Abstract
Respiratory syncytial virus (RSV) is the primary cause of lower respiratory tract infection during childhood and causes severe symptoms in some patients, which may cause hospitalization and death. Mechanisms for differential responses to RSV are unknown. Our objective was to develop an in vitro model of RSV infection to evaluate interindividual variation in response to RSV and identify susceptibility genes. Populations of human-derived HapMap lymphoblastoid cell lines (LCLs) were infected with RSV. Compared with controls, RSV-G mRNA expression varied from ∼1- to 400-fold between LCLs. Basal expression of a number of gene transcripts, including myxovirus (influenza virus) resistance 1 (MX1), significantly correlated with RSV-G expression in HapMap LCLs. Individuals in a case-control population of RSV-infected children who were homozygous (n=94) or heterozygous (n=172) for the predicted deleterious A allele in a missense G/A SNP in MX1 had significantly greater risk for developing severe RSV disease relative to those with the major allele (n=108) (χ2=5.305, P=0.021; OR: 1.750, 95% CI: 1.110, 2.758, P=0.021). We conclude that genetically diverse human LCLs enable identification of susceptibility genes (e.g., MX1) for RSV disease severity in children, providing insight for disease risk.—Ciencewicki, J. M., Wang, X., Marzec, J., Serra, M. E., Bell, D. A., Polack, F. P., Kleeberger, S. R. A genetic model of differential susceptibility to human respiratory syncytial virus (RSV) infection.
Keywords: lymphoblastoid cell lines, HapMap, infant lung disease, gene expression, bronchiolitis, MX1
Respiratory syncytial virus (RSV) is an enveloped, single-stranded RNA virus of the paramyxoviridae family. RSV is the primary cause of lower respiratory tract infection and bronchiolitis during infancy and childhood, with 60–70% of infants being infected within their first year of life and nearly all by 2 yr of age (1). Patients usually present with rhinorrhea, cough, and low-grade fever; however, some susceptible individuals may require hospitalization or die. No RSV vaccine currently exists, and it has been estimated that RSV annually infects 64 million and causes 160,000 deaths (2). Despite efforts to elucidate mechanisms for differential responses to RSV infection, they remain largely unknown.
Previous studies indicated that environmental factors contribute to susceptibility and response to RSV infection, but they cannot account for all observed variation in RSV-related phenotypes (3–6). Differential RSV infection across inbred mouse strains suggests that susceptibility is influenced by genetic background (7, 8). Furthermore, epidemiological studies have found RSV disease severity associated with polymorphisms in genes, including Toll-like receptor 4 (TLR4), cluster of differentiation 14 (CD14), various surfactant proteins, cytokines, and chemokines (9–15). Increased concordance of severe RSV infection in identical twins has also been found (16). However, these studies are cost- and time-intensive, and thus there is a need for a more rapid and cost-effective means to evaluate genome-wide contributions to differential susceptibility to human RSV infection and disease.
The objective of this study was to develop a high-throughput, in vitro cell model of RSV disease to investigate the role of genetic factors in response to RSV and to identify functionally relevant candidate susceptibility genes that can be tested in a human population. Human-derived epithelial cell lines have been used to investigate mechanisms of RSV infection, but there are relatively few distinct lines. Furthermore, these have little genetic or genomic information available and thus are not very useful for investigating the genetic contribution to disease phenotypes. To circumvent these challenges, human lymphoblastoid cell lines (LCLs) have been used to examine genetic components involved in human disease (17–19).
Using the human variation collection from the Coriell Institute (Camden, NJ, USA), which consists of human LCLs established from donors from various ethnic and racial groups, as well as multigenerational families, we found that RSV infectivity differed significantly among individuals and between ethnic groups. We also identified a panel of genes whose basal expression was significantly and reproducibly correlated with RSV infectivity in LCLs. One of these differentially expressed genes, myxovirus (influenza virus) resistance 1 (MX1), contributes to host defense against influenza and other viruses (20–22). Moreover, a functional single-nucleotide polymorphism (SNP) in MX1 was significantly associated with RSV-G expression in the LCLs and RSV disease severity in a human neonatal population. Results of the association study supported findings of the LCL model and indicate the value of LCLs for understanding mechanisms of differential susceptibility to RSV infection.
MATERIALS AND METHODS
Cell lines and culture conditions
Human LCLs were obtained from the Coriell Institute's cell repositories. LCLs were established by Epstein-Barr virus transformation of peripheral blood mononuclear cells using phytohemagluttinin as a mitogen. Cells were cultured in RPMI 1640 (Life Technologies, Grand Island, NY, USA) with 15% FBS and 1% penicillin/streptomycin in a humidified incubator at 37°C with 5% carbon dioxide. To calculate the growth rate of each cell line, cells were initially cultured at a concentration of 0.5 × 106 cells/ml medium. After 3 d, cells were counted to measure the percentage change from the initial concentration, then centrifuged and resuspended in fresh medium at a concentration of 0.5 × 106 cells/ml. The mean of 3 separate measurements was then used to calculate growth rate for each cell line. Growth rate varied widely among LCLs, with a range of 5–388% and mean ± sem of 124.4 ± 13.3%. We asked whether growth rate affected RSV-G mRNA expression (an indicator of viral load) and found no correlation between the percentage growth of individual cell lines and RSV-G mRNA (r2=0.064, P>0.05).
RSV infection
Cells were infected with RSV A2 at a moiety of infection of 4. Cells were concentrated to 5 × 106 cells/ml in serum-free medium, and virus was added to the medium. After 1 h medium with 5% FBS was added to achieve a cell concentration of 5 × 105 cells/ml.
Real-time reverse transcriptase polymerase chain reaction (RT-PCR)
Total RNA was isolated using a Qiagen RNeasy kit (Qiagen, Valencia, CA, USA) as per the supplier's instructions and reverse transcribed using MMLV-reverse transcriptase. First-strand cDNA was amplified by real-time PCR using gene-specific primers and probes and TaqMan Universal PCR mastermix from Applied Biosystems (Foster City, CA, USA) using the Applied Biosystems StepOne Plus Sequence Detector System. For RSV-G gene expression, gene-specific primers and probes (forward:CGCACCGCTAAGACATTAGA; reverse: GTGGATTGCAGGGTTGACTT) were obtained from Invitrogen (Carlsbad, CA, USA) and Power SYBR Green PCR mastermix from Applied Biosystems was used.
Human RSV disease population
A prospective case-control study was conducted in Buenos Aires, Argentina between 2003 and 2006. Participating hospitals included Hospital Francés, Hospital Nacional Dr. Alejandro Posadas, Hospital Evita Pueblo de Berazategui, and Hospital Mi Pueblo de Florencio Varela. Invited participants were previously healthy full-term infants younger than 1 yr of age and born after September 15 of the previous year (15 d after the end of RSV season in Buenos Aires; ref. 23), with signs and symptoms of bronchiolitis for the first time in their lives. Bronchiolitis was defined by clinical signs, including wheezing with or without cough, rales, dyspnea, and increased respiratory rate and retractions of the respiratory muscles. Diagnosis was performed by trained pediatricians. Written, witnessed, informed consent was obtained from all parents or guardians of the infants. The Institutional Review Boards of both the French Hospital (Buenos Aires) and the network of Hospital Materno Infantil de San Isidro (which encompasses the 3 hospitals in the northeastern region of Buenos Aires), as well as the Institutional Review Board of Johns Hopkins University (Baltimore, MD, USA), approved the protocol.
Selection of criteria for RSV disease severity was based on the need for oxygen supplementation and consequent hospitalization. These criteria enabled comparison with previous investigations (e.g., ref. 24). Infants with bronchiolitis were recruited as cases when their oxygen saturation on enrollment was <93% in room air. Controls were infants with oxygen saturation ≥93% while breathing room air.
Exclusion criteria included known or suspected impairment of immunological function, major congenital oral malformations, chronic lung disease, cardiac disease, prematurity (gestational age <37 wk), neuromuscular disorders affecting swallowing, and known or suspected coagulation disorders or bleeding tendency.
Demographic information included age, gender, breastfeeding on enrollment, smoking at home, day care attendance, and presence of siblings younger than 14 yr of age in the household (Table 1). Clinical information included need for and duration of oxygen supplementation, need for and duration of hospitalization, need for intensive care, and death. Investigators monitored the clinical evolution of participating infants through phone calls and/or hospital visits for 7 d after enrollment, and any change in their clinical status was recorded (i.e., worsening clinical condition converting a control participant into a case subject). A second, independent population of infants was recruited in Buenos Aires from 2010 to 2013 using the same criteria as in 2003–2006. The infants were recruited from hospitals [Swiss Medical Center, Centro de Educación Médica e Investigaciones Clínicas (CEMIC), Hospital Español] in the central region of Buenos Aires and public institutions (Hospital Pedro de Elizalde and Hospital Lucio Melendez) in the southern region of Buenos Aires. Genotyping of these infants was performed using nasal aspirates.
Table 1.
Demographic characteristics of infants in the first population
| Characteristic | RSV-positive | RSV-negative |
|---|---|---|
| Number (n) | 370 | 305 |
| Males [n (%)] | 205 (55.4) | 175 (57.4) |
| Breastfed [n (%)] | 306 (82.7) | 239 (78.4) |
| Smokers at home [n (%)] | 216 (58.4) | 166 (54.4) |
| Day care [n (%)] | 13 (3.5) | 17 (5.6) |
| Low socioeconomic status [n (%)] | 255 (68.9) | 244 (80.0) |
| Median sibs <14 yr (n) | 3 | 3 |
| Median age (mo) | 4.0 ± 2.0 | 4.8 ± 1.9 |
RSV viral titer
Viral titers were quantified in nasal aspirates using RT-PCR. RNA was extracted using RNeasy Minikits (Qiagen). RT-PCR was performed using an RSV TaqMan probe with the sequence: 6FAM-CAATGATCATGATTTACCTATTG-MGBNFQ. Viral titers were estimated using a standard curve with known concentrations of RSV.
Allelic discrimination genotyping for MX1 V379I rs469390 SNP (G/A)
Genomic DNA was isolated from whole-blood samples using the Gentra Puregene kit (Gentra Systems, Minneapolis, MN, USA) and was characterized for purity and concentration on a Beckman Coulter DU640 spectrophotometer (Beckman Coulter, Fullerton, CA, USA). Low-yield samples were whole genome amplified with the Qiagen REPLI-g kit. The MX1 V379I substitution (rs469390) in exon 9 was assessed by allelic discrimination with a predesigned SNP genotyping assay from Applied Biosystems. RT-PCR was performed using 20-μl reactions with 5 ng of genomic DNA, 10 μl of TaqMan 2× PCR Master Mix, and 1.0 μl of 20× preoptimized assay mix (C_2274997_10). The 20× mix consisted of 18 μM forward and reverse primers and 8 μM of each allele-specific fluorescently labeled (VIC or FAM) TaqMan MGB probe. Standard PCR cycling conditions were used, with initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, and a post-PCR read at 60°C for 30 s. Allele-specific PCR products were detected on an Applied Biosystems StepOnePlus real-time PCR system, and clustered by genotype using StepOnePlus software. Ambiguous samples were clustered manually and verified by sequencing. Control samples with known genotype as verified by DNA sequencing, were run with each plate. Five percent of the samples were repeated for quality control, and we found 100% concordance. In addition, because of potential ethnic diversity in our study population, 33 ethnic-specific genomic markers were evaluated on the Sequenom iPlex platform by BioServe (Beltsville, MD, USA) to test for admixture.
Statistics
Individual LCL values are presented as means ± sem of ≥3 separate experiments. Group data are presented as means ± sem of LCLs for each ethnic group. Because of heteroscedasticity, Kruskal-Wallis 1-way ANOVA on ranks with Dunn's pairwise multiple-comparison test was used to test for differences in basal TLR3 and TLR4 expression (SigmaPlot 12.0; Systat Software, San Jose, CA, USA). For all other comparisons, parametric 1-way ANOVA was used to determine whether means were significantly different from each other with Student-Newman-Keuls post hoc test to compare donors. Associations between gene expression and genotypes were tested using simple linear regression. Associations between individual MX1 genotypes and RSV disease severity were tested using χ2 and multiple logistic regression methods (SigmaPlot 12.0). We used multiple logistic regression to assess potential confounding of demographic variables in the relationship between RSV disease and MX1 genotype. Where necessary, Bonferroni correction for multiple comparisons was applied. Values of P < 0.05 were considered statistically significant.
RESULTS
Ethnic panel LCL responses to RSV infection
Because TLR3, TLR4, and eukaryotic translation initiation factor 2-α kinase 2 [EIF2AK2; or protein kinase, RNA-activated (PKR)] have reported roles in RSV susceptibility and response (25, 26), we quantified their basal expression in LCLs from 3 human variation panels. Differential mRNA expression of all three genes was found between individuals within each group (data not shown) and across different groups (Fig. 1). Baseline expression of TLR3 was highest in Japanese LCLs compared to the other two panels, expression of TLR4 was highest among Northern Europeans, and expression of EIF2AK2 was significantly higher in African Americans. Considered across all individuals, TLR3 mRNA expression and EIF2AK2 mRNA expression were significantly correlated (r2=0.315, P=0.003; data not shown). No other pairwise correlation between TLR3, TLR4, and EIF2AK2 expression was statistically significant.
Figure 1.
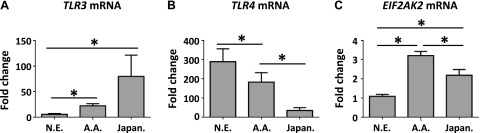
Baseline mRNA expression of TLR3 (A), TLR4 (B), and EIF2AK2 (C) in Northern European (N.E.), African-American (A.A.), and Japanese (Japan.) ethnic panels. Baseline mRNA was quantified in uninfected samples using real-time RT-PCR. Values are normalized to β-actin and expressed as mean ± sem fold induction over a common reference sample. *P < 0.05.
In a pilot investigation with 3 LCLs, RSV-G mRNA was quantified at 24, 48, and 72 h postinfection (hpi). The greatest amount of RSV-G expression in these lines was found at 48–72 hpi (data not shown). RSV-G mRNA expression was subsequently evaluated in individual LCLs from the ethnic panels at 48 hpi, and differences in interindividual response were found for RSV-induced effects (Fig. 2). For example, although RSV-G mRNA expression increased in all African American LCLs at 48 hpi relative to respective vehicles, increases were significantly greater in LCL 37 than in LCL 32 (Fig. 2). Across panels, mean ± sem fold change in RSV-G expression ranged from 6.4 ± 1.2 (Northern European) to 16 ± 9 (Japanese) (Fig. 2). Although mean levels in Japanese LCLs were more than double those in Northern European and African American panels, no significant differences in mean RSV-G expression were found between the groups, likely due to large interindividual variation within groups (Fig. 2). RSV-G mRNA expression in LCLs did not correlate with basal expression of TLR3, TLR4, or EIF2AK2.
Figure 2.

RSV-G mRNA expression at 48 hpi in individual cell lines from the African American (A), Northern European (B), and Japanese (C) ethnic panels, as well as mean values from each panel (D). mRNA was quantified using real-time RT-PCR. Values are normalized to β-actin and expressed as mean ± sem fold change over a common reference sample. *P < 0.05 vs. lowest-responding donor; +P < 0.05 vs. all other individuals.
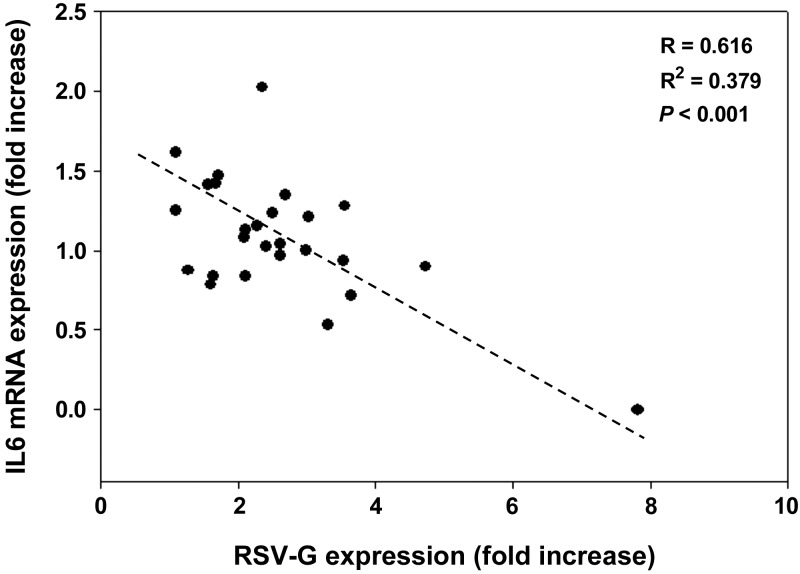
A role for interferon α (IFN-α), IFN-β, and IFN-γ in the innate immune response to RSV has been demonstrated by previous studies (27–29). Among Northern Europeans, small but statistically significant increases in mean IFN-α (IFNA) mRNA were found and differed between lines (e.g., LCL 5 vs. LCL 6; Supplemental Fig. S1). Mean IFNA mRNA expression was significantly higher in Northern Europeans compared with African Americans at 48 hpi (Supplemental Fig. S1), but no significant differences were found between other panels or individuals within panels (data not shown). Mean IFN-β (IFNB) and IFN-γ (IFNG) mRNA expression was not significantly different between panels. Cytokines interleukin 6 (IL-6) and tumor necrosis factor α (TNF-α) have also been implicated in response to RSV (30), but mean mRNA expression did not differ between panels (data not shown). However, a significant inverse correlation was found between RSV-g expression and IL-6 expression among all LCLs at48 hpi (Fig. 3).
Figure 3.
Correlation between mean RSV-G mRNA expression and mean IL-6 mRNA expression at 48 hpi in 26 lymphoblastoid cell lines from all ethnic panels. Points represent ≥3 biological replicates for each cell line. mRNA was quantified using real-time RT-PCR. Values are normalized to β-actin and expressed as fold change over the respective uninfected control.
Responses to RSV in HapMap LCLs
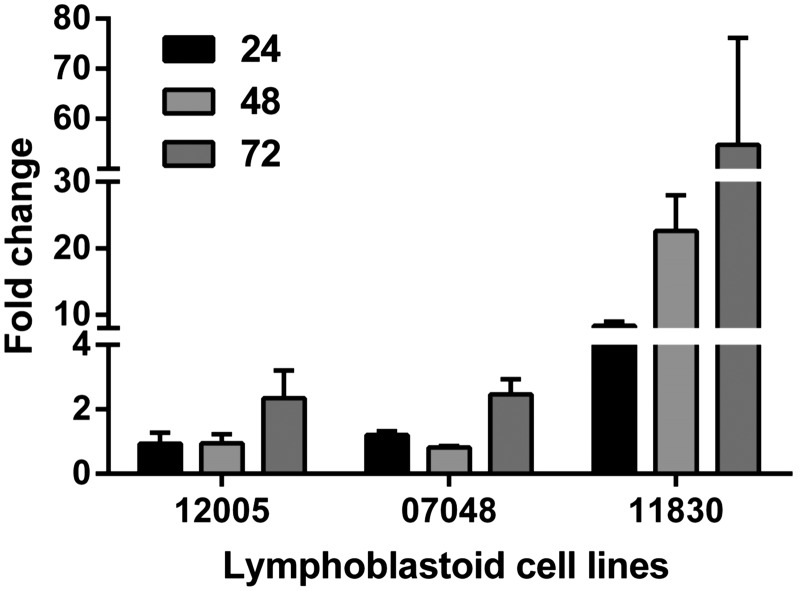
We next sought insight into mechanisms of differential responsiveness to RSV in LCLs by infecting 42 HapMap CEU LCLs to leverage genetic and genomic information available for the lines (31–35). As above, we found significant interindividual variation in viral RSV-G expression following infection with RSV (Fig. 4). With these additional LCLs, we found that RSV-G mRNA expression was consistently greatest at 72 hpi, and HapMap LCLs were phenotyped at this time (Fig. 5).
Figure 4.
RSV-G mRNA expression in 3 CEU HapMap lymphoblastoid cell lines at 24, 48, and 72 hpi. mRNA was quantified using real-time RT-PCR. Values are normalized to β-actin and expressed as mean ± sem fold change over a common reference sample.
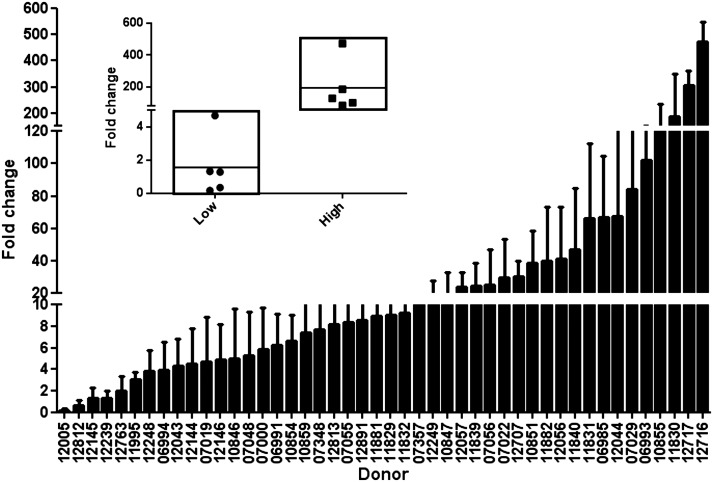
Figure 5.
RSV-G mRNA expression at 72 hpi in 46 CEU HapMap lymphoblastoid cell lines. Data are presented as means ± sem. Inset: box plot of the 5 lowest and 5 highest responding cell lines. mRNA was quantified using real-time RT-PCR. Values are normalized to β-actin and expressed as fold change over a common reference sample.
To identify additional genes whose basal (unstimulated) expression was correlated with response to RSV infection, we used a microarray data set (unpublished results) and obtained 5 published microarray data sets (GSE16778, ref. 34; GSE6536, ref. 35; GSE7761, ref. 36; GSE9703, ref. 37; and GSE11582, ref. 31) from the U.S. National Center for Biotechnology Information (NCBI; Bethesda, MD, USA) Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) for the LCLs. We then used a 3-step approach: first, we performed linear regression of log2-transformed RSV-G expression on log2-transformed gene expression values of every detectable gene; second, we compared the average gene expression levels for the 5 individuals with the highest RSV-G expression against that for the 5 individuals with the lowest RSV-G expression using t test (Fig. 5, inset); last, we selected genes that were associated with RSV-G expression and differential gene expression in common to ≥3 data sets.
Therefore, a significant gene or transcript was associated with RSV-G expression if the expression and RSV levels were significantly correlated (nominal value of P≤0.01) and if the gene expression fold change was ≤0.8 or ≥1.2 in ≥3 of the tested gene expression data sets. These analyses generated a list of 61 transcripts in which basal mRNA expression was significantly and reproducibly correlated with LCL RSV-G expression. One of the genes was MX1, which encodes a protein with antiviral properties (refs. 20, 21, 36 and Fig. 6). Based on these results and the potential antiviral role for MX1, we hypothesized that MX1 is a gene candidate for differential susceptibility to RSV. The remaining transcripts have not been evaluated further in this model.
Figure 6.
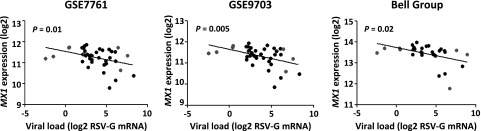
MX1 mRNA expression in 42 lymphoblastoid cell lines observed in 3 different microarray datasets (y axes) plotted against RSV-G mRNA expression observed in the corresponding cell lines from our study (x axes). Values are expressed as log2 transformed. Each data point represents a particular cell line; gray points represent cell lines with high RSV-G expression (right), and cell lines with low RSV-G expression (left) (see Fig. 5 inset).
Role of MX1 Val379Ile in RSV disease severity
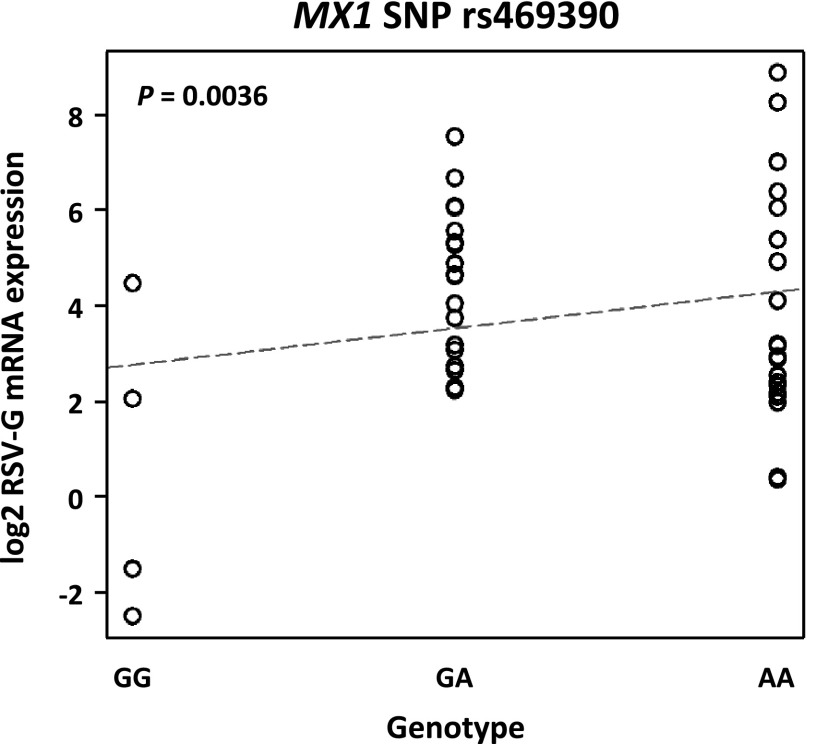
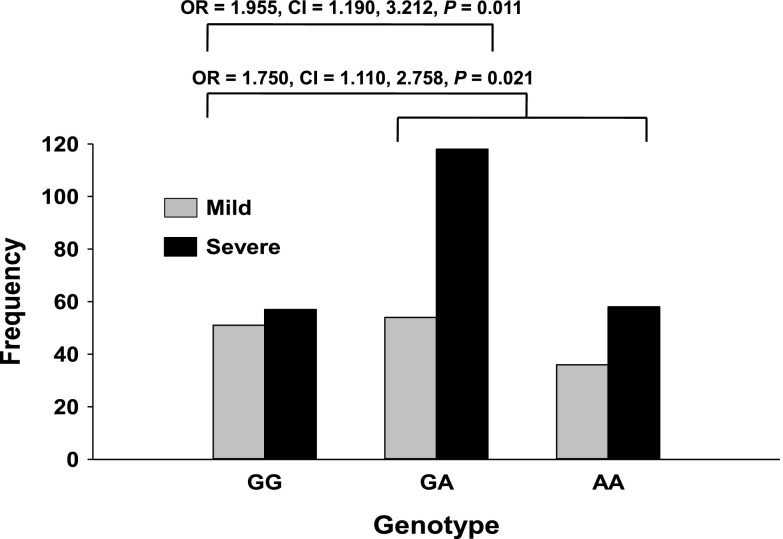
We queried dbSNP (build 129, 2008) to identify potentially relevant SNPs in MX1 based on functional annotations and identified a missense SNP (rs469390, G/A) in exon 9 of NM_002462:19777. On the basis of the 1000 genomes project phase 1 (http://www.1000genomes.org), the alternative allele A frequency is 0.56 for Europeans, 0.23 for Asians, and 0.44 for Africans. This nonsynonymous coding polymorphism causes a substitution of the valine residue with a isoleucine residue at position 379 (Val379Ile; http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?type=rs), and is predicted by PANTHER (37) to have a mild deleterious effect on protein function (subPSEC, −3.25946; P deleterious, 0.5645). In phenotyped LCLs, we used linear regression to determine the relationship between log2 RSV-G mRNA expression and rs469390 genotype as previously described (38, 39). RSV-G mRNA was significantly correlated with rs469390 genotype (P=0.0036; Fig. 7), consistent with the deleterious A allele conferring enhanced risk of RSV load. We next asked whether rs469390 increased the risk of severe RSV disease in infected infants from our prospective case-control study. We genotyped infants with mild or severe RSV infection, and combined allelic frequencies (Table 2) were similar to those published for other populations (http://www.1000genomes.org/). Allelic frequencies satisfied Hardy-Weinberg equilibrium (P>0.05), and no evidence of admixture in the populations was found after analysis of ethnic-specific genomic markers (not shown). χ2 analysis of RSV disease severity among homozygous major allele, heterozygous, and homozygous variants indicated the rs469390 G/A polymorphism was associated with increased risk of severe RSV disease (χ2=7.095, P=0.029). This association was driven largely by the GA genotype risk effect. Infants with ≥1 A allele were at significantly greater risk of developing severe RSV disease (χ2=5.305, P=0.021; OR: 1.750, 95% CI: 1.110, 2.758, P=0.021; Fig. 8), and those infants heterozygous for the polymorphism were at significantly greater risk of severe disease (χ2=6.431, P=0.011; OR: 1.955, 95% CI: 1.190, 3.212, P=0.011; Fig. 8). In the second genotyped population, infants with ≥1 rs469390 A allele (χ2=3.772, P=0.052; OR: 2.013, 95% CI: 1.043, 3.886) or heterozygous (χ2=3.195, P=0.074; OR: 1.981, 95% CI: 0.995, 3.964) were also at greater risk of developing severe RSV disease though the associations did not reach statistical significance likely due to the smaller number of study participants compared to the first population (Supplemental Table S1). Notably, when the two populations were combined (Supplemental Table S2), the strength of the association of ≥1 rs469390 A allele (χ2=8.205, P=0.004; OR: 1.740, 95% CI: 1.203, 2.515) or the heterozygotes (χ2=8.447, P=0.004; OR: 1.827, 95% CI: 1.231, 2.711) was stronger than either population alone.
Figure 7.
Plot of linear regression of mean RSV-G mRNA expression in 42 HapMap LCLs with genotypes for the MX1 rs469390 SNP.
Table 2.
Genotype and allele frequencies for the MX1 rs469390 SNP in RSV-positive and RSV-negative patients (first population)
| Parameter | RSV-positive |
RSV-negative |
||||
|---|---|---|---|---|---|---|
| Mild disease | Severe disease | Total | Mild disease | Severe disease | Total | |
| GG genotype [n (%)] | 51 (36.2) | 57 (24.5) | 108 (28.9) | 40 (27.4) | 51 (32.1) | 91 (29.8) |
| AG genotype [n (%)] | 54 (38.3) | 118 (50.6) | 172 (46.0) | 69 (47.3) | 68 (42.8) | 137 (44.9) |
| AA genotype [n (%)] | 36 (25.5) | 58 (24.9) | 94 (25.1) | 37 (25.3) | 40 (25.1) | 77 (25.2) |
| Total | 141 | 233 | 374 | 146 | 159 | 305 |
| G allele frequency | 0.553 | 0.498 | 0.519 | 0.510 | 0.535 | 0.523 |
| A allele frequency | 0.447 | 0.502 | 0.481 | 0.490 | 0.465 | 0.477 |
Figure 8.
Role of MX1 SNP rs469390 in disease severity among infants with RSV infection. DNA was isolated from whole blood, and infants with mild or severe RSV infection were genotyped for the MX1 SNP (Val379Ile) and were classified as homozygous for the major or minor allele, or heterozygous. χ2 analysis of RSV disease severity among homozygous wild-type, heterozygous, and homozygous variants was used to determine association of genotype with increased disease severity.
Logistic regression analyses of the first population also found that the rs469390 G/A MX1 SNP was associated with increased risk of severe disease among those infected with RSV (unadjusted OR: 2.247, 95% CI: 1.205, 4.191). After adjustment for gender, socioeconomic status, and breastfeeding, the rs469390 G/A Mx1 SNP remained associated with increased risk of severe disease in infected children (OR: 2.476, 95% CI: 1.094, 5.600). Similarly, logistic regression analyses of the combined populations found the rs469390 G/A MX1 SNP was associated with increased risk of severe disease, although the OR was smaller (unadjusted OR: 1.271, 95% CI: 1.010, 1.599). Adjusting for gender, socioeconomic status, and breastfeeding did not significantly change the association.
DISCUSSION
RSV disease affects nearly every infant by 2 yr of age, and, in the absence of a vaccine, it will remain an important public health concern. However, the mechanisms of interindividual susceptibility to RSV disease severity are not understood. We have developed a novel in vitro model of RSV disease using LCLs that have been well characterized and used to evaluate the genetic contribution to environmental stimuli (e.g., 38).
We initially found differential baseline expression of selected innate immunity genes in human variation panel cell lines (i.e., TLR3, TLR4, and EIF2AK2), demonstrating interindividual variation exists between different racial and ethnic groups, and also within members of a particular group. Interestingly, when all LCLs were considered, mRNA expression of TLR3 and EIF2AK2 were significantly correlated. The potential interaction between TLR3 and EIF2AK2 in response to virus infection has been reported (25), and it was hypothesized that virus-induced up-regulation of TLRs and EIF2AK2 may sensitize cells to subsequent viral and bacterial exposures (25). However, we found no correlation between RSV titers and either EIF2AK2 or TLR expression and, at least in the LCLs, differential expression of these antiviral genes did not contribute to interindividual variation in RSV-G mRNA expression after infection.
Differences in RSV-G mRNA levels following infection in LCLs within and between ethnic groups ranged from ∼2- to 60-fold, and from ∼1- to >400-fold in HapMap LCLs. While viral load is not the only measure of disease severity, it is a good indication of the degree of infection. Viral load does not always correlate well with response phenotypes, although a recent investigation in 35 healthy human volunteers found that viral load was associated with disease and that viral load and disease can track together (39). While in vitro infection of LCLs has not been used to predict those individuals who will respond severely irrespective of viral load, it may offer predictive information about which individuals will be more likely to present with higher viral loads on infection, rendering them more susceptible to developing severe RSV disease.
Circulating levels of a host of proinflammatory and anti-inflammatory mediators have been found in the blood of infected humans and mice (e.g., 30, 40). While plasma levels of IL-6 and other cytokines have been used as indicators of RSV disease, results across human studies have been inconsistent (40). In an in vitro system, Dyer et al. (41) found cytokines released from RSV-infected eosinophils at 8 d postinfection and correlated with the amount of virus titer in the cells. In the present study, we found that IL6 mRNA expression was significantly and negatively correlated with RSV-G expression among all LCLs in the ethnic panels. Our results are consistent with Mella et al. (40), who found that serum levels of innate immune cytokines TNF-α, IL-8, and IL-6 were modestly but significantly higher in infants with severe RSV bronchiolitis relative to healthy infants. However, they also found that TNF-α, IL-8, and IL-6 production capacity of whole blood cells was significantly diminished in severe disease infants compared to controls. Similarly, Roberts (42) found that in vitro exposure of human lymphocytes to RSV depressed cell-mediated immune function. Our findings also illustrate one of the advantages to using a battery of LCLs. The relationship between IL6 and RSV-G mRNA expression could only have been identified by comparing many lines.
To test the hypothesis that differential gene expression may have a role in susceptibility and response to RSV infection observed in human populations, we utilized publically available microarray gene expression profiles for HapMap LCLs to identify reproducible differential expression patterns between high and low responding individuals. We found 61 genes significantly correlated with RSV-G mRNA expression among 46 HapMap cell lines across multiple expression datasets. One of the genes was MX1, and because of biological plausibility, we selected it for further investigation. MX1 codes for MXA protein and interferes with viral replication by binding the polymerase subunits (PB2 and NP) of influenza virus and also binds the nucleocapsid of thogoto virus (20, 43). Increased MX1 mRNA expression was found in blood from infants with acute RSV bronchiolitis (44), while overexpression of Bos taurus MX1 significantly reduced RSV disease (45), and deletion of Mx1 (46) significantly enhanced influenza-induced disease in mice.
We hypothesized that polymorphisms in MX1 would predispose infants to severe RSV disease. To address this hypothesis, we identified a nonsynonymous missense MX1-coding SNP that leads to a Val379Ile amino acid substitution. Notably, we found that the predicted deleterious A allele was associated with increased RSV-G expression in the population of LCLs and increased the risk of RSV disease severity in another population, the Argentinian infant case-control cohort. Moreover, when we added a second, independent population to the first population, the strength of the association of the A allele with disease severity was enhanced. Our association results remained robust after adjusting for potentially confounding effects of clinical and epidemiological variables. Together with the LCL findings, this suggested a role for MX1 in RSV disease. The mechanism through which MX1 confers protection against human RSV disease is not completely understood. The amino acid substitution that we investigated occurs in the middle domain in the stalk of the MX1 protein (47), and deleterious effect on protein function may be hypothesized to occur by interfering with oligomerization of the protein and thus influence GTPase activity essential to antiviral activity (48). Additional investigations are needed to elucidate how the Val379Ile amino acid substitution in MXA is involved in host defense against RSV.
Another advantage of this model is the ability to select cell lines that contain SNPs of particular interest for in vitro screening. A bioinformatics approach integrating SNP, genotype, and cell-line selection was used to identify a functionally relevant SNP in a candidate gene. Using this approach, we found it is possible to identify several cell lines with SNPs in genes of interest that could be examined in vitro. It is also possible to take advantage of the multiple trios in the HapMap LCL collection to evaluate expression quantitative trait loci (eQTLs) in the model under investigation as has been done previously (e.g., 17, 31).
While RSV is primarily a respiratory disease, the virus is not restricted to the lung after infection. RSV RNA has been detected in extrapulmonary tissues in infected individuals, including human bone marrow stromal cells, the central nervous system, heart, and liver (49). RSV has also been detected in blood monocytes from Balb/c mice, and in human eosinophils (41), monocytes-macrophages (50), and lymphocytes (51). RSV load in peripheral blood of mice correlated with disease severity (52). Collectively, these studies suggest that RSV is capable of replicating and producing infective virus in multiple blood cell types, though the role they have in RSV disease progression remains unclear (41).
In summary, the combination of in vitro screening for genetic variation in response to RSV and differential gene expression with genotyping of individuals in a human cohort enabled us to make an informed decision about a gene (MX1) that has a role in RSV disease. This translational approach can ultimately be used to develop a set of genes with predictive capability about which individuals are at greater risk for developing severe RSV disease upon infection.
Supplementary Material
Acknowledgments
This research was supported (in part) by a Director's Challenge Program and the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences, U.S. Department of Health and Human Services.
The authors thank Dr. Patricio Acosta for all of his work to obtain the second population of RSV-infected children and his genotyping analyses. The authors also thank Dr. Michael Fessler and Dr. Donald Cook for thoughtful comments on the manuscript.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- CD14
- cluster of differentiation 14
- EIF2AK2
- eukaryotic translation initiation factor 2-α kinase 2
- hpi
- hours postinfection
- IFN
- interferon
- IFNA
- interferon α
- IFNB
- interferon β
- IFNG
- interferon γ
- IL-6
- interleukin 6
- LCL
- lymphoblastoid cell line
- MX1
- myxovirus resistance 1
- PKR
- protein kinase RNA-activated
- RSV
- respiratory syncytial virus
- RT-PCR
- reverse transcriptase polymerase chain reaction
- SNP
- single-nucleotide polymorphism
- TLR
- Toll-like receptor
- TNF-α
- tumor necrosis factor α
REFERENCES
- 1. Glezen W. P., Taber L. H., Frank A. L., Kasel J. A. (1986) Risk of primary infection and reinfection with respiratory syncytial virus. Am. J. Dis. Child. 140, 543–546 [DOI] [PubMed] [Google Scholar]
- 2. Nair H., Nokes D. J., Gessner B. D., Dherani M., Madhi S. A., Singleton R. J., O'Brien K. L., Roca A., Wright P. F., Bruce N., Chandran A., Theodoratou E., Sutanto A., Sedyaningsih E. R., Ngama M., Munywoki P. K., Kartasasmita C., Simoes E. A., Rudan I., Weber M. W., Campbell H. (2010) Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375, 1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lanari M., Giovannini M., Giuffre L., Marini A., Rondini G., Rossi G. A., Merolla R., Zuccotti G. V., Salvioli G. P. (2002) Prevalence of respiratory syncytial virus infection in Italian infants hospitalized for acute lower respiratory tract infections, and association between respiratory syncytial virus infection risk factors and disease severity. Pediatr. Pulmonol. 33, 458–465 [DOI] [PubMed] [Google Scholar]
- 4. Kaan P. M., Hegele R. G. (2003) Interaction between respiratory syncytial virus and particulate matter in guinea pig alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 28, 697–704 [DOI] [PubMed] [Google Scholar]
- 5. Harrod K. S., Jaramillo R. J., Rosenberger C. L., Wang S. Z., Berger J. A., McDonald J. D., Reed M. D. (2003) Increased susceptibility to RSV infection by exposure to inhaled diesel engine emissions. Am. J. Respir. Cell Mol. Biol. 28, 451–463 [DOI] [PubMed] [Google Scholar]
- 6. Bradley J. P., Bacharier L. B., Bonfiglio J., Schechtman K. B., Strunk R., Storch G., Castro M. (2005) Severity of respiratory syncytial virus bronchiolitis is affected by cigarette smoke exposure and atopy. Pediatrics 115, e7–14 [DOI] [PubMed] [Google Scholar]
- 7. Anh D. B., Faisca P., Desmecht D. J. (2006) Differential resistance/susceptibility patterns to pneumovirus infection among inbred mouse strains. Am. J. Physiol. Lung Cell. Mol. Physiol. 291, L426–L435 [DOI] [PubMed] [Google Scholar]
- 8. Stark J. M., McDowell S. A., Koenigsknecht V., Prows D. R., Leikauf J. E., Le Vine A. M., Leikauf G. D. (2002) Genetic susceptibility to respiratory syncytial virus infection in inbred mice. J. Med. Virol. 67, 92–100 [DOI] [PubMed] [Google Scholar]
- 9. Awomoyi A. A., Rallabhandi P., Pollin T. I., Lorenz E., Sztein M. B., Boukhvalova M. S., Hemming V. G., Blanco J. C., Vogel S. N. (2007) Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high-risk infants and young children. J. Immunol. 179, 3171–3177 [DOI] [PubMed] [Google Scholar]
- 10. Gentile D. A., Doyle W. J., Zeevi A., Howe-Adams J., Kapadia S., Trecki J., Skoner D. P. (2003) Cytokine gene polymorphisms moderate illness severity in infants with respiratory syncytial virus infection. Hum. Immunol. 64, 338–344 [DOI] [PubMed] [Google Scholar]
- 11. Paulus S. C., Hirschfeld A. F., Victor R. E., Brunstein J., Thomas E., Turvey S. E. (2007) Common human Toll-like receptor 4 polymorphisms—role in susceptibility to respiratory syncytial virus infection and functional immunological relevance. Clin. Immunol. 123, 252–257 [DOI] [PubMed] [Google Scholar]
- 12. Puthothu B., Forster J., Heinze J., Heinzmann A., Krueger M. (2007) Surfactant protein B polymorphisms are associated with severe respiratory syncytial virus infection, but not with asthma. BMC Pulm. Med. 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puthothu B., Forster J., Heinzmann A., Krueger M. (2006) TLR-4 and CD14 polymorphisms in respiratory syncytial virus-associated disease. Dis. Markers 22, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Puthothu B., Krueger M., Forster J., Heinze J., Weckmann M., Heinzmann A. (2007) Interleukin (IL)-18 polymorphism 133C/G is associated with severe respiratory syncytial virus infection. Pediatr. Infect. Dis. J. 26, 1094–1098 [DOI] [PubMed] [Google Scholar]
- 15. Puthothu B., Krueger M., Heinze J., Forster J., Heinzmann A. (2006) Impact of IL8 and IL8-receptor alpha polymorphisms on the genetics of bronchial asthma and severe RSV infections. Clin. Mol. Allergy 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thomsen S. F., Stensballe L. G., Skytthe A., Kyvik K. O., Backer V., Bisgaard H. (2008) Increased concordance of severe respiratory syncytial virus infection in identical twins. Pediatrics 121, 493–496 [DOI] [PubMed] [Google Scholar]
- 17. Cheung V. G., Conlin L. K., Weber T. M., Arcaro M., Jen K. Y., Morley M., Spielman R. S. (2003) Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 33, 422–425 [DOI] [PubMed] [Google Scholar]
- 18. Correa C. R., Cheung V. G. (2004) Genetic variation in radiation-induced expression phenotypes. Am. J. Hum. Genet. 75, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jen K. Y., Cheung V. G. (2003) Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res. 13, 2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haller O., Frese M., Rost D., Nuttall P. A., Kochs G. (1995) Tick-borne thogoto virus infection in mice is inhibited by the orthomyxovirus resistance gene product Mx1. J. Virol. 69, 2596–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pavlovic J., Zurcher T., Haller O., Staeheli P. (1990) Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 64, 3370–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Staeheli P., Haller O., Boll W., Lindenmann J., Weissmann C. (1986) Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44, 147–158 [DOI] [PubMed] [Google Scholar]
- 23. Laham F. R., Israele V., Casellas J. M., Garcia A. M., Lac Prugent C. M., Hoffman S. J., Hauer D., Thumar B., Name M. I., Pascual A., Taratutto N., Ishida M. T., Balduzzi M., Maccarone M., Jackli S., Passarino R., Gaivironsky R. A., Karron R. A., Polack N. R., Polack F. P. (2004) Differential production of inflammatory cytokines in primary infection with human metapneumovirus and with other common respiratory viruses of infancy. J. Infect. Dis. 189, 2047–2056 [DOI] [PubMed] [Google Scholar]
- 24. Inoue Y., Shimojo N., Suzuki Y., Campos Alberto E. J., Yamaide A., Suzuki S., Arima T., Matsuura T., Tomiita M., Aoyagi M., Hoshioka A., Honda A., Hata A., Kohno Y. (2007) CD14-550 C/T, which is related to the serum level of soluble CD14, is associated with the development of respiratory syncytial virus bronchiolitis in the Japanese population. J. Infect. Dis. 195, 1618–1624 [DOI] [PubMed] [Google Scholar]
- 25. Groskreutz D. J., Monick M. M., Powers L. S., Yarovinsky T. O., Look D. C., Hunninghake G. W. (2006) Respiratory syncytial virus induces TLR3 protein and protein kinase R, leading to increased double-stranded RNA responsiveness in airway epithelial cells. J. Immunol. 176, 1733–1740 [DOI] [PubMed] [Google Scholar]
- 26. Klein Klouwenberg P., Tan L., Werkman W., van Bleek G. M., Coenjaerts F. (2009) The role of Toll-like receptors in regulating the immune response against respiratory syncytial virus. Crit. Rev. Immunol. 29, 531–550 [DOI] [PubMed] [Google Scholar]
- 27. Isaacs D. (1989) Production of interferon in respiratory syncytial virus bronchiolitis. Arch. Dis. Childhood 64, 92–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merolla R., Rebert N. A., Tsiviste P. T., Hoffmann S. P., Panuska J. R. (1995) Respiratory syncytial virus replication in human lung epithelial cells: inhibition by tumor necrosis factor alpha and interferon beta. Am. J. Respir. Crit. Care Med. 152, 1358–1366 [DOI] [PubMed] [Google Scholar]
- 29. Tregoning J. S., Wang B. L., McDonald J. U., Yamaguchi Y., Harker J. A., Goritzka M., Johansson C., Bukreyev A., Collins P. L., Openshaw P. J. (2013) Neonatal antibody responses are attenuated by interferon-γ produced by NK and T cells during RSV infection. Proc. Natl. Acad. Sci. U. S. A. 110, 5576–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diaz P. V., Pinto R. A., Mamani R., Uasapud P. A., Bono M. R., Gaggero A. A., Guerrero J., Goecke A. (2012) Increased expression of the glucocorticoid receptor beta in infants with RSV bronchiolitis. Pediatrics 130, e804–e811 [DOI] [PubMed] [Google Scholar]
- 31. Cheung V. G., Ewens W. J. (2006) Heterozygous carriers of Nijmegen Breakage Syndrome have a distinct gene expression phenotype. Genome Res. 16, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choy E., Yelensky R., Bonakdar S., Plenge R. M., Saxena R., De Jager P. L., Shaw S. Y., Wolfish C. S., Slavik J. M., Cotsapas C., Rivas M., Dermitzakis E. T., Cahir-McFarland E., Kieff E., Hafler D., Daly M. J., Altshuler D. (2008) Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 4, e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Min J. L., Taylor J. M., Richards J. B., Watts T., Pettersson F. H., Broxholme J., Ahmadi K. R., Surdulescu G. L., Lowy E., Gieger C., Newton-Cheh C., Perola M., Soranzo N., Surakka I., Lindgren C. M., Ragoussis J., Morris A. P., Cardon L. R., Spector T. D., Zondervan K. T. (2011) The use of genome-wide eQTL associations in lymphoblastoid cell lines to identify novel genetic pathways involved in complex traits. PLoS One 6, e22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smirnov D. A., Morley M., Shin E., Spielman R. S., Cheung V. G. (2009) Genetic analysis of radiation-induced changes in human gene expression. Nature 459, 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X., Tomso D. J., Chorley B. N., Cho H. Y., Cheung V. G., Kleeberger S. R., Bell D. A. (2007) Identification of polymorphic antioxidant response elements in the human genome. Hum. Mol. Genet. 16, 1188–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pavlovic J., Arzet H. A., Hefti H. P., Frese M., Rost D., Ernst B., Kolb E., Staeheli P., Haller O. (1995) Enhanced virus resistance of transgenic mice expressing the human MxA protein. J. Virol. 69, 4506–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lock E. F., Abdo N., Huang R., Xia M., Kosyk O., O'Shea S. H., Zhou Y. H., Sedykh A., Tropsha A., Austin C. P., Tice R. R., Wright F. A., Rusyn I. (2012) Quantitative high-throughput screening for chemical toxicity in a population-based in vitro model. Toxicol. Sci. 126, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeVincenzo J. P., Wilkinson T., Vaishnaw A., Cehelsky J., Meyers R., Nochur S., Harrison L., Meeking P., Mann A., Moane E., Oxford J., Pareek R., Moore R., Walsh E., Studholme R., Dorsett P., Alvarez R., Lambkin-Williams R. (2010) Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 182, 1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mella C., Suarez-Arrabal M. C., Lopez S., Stephens J., Fernandez S., Hall M. W., Ramilo O., Mejias A. (2012) Innate immune dysfunction is associated with enhanced disease severity in infants with severe respiratory syncytial virus bronchiolitis. J. Infect. Dis. 207, 564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dyer K. D., Percopo C. M., Fischer E. R., Gabryszewski S. J., Rosenberg H. F. (2009) Pneumoviruses infect eosinophils and elicit MyD88-dependent release of chemoattractant cytokines and interleukin-6. Blood 114, 2649–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roberts N. J., Jr. (1982) Different effects of influenza virus, respiratory syncytial virus, and Sendai virus on human lymphocytes and macrophages. Infect. Immun. 35, 1142–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Verhelst J., Parthoens E., Schepens B., Fiers W., Saelens X. (2012) Interferon-inducible protein mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 86, 13445–13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bucasas K. L., Mian A. I., Demmler-Harrison G. J., Caviness A. C., Piedra P. A., Franco L. M., Shaw C. A., Zhai Y., Wang X., Bray M. S., Couch R. B., Belmont J. W. (2012) Global gene expression profiling in infants with acute respiratory syncytial virus broncholitis demonstrates systemic activation of interferon signaling networks. Pediatr. Infect. Dis. J. 32, e68–e76 [DOI] [PubMed] [Google Scholar]
- 45. Dermine M., Desmecht D. (2012) In vivo modulation of the innate response to pneumovirus by type-I and -III interferon-induced Bos taurus Mx1. J. Interferon Cytokine Res. 32, 332–337 [DOI] [PubMed] [Google Scholar]
- 46. Staeheli P., Grob R., Meier E., Sutcliffe J. G., Haller O. (1988) Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8, 4518–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Von der Malsburg A., Abutbul-Ionita I., Haller O., Kochs G., Danino D. (2011) Stalk domain of the dynamin-like MxA GTPase protein mediates membrane binding and liposome tubulation via the unstructured L4 loop. J. Biol. Chem. 286, 37858–37865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao S., von der Malsburg A., Dick A., Faelber K., Schroder G. F., Haller O., Kochs G., Daumke O. (2011) Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35, 514–525 [DOI] [PubMed] [Google Scholar]
- 49. Eisenhut M. (2006) Extrapulmonary manifestations of severe respiratory syncytial virus infection—a systematic review. Crit. Care 10, R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rohwedder A., Keminer O., Forster J., Schneider K., Schneider E., Werchau H. (1998) Detection of respiratory syncytial virus RNA in blood of neonates by polymerase chain reaction. J. Med. Virol. 54, 320–327 [DOI] [PubMed] [Google Scholar]
- 51. Domurat F., Roberts N. J., Jr., Walsh E. E., Dagan R. (1985) Respiratory syncytial virus infection of human mononuclear leukocytes in vitro and in vivo. J. Infect. Dis. 152, 895–902 [DOI] [PubMed] [Google Scholar]
- 52. Torres J. P., Gomez A. M., Khokhar S., Bhoj V. G., Tagliabue C., Chang M. L., Kiener P. A., Revell P. A., Ramilo O., Mejias A. (2010) Respiratory syncytial virus (RSV) RNA loads in peripheral blood correlates with disease severity in mice. Respir. Res. 11, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.