs15280g. [DOI] [PubMed] [Google Scholar]
- 36.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 37.Niemz A, et al. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam B, et al. Solution-based circuits enable rapid and multiplexed pathogen detection. Nat Commun. 2013;4:2001. doi: 10.1038/ncomms3001. [DOI] [PubMed] [Google Scholar]
- 39.Liong M, et al. Magnetic barcode assay for genetic detection of pathogens. Nat Commun. 2013;4:1752. doi: 10.1038/ncomms2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gahan PB, Swaminathan R. Circulating nucleic acids in plasma and serum - Recent developments. Ann N Y Acad Sci. 2008;1137:1–6. doi: 10.1196/annals.1448.050. [DOI] [PubMed] [Google Scholar]
- 41.Anker P, et al. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metast Rev. 1999;18:65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- 42.Stroun M, et al. About the possible origin and mechanism of circulating DNA - Apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–142. doi: 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 43.Root BE, et al. Purification of HIV RNA from Serum Using a Polymer Capture Matrix in a Microfluidic Device. Anal Chem. 2011;83:982–988. doi: 10.1021/ac102736g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu KJ, et al. Decoding Circulating Nucleic Acids in Human Serum Using Microfluidic Single Molecule Spectroscopy. J Am Chem Soc. 2010;132:5793–5798. doi: 10.1021/ja100342q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forshew T, et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 46.Yang Q, et al. Detection and differential diagnosis of colon cancer by a cumulative analysis of promoter methylation. Nat Commun. 2012;3 doi: 10.1038/ncomms2209. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 48.Labib M, et al. Three-Mode Electrochemical Sensing of Ultralow MicroRNA Levels. J Am Chem Soc. 2013;135:3027–3038. doi: 10.1021/ja308216z. [DOI] [PubMed] [Google Scholar]
- 49.Dong H, et al. MicroRNA: Function, Detection, and Bioanalysis. Chem Rev. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 50.Wen Y, et al. DNA Nanostructure-based Interfacial engineering for PCR-free ultrasensitive electrochemical analysis of microRNA. Sci Rep. 2012;2:867. doi: 10.1038/srep00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, et al. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat nanotechnol. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song YJ, et al. Graphene Oxide: Intrinsic Peroxidase Catalytic Activity and Its Application to Glucose Detection. Adv Mater. 2010;22:2206–2210. doi: 10.1002/adma.200903783. [DOI] [PubMed] [Google Scholar]
- 53.Song YJ, et al. Label-free colorimetric detection of single nucleotide polymorphism by using single-walled carbon nanotube intrinsic peroxidase-like activity. Chem Eur J. 2010;16:3617–3621. doi: 10.1002/chem.200902643. [DOI] [PubMed] [Google Scholar]
- 54.Song YJ, et al. Light regulation of peroxidase activity by spiropyran functionalized carbon nanotubes used for label-free colorimetric detection of lysozyme. Chem Commun. 2011;47:9083–9085. doi: 10.1039/c1cc13279b. [DOI] [PubMed] [Google Scholar]
- 55.Song Y, et al. Visual and quantitative detection of copper ions using magnetic silica nanoparticles clicked on multiwalled carbon nanotubes. Chem Commun. 2010;46:6572–6574. doi: 10.1039/c0cc01593h. [DOI] [PubMed] [Google Scholar]
- 56.Xiang Y, Lu Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mousa NA, et al. Droplet-Scale Estrogen Assays in Breast Tissue, Blood, and Serum. Sci Transl Med. 2009;1:1ra2. doi: 10.1126/scitranslmed.3000105. [DOI] [PubMed] [Google Scholar]
- 58.Jebrail MJ, et al. A digital microfluidic method for dried blood spot analysis. Lab chip. 2012;11:3218–3224. doi: 10.1039/c1lc20524b. [DOI] [PubMed] [Google Scholar]
- 59.Shih SC, et al. Dried blood spot analysis by digital microfluidics coupled to nanoelectrospray ionization mass spectrometry. Anal Chem. 2012;84:3731–3738. doi: 10.1021/ac300305s. [DOI] [PubMed] [Google Scholar]
- 60.Hunter Rebecca A, BJP, Hampton Henley W, Breed Elise R, Liang Zhe, Mittal Rohit, Yoseph Benyam P, McDunn Jonathan E, Burd Eileen M, Coopersmith Craig M, Michael Ramsey J, Schoenfisch Mark H. Microfluidic amperometric sensor for analysis of nitric oxide in whole blood. Anal Chem. 2013;85:6066–6072. doi: 10.1021/ac400932s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller MC, et al. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J Oncol. 2012;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang YY, et al. Immunomagnetic nanoscreening of circulating tumor cells with a motion controlled microfluidic system. Biomed Microdevices. 2013;15:673–681. doi: 10.1007/s10544-012-9718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoshino K, et al. Computational analysis of microfluidic immunomagnetic rare cell separation from a particulate blood flow. Anal Chem. 2012;84:4292–4299. doi: 10.1021/ac2032386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoshino K, et al. Microchip-based immunomagnetic detection of circulating tumor cells. Lab chip. 2011;11:3449–3457. doi: 10.1039/c1lc20270g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozkumur E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen P, et al. Multiscale immunomagnetic enrichment of circulating tumor cells: from tubes to microchips. Lab chip. 2014;14:446–458. doi: 10.1039/c3lc51107c. [DOI] [PubMed] [Google Scholar]
- 67.Myung JH, et al. Dendrimer-mediated multivalent binding for the enhanced capture of tumor cells. Angew Chem Int Ed. 2011;50:11769–11772. doi: 10.1002/anie.201105508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao L, et al. High-Purity Prostate Circulating Tumor Cell Isolation by a Polymer Nanofiber-Embedded Microchip for Whole Exome Sequencing. Adv Mater. 2013;25:2897–2902. doi: 10.1002/adma.201205237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoon HJ, et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat Nanotechnol. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S, et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew Chem Int Ed. 2009;48:8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang W, et al. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc Nat Acad Sci USA. 2012;109:18707–18712. doi: 10.1073/pnas.1209893109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng S, et al. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 2011;13:203–213. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gascoyne PR, et al. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis. 2009;30:1388–1398. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Nat Acad Sci USA. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lim LS, et al. Microsieve lab-chip device for rapid enumeration and fluorescence in situ hybridization of circulating tumor cells. Lab chip. 2012;12:4388–4396. doi: 10.1039/c2lc20750h. [DOI] [PubMed] [Google Scholar]
- 77.Punnoose EA, et al. Molecular biomarker analyses using circulating tumor cells. PloS one. 2010;5:e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]