Abstract
NF-κB has long been known to play an important role in autoimmune diseases such as rheumatoid arthritis (RA). Indeed, as our understanding of how NF-κB is utilized has increased, we have been hard put to find a process not associated with this transcription factor family in some way. However, new data originating, in part, from genome-wide association studies have demonstrated that very specific alterations in components of the NF-κB pathway are sufficient to confer increased risk of developing disease. Here we review the data which have identified specific components of the NF-κB pathway, and consider what is known of their mechanisms of action and how these mechanisms might play into the disease process. In addition, the use of genetic information to predict RA is considered.
Keywords: NF-κB, ubiquitin, A20, TNFAIP3, TRAF, REL, single nucleotide polymorphism (SNP), genome-wide association study (GWAS), rheumatoid arthritis (RA)
I. INTRODUCTION
Rheumatoid arthritis (RA) is a chronic autoimmune disease of the joints in which synovial cells proliferate and intermix with infiltrating immune cells to form a pathological tissue termed “pannus.”1 A complex network of cytokines including TNFα, IL-1, IL-6, IL-17, and others is produced leading to a state of inflammation and the secretion of reactive compounds and destructive enzymes such as matrix metalloproteinases (MMPs) and collagenases. These molecules work together to degrade cartilage and extracellular matrix and expose bone cells to the inflammatory environment. Osteoclasts become activated and begin to mobilize hydroxyapatite thus removing bone from the joint. Osteoblasts respond by rebuilding the bone; however, as patterning information has become corrupted, the process leads to the progressive malformation of the joint and crippling of the patient. The cytokine network plays a major role in disease progression. Inflammatory cytokines program the cells comprising pannus to assume their pathological character. Macrophages are considered one of the major contributors of cytokines to this process, producing TNFα, IL-1, and IL-6 in large quantities. Programmed by IL-23, T cells differentiate into Th17 cells which secrete IL-17. IL-17, in turn, promotes neutrophil migration and activation. B cells secrete autoantibodies such as rheumatoid factor and anti-cyclic citrullinated protein (anti-CCP) antibodies. Anti-CCP antibodies are currently of great interest as they are both quite predictive of disease and mark a specific subset of RA.2 In addition to secreting autoantibodies, B cells are extremely efficient antigen presenting cells and so act to continue activating T cells. Fibroblast-like synoviocytes as well as other cells secrete MMPs and RANKL which activate osteoclasts, leading ultimately to the bone malformations that characterize late stage RA.
NF-κB has long been appreciated as a central player in many of the processes underlying the progression of RA. This is not surprising as NF-κB plays a central role in the mediation of inflammation and the immune response.3 NF-κB is comprised of a family of five DNA-binding proteins; relA (p65), relB, c-rel, p50 (derived from the p105 precursor), and p52 (derived from the p100 precursor), which can mix and match to form a large number of dimeric transcription factors. These dimers are held by cytoplasmic inhibitors until their release via signal-mediated degradation of those inhibitors. In the most canonical example, p50/relA dimers are held in the cytoplasm by IκBα. An inflammatory signal such as TNFα. induces the activation of the IκB kinase (IKK) complex to phosphorylate members of the IκB family. Phosphorylated IκB becomes ubiquitinated and is then targeted for degradation by the proteasome. The NF-κB dimers can then translocate to the nucleus and activate the transcription and/or repression of genes. NF-κB has been implicated in cytokine release, activation of virtually all immune cells, osteoclast activation, autoantibody production, cellular proliferation, inhibition of apoptosis, and numerous other processes associated with RA. Tremendous energy has been expended on the generation of NF-κB inhibitors and, indeed, numerous inhibitors have been (and are continuing to be) synthesized. Unfortunately, most have toxicities that render them unusable in the clinic. One reason for this is that NF-κB plays a role in differentiation, metabolism, and certain aspects of CNS function in addition to its role as a mediator of inflammation (for review see 4,5). A more nuanced approach, which is now being pursued, involves the targeting of subsets of NF-κB activities which, hopefully, will target pathogenic processes while leaving physiological processes largely intact. The depth and complexity of regulatory nodes within the NF-κB system is amenable to this strategy.
In some ways, the discussion above paints NF-κB as part of the necessary substratum of circuitry linking signals to responses and making possible all of the processes we collectively think of as RA. However, new data emerging over the past decade have demonstrated that subtle changes in the NF-κB circuitry can have measurable effects on RA susceptibility as well as disease progression. These data derive, in part, from the advances that have been made in high throughput screening of genetic sequences along with the creation of large collections of genetic material from RA patients throughout the world. Additionally, the creation of new statistical tools with which these data may be analyzed has been essential to this effort. Single nucleotide polymorphisms (SNPs) serve as a basic unit of human genetic variation.6 These are high frequency events with (for example) 75% of alleles within the entire human population at some position having a G residue and with 25% of alleles having an A residue at that same location. In contrast, a rare event such as an allele that confers retinitis pigmentosa would be referred to as a mutation rather than a SNP. While SNPs can have functional consequences such as changing the coding sequence of a gene or an important enhancer within a promoter, more often they serve as markers of a genomic region. SNPs, which are found more often than statistics would predict in patients with a particular disease, are said to be associated with that disease. Healthy people who carry that SNP are at an increased risk for developing that disease. The interpretation is that SNPs lie close enough to some unidentified functional change in DNA sequence such that during recombination they almost always segregate with that sequence. Currently, several hundred thousand SNPs have been catalogued and many have been placed on arrays such as the Affymetrix platform, allowing the entire genome to be assessed in one measure. This has spawned genome-wide association studies (GWAS) which are making great strides in the understanding of the genetic basis for human diseases.7 Indeed, as will be described below, several GWAS in recent years have identified components of the NF-κB circuitry as risk factors in RA. Understanding exactly how these changes in certain NF-κB circuits are manifest and how they contribute to RA will both increase our understanding of who is at risk as well as our understanding of how RA develops. Given our need to identify methods of inhibiting subsets of NF-κB circuits, the identification of the specific circuits which place people at risk may identify therapeutic targets as well. Here, we will consider some of the direct components of the NF-κB regulatory circuitry that have been identified by GWAS and review what is currently known about their function and their role in RA.
II. REGULATION OF NF-κB ACTIVATION
Two major pathways of NF-κB activation have been delineated (for recent reviews see 3,8–12). In the classical pathway, relA/p50 dimers (for example) are held in the cytoplasm by a member of the IκB family. IκB is phosphorylated on two serines to create an ubiquitination signal which targets IκB for degradation via the proteasome. The degradation of IκB unmasks a nuclear localization signal on relA/p50, allowing it to translocate to the nucleus and regulate the transcription of genes. Phosphorylation of IκB is mediated by a protein complex referred to as IKK. IKK in turn is associated with a number of different upstream pathways, allowing NF-κB to be activated by a rather large number of different signal transduction events. Examples of signals that activate NF-κB via the classical pathway include antigen receptors, cytokine receptors such as TNFR1 and IL-1R, and Toll-like receptors (TLR). The relA subunit is often part of the classically activated NF-κB dimer. The alternative (noncanonical) pathway of NF-κB activation involves complexes of NF-κB subunits associated with p100 or p105. P100 and p105 are precursors to p52 and p50 subunits, respectively. The C-termini of these proteins contain an IκB-like domain which is cleaved in the creation of p52 and p50 but when intact, can serve the same function as IκB, folding over and masking the nuclear localization sequence. Processing of these precursors is regulated by the NF-κB inducing kinase (NIK). NIK, in turn, is activated by the IKK complex. Examples of signals that activate NF-κB via the alternative pathway include the TNFR superfamily members such as CD40, the lymphotoxin-β receptor, and BAFFR. The IKK complex consists of IKKα and IKKβ associated with the scaffold protein, IKKγ/Nemo, an essential regulatory subunit of the IKK complex.13 It is IKKβ which targets members of the IκB family while it is IKKα which targets NIK. In this fashion, the IKK complex serves as an important regulatory switch. Given that NF-κB is associated with so many different processes, the question of specificity immediately arises. In part, specificity is achieved by the different homo- and heterodimers of NF-κB subunits that can be created, each of which binds to partially overlapping but ultimately different populations of DNA-binding sites.14 However, it is becoming apparent that specificity also comes from chromatin states, exposing some sites while hiding others in a cell-and context-specific fashion.15,16
Ubiquitin, a small protein expressed in all eukaryotic cells, plays a major role in the regulation of NF-κB activation. Its most celebrated role is to be attached to proteins as a post-translational modification. Attachment is often a three-step process (for reviews see 17,18). Ubiquitin is activated via an E1 enzyme, conjugated to an E2 enzyme, and then finally targeted for attachment to a specific protein by an E3 ubiquitin ligase. This is not necessarily a permanent alteration. There also exist de-ubiquitinating enzymes (DUBs) which remove the ubiquitin modification. Ubiquitin may attach as a monomer or it may be concatenated into long chains. In this regard, ubiquitin has several internal lysine residues which serve as connection points (K48 and K63, for example). The C-terminus of one ubiquitin is attached to the lysine of a second ubiquitin which then through multiple iterations creates chains of varying length which are finally attached to the target protein via the E3 ligase. Additionally, linear ubiquitin chains are concatenated by the E3 complex; the linear ubiquitin chain assembly is complex19 and appears to play an important role in NF-κB activation by a variety of signal transduction pathways. Different chains confer different signals, thus allowing ubiquitin to be used for many different purposes within the cell including cellular trafficking, regulation of transcription, responding to DNA damage, and of course, targeting proteins for degradation.20
Activation of NF-κB via engagement of TNFR1 has been extensively analyzed and provides several important examples of ubiquitin utilization. Engagement of TNFR1 by TNFα causes trimerization and the assembly of a receptor complex including TRADD, TRAF2, RIP, and cIAP1/2. RIP is ubiquitinated by cIAP1/2 which has E3 ligase activity. TRAF2 also has E3 ligase activity and may serve to facilitate cIAP1/2. The ubiquitination of RIP (which occurs via K63 linkages) creates a scaffold to which additional proteins bind. TAK1 interacts with ubiquitinated RIP via ubiquitin-binding adaptors, TAB2 and TAB3. IKK is also recruited via the ability of IKKγ/ Nemo’s ubiquitin-binding domain to interact with the RIP ubiquitin chains. The complex is additionally stabilized through linear ubiquitination of IKKγ/Nemo by linear ubiquitin chain assembly complex, which appears to be necessary for efficient activation of NF-κB. TAK1 can then phosphorylate IKKβ leading to the activation of IKK and the phosphorylation of IκB. Phosphorylated IκB is then ubiquitinated by a different E3 ligase leading to its targeting for proteasomal degradation.
III. GENETIC LOCI AFFECTING RA: EARLY WORK
While the etiology of RA remains elusive, it is likely that the disease arises through an interaction between environmental exposures and the genetics of the exposed individuals. Family studies generated the earliest data identifying a genetic component contributing to RA which was mapped via linkage analysis to the HLA-DRB1 locus.21 Subsequent work overcame the significant linkage disequilibrium characteristic of this region to further dissect this region and reveal non-DRB1 HLA associations with anti-CCP positive RA.22–24 With the development of large SNP databases, consortia of laboratories were able to amass sufficiently large populations to identify non-MHC loci such as PTPN22.25 PTPN22 encodes a protein tyrosine phosphatase which negatively regulates T cell function and is probably a general autoimmunity locus as SNPs within this locus associate with multiple autoimmune diseases.26 These two loci represent the low hanging fruit of association studies with high levels of statistical significance.
IV. THE TNFAIP3 LOCUS
The identification of single nucleotide polymorphisms (SNPs) and the development of technology to allow high throughput analysis of thousands of SNPs paved the way for genome-wide association studies (GWAS).7 One of the first large scale GWAS was performed by the Wellcome Trust Case Control Consortium (WTCCC) and examined 1,860 patients suffering from one of seven major diseases (of which one was RA) and compared them to 2,938 controls.27 Of the many associations found, several SNPs were identified on chromosome 6q23 that had a moderate association with RA. This was validated in an independent cohort of 5063 RA patients and 3849 controls.28 An independent group examined 397 RA patients and compared this cohort to individuals from the Framingham Heart Study as a control group and identified two independent SNPs in the same region.29 The 6q23 region is intergenic; lying between the genes encoding the oligodendrocyte lineage transcription factor 3 and the tumor necrosis factor-α-induced protein 3 (TNFAIP3, also known as A20).28 It was appreciated from the beginning that these SNPs were not causal, but rather were tightly linked genetically with one or more causal mutations that were not yet identified. As with PTPN22, the TNFAIP3/ oligodendrocyte lineage transcription factor 3 locus appears to be associated with multiple autoimmune diseases such as type I diabetes, Sjogren’s syndrome, and systemic lupus erythematosus among others.30–32 Additional GWAS have replicated the association of the TNFAIP3/oligodendrocyte lineage transcription factor 3 locus with autoimmune diseases in a variety of specific ethnic groups around the world, further strengthening the ability of this locus to affect disease in a variety of genetic backgrounds.33–35
TNFAIP3 is a negative regulator of NF-κB signaling associated with several important signal transduction pathways including TNFR1, TLR4, and CD40 among others. It was originally identified by virtue of its upregulation by TNFα and it became known as an inhibitor of TNF signaling because high expression levels were associated with resistance to TNFα-mediated apoptosis.36 It was clear from the knockout that TNFAIP3 was a critical regulator of NF-κB activity as TNFAIP3−/− mice rapidly died due to systemic inflammation and MEFs derived from these mice had spontaneously activated NF-κB.37 More recently, a myeloid-specific knockout of TNFAIP3 was constructed.38 The mice developed a spontaneous polyarthritis similar to rheumatoid arthritis strongly implicating the TNFAIP3 gene as the conveyer of risk identified in GWAS.
Using a combination of SNP analysis, genomic sequence data from the 1000 Genomes Project,6 and resequencing of TNFAIP3 regions from nine individuals with SLE, one group identified a TT>A polymorphic dinucleotide variant in a conserved extragenic region likely to contain regulatory sequences.39 They tested this region for DNA-binding factors and found reduced avidity for NF-κB binding. Others have examined these data and consider it likely that the actual disruption is in a BCL-binding motif which, in turn, acts to decrease NF-κB binding.40 This study is of note as it uses the TNFAIP3 GWAS data as a test for a database annotation system for genomic data (RegulomeDB) which allows for the generation of testable hypotheses from large data sets. Sequencing of the TNFAIP3 gene region in patients with multiple autoimmune diseases has also found polymorphisms within the TNFAIP3 gene itself.41 The authors reported 14 exon polymorphisms and 18 intron polymorphisms; however, no expression analysis or functional testing was performed. It will be of great interest to see exactly how these mutations affect TNFAIP3 function.
TNFAIP3 was found to interact with TRAF242 and with TRAF6,43 a mediator of IL-1 activation of NF-κB. Remarkably, it was found that TNFAIP3 had both DUB activity and E3 ligase activity via different domains.44,45 Kinetic analysis demonstrated that TNFAIP3 first removes the K63 ubiquitin chains from RIP1 within minutes of activation and then some time later (3–6 hours) TNFAIP3 uses its E3 ligase activity to place K48 ubiquitin chains on RIP1 resulting in RIP1 degradation via the proteasome (Fig. 1). This mechanism has recently been further elucidated with the construction of gene targeted mice containing mutant forms of TNFAIP3 which have either lost their E3 ligase or their DUB activity.46 The authors found that both activities were necessary for efficient regulation of NF-κB. How-ever, unlike the TNFAIP3 knockout mouse, these mice did not develop any spontaneous inflammatory disease. Importantly, they found that the E3 ligase domain appeared to be necessary for recruiting TNFAIP3 to ubiq-uitinated RIP1 and that its loss resulted in increased RIP1 ubiquitination. Interestingly, the two mutants, when co-expressed, could complement each other through dimerization. These data suggest that the coordination of different functions of TNFAIP3 occur within the context of higher order complexes rather than in single molecules.
FIGURE 1.
TNFAIP3/A20 regulates NF-κB through both deubiquitinase and E3 ligase activities. (A) Upon engagement of TNFR1, clAP1/2 and TRAF2 promote the K63 ubiquitination of RIP1 while LUBAC promotes the linear ubiquitination of the IKK complex. These are brought together, in part, through the actions of the ubiquitin-binding adaptors TAB1 and TAB2. TAK interacts with TAB1 and TAB2 and phosphorylates IKKβ thus activating the IKK complex and initiating the NF-κB activation process. (B) TNFAIP3 (labeled as A20), functioning as a deubiquitinase, removes the K63 ubiquitin chain from RIP1 thus destabilizing the complex and blocking the ability of TAK to phosphorylation IKKβ. (C) Subsequently, TNFAIP3/A20, functioning as an E3 ligase, places K48 ubiquitin chains on RIP1 thus targeting it for proteasomal degradation. In addition, TNFAIP3/A20 escorts TRAF2 to the lysosome (in an ubiquitin-independent manner) where it is degraded as well.
In addition to TNFAIP3 modulating ubiquitin others have found that TNFAIP3 can also inhibit IKK complex activity through a mechanism that is independent of ubiquitin. One group found, upon overexpression of TNFAIP3 in an HEK293 cell line that also overexpressed the IL-1R, that TNFR1 complexes still contained polyubiquitinated RIP1 upon TNFα binding even though NF-κB was strongly inhibited.47 They ultimately found that TNFAIP3 was physically interacting with an N-terminal domain within IKKγ/Nemo. This was occurring through the seventh zinc finger domain (ZnF7) of TNFAIP3 and required the polyubiquitination of IKKγ/Nemo. In the presence of TNFAIP3 bound to IKKγ/Nemo, TAK1 was unable to phosphorylate IKKβ in an in vitro system. The authors speculated that their results differed from those of Wertz et al.45 due to their use of a different cell type implying that TNFAIP3 might inhibit NF-κB differently in different tissues. This work was repeated and extended by several groups to show that the polyubiquitin chains of IKKγ/ Nemo are linear in nature and thus mediated by linear ubiquitin chain assembly complex.48,49
The model which then emerges is one in which TNFAIP3 is a pleiotropic inhibitor of NF-κB that can interact with numerous other proteins, enzymatically modulates ubiquitin chains, and oligomerizes into higher order structures. It will be important to determine if altered risk for RA is mediated primarily by downregulation of expression of fully functional TNFAIP3 or if there exist mutations which selectively alter some subsets of TNFAIP3 functions.
V. TRAF LOCI
A GWAS performed by the North American RA Consortium (NARAC) in collaboration with the Swedish Epidemiologic Investigation of RA (EIRA) examined >300,000 SNPs within 1522 RA patients scoring positive for the presence of anti-CCP antibodies and 1850 controls.50 In addition to PTPN22 and HLA-DRB1 the authors found a SNP with an odds ratio of 1.32 on chromosome 9, lying between the TNF receptor associated factor 1 (TRAF1) and complement component 5. This work was extended by others, finding associations throughout the TRAF1 gene region into the TRAF1-complement component 5 intergenic space but excluding the complement component 5 coding region,51 thus identifying TRAF1 as the most likely disease modifying candidate. As with the TNFAIP3 locus, this association has been replicated in different ethnic groups.52–55 Interestingly, several groups have begun dissecting various aspects of RA and querying if the TRAF1 susceptibility locus associates with isolated disease components. Thus, for example, SNPs within the TRAF1 locus are associated with radiological severity of disease and may serve as a predictor56,57 but are not associated with increased cardiovascular risk.58 As of yet, no work has been published examining TRAF1 expression levels in these patients.
TRAF1 was originally identified as an adaptor protein associated with TNF receptor 2 59 where it binds in combination with TRAF2. Subsequently, TRAF1 was found to interact with other TNFR superfamily members including CD40, receptor activator of NF-κB, 4-1BB, and others.60 Unlike TRAFs 2–6 which encode an E3 ligase activity within the RING domain, TRAF1 is missing this function, suggesting that it should function somewhat like a dominant negative TRAF2. Its role in the regulation of NF-κB has turned out to be complex and is both context and tissue dependent. Early work found that overexpression of TRAF1 in human embryonic kidney cells resulted in the inhibition of TNFa-mediated NF-κB activation while overexpression in HeLa cells resulted in potentiation of TNFα.61,62 Knockout of TRAF1 resulted in a T cell population which responded more vigorously to anti-CD3 stimulation than wild-type T cells and responded to TNFα by proliferating, a response not seen in controls.63 However, examination of TRAF1 and TRAF2 knockout B cell lines indicated that TRAF1 and TRAF2 cooperate to promote NF-κB activation upon CD40 stimulation.64 When CD40 is activated, TRAF2 translocates into lipid rafts in a RING domain-dependent process which appears to be necessary for kinase activation. In a subcellular localization study, TRAF1 was found to displace CD40/TRAF2 from lipid rafts but appeared to be necessary for sustained activation.65 Examination of IKKβ interactions using a two-hybrid screen uncovered an interaction with TRAF1.66 TRAF1 was found to both activate and inhibit the IKK complex depending on dose; low amounts of TRAF1 were stimulatory while high levels were inhibitory. Others have found that TRAF1 can interact differentially with components of the classical and alternative NF-κB pathways. In a study of CD8 T cell survival and the role of TRAF1 in the function of the co-stimulatory receptor 4-1BB, it was found that TRAF1 was required for maximal stimulation via the classical pathway but acted to repress NIK-mediated activation.67 Conversely, another group, studying pulmonary epithelial cells, found that TRAF1 interacted with NIK and stabilized it from TRAF2/cIAP-mediated degradation.68 Clearly, the role of TRAF1 will need to be carefully dissected in each tissue before we have a clear idea of how exactly it functions during RA. The identification of structural TRAF1 mutations affecting RA risk will be especially useful.
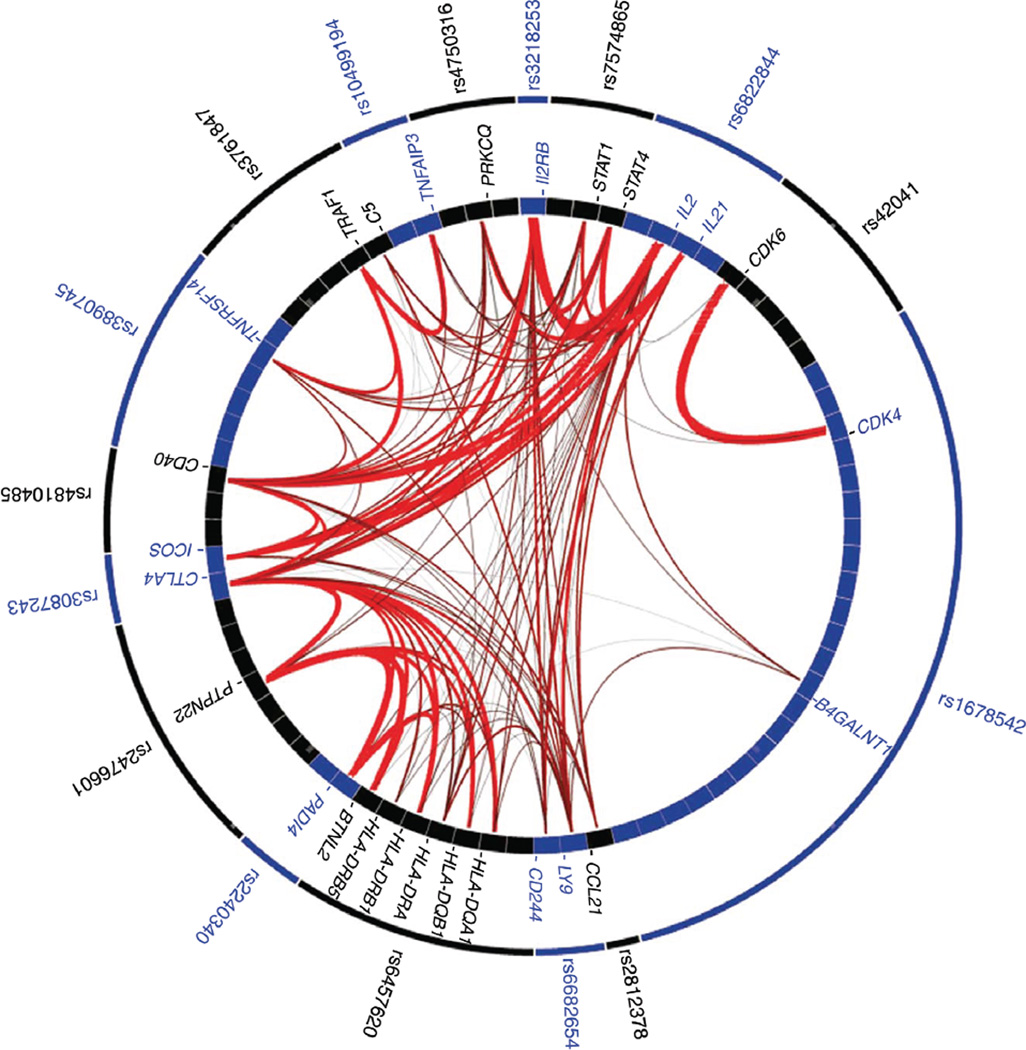
Other TRAF loci have been associated with RA. Using a more directed analysis of just SNPs associated with TRAF loci, a screen of 1273 RA cases with 2463 healthy controls identified a single SNP which mapped upstream of TRAF5 and was placed within a putative enhancer sequence.69 A recently developed functional genomics tool, GRAIL (gene relationships across implicated loci), allows SNPs with association P values that are suggestive to be further analyzed70 and this has resulted in the identification of several new loci to be associated with RA including TRAF6.71 The tool is novel enough that it is worth considering it in more detail. GRAIL uses established statistical text mining methods to query a database of 250,000 published abstracts and create a connectivity or relatedness map between genes (Fig. 2). Known disease loci are fed into the program (the seed regions) and unknown loci are then supplied as the queries. GRAIL returns a P value based on the functional connectedness of the unknown loci to the known loci. The authors tested their tool on the ability to identify validated loci in RA by taking a subset of known RA-associated genetic loci as seeds and querying with other validated loci. GRAIL returned a significant P value 83% of the time. When a large number of irrelevant loci were provided as seeds, approximately 5% returned significant P values, thus, establishing the false positive rate for the process. The authors then used as queries 179 SNPs from their previous GWAS that were close to significance, but had not made the cutoff. The program returned significant P values for 22 of these SNPs (the false positive rate predicted a return of 9–10 SNPs as significant). They then genotyped the 22 SNPs in an independent study involving 7957 RA patients and 11,958 controls and found that seven of the SNPs showed a significant association with RA; CD2-CD58, CD28, PRDM1, TAGAP, PTPRC, TRAF6-RAG1, and FCGR2A.
FIGURE 2.
GRAIL text mining shows functional connectivity among genes known to be associated with RA. Upon input of SNPs known to be associated with RA, the GRAIL utility returns a list of genes near those SNP locations and their connection strengths as measured by mining the abstract database. SNPs known to be associated with RA are shown in the outer ring. Genes which are near the SNPs are represented in the inner ring. For example, CD40 shows strong associations with TRAF1, IL2, and IL21 and a somewhat weaker association with TNFAIP3. PTPN22 shows strong associations with HLA loci, PADI4, and BTNL2. Figure originally published in Nature Genetics; Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. 2009;41:1313-18.71
TRAF6 is of interest in regards to osteoclast function. The main osteoclast activating receptor is called the receptor activator of NF-κB. Upon engagement of its ligand, RANKL, TRAF6 is recruited to the cytoplasmic domain of receptor activator of NF-κB where it functions as an E3 ubiquitin ligase. The importance of TRAF6 in osteoclast function was underscored by the phenotype of TRAF6 knockout mice.72 Osteoclasts from these mice were defective in NF-κB activation and developed bone that was abnormally dense and prone to breakage. TRAF6 associates with p62, a scaffold for the atypical PKCs.73 This complex then contributes to IKK activation. TRAF6 also interacts with IRAK1 to mediate the activation of certain Toll-like receptors (TLRs).74 TLR signaling has been implicated in RA. While a pathogenic cause of RA remains controversial, it is clear that endogenous danger signals (DAMPs) are present in the joints of RA patients and TLRs are activated.75 Association studies have provided mixed results. A GWAS of a Dutch population found no association with TLR loci76 while a meta-analysis including both European and Asian patients found an association with TLR2 and TLR9 but not TLR4.77
VI. THE CD40 LOCUS
The CD40 locus was associated with RA through a meta-analysis of two previous GWAS involving 3393 RA cases and 12,462 controls.78 The associations were then tested against independent populations of 3929 autoantibody positive RA cases and 5807 matched controls. While this association did not replicate in a cohort of 1128 Korean RA patients,79 it was validated in a study of 3962 British RA patients80 and in a Northern Indian RA patient population.81 The CD40 risk allele was found to associate with more severe disease as measured by the degree of joint destruction.82 Extending these results with dense SNP analysis and exonic sequencing in a large cohort of RA patients it was found that the risk allele corresponds to a gain of function mutation resulting in an increase of approximately 33% in B cell expression of CD40.83
CD40, a member of the TNFR superfamily, is a receptor found on lymphoid cells and synovial fibroblasts.84 CD40 engagement by its ligand, CD154, directly activates B cells and can induce expression of autoantibodies including anti-CCP autoantibodies. On T cells, activation of CD40 can provide a costimulatory function.85 CD40L is upregulated in T cells of RA patients86,87 and promotes the downregulation of the inhibitory receptor, CD32B, on B cells via CD40 signaling. Synovial fibroblasts, cultured with activated T cells, secrete cytokines in a CD40-dependent manner as well as RANKL. CD40 must recruit a signaling complex to function. The cytoplasmic region of CD40 contains a site for the binding of TRAF6 and another site for the binding of either TRAF2, 3, or 5. When the CD40 receptor is activated, TRAF1 will either dimerize with TRAF2 or associate with the TRAF2/3/5 site, displacing the activating TRAF.65
The identification of CD40 as an RA risk allele has invoked the concept of a genetically delineated full signaling circuit. Both TRAF1 and TNFAIP3 function to modulate CD40 signaling. Antibodies targeting either CD40 or CD40L were found to ameliorate arthritis when administered before disease in the collagen induced arthritis (CIA) model and in the K/BxN model.88–90 However, treatment with the anti-CD40L antibody after disease was established had little to no effect in either model.88,89 This could argue that the CD40 pathway may be important in the early stages of human RA but less so in later stages. In this case, the genetic data described above have delineated an important mechanism by which the dysregulation of an NF-κB pathway leads to an autoimmune disease. However, others are pursuing the concept that modulation of CD40 signaling in human RA patients will be therapeutically efficacious. Using a high throughput screen followed by testing with human B cells, two novel compounds were identified which, presumably, will move towards clinical testing in the near future.83
VII. THE REL LOCUS
The next component of the NF-κB pathway to be identified by GWAS was REL itself.91 This work underscores the importance of large sample sizes, improved mapping technology, and more robust statistical filters. A previous GWAS (described above50) was reanalyzed using the Illumina HapMap370 Bead Array typing platform with new cases added. In total, 2418 cases and 4504 controls were examined and the data replicated in a case-control data set consisting of 2604 cases and 2882 controls. Clusters of cases and controls were matched to create structured associations. This association was confirmed in a study of an RA patient population from the United Kingdom.92
Activation of relA-containing dimers has been observed to be rapid and transient while activation of c-rel-containing dimers is somewhat delayed and more persistent.93 In T cells, c-rel activation is critically dependent on CD28 co-stimulation, unlike relA-containing dimers which are activated by a variety of costimulatory signals.94 It has been appreciated for some time that different NF-κB subunits have different functions. In a classic study, relA, p50, and c-rel knockout mice were examined for dendritic cell properties.95 Mice doubly deficient in relA and p50 were significantly impaired in their ability to generate dendritic cells while mice doubly deficient in c-rel and p50 generated dendritic cells which were deficient in CD40 signaling. This is especially interesting given the observation, described above, that certain CD40 alleles can confer an increased risk of RA. C-rel is also required for the expression of FoxP3, making c-rel essential for T regulatory cell development.96 Consistent with these data, it was found that the E3 ubiquitin ligase, Peli1, negatively regulates c-rel and in Peli1 knockout mice there develops a spontaneous autoimmune disease in which c-rel accumulates in the nucleus.97 Thus, one could imagine that perturbations of c-rel in either direction could play a role in RA.
VIII. PREDICTIONS OF THERAPEUTIC EFFICACY
The class of therapeutics termed Biologicals has had great success in treating RA. These are protein therapeutics which target either specific cytokines, T cells, or B cells. In many cases, these therapeutics indirectly induce a decrease in the activity of various NF-κB circuits via the blockade of upstream ligands or receptors. Therapeutic success is far from complete, however. While many patients respond, only a subset of patients responds significantly. For example, in a study of the TNFα blocker, adalimumab, it was found that after 1 year only 41% of patients improved by 50% or better.98 Addition of methotrexate increased this to 62% of patients. Given the high cost of these therapeutics along with the possibility of serious adverse effects much effort has gone into attempting to predict response. Numerous potential biomarkers have been examined including clinical parameters, radiological parameters, serum proteins such as autoantibodies or cytokine signatures, as well as the genetics of the patient.99 Focusing here on anti-TNFα therapeutics, numerous studies have found predictive biomarkers; however, they seem to vary from study to study. In one such study that examined 617 RA patients’ response to either etanercept, adalimumab, or infliximab it was found that low disease activity, higher levels of education, not smoking, lack of anti-CCP antibodies, and no glucocorticoid use were predictive of response.100 In another study involving 2625 RA patients’ response to adalimumab it was found that high disease activity along with good functional ability and young age was predictive of response.101 Additionally, a pilot study found high levels of anti-CCP antibodies a good predictor of efficacy for patients taking infliximab.102 These results show a clear need for standardization of disease as well as a careful assessment of patient characteristics. Examination of SNPs has also generated some small successes; however, the associations are modest and hard to reproduce. GWAS data from the Wellcome Trust Case Control Consortium27 were utilized to analyze anti-TNF treatments in 566 RA patients. They found seven SNPs which showed association with response to treatment; however, no obvious connections could be made between SNP locations and RA pathology. Another GWAS study made use of 13 different patient collections, analyzing 2706 RA patients treated with either etanercept, infliximab, or adalimumab.103 They found an association with CD84 with response to etanercept but not infliximab or adalimumab. CD84 is a receptor expressed on T cell and B cells and plays an immunomodulatory role.104 Hypothesis driven genetic studies have also shown some limited success. One group found SNPs associated with the p38 MPAK pathway in an analysis of 1102 RA patients taking either etanercept, infliximab, or adalimumab.105 Another group examined the TNFα promoter in 2127 RA patients taking either etanercept, infliximab, or adalimumab and found that a G at position −308 conferred a better response.106 Thus, while some progress has been made in identifying potential biomarkers, clearly combinations of biomarkers will need to be identified to provide a robust form of personalized medicine capable of predicting response with some accuracy.
CONCLUSION
Genomic analysis has made enormous advances over the past decade. As the cost of genomic sequencing continues to fall we will eventually have large data sets of complete genomes along with medical histories. From this wealth of information, once suitable analytic tools are developed to effectively mine these data, we will truly begin to understand how the alteration of signaling circuits contributes to disease. The current form of GWAS may be considered an intermediate stage in this process. Nevertheless, GWAS in its current form is a powerful tool. Through GWAS, NF-κB pathways have emerged as potential risk factors for RA. While we have appreciated for decades that NF-κB mediates many of the processes involved in disease progression, we now can see that specific perturbations in a subset of NF-κB pathways may themselves play a causative role. Much yet needs to be done. There most certainly will be structural mutants which, when found, may shed important light on how subtle perturbations in the NF-κB network may lead to disease. Additionally, it is likely that these genes do not work alone. Gene interaction analysis will need to be performed and may give us an unparalleled ability to predict both disease risk and potential response to therapeutics. Finally, we may unearth important new targets that will further advance our ability to help those suffering from the crippling end point of RA.
ACKNOWLEDGMENTS
Robert Scheinman is supported by the University of Colorado School of Pharmacy and NIH 5R01EY017533-05.
ABBREVIATIONS
- anti-CCP
anti-cyclic citrullinated protein
- GRAIL
gene relationships across implicated loci
- GWAS
genome-wide association studies
- NIK
NF-κB inducing kinase
- RA
rheumatoid arthritis
- SNPs
single nucleotide polymorphisms
- TLR
Toll-like receptors
- TNFAIP3
tumor necrosis factor-α-induced protein 3
- TRAF1
TNF receptor associated factor 1
REFERENCES
- 1.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 2.van Venrooij WJ, van Beers JJ, Pruijn GJ. Anti-CCP antibodies: The past, the present and the future. Nat Rev Rheumatol. 2011;7:391–398. doi: 10.1038/nrrheum.2011.76. [DOI] [PubMed] [Google Scholar]
- 3.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 4.Kaltschmidt B, Kaltschmidt C. NF-kappaB in the nervous system. Cold Spring Harb Perspect Biol. 2009;1:a001271. doi: 10.1101/cshperspect.a001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tornatore L, Thotakura AK, Bennett J, Moretti M, Franzoso G. The nuclear factor kappa B signaling pathway: Integrating metabolism with inflammation. Trends Cell Biol. 2012;22:557–566. doi: 10.1016/j.tcb.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from populationscale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush WS, Moore JH. Chapter 11: Genomewide association studies. PLoS Comput Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown KD, Claudio E, Siebenlist U. The roles of the classical and alternative nuclear factor-kappaB pathways: Potential implications for autoimmunity and rheumatoid arthritis. Arthritis Res Ther. 2008;10:212. doi: 10.1186/ar2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 10.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 11.Razani B, Reichardt AD, Cheng G. Non-canonical NF-kappaB signaling activation and regulation: Principles and perspectives. Immunol Rev. 2011;244:44–54. doi: 10.1111/j.1600-065X.2011.01059.x. [DOI] [PubMed] [Google Scholar]
- 12.Sun SC. The noncanonical NF-kappaB pathway. Immunol Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu F, Xia Y, Parker AS, Verma IM. IKK biology. Immunol Rev. 2012;246:239–253. doi: 10.1111/j.1600-065X.2012.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smale ST. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunol Rev. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 15.Natoli G. NF-kappaB and chromatin: Ten years on the path from basic mechanisms to candidate drugs. Immunol Rev. 2012;246:183–192. doi: 10.1111/j.1600-065X.2012.01103.x. [DOI] [PubMed] [Google Scholar]
- 16.Smale ST. Hierarchies of NF-kappaB target-gene regulation. Nat Immunol. 2011;12:689–694. doi: 10.1038/ni.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda F, Dikic I. Atypical ubiquitin chains: New molecular signals. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emmerich CH, Schmukle AC, Walczak H. The emerging role of linear ubiquitination in cell signaling. Sci Signal. 2011;4:re5. doi: 10.1126/scisignal.2002187. [DOI] [PubMed] [Google Scholar]
- 20.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis: An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 22.Ding B, Padyukov L, Lundstrom E, Seielstad M, Plenge RM, Oksenberg JR, Gregersen PK, Alfredsson L, Klareskog L. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–38. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HS, Lee AT, Criswell LA, Seldin MF, Amos CI, Carulli JP, Navarrete C, Remmers EF, Kastner DL, Plenge RM, Li W, Gregersen PK. Several regions in the major histocompatibility complex confer risk for anti-CCP-antibody positive rheumatoid arthritis, independent of the DRB1 locus. Mol Med. 2008;14:293–300. doi: 10.2119/2007-00123.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vignal C, Bansal AT, Balding DJ, Binks MH, Dickson MC, Montgomery DS, Wilson AG. Genetic association of the major histocompatibility complex with rheumatoid arthritis implicates two non-DRB1 loci. Arthritis Rheum. 2009;60:53–62. doi: 10.1002/art.24138. [DOI] [PubMed] [Google Scholar]
- 25.Gregersen PK, Batliwalla F. PTPN22 and rheumatoid arthritis: gratifying replication. Arthritis Rheum. 2005;52:1952–1955. doi: 10.1002/art.21125. [DOI] [PubMed] [Google Scholar]
- 26.Gregersen PK, Lee HS, Batliwalla F, Begovich AB. PTPN22: Setting thresholds for autoimmunity. Semin Immunol. 2006;18:214–223. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Wellcome-Trust-Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, Donn R, Symmons D, Hider S, Bruce IN, Wellcome Trust Case Control Consortium. Wilson AG, Marinou I, Morgan A, Emery P, YEAR Consortium. Carter A, Steer S, Hocking L, Reid DM, Wordsworth P, Harrison P, Strachan D, Worthington J. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, Pe’er I, Burtt NP, Blumenstiel B, DeFelice M, Parkin M, Barry R, Winslow W, Healy C, Graham RR, Neale BM, Izmailova E, Roubenoff R, Parker AN, Glass R, Karlson EW, Maher N, Hafler DA, Lee DM, Seldin MF, Remmers EF, Lee AT, Padyukov L, Alfredsson L, Coblyn J, Weinblatt ME, Gabriel SB, Purcell S, Klareskog L, Gregersen PK, Shadick NA, Daly MJ, Altshuler D. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun F, Li P, Chen H, Wu Z, Xu J, Shen M, Leng X, Shi Q, Zhang W, Tian X, Li Y, Zhang F. Association studies of TNFSF4, TNFAIP3 and FAM167A-BLK polymorphisms with primary Sjogren’s syndrome in Han Chinese. J Hum Genet. 2013;58(7):475–479. doi: 10.1038/jhg.2013.26. [DOI] [PubMed] [Google Scholar]
- 31.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, Plenge RM, Behrens TW, Wither JE, Rioux JD, Fortin PR, Graham DC, Wong AK, Vyse TJ, Daly MJ, Altshuler D, Moser KL, Gaffney PM. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fung EY, Smyth DJ, Howson JM, Cooper JD, Walker NM, Stevens H, Wicker LS, Todd JA. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/ TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10:188–191. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 33.Ben Hamad M, Cornelis F, Maalej A, Petit-Teixeira E. A Tunisian case-control association study of a 6q polymorphism in rheumatoid arthritis. Rheumatol Int. 2012;32:1849–1850. doi: 10.1007/s00296-011-1996-6. [DOI] [PubMed] [Google Scholar]
- 34.Dieguez-Gonzalez R, Calaza M, Perez-Pampin E, Balsa A, Blanco FJ, Cañete JD, Caliz R, Carreño L, de la Serna AR, Fernandez-Gutierrez B, Ortiz AM, Herrero-Beaumont G, Pablos JL, Narvaez J, Navarro F, Marenco JL, Gomez-Reino JJ, Gonzalez A. Analysis of TNFAIP3, a feedback inhibitor of nuclear factor-kappaB and the neighbor intergenic 6q23 region in rheumatoid arthritis susceptibility. Arthritis Res Ther. 2009;11:R42. doi: 10.1186/ar2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawasaki A, Ito I, Ito S, Hayashi T, Goto D, Matsumoto I, Takasaki Y, Hashimoto H, Sumida T, Tsuchiya N. Association of TNFAIP3 polymorphism with susceptibility to systemic lupus erythematosus in a Japanese population. J Biomed Biotechnol. 2010;2010:207578. doi: 10.1155/2010/207578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opipari AW, Jr, HM Hu, Yabkowitz R, Dixit VM. The A20 zinc finger protein protects cells from tumor necrosis factor cytotoxicity. J Biol Chem. 1992;267:12424–12427. [PubMed] [Google Scholar]
- 37.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matmati M, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, Geboes L, Louagie E, McGuire C, Vereecke L, Chu Y, Boon L, Staelens S, Matthys P, Lambrecht BN, Schmidt-Supprian M, Pasparakis M, Elewaut D, Beyaert R, van Loo G. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- 39.Adrianto I, Wen F, Templeton A, Wiley G, King JB, Lessard CJ, Bates JS, Hu Y, Kelly JA, Kaufman KM, Guthridge JM, Alarcón-Riquelme M, BIOLUPUS and GENLES Networks. Anaya JM, Bae SC, Bang SY, Boackle SA, Brown EE, Petri MA, Gallant C, Ramsey-Goldman R, Reveille JD, Vila LM, Criswell LA, Edberg JC, Freedman BI, Gregersen PK, Gilkeson GS, Jacob CO, James JA, Kamen DL, Kimberly RP, Martin J, Merrill JT, Niewold TB, Park SY, Pons-Estel BA, Scofield RH, Stevens AM, Tsao BP, Vyse TJ, Langefeld CD, Harley JB, Moser KL, Webb CF, Humphrey MB, Montgomery CG, Gaffney PM. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using Regulome DB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musone SL, Taylor KE, Nititham J, Chu C, Poon A, Liao W, Lam ET, Ma A, Kwok PY, Criswell LA. Sequencing of TNFAIP3 and association of variants with multiple autoimmune diseases. Genes Immun. 2011;12:176–182. doi: 10.1038/gene.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappaB activation. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heyninck K, Beyaert R. The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-kappaB activation at the level of TRAF6. FEBS Lett. 1999;442:147–150. doi: 10.1016/s0014-5793(98)01645-7. [DOI] [PubMed] [Google Scholar]
- 44.Evans PC, Ovaa H, Hamon M, Kilshaw PJ, Hamm S, Bauer S, Ploegh HL, Smith TS. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem J. 2004;378:727–734. doi: 10.1042/BJ20031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 46.Lu TT, Onizawa M, Hammer GE, Turer EE, Yin Q, Damko E, Agelidis A, Shifrin N, Advincula R, Barrera J, Malynn BA, Wu H, Ma A. Dimerization and ubiquitin mediated recruitment of a20, a complex deubiquitinating enzyme. Immunity. 2013;38:896–905. doi: 10.1016/j.immuni.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tokunaga F, Nishimasu H, Ishitani R, Goto E, Noguchi T, Mio K, Kamei K, Ma A, Iwai K, Nureki O. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation. Embo J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verhelst K, Carpentier I, Kreike M, Meloni L, Verstrepen L, Kensche T, Dikic I, Beyaert R. A20 inhibits LUBAC-mediated NF-kappaB activation by binding linear polyubiquitin chains via its zinc finger 7. Embo J. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plenge EM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR, Li W, Tan AK, Bonnard C, Ong RT, Thalamuthu A, Pettersson S, Liu C, Tian C, Chen WV, Carulli JP, Beckman EM, Altshuler D, Alfredsson L, Criswell LA, Amos CI, Seldin MF, Kastner DL, Klareskog L, Gregersen PK. TRAF1-C5 as a risk locus for rheumatoid arthritis—A genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang M, Rowland CM, Garcia VE, Schrodi SJ, Catanese JJ, van der Helm-van Mil AH, Ardlie KG, Amos CI, Criswell LA, Kastner DL, Gregersen PK, Kurreeman FA, Toes RE, Huizinga TW, Seldin MF, Begovich AB. A large-scale rheumatoid arthritis genetic study identifies association at chromosome 9q33.2. PLoS Genet. 2008;4:e1000107. doi: 10.1371/journal.pgen.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fakhfakh Karray E, Chalbi H, Ben Dhifallah I, Zakraoui L, Hamzaoui K. Association study of TRAF1-C5 polymorphism with susceptibility to rheumatoid arthritis in Tunisian population. Joint Bone Spine. 2012;79:331–332. doi: 10.1016/j.jbspin.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Han TU, Bang SY, Kang C, Bae SC. TRAF1 polymorphisms associated with rheumatoid arthritis susceptibility in Asians and in Caucasians. Arthritis Rheum. 2009;60:2577–2584. doi: 10.1002/art.24759. [DOI] [PubMed] [Google Scholar]
- 54.Mohamed RH, Pasha HF, El-Shahawy EE. Influence of TRAF1/C5 and STAT4 genes polymorphisms on susceptibility and severity of rheumatoid arthritis in Egyptian population. Cell Immunol. 2012;273:67–72. doi: 10.1016/j.cellimm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Zhu J, Zhang D, Wu F, He F, Liu X, Wu L, Zhou B, Liu J, Lu F, Liu J, Luo R, Long W, Yang M, Ma S, Wu X, Shi Y, Wu T, Lin Y, Yang J, Yuan G, Yang Z. Single nucleotide polymorphisms at the TRAF1/C5 locus are associated with rheumatoid arthritis in a Han Chinese population. BMC Med Genet. 2011;12:53. doi: 10.1186/1471-2350-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plant D, Thomson W, Lunt M, Flynn E, Martin P, Eyre S, Farragher T, Bunn D, Worthington J, Symmons D, Barton A. The role of rheumatoid arthritis genetic susceptibility markers in the prediction of erosive disease in patients with early inflammatory polyarthritis: Results from the Norfolk Arthritis Register. Rheumatology (Oxford) 2011;50:78–84. doi: 10.1093/rheumatology/keq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viatte S, Plant D, Lunt M, Fu B, Flynn E, Parker BJ, Galloway J, Solymossy C, Worthington J, Symmons DP, Dixey JJ, Young A, Barton A. Investigation of rheumatoid arthritis genetic susceptibility markers in the early rheumatoid arthritis study further replicates the TRAF1 association with radiological damage. J Rheumatol. 2013;40:144–156. doi: 10.3899/jrheum.121034. [DOI] [PubMed] [Google Scholar]
- 58.Palomino-Morales R, Gonzalez-Juanatey C, Vazquez-Rodriguez TR, Rodriguez L, Miranda-Filloy JA, Pascual-Salcedo D, Balsa A, Fernandez-Gutierrez B, Llorca J, Martin J, Gonzalez-Gay MA. Lack of association of PTPN22, STAT4 and TRAF1/ C5 gene polymorphisms with cardiovascular risk in rheumatoid arthritis. Clin Exp Rheumatol. 2010;28:695–701. [PubMed] [Google Scholar]
- 59.Rothe M, Wong SC, Henzel WJ, Goeddel DV. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 60.Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 61.Carpentier I, Beyaert R. TRAF1 is a TNF inducible regulator of NF-kappaB activation. FEBS Lett. 1999;460:246–250. doi: 10.1016/s0014-5793(99)01356-3. [DOI] [PubMed] [Google Scholar]
- 62.Schwenzer R, Siemienski K, Liptay S, Schubert G, Peters N, Scheurich P, Schmid RM, Wajant H. The human tumor necrosis factor (TNF) receptor-associated factor 1 gene (TRAF1) is up-regulated by cytokines of the TNF ligand family and modulates TNF-induced activation of NF-kappaB and c-Jun N-terminal kinase. J Biol Chem. 1999;274:19368–19374. doi: 10.1074/jbc.274.27.19368. [DOI] [PubMed] [Google Scholar]
- 63.Tsitsikov EN, Laouini D, Dunn IF, Sannikova TY, Davidson L, Alt FW, Geha RS. TRAF1 is a negative regulator of TNF signaling, enhanced TNF signaling in TRAF1-deficient mice. Immunity. 2001;15:647–657. doi: 10.1016/s1074-7613(01)00207-2. [DOI] [PubMed] [Google Scholar]
- 64.Xie P, Hostager BS, Munroe ME, Moore CR, Bishop GA. Cooperation between TNF receptor-associated factors 1 and 2 in CD40 signaling. J Immunol. 2006;176:5388–5400. doi: 10.4049/jimmunol.176.9.5388. [DOI] [PubMed] [Google Scholar]
- 65.Arron JR, Pewzner-Jung Y, Walsh MC, Kobayashi T, Choi Y. Regulation of the subcellular localization of tumor necrosis factor receptor-associated factor (TRAF)2 by TRAF1 reveals mechanisms of TRAF2 signaling. J Exp Med. 2002;196:923–934. doi: 10.1084/jem.20020774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sughra K, Birbach A, de Martin R, Schmid JA. Interaction of the TNFR-receptor associated factor TRAF1 with I-kappa B kinase-2 and TRAF2 indicates a regulatory function for NF-kappa B signaling. PLoS One. 2010;5:e12683. doi: 10.1371/journal.pone.0012683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McPherson AJ, Snell LM, Mak TW, Watts TH. Opposing roles for TRAF1 in the alternative versus classical NF-kappaB pathway in T cells. J Biol Chem. 2012;287:23010–23019. doi: 10.1074/jbc.M112.350538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choudhary S, Kalita M, Fang L, Patel KV, Tian B, Zhao Y, Edeh CB, Brasier AR. Inducible tumor necrosis factor (TNF) receptor-associated factor-1 expression couples the canonical to the non-canonical NF-kappaB pathway in TNF stimulation. J Biol Chem. 2013;288:14612–14623. doi: 10.1074/jbc.M113.464081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potter C, Eyre S, Cope A, Worthington J, Barton A. Investigation of association between the TRAF family genes and RA susceptibility. Ann Rheum Dis. 2007;66:1322–1326. doi: 10.1136/ard.2006.065706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, International Schizophrenia Consortium. Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: Predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, Catanese JJ, Xie G, Stahl EA, Chen R, Alfredsson L, Amos CI, Ardlie KG, BIRAC Consortium. Barton A, Bowes J, Burtt NP, Chang M, Coblyn J, Costenbader KH, Criswell LA, Crusius JB, Cui J, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Kurreeman FA, Lee AT, Liu X, Li Y, Martin P, Morgan AW, Padyukov L, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth P, YEAR Consortium. Altshuler D, Karlson EW, Toes RE, de Vries N, Begovich AB, Siminovitch KA, Worthington J, Klareskog L, Gregersen PK, Daly MJ, Plenge RM. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger JM, Paige CJ, Lacey DL, Dunstan CR, Boyle WJ, Goeddel DV, Mak TW. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 74.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 75.Goh FG, Midwood KS. Intrinsic danger: Activation of Toll-like receptors in rheumatoid arthritis. Rheumatology (Oxford) 2012;51:7–23. doi: 10.1093/rheumatology/ker257. [DOI] [PubMed] [Google Scholar]
- 76.Coenen MJ, Enevold C, Barrera P, Schijvenaars MM, Toonen EJ, Scheffer H, Padyukov L, Kastbom A, Klareskog L, Barton A, Kievit W, Rood MJ, Jansen TL, Swinkels D, van Riel PL, Franke B, Bendtzen K, Radstake TR. Genetic variants in toll-like receptors are not associated with rheumatoid arthritis susceptibility or anti-tumour necrosis factor treatment outcome. PLoS One. 2010;5:el4326. doi: 10.1371/journal.pone.0014326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee YH, Bae SC, Song GG. Meta-analysis demonstrates association between TLR polymorphisms and rheumatoid arthritis. Genet Mol Res. 12(1):328–334. doi: 10.4238/2013.February.7.2. [DOI] [PubMed] [Google Scholar]
- 78.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, Gianniny L, Korman BD, Padyukov L, Kurreeman FA, Chang M, Catanese JJ, Ding B, Wong S, van der Helm-van Mil AH, Neale BM, Coblyn J, Cui J, Tak PP, Wolbink GJ, Crusius JB, van der Horst-Bruinsma IE, Criswell LA, Amos CI, Seldin MF, Kastner DL, Ardlie KG, Alfredsson L, Costenbader KH, Altshuler D, Huizinga TW, Shadick NA, Weinblatt ME, de Vries N, Worthington J, Seielstad M, Toes RE, Karlson EW, Begovich AB, Klareskog L, Gregersen PK, Daly MJ, Plenge RM. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee HS, Korman BD, Le JM, Kastner DL, Remmers EF, Gregersen PK, Bae SC. Genetic risk factors for rheumatoid arthritis differ in Caucasian and Korean populations. Arthritis Rheum. 2009;60:364–371. doi: 10.1002/art.24245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orozco G, Eyre S, Hinks A, Ke X, Wellcome Trust Case Control consortium YEAR Consortium. Wilson AG, Bax DE, Morgan AW, Emery P, Steer S, Hocking L, Reid DM, Wordsworth P, Harrison P, Thomson W, Barton A, Worthington J. Association of CD40 with rheumatoid arthritis confirmed in a large UK case-control study. Ann Rheum Dis. 2010;69:813–816. doi: 10.1136/ard.2009.109579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prasad P, Kumar A, Gupta R, Juyal RC, Thelma BK. Caucasian and Asian specific rheumatoid arthritis risk loci reveal limited replication and apparent allelic heterogeneity in north Indians. PLoS One. 2012;7:e31584. doi: 10.1371/journal.pone.0031584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Linden MP, Feitsma AL, le Cessie S, Kern M, Olsson LM, Raychaudhuri S, Begovich AB, Chang M, Catanese JJ, Kurreeman FA, van Nies J, van der Heijde DM, Gregersen PK, Huizinga TW, Toes RE, van der Helm-Van Mil AH. Association of a single-nucleotide polymorphism in CD40 with the rate of joint destruction in rheumatoid arthritis. Arthritis Rheum. 2009;60:2242–2247. doi: 10.1002/art.24721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li G, Diogo D, Wu D, Spoonamore J, Dancik V, Franke L, Kurreeman F, Rossin EJ, Duclos G, Hartland C, Zhou X, Li K, Liu J, De Jager PL, Siminovitch KA, Zhernakova A, Raychaudhuri S, Bowes J, Eyre S, Padyukov L, Gregersen PK, Worthington J, Rheumatoid Arthritis Consortium International (RACI) Gupta N, Clemons PA, Stahl E, Tolliday N, Plenge RM. Human genetics in rheumatoid arthritis guides a high-throughput drug screen of the CD40 signaling pathway. PLoS Genet. 2013;9:e1003487. doi: 10.1371/journal.pgen.1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peters AL, Stunz LL, Bishop GA. CD40 and autoimmunity: The dark side of a great activator. Semin Immunol. 2009;21:293–300. doi: 10.1016/j.smim.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Munroe ME, Bishop GA. A costimulatory function for T cell CD40. J Immunol. 2007;178:671–682. doi: 10.4049/jimmunol.178.2.671. [DOI] [PubMed] [Google Scholar]
- 86.Berner B, Wolf G, Hummel KM, Muller GA, Reuss-Borst MA. Increased expression of CD40 ligand (CD154) on CD4+ T cells as a marker of disease activity in rheumatoid arthritis. Ann Rheum Dis. 2000;59:190–195. doi: 10.1136/ard.59.3.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Burch E, Cai L, So E, Hubbard F, Matteson EL, Strome SE. CD40 mediates down-regulation of CD32B on specific memory B Cell populations in rheumatoid arthritis. J Immunol. 2013;190:6015–6022. doi: 10.4049/jimmunol.1203366. [DOI] [PubMed] [Google Scholar]
- 88.Durie FH, Fava RA, Foy TM, Aruffo A, Ledbetter JA, Noelle RJ. Prevention of collagen-induced arthritis with an antibody to gp39, the ligand for CD40. Science. 1993;261:1328–1330. doi: 10.1126/science.7689748. [DOI] [PubMed] [Google Scholar]
- 89.Kyburz D, Carson DA, Corr M. The role of CD40 ligand and tumor necrosis factor alpha signaling in the transgenic K/BxN mouse model of rheumatoid arthritis. Arthritis Rheum. 2000;43:2571–2577. doi: 10.1002/1529-0131(200011)43:11<2571::AID-ANR26>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 90.Mauri C, Mars LT, Londei M. Therapeutic activity of agonistic monoclonal antibodies against CD40 in a chronic autoimmune inflammatory process. Nat Med. 2000;6:673–679. doi: 10.1038/76251. [DOI] [PubMed] [Google Scholar]
- 91.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, Seldin MF, Criswell LA, Plenge RM, Holers VM, Mikuls TR, Sokka T, Moreland LW, Bridges SL, Jr, Xie G, Begovich AB, Siminovitch KA. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eyre S, Hinks A, Flynn E, Martin P, Wilson AG, Maxwell JR, Morgan AW, Emery P, Steer S, Hocking LJ, Reid DM, Harrison P, Wordsworth P, Thomson W, Worthington J, Barton A. Confirmation of association of the REL locus with rheumatoid arthritis susceptibility in the UK population. Ann Rheum Dis. 2010;69:1572–1573. doi: 10.1136/ard.2009.122887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sen R, Smale ST. Selectivity of the NF-{kappa} B response. Cold Spring Harb Perspect Biol. 2009;2:a000257. doi: 10.1101/cshperspect.a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou XY, Yashiro-Ohtani Y, Nakahira M, Park WR, Abe R, Hamaoka T, Naramura M, Gu H, Fujiwara H. Molecular mechanisms underlying differential contribution of CD28 versus non-CD28 costimulatory molecules to IL-2 promoter activation. J Immunol. 2002;168:3847–3854. doi: 10.4049/jimmunol.168.8.3847. [DOI] [PubMed] [Google Scholar]
- 95.Ouaaz F, Arron J, Zheng Y, Choi Y, Beg AA. Dendritic cell development and survival require distinct NF-kappaB subunits. Immunity. 2002;16:257–270. doi: 10.1016/s1074-7613(02)00272-8. [DOI] [PubMed] [Google Scholar]
- 96.Lio CW, Hsieh CS. Becoming self-aware: The thymic education of regulatory T cells. Curr Opin Immunol. 2011;23:213–219. doi: 10.1016/j.coi.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chang M, Jin W, Chang JH, Xiao Y, Brittain GC, Yu J, Zhou X, Wang YH, Cheng X, Li P, Rabinovich BA, Hwu P, Sun SC. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat Immunol. 2011;12:1002–1009. doi: 10.1038/ni.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Breedveld FC, Weisman MH, Kavanaugh AF, Cohen SB, Pavelka K, van Vollenhoven R, Sharp J, Perez JL, Spencer-Green GT. The PREMIER study: A multicenter, randomized, double-blind clinical trial of combination therapy with adali-mumab plus methotrexate versus methotrexate alone or adalimumab alone in patients with early, aggressive rheumatoid arthritis who had not had previous methotrexate treatment. Arthritis Rheum. 2006;54:26–37. doi: 10.1002/art.21519. [DOI] [PubMed] [Google Scholar]
- 99.van den Broek M, Visser K, Allaart CF, Huizinga TW. Personalized medicine: Predicting responses to therapy in patients with RA. Curr Opin Pharmacol. 2013;13:463–469. doi: 10.1016/j.coph.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 100.Canhao H, Rodrigues AM, Mourao AF, Martins F, Santos MJ, Canas-Silva J, Polido-Pereira J, Pereira Silva JA, Costa JA, Araújo D, Silva C, Santos H, Duarte C, da Silva JA, Pimentel-Santos FM, Branco JC, Karlson EW, Fonseca JE, Solomon DH. Comparative effectiveness and predictors of response to tumour necrosis factor inhibitor therapies in rheumatoid arthritis. Rheumatology (Oxford) 2012;51:2020–2026. doi: 10.1093/rheumatology/kes184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kleinert S, Tony HP, Krause A, Feuchtenberger M, Wassenberg S, Richter C, Röther E, Spieler W, Gnann H, Wittig BM. Impact of patient and disease characteristics on therapeutic success during adalimumab treatment of patients with rheumatoid arthritis: Data from a German noninterventional observational study. Rheumatol Int. 2012;32:2759–2767. doi: 10.1007/s00296-011-2033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Klaasen R, Cantaert T, Wijbrandts CA, Teitsma C, Gerlag DM, Out TA, de Nooijer MJ, Baeten D, Tak PP. The value of rheumatoid factor and anti-citrullinated protein antibodies as predictors of response to infliximab in rheumatoid arthritis: An exploratory study. Rheumatology (Oxford) 2011;50:1487–1493. doi: 10.1093/rheumatology/ker010. [DOI] [PubMed] [Google Scholar]
- 103.Cui J, Stahl EA, Saevarsdottir S, Miceli C, Diogo D, Trynka G, Raj T, Mirkov MU, Canhao H, Ikari K, Terao C, Okada Y, Wedrén S, Askling J, Yamanaka H, Momohara S, Taniguchi A, Ohmura K, Matsuda F, Mimori T, Gupta N, Kuchroo M, Morgan AW, Isaacs JD, Wilson AG, Hyrich KL, Herenius M, Doorenspleet ME, Tak PP, Crusius JB, van der Horst-Bruinsma IE, Wolbink GJ, van Riel PL, van de Laar M, Guchelaar HJ, Shadick NA, Allaart CF, Huizinga TW, Toes RE, Kimberly RP, Bridges SL, Jr, Criswell LA, Moreland LW, Fonseca JE, de Vries N, Stranger BE, De Jager PL, Raychaudhuri S, Weinblatt ME, Gregersen PK, Mariette X, Barton A, Padyukov L, Coenen MJ, Karlson EW, Plenge RM. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 2013;9:e1003394. doi: 10.1371/journal.pgen.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 105.Coulthard LR, Taylor JC, Eyre S, Biologics in Rheumatoid Arthritis Genetics and Genomics. Robinson JI, Wilson AG, Isaacs JD, Hyrich K, Emery P, Barton A, Barrett JH, Morgan AW, McDermott MF. Genetic variants within the MAP kinase signalling network and anti-TNF treatment response in rheumatoid arthritis patients. Ann Rheum Dis. 2012;70:98–103. doi: 10.1136/ard.2010.133249. [DOI] [PubMed] [Google Scholar]
- 106.Zeng Z, Duan Z, Zhang T, Wang S, Li G, Gao J, Ye D, Xu S, Xu J, Zhang L, Pan F. Association between tumor necrosis factor-alpha (TNF-alpha) promoter −308 G/A and response to TNF-alpha blockers in rheumatoid arthritis: A meta-analysis. Mod Rheumatol. 2013;23:489–495. doi: 10.1007/s10165-012-0699-5. [DOI] [PubMed] [Google Scholar]