Abstract
Cervicovaginal human papillomavirus (HPV) viral load has been purported as a potential marker for the detection of high-grade cervical intraepithelial neoplasia or cancer (≥CIN2). To examine disease association with type-specific viral load for the full-range of anogenital HPV infections, we conducted cross-sectional and prospective analyses of ∼2,000 HPV-infected women from a 10,000-woman population-based study in Guanacaste, Costa Rica with 7 years of follow-up. Cervical specimens were tested for >40 HPV types using a MY09/MY11 L1 consensus primer PCR method with type-specific dot blot hybridization and PCR signal intensity as a measure of viral load. A positive association was observed between prevalent ≥CIN2 and high viral load compared to low viral load for women with baseline single HPV16 infections (OR = 19.2, 95% CI = 4.4–83.2) and single non-16 carcinogenic infections (OR = 9.2, 95% CI = 2.1–39.9). Inclusion of women with multiple HPV types did not substantially change these associations. In prospective follow-up, only women infected with HPV16 alone (OR = 27.2, 95% = 3.5-213.5) had a strong association between high viral load and incident ≥CIN2; non-16 carcinogenic high viral load was not associated with incident ≥CIN2 (OR = 0.7, 95% CI = 0.2–1.9). Single noncarcinogenic type viral load was not associated with increased risk of prevalent or incident ≥CIN2 (OR = 1.2 and 1.1, respectively). In conclusion, carcinogenic high viral load was associated with prevalent ≥CIN2; however HPV16 was uniquely associated with incident ≥CIN2. The extent to which these observations can be translated into clinical practice must be rigorously examined in the context of the method of viral load measurement and the type-specific differences observed for incident ≥CIN2.
Keywords: human papillomavirus, viral load, genotype, screening
Human papillomaviruses (HPV) are the causal agent of invasive cervical cancer.1 Current screening guideline in the US incorporate HPV DNA testing of cervical samples to clarify equivocal Pap tests and as an adjunct to cytology in primary screening among women over age 30 years. A negative HPV test greatly reduces the risk of a cancer precursor. This allows women with equivocal cytology to return to the general screening pool and women 30-years and older with normal cytology and no high-risk HPV to extend their screening interval to 3 years. Although HPV testing is clinically useful, many women with self-limited carcinogenic HPV infections test positive and are subjected to unnecessary procedures. Therefore, identifying markers that indicate which HPV infections will persist and result in progression to cancer precursors is an important research objective.
High concentration of HPV DNA in cervical specimens (HPV viral load)2–16 has been evaluated as a marker for the presence of prevalent cervical cancer precursors (defined in the United States for treatment purposes as CIN2 or worse, ≥CIN2) or incident ≥CIN2. Following a typical infectious disease model, viral load as a measure of “dosage” (i.e., HPV copy number) might be predictive of ≥CIN2 or the duration of infection. However measuring viral load in routinely collected cervical samples is limited by potential bias in specimen sampling, the complexity of interpreting viral load measures in the context of multiple concurrent infections, and ambiguities in determining viral copy number per cell. Methods employed to measure HPV load vary in the ability to address these limitations14 and this variability could explain the inconsistencies in studies of HPV load and ≥CIN2.
We therefore conducted detailed analyses of HPV DNA load and risk of prevalent and incident histologically-confirmed ≥CIN2 using enrollment samples from a large, population-based natural history study conducted in Guanacaste, Costa Rica with up to 7 years of follow-up. By using a qualitative viral load estimate based on type-specific probe hybridization intensity, we were able to evaluate the association between HPV viral loads across the genotype spectrum.
Material and methods
Study population
The Proyecto Epidemiológico Guanacaste is a population-based study in a province of Costa Rica where rates of cervical cancer are historically high. Study participants were enrolled between June 1993 and December 1994 with the approval of the National Cancer Institute (NCI) and Costa Rican institutional review boards.17,18 Of the 11,742 potentially eligible subjects, 10,049 (93.4%) women provided written informed consent. Detailed methods of recruitment, screening and follow-up were previously published.19
For both the cross-sectional (enrollment/prevalent) and prospective (incident) parts of this analysis, we excluded women with prior hysterectomies (n = 630), who were virgins (n = 583), or who refused a pelvic exam (n = 291) resulting in a baseline analytic group of 8,545.19 When examining disease associations with qualitative viral load (PCR signal intensity), women who at enrollment were found to be positive only for a combination of rare HPV types (dot blot mix) (n = 8) or uncharacterized types (n = 250) or missing PCR (n = 32) or HPV negative (n = 6,263) were excluded, resulting in 1,296 women with single HPV infections and 696 women with multiple HPV infections in the final analysis. Women positive for the rare HPV types or for uncharacterized types were excluded as we could not definitively ascertain whether these women had single or multiple HPV infections nor could we accurately categorize these types in specific alpha genotype groups and/or risk groups. For the incident disease analysis, the final analysis group excluded women diagnosed with ≥CIN2 at enrollment (n = 63) or women with missing follow-up data (n = 142).
HPV DNA testing and viral load determination
During a single pelvic examination at baseline, 2 exfoliative cervical specimens were collected. Using a Cervex brush, the first specimen was employed to prepare both a conventional Pap smear and thin-layer cytology slides (ThinPrep, Cytyc Corporation)19 and a second cervical specimen was collected using a Dacron swab, placed into Digene specimen transport medium (Digene Corporation, Gaithersburg, MD), and stored at −70°C for future testing.
DNA was partially purified using standard protocols and PCR amplified using a MY09/M11 L1 consensus primer PCR (MY09/11 PCR) method with TaqGold polymerase as previously described.20 HPV type-specific infections were detected by dot blot hybridization of PCR products using type-specific oligonucleotide probes for HPV types 2, 6, 11, 13, 16, 18, 26, 31–35, 39, 40, 42–45, 51–59, 61, 62, 64, 66–74, 81, 82 (AE2 and W13B), 83–85 and 89.21 Probes for HPV Types 2, 13, 34, 42–4, 57, 64, 69, 74, 82 (AE2 and W13B) and 54 (AE9) were pooled in dot blot hybridizations for detection of rare types (dot blot mix). For these analyses, HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 6822 plus HPV6623 were considered carcinogenic. Specimens that tested HPV positive based on a radiolabeled generic probe mix, but were not positive for any type specific probe, were considered to be positive for uncharacterized HPV types. We also examined associations with two alpha “species groups.”24 Group 1 contained mostly carcinogenic types: α9/ α11: HPV types 16, 31, 33, 34, 35, 52, 58, 64, 67 and 73; α7: HPV types 18, 39, 45, 59, 68, 70 and 85; and α5/α6: HPV types 26, 51, 53, 56, 66, 69 and 82. Group 2 contained exclusively noncarcinogenic HPV types: α3/α15: HPV types 61, 62, 71, 72, 81, 83, 84 and 89; and α1/α8/α10/α13: HPV types 6, 11, 32, 40, 42, 54, 55 and 74.
To determine HPV PCR signal strength, three experienced HPV investigators interpreted type-specific dot blot results and discrepancies were resolved by consensus. Dot blot oligonucleotide hybridization signal intensity, a measure of viral load, was evaluated by two researchers using a qualitative index on a scale of 1– 5 (weakest = 1 and strongest = 5). The index represents the strength of the hybridization signal established by observing the density and diameter of the PCR product on the autoradiogram. For the current analyses, viral load findings were collapsed into dichotomous categories of low (PCR signal strength index of 1–3) and high (PCR signal strength index of 4–5) viral load. We also explored an alternative categorization of <3 versus ≥3. The overall trends were similar when the PCR signal strength of 3 was included in the low or high viral load definition; therefore only one analysis is presented. A detailed description of the HPV16 and HPV 18 TaqMan PCR methods and testing in this population have been previously described.14,25
Statistical analysis
On the basis of masked histopathology reviews performed in the US, final diagnoses were assigned by algorithm: invasive cancer, CIN3, CIN2 and <CIN2.26 Unconditional logistic regression was used to calculate odds ratios (ORs) with 95% CIs as an estimate of the relative risk of ≥CIN2. We examined separately the association of type-specific qualitative viral load with CIN2 and CIN3 or worse. As our patterns for CIN2 and CIN3 or worse did not substantially differ, only results for ≥CIN2 are shown. When estimating the viral load association with incident ≥CIN2, we repeated all analyses restricting to (i) ≥CIN2 cases occurring within 2 years of the enrollment viral load measurement and (ii) ≥CIN2 cases occurring at least 2 years after the enrollment viral load measurement.
Viral load exposure was defined in three ways: (i) for each individual type restricted to single infections, (ii) hierarchical type-specific classification in analyses including multiple genotype infections and (iii) summed viral load across all genotypes. In analyses restricted to single infections, the PCR signal strength from the type-specific oligonucleotide probe hybridization was used. In the second analysis, when a woman was coinfected with more than 1 HPV genotype (and thus had more than one viral load exposure), we hierarchically selected the highest single carcinogenic viral load signal strength or the highest single viral load from a noncarcinogenic type if there were no carcinogenic coinfections. Thus, in women coinfected with carcinogenic and noncarcinogenic types, the carcinogenic type with the highest viral load was selected even if a coinfecting noncarcinogenic type had a higher viral load, because noncarcinogenic types only rarely produce CIN2 or CIN3.
In the third analysis, we simply summed the type-specific viral load signal strength across all coinfections for a cumulative measure of viral burden and categorized this measure according to tertile distribution. In these grouped analyses, we estimated the association between viral load and risk of ≥CIN2 for all HPV types and in subgroups of types defined by carcinogenicity and by phylogenetic alpha species.24
The distribution of absolute viral load as determined by TaqMan PCR for HPV16 and HPV18 in categories of PCR signal strength were evaluated by box and whiskers plots. PCR signal intensity and TaqMan PCR viral load were compared using Spearman rank correlation.
Results
Association of viral load with prevalent ≥CIN2
To validate the use of the PCR signal strength index as a surrogate viral load measure, type-specific signal strength intensity in women testing positive for HPV16 and/or HPV18 was compared to real-time PCR. Figure 1 indicates that in women testing positive for either HPV16 or HPV18, the PCR signal intensity was highly correlated to real-time PCR [Spearman's ρ (rho)] = 0.70 and 0.63, respectively). These data suggest that stratification of lower and higher viral load by hybridization signal intensity is a valid method for assessment of the relative viral load.
Figure 1.
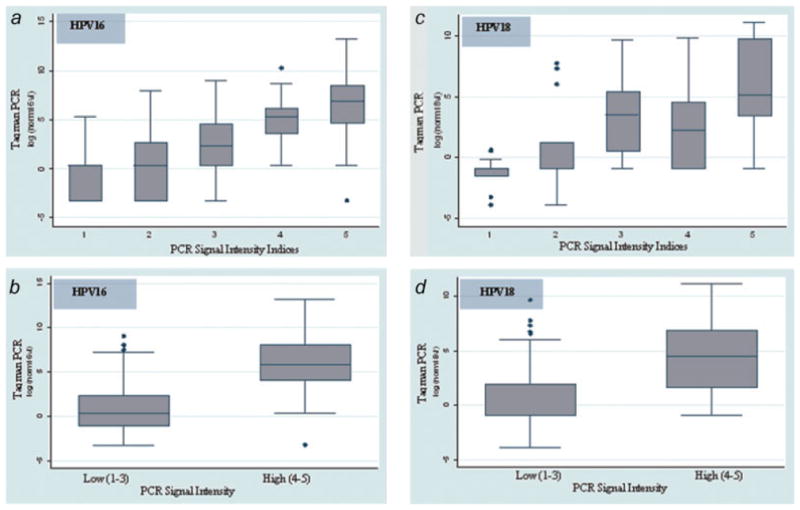
Box plots of log10-transformed HPV16 (a and b) and HPV18 (c and d) normalized viral load (viral load per 20,000 GAPDH copies) by oligonucleotide hybridization signal intensity in the original 5 (a and c) and collapsed 2 (b and d) categories. The median crude HPV copy number (and interquartile range) by hybridization signal intensity, HPV 16: 1 = 10 (1–48), 2 = 6 (1–240), 3 = 144 (27–1661), 4 = 2538 (294–12,610), 5 = 14,382 (2988–61,936); HPV 18: 1 = 2 (1–7), 2 = 221 (1–5682), 3 = 745 (154–6137), 4 = 1387 (172–4881), 5 = 5001 (1803–175,651). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Using the type-specific dichotomous viral load signal strength index, we found a strong relationship with prevalent ≥CIN2 and high viral load in women with any single HPV type (OR = 10.4, 95% CI = 4.5–24.4) and in women with single and multiple HPV types combined (OR = 13.0, 95% CI = 6.3–26.8) (Table I). We found that HPV16 positive women with no other HPV type had the strongest relationship (OR = 19.2, 95% CI = 4.4–83.2). Inclusion of women with multiple HPV types in addition to HPV16; however, did not strengthen this association (OR = 12.3, 95% CI = 4.3–34.8). To limit the possibility that the combined carcinogenic viral load association was driven mostly by inclusion of HPV16, we reestimated the relative odds after exclusion of the women having HPV16 and found only a modest attenuation of the risk estimate (OR = 13.6 vs. 9.2 among single carcinogenic HPV types with and without HPV16, respectively). Whereas low prevalence of individual HPV types precluded a type-specific analysis, we were able to examine disease associations in women with single HPV types restricted to each of the 3 carcinogenic-containing species groups (α9/11, α7 and a5/6). Similar increased odds of ≥CIN2 among women with high viral load in each species (data not shown) indicated that the association between prevalent ≥CIN2 and high viral load was not unique to HPV16 or HPV16-related genotypes.
Table I. Association of HPV Viral Load with Prevalent ≥Cin2.
| HPV risk group | Qualitative viral load1 | Single HPV infections | Single and multiple HPV infections2 | ||
|---|---|---|---|---|---|
|
|
|
||||
| N (cases/controls)3 | OR (95% CI) | N (cases/controls) | OR (95% CI) | ||
| All infections | Low (1–3) | 6/646 | ref. | 8/893 | ref. |
| High (4–5) | 57/587 | 10.4 (4.5–24.4) | 114/977 | 13.0 (6.3–26.8) | |
| Carcinogenic4 | Low (1–3) | 4/269 | ref. | 6/451 | ref. |
| High (4–5) | 55/272 | 13.6 (4.9–38.0) | 108/558 | 14.5 (6.3–33.4) | |
| HPV16+ | Low (1–3) | 2/64 | ref. | 4/110 | ref. |
| High (4–5) | 36/60 | 19.2 (4.4–83.2) | 58/130 | 12.3 (4.3–34.8) | |
| Carcinogenic, HPV16- | Low (1–3) | 2/205 | ref. | 3/353 | ref. |
| High (4–5) | 19/212 | 9.2 (2.1–39.9) | 49/416 | 13.9 (4.3–44.8) | |
| Noncarcinogenic | Low (1–3) | 2/377 | ref. | 2/442 | ref. |
| High (4–5) | 2/315 | 1.2 (0.2–8.5) | 6/419 | 3.2 (0.6–15.8) | |
HPV, human papillomavirus; OR, odds ratio; 95% CI, 95% confidence interval.
Viral load for HPV groups and species reflects type-specific HPV PCR probes. Parentheses indicate range of PCR signal strength indices.
For multiple infections, we utilized the maximum signal intensity value of any carcinogenic HPV type, if such a type was present and the maximum signal intensity value among the noncarcinogenic types for women with noncarcinogenic types only.
Cases were women with prevalent CIN2 or worse at enrollment. Controls were women with prevalent CIN1 or better at enrollment.
For women with multiple HPV types, women had to have had at least one carcinogenic HPV type to be categorized as ‘Carcinogenic.’
As others have attempted to utilize Hybrid Capture 2 (HC2 RLU/CO; Digene Corporation, Gaithersburg, MD), a measure of cumulative viral burden, as a surrogate of viral load, we too attempted to evaluate associations of a summation of our qualitative viral load measure, again for risk of prevalent ≥CIN2. We found that women with mid-level (summed PCR signal strength indices ranging from 4–5) and the highest level (summed signal strength indices ≥6) of any HPV type were more likely to have prevalent ≥CIN2 (OR = 10.0 and 13.7, respectively) compared to women with the lowest values (summed signal strength indices ranging from 1–3) (Table II). The highest summed viral load category (≥6) represented samples testing positive for multiple HPV types relative to the lower viral load categories.
Table II. Association of the Sum of HPV Viral Load with Prevalent >Cin2.
| HPV risk group | Qualitative viral load1 | N (cases/controls)2 | OR (95% CI) |
|---|---|---|---|
| All infections | Low (1–3) | 6/698 | ref. |
| Mid (4–5) | 60/697 | 10.0 (4.3–23.3) | |
| High (6+) | 56/475 | 13.7 (5.9–32.1) | |
| Carcinogenic3 | Low (1–3) | 4/297 | ref. |
| Mid (4–5) | 56/337 | 12.3 (4.4–34.4) | |
| High (6+) | 54/375 | 10.7 (3.8–29.9) | |
| HPV16+ | Low (1–3) | 2/69 | ref. |
| Mid (4–5) | 36/73 | 17.0 (3.9–73.4) | |
| High (6+) | 24/98 | 8.4 (1.9–36.9) | |
| Carcinogenic, HPV16− | Low (1–3) | 2/228 | ref. |
| Mid (4–5) | 20/264 | 8.6 (2.0–37.3) | |
| High (6+) | 30/277 | 12.3 (2.9–52.2) | |
| Noncarcinogenic | Low (1–3) | 2/401 | ref. |
| Mid (4–5) | 4/360 | 2.2 (0.4–12.2) | |
| High (6+) | 2/100 | 4.0 (0.6–28.8) |
HPV, human papillomavirus; OR, odds ratio; 95% CI, 95% confidence interval.
Viral load for HPV groups and species reflects type-specific HPV PCR probes. Parentheses indicate range of the sums of PCR signal strength indices.
Cases were women with prevalent CIN2 or worse at enrollment. Controls were women with prevalent CIN1 or better at enrollment. Women with both single and multiple HPV infections are included.
For women with multiple HPV types, women had to have had at least one carcinogenic HPV type to be categorized as ‘Carcinogenic’
Association of viral load with incident ≥CIN2
A baseline viral load measure in women without high grade neoplasia (<CIN2) was associated with an increased risk of incident ≥CIN2 over 7 years only in women testing positive for HPV16 alone (OR = 27.2, 95% = 3.5–213.5; Table III). After excluding women with HPV16 positive single infections (i.e., among women with single, non-16, carcinogenic type infection), we observed no increased risk associated with higher viral load (OR = 0.7, 95% CI = 0.2–1.9; Table III), suggesting that the overall association with incident ≥CIN2 was explained by HPV16 viral load. Baseline genotypes for non-HPV16 carcinogenic ≥CIN2 cases are listed in Table IV. When women testing positive for multiple HPV types were included in the analyses along with the single infections, the risk of incident >CIN2 among all women was similar in all cases except in women having HPV16 alone. Among women with HPV 16 plus other carcinogenic coinfections, the risk was significantly attenuated (OR = 2.6, 95% CI = 1.2–5.8), compared to women with only a single HPV16 infection. Viral load was not associated with increased risk of incident ≥CIN2 in women positive for either single or single and multiple noncarcinogenic HPV types combined (OR = 1.2 and 1.1 respectively; Table III). Similar results were obtained when using summed viral load categories, with significant associations observed only among women testing positive for HPV16, with no further increase risk when viral load exceeded 5 (implying multiple infections) (Table V).
Table III. Association of HPV Viral Load with Incident ≥Cin2.
| HPV risk group | Qualitative viral load1 | Single HPV infections | Single and multiple HPV infections2 | ||
|---|---|---|---|---|---|
|
|
|
||||
| N (cases/controls)3 | OR (95% CI) | N (cases/controls) | OR (95% CI) | ||
| All infections | Low (1–3) | 15/560 | ref. | 26/762 | ref. |
| High (4–5) | 28/488 | 2.1 (1.1–4.1) | 53/788 | 2.0 (1.2–3.2) | |
| Carcinogenic4 | Low (1–3) | 10/231 | ref. | 20/375 | ref. |
| High (4–5) | 23/214 | 2.5 (1.2–5.3) | 47/425 | 2.1 (1.2–3.6) | |
| HPV16+ | Low (1–3) | 1/56 | ref. | 10/87 | ref. |
| High (4–5) | 17/35 | 27.2 (3.5–213.5) | 25/83 | 2.6 (1.2–5.8) | |
| Carcinogenic, HPV16− | Low (1–3) | 9/175 | ref. | 14/294 | ref. |
| High (4–5) | 6/179 | 0.7 (0.2–1.9) | 18/336 | 1.1 (0.6–2.3) | |
| Noncarcinogenic | Low (1–3) | 5/329 | ref. | 6/387 | ref. |
| High (4–5) | 5/274 | 1.2 (0.3–4.2) | 6/363 | 1.1 (0.3–3.3) | |
HPV, human papillomavirus; OR, odds ratio; 95% CI, 95% confidence interval.
Viral load for HPV groups and species reflects type-specific HPV PCR probes. Parentheses indicate range of PCR signal strength indices.
For multiple infections, we utilized the maximum signal intensity value of any carcinogenic HPV type, if such a type was present and the maximum signal intensity value among the noncarcinogenic types for women with noncarcinogenic types only.
Cases were women with incident CIN2 or worse during follow-up after excluding women with CIN2 or worse at enrollment based on both cytology and histology. Controls were women without incident CIN2 or worse at enrollment and during follow-up.
For women with multiple HPV types, women had to have had at least one carcinogenic HPV type to be categorized as ‘Carcinogenic’
Table IV. Baseline Genotype and Viral Load Among the 15 Women with Single, Non- HPV 16 Carcinogenic HPV Infection and Incident ≥Cin2 During 7 Years of Follow-Up.
| HPV genotype | Hybridization Signal strength | Enrollment diagnosis | Final diagnosis |
|---|---|---|---|
| HPV 31 | 2 | Normal | CIN 2 |
| HPV 31 | 2 | Normal | CIN 2 |
| HPV 33 | 4 | Normal | CIN 2 |
| HPV 18 | 1 | Normal | CIN 3 |
| HPV 31 | 5 | Normal | CIN 3 |
| HPV 51 | 1 | Normal | CIN 3 |
| HPV 18 | 3 | Normal | CIN 3 |
| HPV 52 | 4 | Normal | CIN 3 |
| HPV 18 | 2 | Normal | cancer |
| HPV 33 | 5 | Equivocal | CIN 2 |
| HPV 31 | 2 | Equivocal | CIN 2 |
| HPV 33 | 4 | Equivocal | CIN 3 |
| HPV 33 | 3 | Equivocal | CIN 3 |
| HPV 35 | 5 | Equivocal | CIN 3 |
| HPV 35 | 2 | LSIL | CIN 3 |
Genotype and hybridization signal strength was measured at enrollment.
Table V. Association of the Sum of HPV Viral Load with Incident >Cin2.
| HPV risk group | Qualitative Viral Load1 | N (cases/controls)2 | OR (95% CI) |
|---|---|---|---|
| All infections | Low (1–3) | 15/605 | ref. |
| Mid (4–5) | 34/578 | 2.4 (1.3–4.4) | |
| High (6+) | 30/367 | 3.3 (1.8–6.2) | |
| Carcinogenic3 | Low (1–3) | 10/254 | ref. |
| Mid (4–5) | 28/267 | 2.7 (1.3–5.6) | |
| High (6+) | 29/279 | 2.6 (1.3–5.5) | |
| HPV16+ | Low (1–3) | 1/61 | ref. |
| Mid (4–5) | 19/45 | 25.8 (3.3–199.5) | |
| High (6+) | 15/64 | 14.3 (1.8–111.5) | |
| Carcinogenic, HPV16− | Low (1–3) | 9/193 | ref. |
| Mid (4–5) | 9/222 | 0.9 (0.3–2.2) | |
| High (6+) | 14/215 | 1.4 (0.6–3.3) | |
| Noncarcinogenic | Low (1–3) | 5/351 | ref. |
| Mid (4–5) | 6/311 | 1.4 (0.4–4.5) | |
| High (6+) | 1/88 | 0.8 (0.09–6.9) |
HPV, human papillomavirus; OR, odds ratio; 95% CI, 95% confidence interval.
Viral load for HPV groups and species reflects type-specific HPV PCR probes. Parentheses indicate range of sums of PCR signal strength indices.
Cases were women with incident CIN2 or worse during folllow-up after excluding women with CIN2 or worse at enrollment based on both cytology and histology. Controls were women without incident CIN2 or worse at enrollment and during follow-up. Women with both single and multiple HPV infections are included.
For women with multiple HPV types, women had to have had at least one carcinogenic HPV type to be categorized as ‘Carcinogenic’
Because of the known limitations of colposcopy as a reference standard,27 we repeated the analyses based on the time of incident ≥CIN2 diagnosis to evaluate the potential bias resulting from inclusion of missed prevalent lesions. A similar proportion of incident cases associated with HPV16 infection occurred within 2 years of enrollment (5/18, 27.8%) as incident cases associated with non-HPV16-carcinogenic infection (3/15, 20%; p = 0.6), suggesting that the uniquely increased risk of incident ≥CIN2 for high HPV 16 viral load was not a result of more missed prevalent lesions among HPV16-positive women. Indeed, the inferences were unchanged when alternately restricting the analysis to cases occurring within 2 years of enrollment, and cases occurring after 2 years of enrollment (data not shown).
Discussion
In a population-based study, we found an association between prevalent, histologically confirmed ≥CIN2 and high viral load in women with single HPV16 infections14 using a qualitative viral load measure based on oligonucleotide probe hybridization signal intensity. We also detected an association between carcinogenic, but not noncarcinogenic, high viral load and prevalent ≥CIN2. This effect was retained even after exclusion of women with HPV16, suggesting that carcinogenic high viral load in prevalent ≥CIN2 is not uniquely associated with HPV16 infection. These data are consistent with our recent analyses examining associations between viral load and cytologic abnormalities.12
Our primary analyses were restricted to women testing positive for single HPV types since it is difficult to attribute viral load to lesion development when women have coinfections. Under the one-virus/one-lesion paradigm, only the viral load for the genotype that gives rise to the neoplastic lesion is thought to be relevant to disease risk.28 As 35% of women with characterized HPV types in this population have multiple HPV types, we evaluated viral load-disease relationships in all women with multiple HPV types by assigning a viral load measure as the highest qualitative viral load among the multiple carcinogenic HPV types. Using this approach, we did not observe a substantial difference from what we observed for single infections, suggesting that exclusion of women with multiple infections did not substantially bias our primary results.
It is important to differentiate grouped type-specific viral load estimates such as those used in our analysis from the more commonly used measurement of cumulative viral burden (e.g., HC2 RLU/CO).5,29 By creating a composite viral load score based on the summation signal strength indices from all concomitant infections, we estimated a cumulative viral burden similar to HC2. Unlike HC2 viral load, which is a summation of the total viral burden from any carcinogenic HPV in the sample (and thus inherently inclusive of HPV16), our summed viral load measure allows examination of disease-viral burden associations after removing the dominating effect of HPV16. When we examined women that had tested positive for both single and multiple HPV types, we found that women with higher carcinogenic viral loads, with or without including HPV16, have greater risk of ≥CIN2. This finding was not dissimilar to what would be detected with HC2 and further suggests that increasing viral load because of increasing number of infecting HPV types (signal strength index ≥6) is associated with a modest increase in odds of prevalent ≥CIN2 among women with non-HPV16 carcinogenic types. Among women infected with HPV16; however, testing positive for other HPV types did not increase risk. HPV16, found alone at high viral load, was the greatest risk predictor that we observed, and might represent an enlarging clone of HPV-infected cells facilitating diagnosis of ≥CIN2. These data are consistent with previous reports from this population.30
When evaluating the prognostic association of a baseline HPV viral load with risk of incident ≥CIN2, we confirmed earlier reports of a strong association with HPV16 viral load.9 We failed to detect an association for pooled non-HPV16 carcinogenic types and incident ≥CIN2 (Table III). Small numbers of cases of individual non-16 types do not allow us to exclude the possibility that one or more of the non-HPV16 carcinogenic types have a viral load association with incident ≥CIN2. Larger studies are required for more complete evaluation of non-16 carcinogenic type-specific viral load associations with incident ≥CIN2.
It is important to consider the complex natural history of HPV infection and CIN diagnosis when interpreting the type-specific risk differences for incident ≥CIN2. First, disease occurring within 2 years of the baseline viral load measure may represent missed prevalent disease; however, we did not observe a significantly higher proportion of HPV16 positive incident ≥CIN2 cases during the first two years of follow-up relative to non-HPV16 carcinogenic cases. We therefore do not believe that missed diagnoses explain the difference in risk observed with HPV16. Second, we are measuring viral genotype and load at a fixed time, following for development of disease without concomitant follow-up of the type-specific viral infection. It is quite feasible that a proportion of incident ≥CIN2 detected over 7 years of follow-up resulted not from the baseline infection, but from a subsequent, unmeasured infection. In these cases, it is not surprising that the baseline type-specific viral load was not predictive of disease progression. To evaluate the possible use of repeated viral load measurements, we are currently looking at longitudinal analyses, trends in viral load, and associated risks of incident ≥CIN2. Finally, in Figure 2, we have represented two possible scenarios of subsequent (and unmeasured in this case) natural history of infection following a high baseline viral load measure. Specifically, any cross-sectional identification of high viral load in the absence of neoplasia may reflect the peak of viral replication prior to immune recognition and clearance (Fig. 2a) or a persistently high viral load (Fig. 2b). The unique predictive value of HPV16 viral load for incident ≥CIN2 could be explained if women with HPV16 are more likely to be detected at cross-section with persistent infection and high viral load (State B, Fig. 2) relative to non-HPV16 carcinogenic infections. The fact that HPV16 accounts for nearly 50% of all cervical cancers worldwide, despite being only marginally more prevalent than other types as an asymptomatic infection in the general population, points to poorly-understood but clearly unique properties of HPV16 relative to the other carcinogenic HPV genotypes. This is also reflected in the present study where the cumulative risk of ≥CIN2 diagnosis was higher for HPV16 (20.6%), compared to women with non-HPV16 carcinogenic types (5.1%) or noncarcinogenic types (1.6%) (data not shown).
Figure 2.
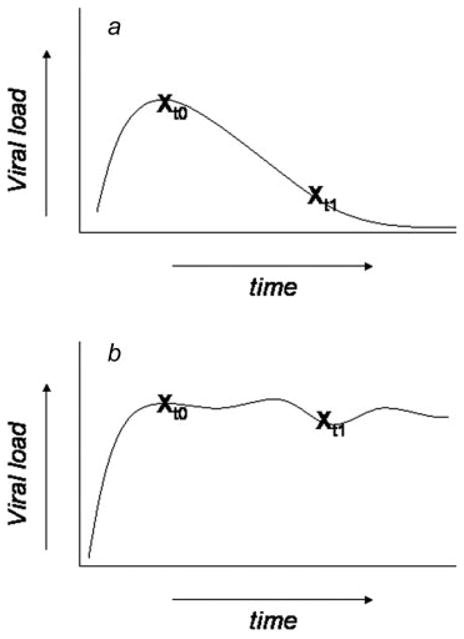
Schematic representation of underlying trajectory of HPV infection from cross-sectional viral load measurements. A similar baseline viral load measurement (t0) is found in subject (a) and subject (b). However, longitudinal characterization of the subsequent natural history of the infection demonstrates that the high viral load could reflect one of two underlying states (a) the peak viral replication prior to immune recognition and clearance, or (b) a persistently high viral load, inclusive of a persistent neoplasia. To the extent that women with HPV 16 may be more likely at cross-section to be in state (b) relative to other genotypes, this could explain the unique predictive value of HPV 16 viral load for incident CIN2+.
In summary, carcinogenic high viral load was associated with prevalent ≥CIN2; however only HPV16 was associated with incident ≥CIN2. Because (i) complex research assays which are not clinically available were used to estimate type-specific viral load and (ii) the limitation of prognostic benefit of viral load testing to HPV16 only, efforts to translate these findings into clinical application should be approached with caution.
Acknowledgments
The authors acknowledge the enthusiastic work of the study staff in Guanacaste and the support of the local health authorities that made this effort possible, and the technical support of the Burk lab personnel.
Grant sponsor: NCI; Grant numbers: P50 CA098252, NO1-CP-21081, NO1-CP-33061, NO1-CP-40542, NO1-CP-50535, NO1-CP-81023; Grant sponsor: FUCODOCSA, Costa Rica; Grant number: CA78527; Grant sponsor: NIH.
References
- 1.Bosch F, Lorincz A, Munoz N, Meijer C, Shah K. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–65. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flores R, Papenfuss M, Klimecki WT, Giuliano AR. Cross-sectional analysis of oncogenic HPV viral load and cervical intraepithelial neoplasia. Int J Cancer. 2006;118:1187–93. doi: 10.1002/ijc.21477. [DOI] [PubMed] [Google Scholar]
- 3.Bigras G, de Marval F. The probability for a Pap test to be abnormal is directly proportional to HPV viral load: results from a Swiss study comparing HPV testing and liquid-based cytology to detect cervical cancer precursors in 13,842 women. Br J Cancer. 2005;93:575–81. doi: 10.1038/sj.bjc.6602728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moberg M, Gustavsson I, Gyllensten U. Type-specific associations of human papillomavirus load with risk of developing cervical carcinoma in situ. Int J Cancer. 2004;112:854–9. doi: 10.1002/ijc.20480. [DOI] [PubMed] [Google Scholar]
- 5.Dalstein V, Riethmuller D, Pretet JL, Le Bail Carval K, Sautiere JL, Carbillet JP, Kantelip B, Schaal JP, Mougin C. Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: a longitudinal French cohort study. Int J Cancer. 2003;106:396–403. doi: 10.1002/ijc.11222. [DOI] [PubMed] [Google Scholar]
- 6.Schlecht NF, Trevisan A, Duarte-Franco E, Rohan TE, Ferenczy A, Villa LL, Franco EL. Viral load as a predictor of the risk of cervical intraepithelial neoplasia. Int J Cancer. 2003;103:519–24. doi: 10.1002/ijc.10846. [DOI] [PubMed] [Google Scholar]
- 7.Beskow AH, Gyllensten UB. Host genetic control of HPV 16 titer in carcinoma in situ of the cervix uteri. Int J Cancer. 2002;101:526–31. doi: 10.1002/ijc.90010. [DOI] [PubMed] [Google Scholar]
- 8.Lillo FB, Lodini S, Ferrari D, Stayton C, Taccagni G, Galli L, Lazzarin A, Uberti-Foppa C. Determination of human papillomavirus (HPV) load and type in high-grade cervical lesions surgically resected from HIV-infected women during follow-up of HPV infection. Clin Infect Dis. 2005;40:451–7. doi: 10.1086/427032. [DOI] [PubMed] [Google Scholar]
- 9.Castle PE, Schiffman M, Scott DR, Sherman ME, Glass AG, Rush BB, Schussler JE, Wacholder S, Lorincz AT. Semiquantitative human papillomavirus type 16 viral load and the prospective risk of cervical precancer and cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1311–14. doi: 10.1158/1055-9965.EPI-04-0799. [DOI] [PubMed] [Google Scholar]
- 10.Snijders PJ, Hogewoning CJ, Hesselink AT, Berkhof J, Voorhorst FJ, Bleeker MC, Meijer CJ. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31 and 33-positive women with normal cytology. Int J Cancer. 2006;119:1102–7. doi: 10.1002/ijc.21956. [DOI] [PubMed] [Google Scholar]
- 11.Moberg M, Gustavsson I, Wilander E, Gyllensten U. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br J Cancer. 2005;92:891–4. doi: 10.1038/sj.bjc.6602436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacic MB, Castle PE, Herrero R, Schiffman M, Sherman ME, Wacholder S, Rodriguez AC, Hutchinson ML, Bratti MC, Hildesheim A, Morales J, Alfaro M, et al. Relationships of human papillomavirus type, qualitative viral load, and age with cytologic abnormality. Cancer Res. 2006;66:10112–19. doi: 10.1158/0008-5472.CAN-06-1812. [DOI] [PubMed] [Google Scholar]
- 13.Sherman ME, Wang SS, Wheeler CM, Rich L, Gravitt PE, Tarone R, Schiffman M. Determinants of human papillomavirus load among women with histological cervical intraepithelial neoplasia 3: dominant impact of surrounding low-grade lesions. Cancer Epidemiol Biomarkers Prev. 2003;12:1038–44. [PubMed] [Google Scholar]
- 14.Gravitt PE, Burk RD, Lorincz A, Herrero R, Hildesheim A, Sherman ME, Bratti MC, Rodriguez AC, Helzlsouer KJ, Schiffman M. A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol Biomarkers Prev. 2003;12:477–84. [PubMed] [Google Scholar]
- 15.Ylitalo N, Josefsson A, Melbye M, Sorensen P, Frisch M, Andersen PK, Sparen P, Gustafsson M, Magnusson P, Ponten J, Gyllensten U, Adami HO. A prospective study showing long-term infection with human papillomavirus 16 before the development of cervical carcinoma in situ. Cancer Res. 2000;60:6027–32. [PubMed] [Google Scholar]
- 16.Terry G, Ho L, Szarewski A, Cuzick J. Semiautomated detection of human papillomavirus DNA of high and low oncogenic potential in cervical smears. Clin Chem. 1994;40:1890–2. [PubMed] [Google Scholar]
- 17.Herrero R, Schiffman MH, Bratti C, Hildesheim A, Balmaceda I, Sherman ME, Greenberg M, Cardenas F, Gomez V, Helgesen K, Morales J, Hutchinson M, et al. Design and methods of a population-based natural history study of cervical neoplasia in a rural province of Costa Rica: the Guanacaste Project. Rev Panam Salud Publica. 1997;1:362–75. doi: 10.1590/s1020-49891997000500005. [DOI] [PubMed] [Google Scholar]
- 18.Herrero R, Hildesheim A, Bratti C, Sherman ME, Hutchinson M, Morales J, Balmaceda I, Greenberg MD, Alfaro M, Burk RD, Wacholder S, Plummer M, et al. Population-based study of human papillomavirus infection and cervical neoplasia in rural Costa Rica. J Natl Cancer Inst. 2000;92:464–74. doi: 10.1093/jnci/92.6.464. [DOI] [PubMed] [Google Scholar]
- 19.Bratti MC, Rodriguez AC, Schiffman M, Hildesheim A, Morales J, Alfaro M, Guillen D, Hutchinson M, Sherman ME, Eklund C, Schussler J, Buckland J, et al. Description of a seven-year prospective study of human papillomavirus infection and cervical neoplasia among 10000 women in Guanacaste, Costa Rica. Rev Panam Salud Publica. 2004;15:75–89. doi: 10.1590/s1020-49892004000200002. [DOI] [PubMed] [Google Scholar]
- 20.Castle PE, Schiffman M, Gravitt PE, Kendall H, Fishman S, Dong H, Hildesheim A, Herrero R, Bratti MC, Sherman ME, Lorincz A, Schussler JE, et al. Comparisons of HPV DNA detection by MY09/11 PCR methods. J Med Virol. 2002;68:417–23. doi: 10.1002/jmv.10220. [DOI] [PubMed] [Google Scholar]
- 21.Qu W, Jiang G, Cruz Y, Chang CJ, Ho GY, Klein RS, Burk RD. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol. 1997;35:1304–10. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 23.Cogliano V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005;6:204. doi: 10.1016/s1470-2045(05)70086-3. [DOI] [PubMed] [Google Scholar]
- 24.Schiffman M, Herrero R, Desalle R, Hildesheim A, Wacholder S, Rodriguez AC, Bratti MC, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, et al. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337:76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Gravitt PE, Peyton C, Wheeler C, Apple R, Higuchi R, Shah KV. Reproducibility of HPV 16 and HPV 18 viral load quantitation using TaqMan real-time PCR assays. J Virol Methods. 2003;112:23–33. doi: 10.1016/s0166-0934(03)00186-1. [DOI] [PubMed] [Google Scholar]
- 26.Schiffman M, Khan MJ, Solomon D, Herrero R, Wacholder S, Hildesheim A, Rodriguez AC, Bratti MC, Wheeler CM, Burk RD. A study of the impact of adding HPV types to cervical cancer screening and triage tests. J Natl Cancer Inst. 2005;97:147–50. doi: 10.1093/jnci/dji014. [DOI] [PubMed] [Google Scholar]
- 27.Gage JC, Hanson VW, Abbey K, Dippery S, Gardner S, Kubota J, Schiffman M, Solomon D, Jeronimo J. Number of cervical biopsies and sensitivity of colposcopy. Obstet Gynecol. 2006;108:264–72. doi: 10.1097/01.AOG.0000220505.18525.85. [DOI] [PubMed] [Google Scholar]
- 28.Park TW, Richart RM, Sun XW, Wright TC., Jr Association between human papillomavirus type and clonal status of cervical squamous intraepithelial lesions. J Natl Cancer Inst. 1996;88:355–8. doi: 10.1093/jnci/88.6.355. [DOI] [PubMed] [Google Scholar]
- 29.Castle PE, Schiffman M, Wheeler CM. Hybrid capture 2 viral load and the 2-year cumulative risk of cervical intraepithelial neoplasia grade 3 or cancer. Am J Obstet Gynecol. 2004;191:1590–7. doi: 10.1016/j.ajog.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Herrero R, Castle PE, Schiffman M, Bratti MC, Hildesheim A, Morales J, Alfaro M, Sherman ME, Wacholder S, Chen S, Rodriguez AC, Burk RD. Epidemiologic profile of type-specific human papillomavirus infection and cervical neoplasia in Guanacaste, Costa Rica. J Infect Dis. 2005;191:1796–807. doi: 10.1086/428850. [DOI] [PubMed] [Google Scholar]


