Abstract
Hypoxia-inducible factors (HIFs) are master regulators of the transcriptional response to hypoxia. To gain insight into the structural and functional evolution of the HIF family, we characterized the HIFα gene from amphioxus, an invertebrate chordate, and identified several alternatively spliced HIFα isoforms. Whereas HIFα Ia, the full-length isoform, contained a complete oxygen-dependent degradation (ODD) domain, the isoforms Ib, Ic, and Id had 1 or 2 deletions in the ODD domain. When tagged with GFP and tested in mammalian cells, the amphioxus HIFα Ia protein level increased in response to hypoxia or CoCl2 treatment, whereas HIFα Ib, Ic, and Id showed reduced or no hypoxia regulation. Deletion of the ODD sequence in HIFα Ia up-regulated the HIFα Ia levels under normoxia. Gene expression analysis revealed HIFα Ic to be the predominant isoform in embryos and larvae, whereas isoform Ia was the most abundant form in the adult stage. The expression levels of Ib and Id were very low. Hypoxia treatment of adults had no effect on the mRNA levels of these HIFα isoforms. Functional analyses in mammalian cells showed all 4 HIFα isoforms capable of entering the nucleus and activating hypoxia response element–dependent reporter gene expression. The functional nuclear location signal (NLS) mapped to 3 clusters of basic residues. 775KKARL functioned as the primary NLS, but 737KRK and 754KK also contributed to the nuclear localization. All amphioxus HIFα isoforms had 2 functional transactivation domains (TADs). Its C-terminal transactivation (C-TAD) shared high sequence identity with the human HIF-1α and HIF-2α C-TAD. This domain contained a conserved asparagine, and its mutation resulted in an increase in transcriptional activity. These findings reveal many ancient features of the HIFα family and provide novel insights into the evolution of the HIFα family.—Gao, S., Lu, L., Bai, Y., Zhang, P., Song, W., Duan, C. Structural and functional analysis of amphioxus HIFα reveals ancient features of the HIFα family.
Keywords: oxygen-dependent degradation, nuclear localization signal, transactivation domain, cephalochordate
Animal cells require oxygen for efficient cellular respiration and energy production. Under hypoxia (low oxygen tension), several genes are activated as an adaptive response. These hypoxia-responsive genes are involved in metabolic changes, erythropoiesis, angiogenesis, cell growth, differentiation, and apoptosis (1–3). They are also associated with many human diseases, such as anemia, ischemia, and cancer (1–3). Hypoxia-inducible factors (HIFs) are master regulators of the transcriptional response to hypoxia (2). HIFs are heterodimeric transcription factors consisting of an α subunit and a β subunit [also known as aryl hydrocarbon receptor nuclear translocator (ARNT)]. HIFα and HIFβ both belong to the basic helix-loop-helix (bHLH)/Per/ARNT/Sim (PAS) superfamily (4). Whereas HIFβ is stable, HIFα is tightly regulated by oxygen levels (2). This disparity is well documented in the case of human HIF-1α. Under normoxia, several prolyl hydroxylase domain (PHD) enzymes hydroxylate HIF-1α at P402 and P564 in its oxygen-dependent degradation (ODD) domain. Hydroxylated HIF-1α is recognized by von Hippel-Lindau tumor suppressor (VHL), polyubiquitinated, and targeted to the proteasome for degradation (5–9). Since oxygen is a cosubstrate for PHDs (6), PHD activities are inhibited under hypoxia, leading to the stabilization of HIF-1α. The stabilized HIF-1α is translocated to the nucleus via its nuclear localization signal (NLS) motif (10, 11). In the nucleus, HIF-1α dimerizes with HIFβ and binds to the hypoxia response elements (HREs; A/GCGTG) on the promoter regions of its target genes, to stimulate gene expression (2). Oxygen availability also regulates HIF-1α activity through another hydroxylation event, where factor-inhibiting HIF (FIH), another hydroxylase, hydroxylates human HIF-1α at N803 in its C-terminal transactivation domain (C-TAD). This asparagine hydroxylation blocks the recruitment of coactivators to HIF-1α and thereby inhibits HIF-1α's transcriptional activity under normoxia (10, 12, 13). In addition to HIF-1α's role in hypoxia, recent studies suggest that it plays an important part in early development (14–16). Furthermore, HIF-1α stability and activity are also regulated by nonhypoxic stimuli, such as hormonal, metabolic, and inflammatory signals (17–21).
Humans and other mammals have 3 HIFα genes: HIF-1α, HIF-2α, and HIF-3α. HIF-1α and HIF-2α share significant sequence identity and similar domain arrangements, both containing 2 TADs (22). HIF-3α shares relatively low sequence identity with HIF-1α/2α and lacks the C-TAD (23). In comparison, only a single HIFα gene is found in the nematode Caenorhabditis elegans (24) and the fruit fly (25). Loenarz et al. (26) surveyed HIF genes in 50 eukaryote genomes and found that all vertebrate genomes have multiple HIFα genes, whereas all invertebrate genomes surveyed contain a single HIFα gene. Moreover, they demonstrated that the HIF pathway is functional in Trichoplax adhaerens (26). Despite much progress, many important questions remain regarding the evolution of the HIF pathway. For example, although it is clear that the multiple vertebrate HIFα genes have evolved from a common ancestor, whether the ancestral HIFα has an HIF-1α/2α- or HIF-3α-like structure is unclear. In addition, the C-TAD asparagine hydroxylation and FIH-like molecule have not been found in T. adhaerens (26) or the fruit fly (25). It is not clear whether the asparagine hydroxylation regulation mechanism evolved prior to the emergence of vertebrates.
The amphioxus is considered the most basal species in the chordate subphylum and shares a common ancestor with modern vertebrate lineage, which dates back >5.2 × 108 yr (27). It has been found to evolve relatively slowly compared to other existent chordates, such as tunicates (28, 29). Its genome shares strong synteny with those of modern vertebrates and thus provides an excellent model for studying chordate gene evolution (30). Comparative genomic studies have shown that the amphioxus did not go through the 2 to 3 rounds of whole-genome duplications seen in vertebrates (28, 31). Therefore, it often contains a single gene of a given vertebrate gene family. Indeed, a single HIFα gene was identified in the amphioxus by database searches (26) and molecular cloning (this study).
In this study, we cloned and characterized the amphioxus HIFα gene and determined its expression and physiological regulation. We performed structural and functional analysis by transfecting amphioxus HIFα in mammalian cells. Our results indicate that amphioxus HIFα shares many similarities with vertebrate HIF-1α/2α. Its C-TAD contains a conserved asparagine that is functional in modulating the transcriptional activity. We further showed that the single amphioxus HIFα gene produces multiple spliced transcripts. Although HIFα Ia contains a complete ODD domain and its protein levels are increased under hypoxia when tested in mammalian cells, HIFα Ib, Ic, and Id all have partial ODD domains, and their protein levels did not increase by hypoxia. Of note, although the hypoxia-insensitive HIFα Ic isoform is the predominant HIFα isoform expressed in embryos and larvae, the hypoxia-inducible isoform Ia is the most abundant isoform in the adult stage.
MATERIALS AND METHODS
All chemicals and reagents were purchased from Fisher Scientific (Pittsburgh, PA, USA) unless stated otherwise. Cell culture media and oligonucleotide primers were purchased from Invitrogen (Carlsbad, CA, USA). Restriction enzymes were purchased from New England Biolabs (Ipswich, MA, USA) or Promega (Madison, WI, USA).
Experimental animals
Amphioxi (Branchiostoma belcheri tsingtauense) were collected from the seashore in Qingdao, China, and were spawned in the marine aquatic animal facility. Embryos and larvae were collected at different developmental stages and frozen in liquid nitrogen for RNA extraction. To determine the effect of reduced oxygen levels on HIFα gene expression, we divided the adult animals (2–3 yr old) into 3 groups and transferred them to normal seawater (dissolved oxygen level, ∼8.3 mg/L), moderately (30% of the normal oxygen level), or severely (10% of the normal oxygen level) hypoxic water. The hypoxic water was generated by bubbling nitrogen gas into the system water (32). Animals (5–6/group) were sampled at 12 and 24 h after the transfer and were frozen in liquid nitrogen for RNA extraction. These experiments were conducted at Ocean University of China with protocols approved by the university's Committee on the Ethics of Animal Experiments.
Molecular cloning and sequence analysis
To identify the amphioxus HIFα and clone it for functional analysis, we performed a TBLASTN search [U.S. National Center for Biotechnology Information (NCBI), Bethesda, MD, USA; http://blast.ncbi.nlm.nih.gov] against the Branchiostoma floridae genome database. A single putative HIFα (BRAFLDRAFT_208408, 336 aa) was identified. This putative protein shows high sequence identity with human HIF1α. 5′-RACE was performed with the SMART RACE kit (Clontech, Mountain View, CA, USA). Total RNA was extracted with TRIzol reagent (Invitrogen) and reverse transcribed with M-MLV reverse transcriptase (Invitrogen) into RACE-ready first-strand cDNA. The 5′ RACE product was subcloned into a pGEM-T Easy Vector (Promega) and sequenced. Based on the open reading frame sequence predicted by GeneScan software (Applied Biosystems, Foster City, CA, USA; ref. 33), 2 polymerase chain reaction (PCR) primers (forward 5′-CGGAACACCTGAACACCTTT-3′, reverse 5′-AGCAGTCCCAAGCATCAAGT-3′) were designed and used to amplify a 2679-bp DNA fragment encoding the full-length amphioxus HIFα with Phusion DNA polymerase (New England Biolabs). The PCR product was subcloned into a pGEM-T Easy Vector (Promega), and >30 colonies were sequenced. As a result, 4 distinct transcripts, termed amphioxus HIFα isoforms a–d (Ia, Ib, Ic, and Id), were identified.
The genomic structure was determined by comparing the full-length cDNA sequence with the available B. floridae genome sequence (v. 2.0). Because the public database has an ambiguous sequence near exon 8 (124 n repeats) and since our RNA was isolated from B. belcheri tsingtauense, the genomic structure was ascertained by genomic PCR using LATaq DNA polymerase (Takara Bio Inc., Shiga, Japan).
For phylogenic analysis, the sequence of known HIFαs was obtained from the GenBank (NCBI; http://www.ncbi.nlm.nih.gov/genbank/) or Ensembl (European Bioinformatics Institute/Wellcome Trust Sanger Institute; http://www.ensembl.org) database. Sequences were aligned by MUSCLE (34), and the ambiguously aligned sites were removed by Gblocks v. 0.91b (35). Maximum-likelihood analysis was performed with PhyML 3.0 (36) with the JTT model (G=1.344) selected by ProTest (37). A neighbor-joining tree was constructed with an amino:Poisson correction model, by using Mega 4 (http://www.megasoftware.net). All the trees were bootstrapped for 1000 replicates. The sequences of human, mouse, and frog ARNT proteins, which belong to the bHLH/PAS superfamily, were used as the outgroups.
Reverse transcription PCR (RT-PCR) and real-time quantitative RT-PCR (qRT-PCR)
RNA isolation and cDNA synthesis were performed as described above. PCR was performed with Taq polymerase in the following conditions: 94°C for 3 min, followed by 38 cycles of 94°C for 20 s, 55°C for 30 s, 72°C for 90 s, and a final extension at 72°C for 10 min. A primer set (forward 5′-AAGAGGGGACTGTGTTGTCG-3′, reverse, 5′-CTCCGCCGTCTGTTTGTATT-3′) was designed to flank the deletions to detect all 4 isoforms simultaneously. Amphioxus HIFα isoform plasmids were included as a positive control. The β-actin primers are 5′-CTCCGGTATGTGCAAGGC-3′ and 5′-GCTGGGCTGTTGAAGGTC-3′.
qRT-PCR was performed with the iCycler iQ5 Multicolor real-time PCR detection system (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. A primer set (forward 5′-GACCAACAAATGAGCGACCAG-3′, reverse 5′-AGGGTGGGTGAAGTCAAAGATAC-3′) was designed targeting an N-terminal sequence that is common in all the isoforms, to amplify them with the same efficiency. qPCR was performed with SYBR Green PCR Master Mix (Applied Biosystems) under the following conditions: 94°C for 3 min, followed by 40 cycles of 94°C for 20 s, 60°C for 30 s, and 72°C for 30 s. β-Actin was used as the internal control (forward 5′-TCTGGCATCATACCTTCTACAA-3′, reverse 5′-TCTGTGTCATCTTTTCCCTGTT-3′).
Plasmid construction
The pCEP4-human HIF-1α and pcDNA 3.1-zebrafish Hif-1α plasmids have been reported (24, 38). To engineer an amphioxus HIFα expression construct (pCS2-AmHIFα-EGFP), we amplified DNA encoding the amphioxus HIFα sequence by PCR and subcloned it into the pCS2-EGFP expression vector. An Myc-tagged amphioxus HIFα expression construct was made by subcloning the amphioxus HIFα DNA into the pcDNA 3.1/Myc-His (−)A vector (Invitrogen). Several truncated HIFα mutants, covering aa 717–881, were generated by PCR, with pCS2-AmHIFα-EGFP used as a template. In addition, several point mutations were generated by site-directed mutagenesis, using PfuTurbo polymerase (Stratagene-Agilent, Palo Alto, CA, USA) according to the manufacturer's instructions. To map and characterize the transcriptional activation domains in amphioxus HIFα, DNA encoding the putative C-TAD (aa 839–881) and N-terminal transactivation domain (N-TAD; aa 425–515) were amplified by PCR and subcloned into the pBIND vector (Promega). Truncated protein, deleting C-TAD (aa 1–838, Δ839–881), was generated to test the transactivation importance of C-TAD. N-TAD deletion mutant (ΔNTAD; aa Δ425–515) was generated by using restriction enzyme-directed inverse PCR (39). To study the ODD domain, we engineered a deletion mutant (Δ397–651) by using the same method. To further study the ODD domain, we constructed a fusion protein plasmid by fusing the amphioxus HIFα ODD domain to the human ARNT-TAD domain. For this, DNA encoding the TAD domain of human ARNT (aa 581–789) was amplified and subcloned into the pBIND vector. The putative ODD domain of amphioxus HIFα was then amplified and subcloned into pBIND-ANRT at the BamHI site, resulting in pBIND-ODD-ANRT. All the primers used in the present study are listed in Supplemental Table S1. All plasmids were verified by DNA sequencing.
Cell culture
Human embryonic kidney (HEK) 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 2% penicillin and streptomycin. Human osteosarcoma U2OS cells were cultured in McCoy's 5A medium supplemented with 10% FBS and 2% penicillin and streptomycin. Hypoxia treatment was conducted in a hypoxic chamber filled with 1% O2 (40).
Transcription activity assay
HEK 293T cells were used because they possess high transfection efficiency. Briefly, subconfluent cells were transfected with the intended plasmid DNA using Opti-MEM and Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA; ref. 40). Luciferase activity was measured with the Dual-Luciferase Reporter Assay (Promega) at 24 h after transfection.
Subcellular localization
U2OS cells, which are large and flat, were used in visualizing the subcellular localization of amphioxus HIFα-EGFP. The cells were transiently transfected with the intended plasmid DNA, with Opti-MEM and Lipofectamine 2000 (40). At 48 h after transfection, the cells were washed with 1× PBS and fixed with 4% paraformaldehyde (PFA) for 1 h. After staining with 4′,6-diamidino-2-phenylindole (DAPI) at 0.5 μg/ml for 10 min, the cells were washed with 1× PBS and mounted. Images were captured with an Eclipse E600 microscope (Nikon, Gamagori, Japan) with a Spot Slider2 camera (model 1.4.0, software v.4.0.9; Diagnostic Instruments, Inc., Sterling Heights, MI, USA).
Western immunoblot analysis
Cells were lysed in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM EGTA, and 0.1% TritonX-100, pH 7.5) containing protease and phosphatase inhibitors (1 μg/ml leupeptin, 1 μg/ml pepstatin, 2 μM PMSF, 2 μg/ml aprotinin, 2.5 μM sodium pyrophosphate, 1 μM sodium fluoride, and 1 μM sodium orthovanadate). Protein concentrations were determined with a BCA Protein Assay kit (Pierce Biotechnology, Rockford, IL, USA). Homogenized samples were mixed with 4× protein loading buffer and 2-mercaptoethnol (final concentration 5%) and boiled for 5 min. Equal amounts of samples were separated by 7.5% SDS-PAGE and subjected to Western blot analysis.
Statistics
All values are expressed as means ± sd, unless otherwise stated. Statistical analyses were performed by Tukey post hoc ANOVA (1-way analysis of variance; Prism 5; GraphPad Software, La Jolla, CA, USA).
RESULTS
Characterization of the amphioxus HIFα gene and identification of multiple amphioxus HIFα transcripts
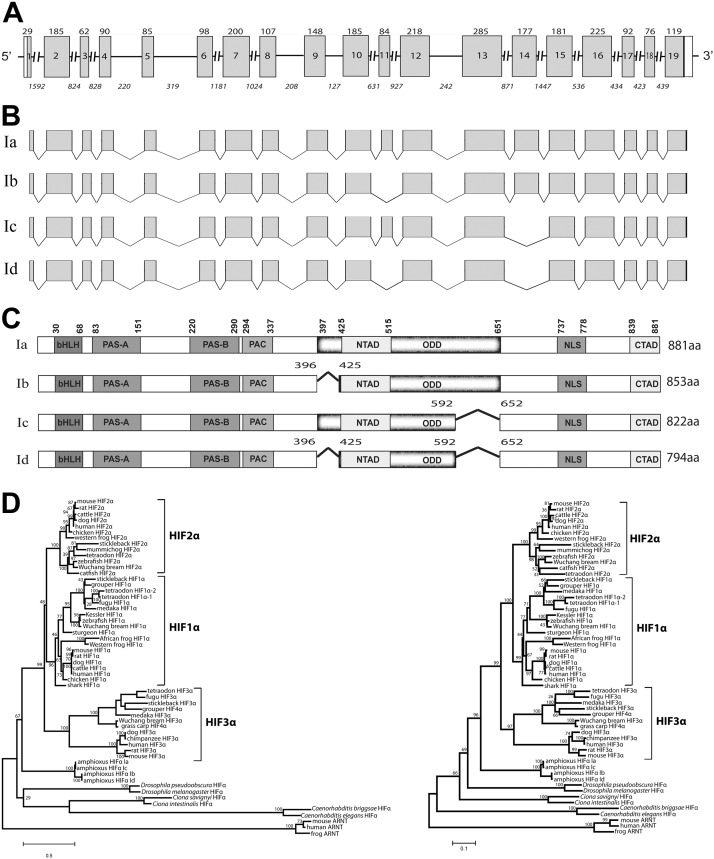
Molecular cloning and sequence analysis indicate that the amphioxus HIFα coding DNA sequence is 2.6 kb in size, and the gene contains 19 exons and 18 introns (Fig. 1A). Four alternatively spliced transcripts, amphioxus HIFα isoforms Ia to Id (HM188448, HM188449, HM188450, and HM188451), were identified by RACE and RT-PCR experiments (Fig. 1B). HIFα Ia, the longest isoform, encodes a protein of 881 aa (Fig. 1C and Supplemental Fig. S1). HIFα Ib lacks aa 397–424, which is encoded by exon 11 (Fig. 1B). Ic lacks an exon 14 sequence (aa 593–651), and Id lacks the sequences of exons 11 and 14 (Fig. 1B). All 4 HIFα isoforms contain a recognizable bHLH, PAS-A, PAS-B, and C-TAD (Fig. 1C). Whereas amphioxus HIFα Ia contains a functional and complete ODD domain (see below), amphioxus HIFα Ib, Ic, and Id have 1 or 2 deletions in this domain (Fig. 1C). Phylogenetic analyses suggest that amphioxus HIFα occupies a basal position corresponding to the common ancestor of vertebrate HIF-1α to -3α (Fig. 1D).
Figure 1.
Characterization of the amphioxus HIFα gene and several alternatively spliced HIFα isoforms. A) Schematic diagram of the amphioxus HIFα gene. Gray boxes represent protein coding regions, open boxes represent untranslated regions, and lines represent introns. Numbers inside the boxes represent the exon number; numbers above each box represent the exon size (bp). Italic numbers below each line represent the intron size (bp). B) Schematic diagram of multiple transcripts identified by RACE and RT-PCR. Note that the exon 11 sequence is not present in transcripts Ib and Id, and exon 14 is absent in Ic and Id. C) Schematic diagram of the amphioxus HIFα isoforms. Numbers in Ib, Ic, and Id represent the position of deletions 1 and 2. Characteristic domains are marked in bold letters according to the amino acid positions of Ia. D) Phylogenetic trees. Full-length amino acid sequences of known chordates and vertebrate HIFα were aligned and analyzed by phyML3.0 with the JTT model for the maximum-likelihood (ML) tree (left). A neighbor-joining (NJ) tree was also constructed with the same dataset by Mega 4 (right). Numbers on the brackets are the bootstrap values with 1000 replications.
Expression and hypoxic regulation of different amphioxus HIFα transcripts in embryonic, larval, and adult amphioxi
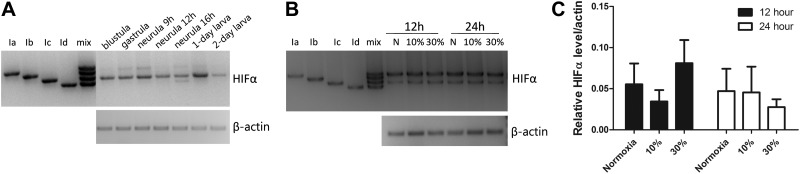
To determine the expression patterns of the HIFα isoforms, we developed RT-PCR assays to detect the multiple HIFα transcripts simultaneously (Fig. 2A). In early development, HIFα Ic mRNA was the most abundantly expressed and easily detected in all early developmental stages, ranging from blastula (3.5 h after fertilization) to 2-d larva (Fig. 2A). HIFα Ia mRNA became detectable at the gastrula stage and remained at lower but detectable levels thereafter. HIFα Id mRNA was detected only at the neurula stage (16 h), and HIFα Ib mRNA was not detected in any of the stages examined (Fig. 2A). In the adult stage, HIFα Ia mRNA was the most abundant transcript (Fig. 2B). HIFα Ic mRNA was also detected, but at lower levels. HIFα Ib and Id mRNA were not detected (Fig. 2B). These data suggest that, although HIFα Ic is the predominant isoform in early development, HIFα Ia becomes the predominant isoform in the adult stage.
Figure 2.
Expression of HIFα Ia to Id mRNAs in amphioxus embryos, larvae, and adults and the effect of hypoxia. A) RT-PCR analysis of HIFα Ia to Id transcripts in early development. Lanes 1–4, positive controls containing cloned HIFα Ia, Ib, Ic, and Id DNA; and lane 5, the mixture of all 4 HIFα plasmid DNAs. Similar results were obtained in 3 independent experiments. B) Expression of HIFα Ia to Id transcripts in the adult amphioxus. Adult amphioxi were kept in seawater with ambient O2 levels (normoxia) or were transferred to moderately (30% of ambient O2 levels) or severely (10% of ambient O2 levels) hypoxic water for the indicated times. RT-PCR assays were performed as described in A. Similar results were obtained in 3 independent experiments. C) Total amphioxus HIFα mRNA levels in the samples described in B were measured by qRT-PCR. Values are expressed relative to β-actin mRNA levels; values are means ± sd (n=3). There was no significant difference among these groups (P>0.05).
We next examined the effect of hypoxia. Adult animals were transferred to seawater containing various levels of dissolved oxygen for 12 and 24 h. Moderate or severe hypoxia (30 and 10% of the ambient O2 level, respectively) did not change the relative levels of HIFα Ia to Id mRNAs (Fig. 2B). The total HIFα mRNA levels were measured by real-time qRT-PCR by using a primer set targeting an N-terminal sequence common to all the isoforms. Again, no significant difference was detected among these groups (Fig. 2C), suggesting that hypoxia does not alter the mRNA expression of HIFα isoforms.
Identification of a functional ODD domain and differential oxygen-dependent regulation of the HIFα isoforms
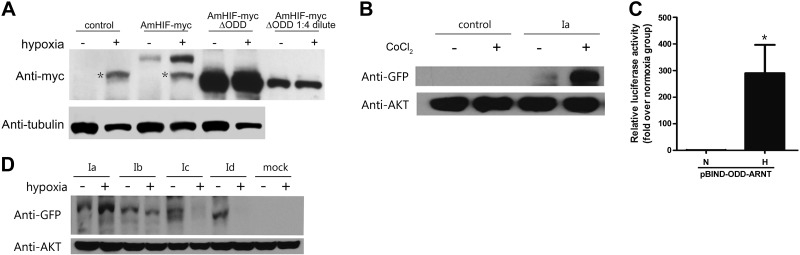
We examined whether changes in oxygen tension regulate amphioxus HIFα at the protein level. Because there is no amphioxus cell line or primary cell culture system available, we used human HEK293T cells instead. The cells were transfected with either a Myc-tagged amphioxus HIFα Ia expression construct or an empty vector and subjected to 24 h hypoxia treatment. In the HIFα Ia-Myc-transfected group, low levels of HIFα Ia-Myc were detected under normoxia. There was a marked increase in the HIFα Ia-Myc levels under hypoxia (Fig. 3A). No amphioxus HIFα Ia-Myc was detected in the empty vector–transfected control group under normoxia or hypoxia (Fig. 3A). In both groups, there was a hypoxia-inducible protein that showed nonspecific immunoreactivity to the Myc antibody used. To further examine the effect of hypoxia, the cells were transfected with an amphioxus HIFα Ia-EGFP expression construct and subjected to hypoxia. Hypoxia markedly increased the HIFαIa-EGFP levels (Fig. 3D, lanes 1 and 2). Next, cells expressing HIFα Ia-EGFP were treated with CoCl2, a small compound that can inhibit vertebrate PHD activity and thus increase vertebrate HIFα stability under normoxia (38, 41). CoCl2 treatment resulted in a robust increase in HIFα Ia-EGFP levels (Fig. 3B). Collectively, these data suggest that amphioxus HIFα Ia is regulated by oxygen-dependent degradation.
Figure 3.
Amphioxus HIFα had a functional ODD domain, and different HIFα isoforms had different responses to hypoxia. A) Effects of hypoxia and the deletion of ODD domain. HEK 293T cells transfected with the indicated amphioxus HIFα constructs or empty vector (control) were subjected to 24 h hypoxia (1% oxygen) or kept under normoxia (ambient air). Cells were lysed and analyzed by Western immunoblot with the indicated antibodies. Asterisks indicate nonspecific bands. B) Effect of CoCl2. Cells transfected with pCS2-EGFP-amHIFaIa or mock-transfected cells were treated with 50 μM CoCl2 for 24 h and analyzed as described above. C) Amphioxus HIFα ODD domain conferred oxygen responsiveness. HEK 293T cells were transfected with pBIND-ODD-ARNT together with pG5luc. Gal4-AmHIF-ODD-ARNT contains the amphioxus HIFα ODD domain fused to the TAD of human ARNT. At 1 d after transfection, cells were switched to hypoxia or kept in normoxia for 24 h and lysed, and luciferase activity was measured. Transactivation activity is expressed as fold increase over the normoxia group. Values are means ± sd (n=3). *P < 0.05. D) Effects of hypoxia on the amphioxus HIFα isoforms. Cells transfected with pCS2-EGFP-amHIFαIa, Ib, Ic, or Id were subjected to 24 h hypoxia (1% oxygen) or normoxia (ambient air) and analyzed by Western immunoblot with the indicated antibodies. Empty transfected cells were used as the control.
A sequence comparison indicated that the sequence between aa 397 and 651 in amphioxus HIFα Ia corresponds to the location of the ODD domain in human HIF-1α. To test whether this region encodes a functional ODD domain, we deleted aa 397–651 in amphioxus HIFα Ia. This deletion resulted in a dramatic increase in the HIFα Ia-Myc levels under normoxia and abolished the hypoxia-induced increase (Fig. 3A), suggesting that this region is indeed necessary for the oxygen-dependent regulation. To test whether this region is sufficient for the oxygen-dependent regulation, we fused it to human HIFβ, resulting in the pBIND-ODD-ARNT construct. As mentioned earlier, HIFβ is not regulated by hypoxia (42). When tested in HEK 293T cells, hypoxia caused a more than 300-fold increase in pG5luc reporter gene activity in cells transfected with the pBIND-ANRT construct (Fig. 3C). These results suggest that amphioxus HIFα Ia contains a functional ODD in the region of aa 397–651.
Among the 4 HIFα isoforms identified in this study, only Ia contains the complete aa 397–651 sequence. Amphioxus HIFα Ib, Ic, and Id had 1 or 2 deletions in this region (see Fig. 1C). We speculated that these different isoforms have different levels of oxygen-dependent regulation. We tested this hypothesis by transfecting them in HEK 293T cells. Under hypoxia, the levels of HIFα Ia showed a marked increase. Hypoxia did not change the levels of HIFα Ib, but it decreased the levels of HIFα Ic and Id (Fig. 3D). This pattern was observed in 3 independent experiments. These data not only support the idea that the aa 397–651 region contains a functional ODD, but also indicate that the 4 amphioxus HIFα isoforms have different degrees of oxygen-dependent regulation.
All 4 amphioxus HIFα isoforms were nuclear proteins capable of activating HRE-dependent gene expression
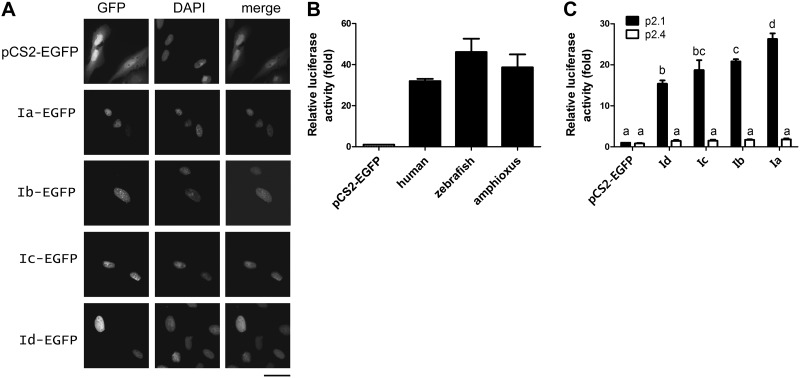
When tagged with EGFP and transfected into cultured U2OS cells, all 4 amphioxus HIFα isoforms were detected in the nucleus (Fig. 4A). U2OS cells were used because they have a flat shape with easily visible nuclei. Their transcriptional activities were examined with the HRE-containing reporter gene p2.1 in HEK 293T cells (43). Cotransfection of pCS2-AmHIFIa-EGFP with p2.1 resulted in a 26 ± 1.4-fold increase in the reporter gene levels. This activity was comparable to those of human and zebrafish HIF-1α (Fig. 4B). When the HRE in the reporter gene was mutated (i.e., p2.4), this induction was abolished (Fig. 4C). Amphioxus HIFα Ib, Ic, and Id all had significant activities, although their activities were lower than that of Ia (Fig. 4C).
Figure 4.
All 4 amphioxus HIFα isoforms were nuclear proteins capable of activating HRE-dependent gene expression. A) Nuclear localization. U2OS cells were transfected with the indicated pCS2 plasmids. Left panels: EGFP signal was visualized 48 h after transfection. Middle panels: corresponding DAPI staining. Right panels: merged images. Scale bar = 50 μm. B) Amphioxus HIFα Ia had transactivation activity comparable to those of human and zebrafish HIF-1α. C) Transactivation activities of amphioxus HIFα isoforms. HEK 293T cells were transfected with pCS2-EGFP or the indicated HIFα expression vectors together with the p2.1 reporter (solid bars) or the p2.4 (open bars) plasmids. pRL-SV40T (Renilla) was cotransfected as an internal control. Luciferase activity was measured at 24 h after the transfection. Transactivation activity is expressed as fold increase over the pCS2-EGFP control group. Values are means ± sd (n=3). Groups with different letters differ significantly from each other (P<0.05).
Identification and mapping of a functional NLS in amphioxus HIFα
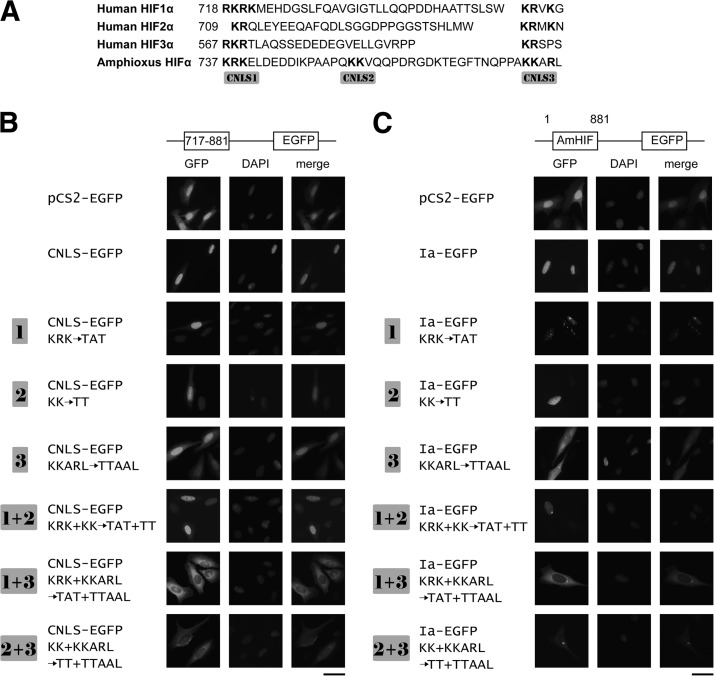
Human HIF-1α has a bipartite NLS located in its C-terminal region (1). Sequence analysis revealed the presence of 3 clusters of basic residues in the corresponding region of amphioxus HIFαs (717–881 aa, labeled as CNLS-1–3 in Fig. 5A). When this region was cloned into pCS2-EGFP (CNLS-EGFP) and tested, the CNLS-EGFP signal was exclusively found in the nucleus (Fig. 5B). To determine which of these motifs is critical, we mutated them individually and in combination. Mutation of CNLS-1 alone (i.e., 737KRK/TAT, mutant 1 in Fig. 5B) did not affect the nuclear localization (Fig. 5B). Likewise, mutation of CNLS-2 (754KK/TT, mutant 2) had little effect. Similar results were obtained when both CNLS-1 and CNLS-2 were mutated, suggesting that these two clusters are not essential. When the basic residues in CNLS-3 were mutated, however, the signal was detected in both the cytoplasm and nucleus (Fig. 5B). When CNLS-3 was mutated together with CNLS-1 or CNLS-2, the signals became exclusively cytoplasmic. Next, the same mutations were introduced to the full-length amphioxus HIFαIa-EGFP background. As shown in Fig. 5C, the wild-type amphioxus HIFαIa-EGFP was detected in the nucleus exclusively. Mutation of CNLS-1 and CNLS-2, individually or in combination, did not change the nuclear localization. When CNLS-3 was mutated, equally strong signals were detected in the nucleus and cytoplasm. In comparison, the GFP signal was found exclusively in cytoplasm in cells transfected with the 1+3 or 2+3 double mutant. These results suggest that these 3 clusters are all involved in the nuclear localization, but their roles are different: 775KKARL is the primary NLS, but 737KRK and 754KK also contribute, and the latter 2 are redundant.
Figure 5.
Several clusters of basic residues in the amphioxus HIFα C-terminal region are critical for its nuclear localization. A) Sequence comparison of the C-terminal region of amphioxus HIFα with those of human HIF-1α to -3α. Bold letters indicate basic amino acid residues. B) Mutational analysis of CNLS. CNLS-EGFP contains the C-terminal region (aa 717–881) fused in frame with EGFP. The 3 clusters of basic residues were mutated individually or in combination, as indicated. C) Same mutations were made in the pCS2-AmHIFIa-EGFP background. U2OS cells were transfected with the indicated plasmid. Left panels: EGFP signal was visualized at 48 h after transfection. Middle panels: corresponding DAPI staining. Right panels: merged images. Scale bars = 50 μm.
Amphioxus HIFαs had 2 functional TADs
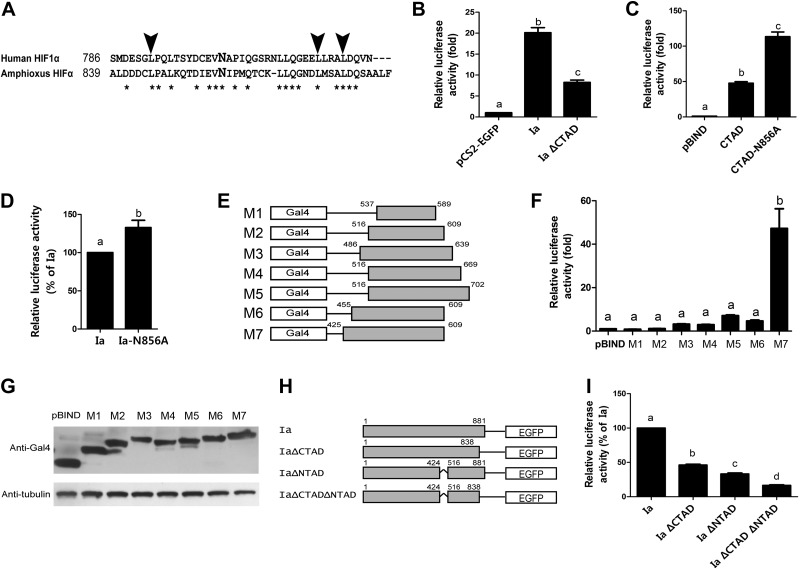
The C-terminal region of amphioxus HIFα shared high sequence identity with human HIF1α C-TAD (Fig. 6A). To test whether this region contains a functional TAD, we deleted (i.e., HIFαIaΔCTAD) and examined it. Whereas expression of amphioxus HIFα Ia resulted in a 20 ± 1.2-fold increase in p2.1 reporter activity, HIFαIaΔCTAD caused only an 8 ± 0.9-fold increase (Fig. 6B). This result suggests that this region is necessary for the full transcriptional activity of amphioxus HIFα Ia. We subcloned the putative C-TAD sequence into the pBIND vector and tested its transactivation activity. Transfection of cells with pBIND-AmHIFαCTAD resulted in a 48 ± 4.2-fold increase (Fig. 6C), indicating that this region indeed contains a functional transactivation domain. Hydroxylation of residue N803 in human HIF1α C-TAD inhibits its transcriptional activity (11, 22, 42). Sequence analysis indicates that this asparagine is conserved in amphioxus HIFα C-TAD (Fig. 6A). Changing this conserved asparagine into alanine resulted in a significant increase in the transactivation activity (Fig. 6C). Likewise, when this mutation was tested in the full-length amphioxus HIFα Ia background, it increased the transactivation activity (Fig. 6D).
Figure 6.
Amphioxus HIFα contained 2 functional TADs. A) Sequence alignment of human HIF-1α C-TAD with the corresponding region of amphioxus HIFα. Asterisks indicate identical residues. Arrowheads mark the conserved leucine residues that are known to be important for human HIF-1α binding to its coactivators. The conserved asparagine residue is marked in a larger, bold letter. B) Deletion of C-TAD in amphioxus HIFα decreased its transactivation activity. HEK 293T cells were transfected with pCS2-EGFP, pCS2-AmHIFIa (Ia), or pCS2-AmHIFIaΔCTAD, together with p2.1 and pRL-SV40T (Renilla). Luciferase activity was measured 24 h after transfection. C) Amphioxus HIFα C-TAD had strong transactivation activity, and mutation of N856 further increased its activity. HEK 293T cells were transfected with pBIND, pBIND-CTAD, or pBIND-CTAD-N856A, together with pG5luc and pRL-SV40T (Renilla). At 24 h after transfection, luciferase activity was measured. Transactivation activity is expressed as fold increase over the control group in all panels. Values are means ± sd (n=3). Groups with different letters differ significantly from each other (P<0.05). D) Mutation of N856 increased full-length amphioxus HIFα activity. HEK 293T cells were transfected with pCS2-AmHIFIa (Ia) or pCS2-AmHIFIa-N856A (Ia-N856A), together with p2.1 and pRL-SV40T. E–G) Identification and mapping of an additional TAD. E) The 7 truncation mutants used to map the additional TAD. Numbers above the gray boxes indicate amino acid positions according to isoform Ia. F) Results of the measurement of their activities. G) Successful expression of the indicated Gal4-fusion proteins was validated by immunoblot analysis. H, I) Both C- and N-TAD are necessary for full transcriptional activity. H) Schematic diagram of the amphioxus HIFα mutants. I) Mutant activities were measured as described above and are shown relative to that of amphioxus HIFα Ia. Values are means ± sd (n=3). Groups with different letters differ significantly from each other (P<0.05).
All known vertebrate HIF-1α and HIF-2α have 2 TADs. In addition to the C-TAD, there is an N-TAD embedded in their ODD domains (5). The middle region differs considerably between amphioxus and human HIF-1α/2α (Supplemental Fig. S1). To determine whether amphioxus HIFα has a second TAD and to map its location, we engineered and tested 7 truncated mutants covering various regions of amphioxus HIFα (Fig. 6E, F). Expression of M7, which covers the sequence between amino acid residues 425 and 609, resulted in a 47 ± 18.0-fold increase in transcriptional activity (Fig. 6F), suggesting that this region contains another functional TAD. In comparison, expression of M1–M6 did not show a significant change (Fig. 6F). The lack of activity of M1–M6 is not due to reduced protein expression and/or increased degradation, because the levels of these mutant proteins were comparable (Fig. 6G). To determine the relative importance of the aa 425–515 region in the full-length amphioxus HIFα background and to clarify its relationship with C-TAD, we deleted this region alone or in combination with ΔCTAD (Fig. 6H). Whereas deletion of the C-TAD resulted in a 54 ± 2.8% reduction in transcriptional activity, deletion of the N-TAD caused a 67 ± 3.3% reduction (Fig. 6I). Deletion of both C- and N-TAD further reduced the transcriptional activity to 16 ± 2.1% (Fig. 6I). Taken together, these results suggest that amphioxus HIFα has 2 functional TADs.
DISCUSSION
The present study revealed that amphioxus HIFα shares many structural and functional similarities with human HIF-1α and HIF-2α. Like human HIF-1α and HIF-2α, amphioxus HIFα has 2 functional TADs in the middle and C-terminal regions and a functional NLS in the C-terminal region. The amphioxus C-TAD, in particular, shares high sequence identity with that of human HIF-1α/2α, and there is a conserved asparagine in this region. Mutation of this asparagine significantly increased transcriptional activity, suggesting that the transcriptional activity of amphioxus HIFα is probably regulated by asparagine hydroxylation. Furthermore, we showed that the single amphioxus HIFα gene produced multiple spliced transcripts. Of note, although these isoforms all encode nuclear proteins capable of activating HRE-dependent gene expression, they differ in their ODD domains and exhibit different degrees of oxygen-dependent regulation. Gene expression analysis results suggest that, although HIFα Ic is the predominant isoform in embryos and larvae, HIF Ia is the most abundant isoform expressed in the adult tissues. Hypoxia treatment has no effect on the mRNA levels of any of these HIFα isoforms.
A key mechanism of HIFα regulation in mammals is its oxygen-dependent degradation. For example, under normoxia, 2 proline residues in the human HIF-1α ODD domain are hydroxylated by PHDs and targeted to the proteasome for degradation (5–9). Our data showed that the full-length amphioxus HIFα Ia is most likely regulated by a similar mechanism. When tested in cultured human cells, hypoxia increased amphioxus HIFα Ia abundance. Further, the addition of CoCl2, a PHD inhibitor, increased amphioxus HIFα Ia levels. We have identified a region in the middle of amphioxus HIFα Ia that is both necessary and sufficient in conferring the oxygen-dependent regulation. Although this region shares low overall sequence identity with the human HIF1α ODD domain (<20%), sequence alignment suggests that there are 2 typical proline hydroxylation motifs (LXXLAP) present in the amphioxus HIFα ODD domain.
Among the 4 HIFα isoforms identified in this study, only HIFα Ia contained the entire ODD domain and showed increased levels under hypoxia and CoCl2 treatments. In comparison, amphioxus HIF isoforms Ib, Ic, and Id had 1 or 2 deletions in the ODD domain. When tested in mammalian cells, hypoxia did not change the levels of HIFα Ib. In fact, hypoxia treatment resulted in a marked decrease in the levels of HIFα Ic and Id. This decrease is probably due to the combined effects of reduced protein synthesis under hypoxia and the rapid turnover rate of HIFα protein in mammalian cells. It is known that mammalian HIFα genes are subjected to complex regulation and produce a large number of mRNA variants due to utilization of different promoters, different transcription initiation sites, and alternative splicing. In the case of human HIF-1α, alternative splicing alone generates 6 different isoforms. Among them, 4 have a partial ODD domain (1). The most complicated case is the human HIF-3α gene, which gives rise to numerous transcripts. One of them, HIF-3α4, does not have an ODD domain or a TAD (44). The mouse HIF-3α spliced variant, termed the inhibitory PAS domain protein (IPAS), has a similar structure (45). Although the existence of several human HIFα isoforms lacking an ODD domain or having a partial ODD domain has been shown (1), it is not known whether they have different degrees of oxygen-dependent regulation. More studies are needed to explore their differential regulation and functions in human and mammalian systems.
When introduced into cultured human cells, amphioxus HIFα Ia to Id were found in the nucleus. The functional NLS is mapped to the C-terminal region shared by all 4 isoforms. We have identified 3 clusters of basic residues in this region that are involved in the nuclear targeting of amphioxus HIFαs. The 775KKARL motif (C-NLS3) is the most critical. Mutation of this motif alone reduced but did not abolish the nuclear localization of amphioxus HIFα. Two additional motifs, 737KRK and 754KK, were also involved. Although mutation of 737KRK or 754KK alone had no effect on nuclear localization, their mutation together with 775KKARL completely eliminated the nuclear location. These findings are different from those reported in vertebrate HIF-1α and HIF-2α. A bipartite NLS in their C-terminal regions of human HIF-1α and HIF-2α is important for their nuclear localization (11). Sima, the fruit fly HIFα homologue, also has a bipartite NLS in its C-terminal region (46). The functional NLS in vertebrate HIF-3α has not been characterized. It is of interest to determine whether HIF-3α has an NLS similar to that of amphioxus HIFα or to that of human HIF-1α/2α.
Our results suggest that all amphioxus HIFα isoforms identified in this study are capable of stimulating HRE-reporter gene expression. In fact, they had transactivation activity comparable to that of human HIF-1α and zebrafish Hif-1α when tested in human cells. These results suggest that amphioxus HIFαs are capable of forming functional heterodimers with human HIFβ and can activate HRE reporter gene expression in human cells despite >5.2 × 108 yr of separation in evolution. This conclusion is also supported by structural analysis results. Studies in mammalian HIFαs have shown that the bHLH domain is responsible for DNA binding, PAS domains facilitate their heterodimerization with HIFβ, and the PAC domain is involved in dimerization and target gene specificity (22). These domains of amphioxus HIFα share high sequence identities with those in human HIF-1α (>60%).
Like human HIF-1α and HIF-2α, amphioxus HIFα protein contains 2 functional TADs. The amphioxus HIFα C-TAD is 44% identical with that of human HIF-1α. This domain alone is sufficient to drive HRE-dependent reporter gene expression when tested in human cells. Deletion of this sequence resulted in a significant reduction in transcriptional activity. In addition to the structurally conserved C-TAD, we identified another TAD and mapped it between amino acid residues 425 and 515. This TAD is analogous to the N-TAD found in human HIF-1α/2α, but its amino acid sequence shares little similarity. The amphioxus N-TAD has strong transactivation activity. Its deletion decreased the transcriptional activity of amphioxus HIFα and this reduction is additive to that of C-TAD deletion. Together, these results suggest that amphioxus HIFα has 2 functional TADs. In this regard, amphioxus HIFα is similar to vertebrate HIF-1α and HIF-2α but different from HIF-3α, which only has 1 TAD (47).
One important mechanism by which oxygen tension regulates vertebrate HIF-1α/2α activity is the asparagine hydroxylation in their C-TADs (13). Intriguingly, this asparagine hydroxylation mechanism is not found in invertebrate species (26). In this study, we found a conserved asparagine hydroxylation site in amphioxus HIFα C-TAD. In its surrounding, there are several leucine residues, which are required for physical interaction with CBP/p300 in HIF-1α and HIF-2α (48). Mutation of the asparagine residue into alanine resulted in increased transcriptional activity. This finding supports the notion that asparagine hydroxylation-mediated regulation of HIFα activity is an evolutionarily ancient mechanism. It is also in agreement with a recent report by Loenarz et al. (26), which demonstrated the presence of a FIH-like molecule in the amphioxus genome through a bioinformatics approach.
Among the 3 mammalian HIFα genes, HIF-1α and HIF-2α are regulated by oxygen tension at post-translational levels. However, the HIF-3α gene is differentially regulated. Hypoxia increases HIF-3α mRNA levels in rat cerebral cortex, hippocampus, and lung, whereas it does not affect the levels of HIF-1α, HIF-2α, and HIF-β mRNA in these tissues (49). Likewise, HIF-3α protein and mRNA levels are increased under hypoxia in cultured human lung epithelial cells, because of increased protein stability as well as transcription (50). IPAS, an alternatively spliced variant of mouse HIF-3α, has been suggested to be a direct HIF-1α target gene (45). This hypoxia-induced increase in hif-3α mRNA expression is conserved in zebrafish (47). In this study, we found that hypoxia increased HIFα Ia protein levels. However, the amphioxus HIFα gene mRNA expression was not regulated by hypoxia. In this regard, the amphioxus HIFα gene is also more similar to the vertebrate HIF-1α/2α genes than to the HIF-3α gene.
Our gene expression analysis revealed that HIFα transcripts are expressed in all stages of amphioxus life cycle. Of note, whereas HIFα Ic is the predominant isoform in embryos and larvae, HIFα Ia is the most abundant isoform in the adult tissues. This switch from a hypoxia-insensitive HIFα isoform to a hypoxia responsive isoform between the larval and adult stages is interesting. In addition to their well-known roles in hypoxia sensing and signaling, HIFs have been suggested to play important roles in embryonic development and organogenesis in invertebrates (14) and mammals (15). A recent study in zebrafish and Xenopus laevis has shown that Hif-1α is critical for neural crest migration (16). These researchers further showed that Hif-1α controls the expression of Twist, which in turn represses E-cadherin during the epithelial-to-mesenchymal transition (EMT) of neural crest cells (16). Although HIFs were identified by their roles in hypoxic response and hypoxia is a key physiological regulator of HIFα stability and activity, recent studies have shown that HIF-1α stabilization and activation are often detected in nonhypoxic conditions. It was reported that NF-κB directly modulates HIF-1α expression under normoxia (21). Growth factors and hormones such as insulin-like growth factor have been shown to stimulate HIF-1α expression and activity via the PI3 kinase/mTOR signaling pathway (17, 20). Likewise, transforming growth factor β1 increases HIF-1α expression and activity and induces MET in renal tubular cells via oxygen-independent mechanisms (18). These findings suggest that HIFs play critical roles in early development via both oxygen-dependent and -independent mechanisms. That the hypoxia-insensitive HIFα Ic is the predominant isoform expressed in amphioxus embryos and larvae strongly supports this notion. Further studies are needed to determine the roles of amphioxus HIFαs in the hypoxic response and in early development and to clarify the underlying mechanisms.
Supplementary Material
Acknowledgments
The authors thank Dr. Xungang Tan (Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China) for providing animals for the gene expression analysis; Drs. Yinlong Qiu, Ya Yang, and Wenfeng Qian (University of Michigan) for advice on the phylogenetic analysis; Dr. Jie Huang (Ocean University of China) for help with the tree computation; Dr. Eric Huang, (U.S. National Cancer Institute) for providing the pANRT construct; and Mr. Nicholas Silva (University of Michigan) for proofreading the manuscript.
This work was supported by U.S. National Science Foundation grants IOB-0543018 and IOS-1051034 to C.D.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- ARNT
- aryl hydrocarbon receptor nuclear translocator
- bHLH
- basic helix-loop-helix
- C-TAD
- C-terminal transactivation domain
- DAPI
- 4′,6-diamidino-2-phenylindole
- FIH
- factor-inhibiting HIF
- HEK
- human embryonic kidney
- HIF
- hypoxia-inducible factor
- HRE
- hypoxia response element
- IPAS
- inhibitory Per/ARNT/Sim
- N-TAD
- N-terminal transactivation domain
- NLS
- nuclear localization signal
- ODD
- oxygen-dependent degradation
- PAS
- Per/ARNT/Sim
- PCR
- polymerase chain reaction
- PHD
- prolyl hydroxylase domain
- qRT-PCR
- quantitative reverse transcription-polymerase chain reaction
- RT-PCR
- reverse transcription-polymerase chain reaction
REFERENCES
- 1. Prabhakar N. R., Semenza G. L. (2012) Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol. Rev. 92, 967–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Semenza G. L. (2012) Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greer S. N., Metcalf J. L., Wang Y., Ohh M. (2012) The updated biology of hypoxia-inducible factor. EMBO J. 31, 2448–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Hypoxia-inducible factor-1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanimoto K., Makino Y., Pereira T., Poellinger L. (2000) Mechanism of regulation of the hypoxia-inducible factor-l alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19, 4298–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bruick R. K., McKnight S. L. (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 [DOI] [PubMed] [Google Scholar]
- 7. Epstein A. C. R., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J.2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 8. Masson N., Willam C., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20, 5197–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275 [DOI] [PubMed] [Google Scholar]
- 10. Kallio P. J., Okamoto K., O'Brien S., Carrero P., Makino Y., Tanaka H., Poellinger L. (1998) Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1 alpha. EMBO J. 17, 6573–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo J. C., Shibuya M. (2001) A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia inducible factors (1 alpha, 2 alpha and 3 alpha). Oncogene 20, 1435–1444 [DOI] [PubMed] [Google Scholar]
- 12. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 14. Pocock R., Hobert O. (2008) Oxygen levels affect axon guidance and neuronal migration in Caenorhabditis elegans. Nat. Neurosci. 11, 894–900 [DOI] [PubMed] [Google Scholar]
- 15. Dunwoodie S. L. (2009) The role of hypoxia in development of the mammalian embryo. Dev. Cell 17, 755–773 [DOI] [PubMed] [Google Scholar]
- 16. Barriga E. H., Maxwell P. H., Reyes A. E., Mayor R. (2013) The hypoxia factor Hif-1α controls neural crest chemotaxis and epithelial to mesenchymal transition. J. Cell Biol. 27, 759–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feldser D., Agani F., Iyer N. V., Pak B., Ferreira G., Semenza G. L. (1999) Reciprocal positive regulation of hypoxia-inducible factor 1alpha and insulin-like growth factor 2. Cancer Res. 59, 3915–3918 [PubMed] [Google Scholar]
- 18. Han W. Q., Zhu Q., Hu J., Li P. L., Zhang F., Li N. (2013) Hypoxia-inducible factor prolyl-hydroxylase-2 mediates transforming growth factor beta 1-induced epithelial-mesenchymal transition in renal tubular cells. Biochim. Biophys. Acta 1833, 1454–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shay J. E., Simon M. C. (2012) Hypoxia-inducible factors: crosstalk between inflammation and metabolism. Semin. Cell Dev. Biol. 23, 389–394 [DOI] [PubMed] [Google Scholar]
- 20. Treins C., Giorgetti-Peraldi S., Murdaca J., Monthouël-Kartmann M. N., Van Obberghen E. (2005) Regulation of hypoxia-inducible factor (HIF)-1 activity and expression of HIF hydroxylases in response to insulin-like growth factor I. Mol. Endocrinol. 19, 1304–1317 [DOI] [PubMed] [Google Scholar]
- 21. Van Uden P., Kenneth N. S., Rocha S. (2008) Regulation of hypoxia-inducible factor-1alpha by NF-kappaB. Biochem. J. 412, 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pugh C. W., O'Rourke J. F., Nagao M., Gleadle J. M., Ratcliffe P. J. (1997) Activation of hypoxia-inducible factor-1: definition of regulatory domains within the alpha subunit. J. Biol. Chem. 272, 11205–11214 [DOI] [PubMed] [Google Scholar]
- 23. Gu Y. Z., Moran S. M., Hogenesch J. B., Wartman L., Bradfield C. A. (1998) Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr. 7, 205–213 [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang B. H., Rue E., Wang G. L., Roe R., Semenza G. L. (1996) Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J. Biol. Chem. 271, 17771–17778 [DOI] [PubMed] [Google Scholar]
- 25. Nambu J. R., Chen W., Hu S., Crews S. T. (1996) The Drosophila melanogaster similar bHLH-PAS gene encodes a protein related to human hypoxia-inducible factor 1 alpha and Drosophila single-minded. Gene 172, 249–254 [DOI] [PubMed] [Google Scholar]
- 26. Loenarz C., Coleman M. L., Boleininger A., Schierwater B., Holland P. H., Ratcliffe P. J., Schofield C. J. (2011) The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 12, 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holland L. Z., Laudet V., Schubert M. (2004) The chordate amphioxus: an emerging model organism for developmental biology Cell. Mol. Life Sci. 61, 2290–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holland L. Z., Albalat R., Azumi K., Benito-Gutierrez E., Blow M. J., Bronner-Fraser M., Brunet F., Butts T., Candiani S., Dishaw L. J., Ferrier D. E., Garcia-Fernàndez J., Gibson-Brown J. J., Gissi C., Godzik A., Hallböök F., Hirose D., Hosomichi K., Ikuta T., Inoko H., Kasahara M., Kasamatsu J., Kawashima T., Kimura A., Kobayashi M., Kozmik Z., Kubokawa K., Laudet V., Litman G. W., McHardy A. C., Meulemans D., Nonaka M., Olinski R. P., Pancer Z., Pennacchio L. A., Pestarino M., Rast J. P., Rigoutsos I., Robinson-Rechavi M., Roch G., Saiga H., Sasakura Y., Satake M., Satou Y., Schubert M., Sherwood N., Shiina T., Takatori N., Tello J., Vopalensky P., Wada S., Xu A., Ye Y., Yoshida K., Yoshizaki F., Yu J. K., Zhang Q., Zmasek C. M., de Jong P. J., Osoegawa K., Putnam N. H., Rokhsar D. S., Satoh N., Holland P. W. (2008) The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1380–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Putnam N. H., Butts T., Ferrier D. E. K., Furlong R. F., Hellsten U., Kawashima T., Robinson-Rechavi M., Shoguchi E., Terry A., Yu J. K., Benito-Gutiérrez E. L., Dubchak I., Garcia-Fernàndez J., Gibson-Brown J. J., Grigoriev I. V., Horton A. C., de Jong P. J., Jurka J., Kapitonov V. V., Kohara Y., Kuroki Y., Lindquist E., Lucas S., Osoegawa K., Pennacchio L. A., Salamov A. A., Satou Y., Sauka-Spengler T., Schmutz J., Shin-I. T., Toyoda A., Bronner-Fraser M., Fujiyama A., Holland L. Z., Holland P. W., Satoh N., Rokhsar D. S. (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1063 [DOI] [PubMed] [Google Scholar]
- 30. Holland L. Z., Sower S. A. (2010) Insights of early chordate genomics: endocrinology and development in amphioxus, tunicates and lampreys: introduction to the symposium. Integr. Comp. Biol. 50, 17–21 [DOI] [PubMed] [Google Scholar]
- 31. Dehal P., Boore J. L. (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3, e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kamei H., Ding Y., Kajimura S., Wells M., Chiang P., Duan C. M. (2011) Role of IGF signaling in catch-up growth and accelerated temporal development in zebrafish embryos in response to oxygen availability. Development 138, 777–786 [DOI] [PubMed] [Google Scholar]
- 33. Burge C., Karlin S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 [DOI] [PubMed] [Google Scholar]
- 34. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 [DOI] [PubMed] [Google Scholar]
- 36. Guindon S., Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 37. Abascal F., Zardoya R., Posada D. (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105 [DOI] [PubMed] [Google Scholar]
- 38. Kajimura S., Aida K., Duan C. M. (2006) Understanding hypoxia-induced gene expression in early development: in vitro and in vivo analysis of hypoxia-inducible factor 1-regulated zebra fish insulin-like growth factor binding protein 1 gene expression. Mol. Cell. Biol. 26, 1142–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williams M., Louw A. I., Birkholtz L. M. (2007) Deletion mutagenesis of large areas in Plasmodium falciparum genes: a comparative study. Malaria J. 6, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ren H. X., Accili D., Duan C. M. (2010) Hypoxia converts the myogenic action of insulin-like growth factors into mitogenic action by differentially regulating multiple signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 107, 5857–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Piret J. P., Mottet D., Raes M., Michiels C. (2002) CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann. N. Y. Acad. Sci. 973, 443–447 [DOI] [PubMed] [Google Scholar]
- 42. Semenza G. L. (2007) Life with oxygen. Science 318, 62–64 [DOI] [PubMed] [Google Scholar]
- 43. Semenza G. L., Jiang B. H., Leung S. W., Passantino R., Concordet J. P., Maire P., Giallongo A. (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 44. Pasanen A., Heikkilä M., Rautavuoma K., Hirsilä M., Kivirikko K. I., Myllyharju J. (2010) Hypoxia-inducible factor (HIF)-3alpha is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int. J. Biochem. Cell Biol. 42, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 45. Makino Y., Cao R. H., Svensson K., Bertilsson G. R., Asman M., Tanaka H., Cao Y. H., Berkenstam A., Poellinger L. (2001) Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 414, 550–554 [DOI] [PubMed] [Google Scholar]
- 46. Romero N. M., Irisarri M., Roth P., Cauerhff A., Samakovlis C., Wappner P. (2008) Regulation of the Drosophila hypoxia-inducible factor alpha Sima by CRM1-dependent nuclear export. Mol. Cell. Biol. 28, 3410–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang P., Zhou J. F., Li Y., Liu Y. Z., Lu L., Yao Q., Duan C. M. (2012) Molecular, functional, and gene expression analysis of zebrafish hypoxia-inducible factor-3α. Am. J. Physiol. Regul. Integr. Comp. Physiol. 303, R1165–R1174 [DOI] [PubMed] [Google Scholar]
- 48. Lando D., Pongratz I., Poellingers L., Whitelaw M. L. (2000) A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1 alpha and the HIF-like factor. J. Biol. Chem. 275, 4618–4627 [DOI] [PubMed] [Google Scholar]
- 49. Heidbreder M., Fröhlich F., Jöhren O., Dendorfer A., Qadri F., Dominiak P. (2003) Hypoxia rapidly activates HIF-3alpha mRNA expression. FASEB J. 17, 1541–1543 [DOI] [PubMed] [Google Scholar]
- 50. Li Q. F., Wang X. R., Yang Y. W., Lin H. (2006) Hypoxia upregulates hypoxia inducible factor (HIF)-3alpha expression in lung epithelial cells: characterization and comparison with HIF-1alpha. Cell Res. 16, 548–558 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.