Abstract
The emergent self-organization of a neuronal network in a developing nervous system is the result of a remarkably orchestrated process involving a multitude of chemical, mechanical and electrical signals. Little is known about the dynamic behavior of a developing network (especially in a human model) primarily due to a lack of practical and non-invasive methods to measure and quantify the process. Here we demonstrate that by using a novel optical interferometric technique, we can non-invasively measure several fundamental properties of neural networks from the sub-cellular to the cell population level. We applied this method to quantify network formation in human stem cell derived neurons and show for the first time, correlations between trends in the growth, transport, and spatial organization of such a system. Quantifying the fundamental behavior of such cell lines without compromising their viability may provide an important new tool in future longitudinal studies.
The emergence of network self-organization in the developing central nervous system is a highly complex and insufficiently understood process1. Neurons use a diverse variety of internal and external guidance cues to direct their organization into functional units. In a developed nervous system the spatial arrangement (i.e., location) of individual neurons is known to be closely linked to specific functional activity, a phenomenon which has been studied extensively in the visual cortex2,3,4. This spatial arrangement ultimately determines how neurons connect to each other to form neural circuits. Once a connection is made, neurons communicate with each other using a complex system of neurotransmitters, neurotrophins, peptides, and cytokines1,5,6,7,8,9,10,11,12,13. The spatiotemporal interaction among individual neurons within a network is the basis of all functional activity in the nervous system and higher level cognitive functions such as sensory perception, memory, and learning. Thus, understanding the formation, stabilization, and behavior of neural networks is a key area of study in neuroscience, developmental and synthetic biology, and regenerative medicine. In order to gain insight into how functional neural networks form and develop, it is necessary to quantify how individual neurons grow and spatially organize with respect to each other, and to understand the dynamics of how mass is transported within connected units, preferably through longitudinal measurements within the same neural cultures.
Neural network formation has been studied by various models, ranging from whole brain animal studies9,14 to in vitro dissociated neuron cultures11,15. However, there are many limitations to each of these models. For example, the analysis of in vitro neuron cultures is limited by the relative lack of data that can be obtained from conventional live-cell imaging techniques such as phase-contrast or differential interference contrast (DIC), which only report on morphology and must be coupled with other methods such as fluorescence to provide functional information. Fluorescence imaging techniques have been used to image vesicle transport and synapse formation16. More recently, fluorescent voltage-sensors have been used to optically measure electrical activity17,18. However, the limitations inherent to fluorophores, such as photo-bleaching and photo-toxicity, do not allow for continuous monitoring of these processes during dynamic neural-network function on the relevant time scales of days to weeks.
Recent advances in micro-fabrication methods have led to several innovative approaches to study in vitro neural cultures. For example, multi-electrode arrays have been extensively used for measuring electrical activity of neural cultures19,20. However, due to their limited spatial resolution they only provide an incomplete electrophysiological picture of the system. Micro-patterning methods have enabled the design of spatial order that generally mimics what is found in live neurons11,21,22,23. Furthermore, recently developed microfluidic approaches are amenable to rapid, high content analysis of various cell properties1,10,12,24,25,26,27,28. Such methods provide important information on how cells behave when spatial cues are provided but are inherently limited in their capabilities to address how spatial order naturally emerges in a culture.
Little is known about the neuron culture in terms of spatial organization, metabolic activity, and mass transport. Our current understanding of these phenomena is limited by the lack of available technologies to quantitatively measure a forming neural network at multiple temporal and spatial scales, in a high throughput and minimally invasive manner. In this work we show that by using a recently developed optical interferometric technique known as spatial light interference microscopy (SLIM)29,30 we can measure several fundamental properties of the developing neural network on a broad range of spatial (sub-cellular to millimeter) and temporal scales (seconds to days) in a completely label-free and non-invasive manner.(i.e., no extrinsic contrast agents or perturbation) and quantify differences in these parameters between a normal and perturbed network. Our results demonstrate that measurements on cell growth, intracellular transport, and scattering structure function are quantitative indicators of neural network emergence and that changes in these metrics are intrinsically linked.
SLIM is a quantitative phase imaging (QPI) modality which measures the pathlength shift distribution of an optical wave front as it passes through a cell31. Due to its common-path geometry, broadband illumination, and ability to interface with existing phase contrast microscopes, SLIM is a highly sensitive and versatile QPI modality32. It has been demonstrated that SLIM has spatial and temporal path length sensitivities of 0.3 nm and 0.03 nm, respectively29. The measured path length information at each pixel is directly related to the dry mass density (fg/μm2) at that point, and thus by integrating over an area the total dry mass or non-aqueous content may be measured33,34,35. When translated into dry mass, SLIM's sensitivity corresponds to changes on the order of femtograms. In combination with SLIM's multimodal capabilities this has allowed for the measurement of single cell dry mass growth in a cell cycle dependent manner35. When this measurement is performed with high temporal resolution it can be used to quantify changes in inter- and intra- cellular mass transport characteristics. This method is known as Dispersion-relation Phase Spectroscopy (DPS)36,37. While the mass growth measurements provide information on the overall metabolic activity of the system, DPS reports on the physical nature of the mass transported within cells. DPS can detect with high sensitivity, a shift between a dominantly diffusive transport (i.e. thermal energy-driven), and directed transport (i.e., using molecular motors). Our results show that in a developing neural network, changes in the overall growth rate, network morphology and mass transport behavior of the system are intrinsically linked. It should be noted that DPS does not provide a direct measurement of how energy is being consumed in the system.
In addition to dry mass growth and transport, the measurement of the complex optical field allows for direct measurements of the angular scattering spectrum from the sample38,39,40,41. It has recently been shown that despite the complexity of the network formation process in vivo, neurons will self-organize in a non-random manner in vitro without the provision of any additional external environmental cues28, that is, relying only on chemical gradients. Previous work on analyzing this organization relied on detecting the location of individual neurons in an image and was, thus, time-consuming and prone to error. The angular scattering data measured by SLIM allows for monitoring how spatial correlations in the culture evolve over time at various spatial scales simultaneously, allowing for a way to quantify organization at all spatial scales without the need for image segmentation. It has been shown that the shape of the angular scattering power spectrum reports on the large scale spatial organization of tissue42 and on the fractal properties of the clusters and cells within the clusters. Performing this measurement on a developing neuronal network revealed that changes in spatial correlations at certain scales are linked to the overall growth behavior of the culture. Specifically, we find that changes in organization across broad spatial scales occur after the mass growth rate stabilizes.
The ability to simultaneously measure the growth, transport characteristics and spatial correlations of an evolving neural network provides a unique and efficient way to quantify the behavior of such systems. In this work we show that changes in the trends of these fundamental properties are intrinsically linked temporally and across broad spatial scales under normal conditions. Furthermore, we show that when normal network formation is disrupted these links break down and the self-organization phenomena is no longer observable. Since all of these parameters are obtained from the same quantitative phase measurement, using a completely non-invasive and label-free method, they allow for monitoring the evolution of an in vitro neural network in an unprecedented manner.
Results
We used human embryonic stem cell derived neurons43,44, which hold therapeutic potential45,46 and express multiple well established neuronal markers including MAP247. Experiments were conducted in both optimal culture conditions (untreated sample) and in the presence of LiCl, an inhibitor of neurite growth48. Imaging was initiated 6 hours after the predifferentiated neurons were replated; this is regarded as t = 0 hours for the remainder of the manuscript. A 0.87 mm × 1.16 mm area of the culture was continuously scanned with SLIM using a 10×/0.3 NA objective at a rate of one scan every 10 minutes. High resolution and high frame rate movies were recorded at several locations at 0 hours and 24 hours using a 40×/0.75 NA objective at a rate of 0.5 Hz (For more details on the cell culture and imaging procedures please refer to the Materials and Methods). The development of the neuronal network was characterized using several analysis techniques with the goals of understanding the dynamic behavior at the single cell level and also to characterize the system at the population level in terms of mass growth, mass transport, and spatial organization. First, the system was characterized in terms of the total dry mass growth within the entire field of view (Fig. 1). This provides a broad view of the metabolic activity of the culture. Second, mass transport in the system was quantified using DPS (Fig. 2). Finally, the angular light scattering spectrum was calculated at each time point in the 24-hour movie to characterize the spatial organization of the culture (Figs. 3 and 4).
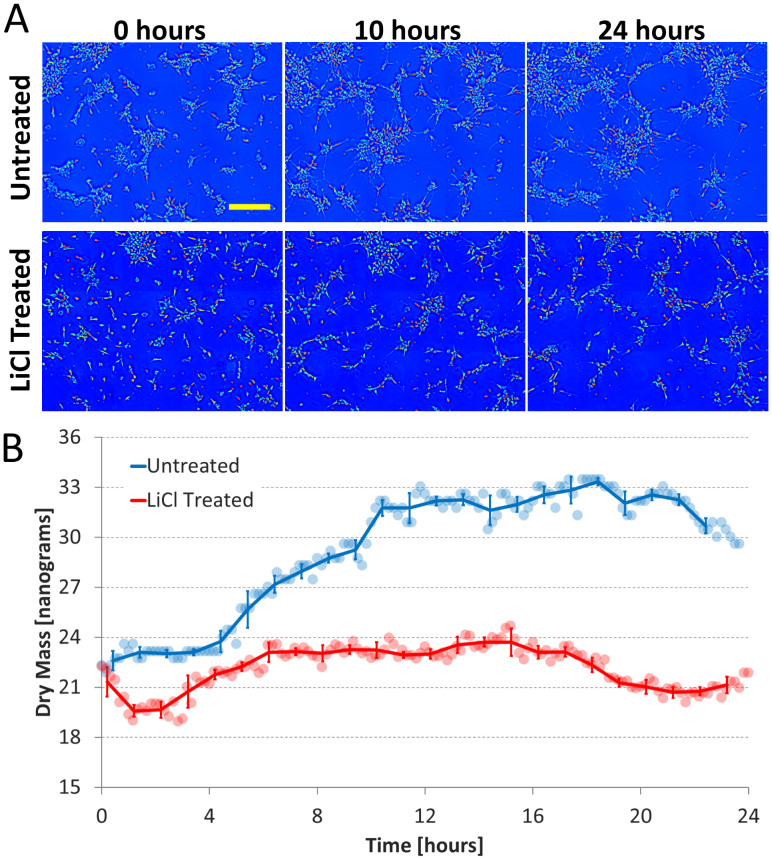
Figure 1.
Dry Mass Growth (A) Dry mass density maps acquired at 0, 10, and 24 hours for both the untreated and LiCl treated cultures; the yellow scale bar corresponds to 200 μm. (B) Total dry mass vs. time of the entire field of view for both conditions. Round markers are raw mass data, solid lines are the average over 1 hour, and error bars indicate standard deviation. For the untreated culture it can be seen that majority of the mass growth occurs between 0 and 10 hours, the time during which the cells are extending processes most actively. In the LiCl treated culture neurite outgrowth is severely retarded and no significant increase in mass is observed during any period.
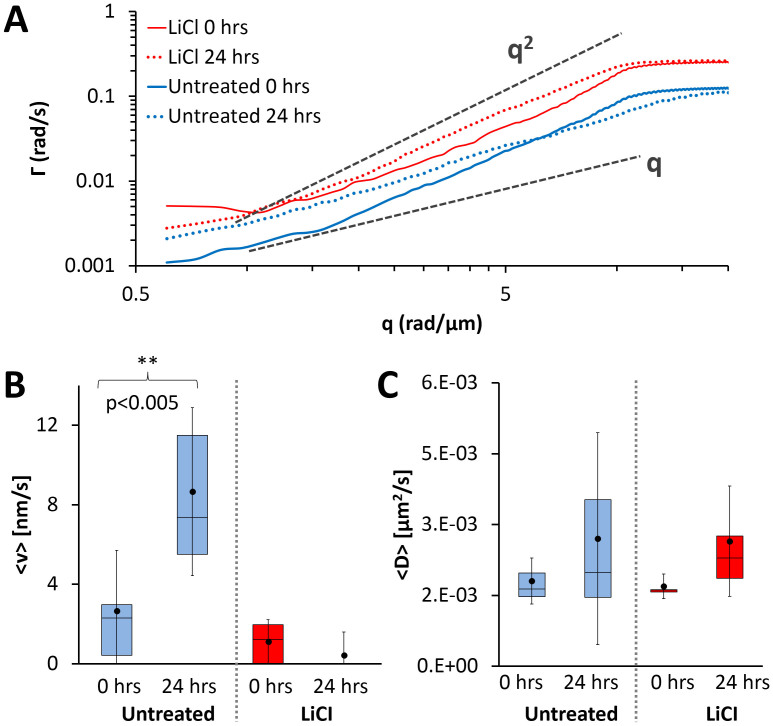
Figure 2.
Mass Transport (A) The average dispersion curve calculated from high resolution movies are shown for both the untreated (blue) and LiCl (red) cultures at 0 (solid lines) and 24 hours (dashed lines); the untreated curves have been offset for clarity. The dashed gray lines indicate a quadratic and linear relationship as labelled. For the untreated culture, a clear shift from a quadratic to a linear relationship can be observed from 0 to 24 hours, whereas for the LiCl culture no shift is observed. (B) Comparison of mean advection velocities obtained from DPS analysis. A statistically significant difference between the mean values at 0 hours and 24 hours is found for the untreated culture, indicating a shift towards more deterministic transport. For the LiCL treated culture there is a slight decrease in the mean advection velocity over 24 hours. (C) Comparison of mean diffusion coefficients obtained from DPS analysis. There is no statistically significant difference between 0 hours and 24 hours for both conditions, however there is an increase in the range of the measured values. The whiskers indicate one standard deviation above and below the mean (black dot).
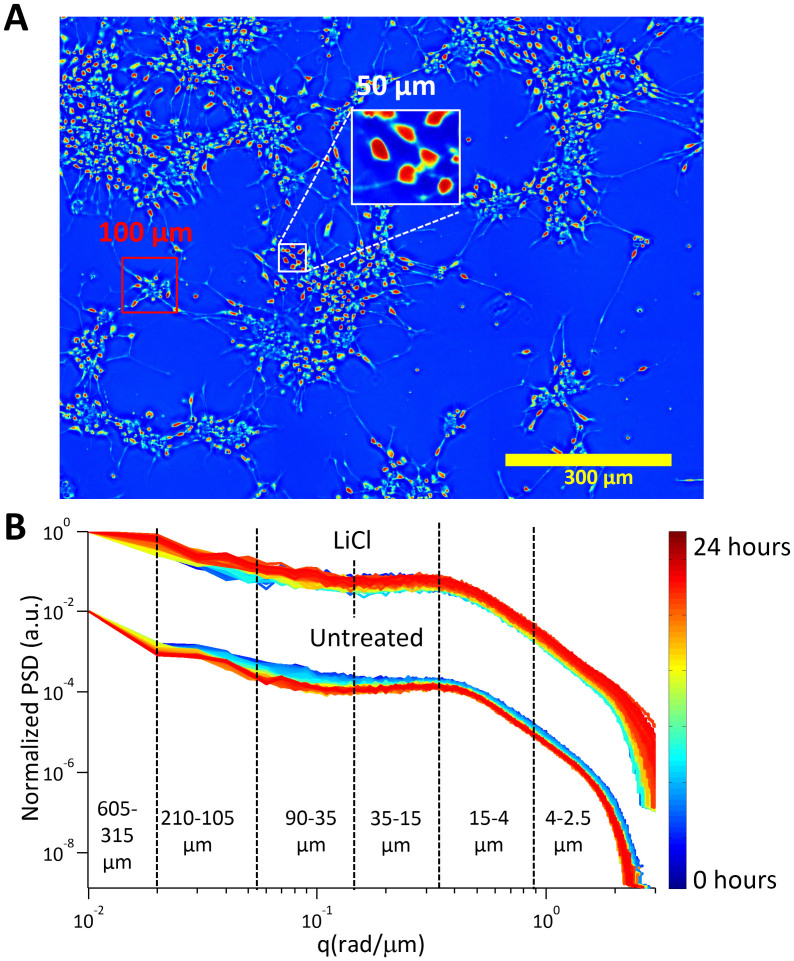
Figure 3.
Angular scattering analysis (A) Dry mass density map of the untreated culture at 24 hours. Various spatial scales have been labeled to aid in the interpretation of the angular scattering maps. (B) Radially averaged profile plots of the angular light scattering map from each time point in the data for both LiCl treated and untreated cultures. The plots are color coded as shown in the color bar. The dashed lines and corresponding labels indicate the various spatial ranges according to which the profiles were split. For the untreated culture, the slopes in each region change with time, with the greatest changes occurring at the spatial scales corresponding to the size of the clusters, the inter cluster distance and inter-soma distance. No such change can be observed in the LiCl treated culture.
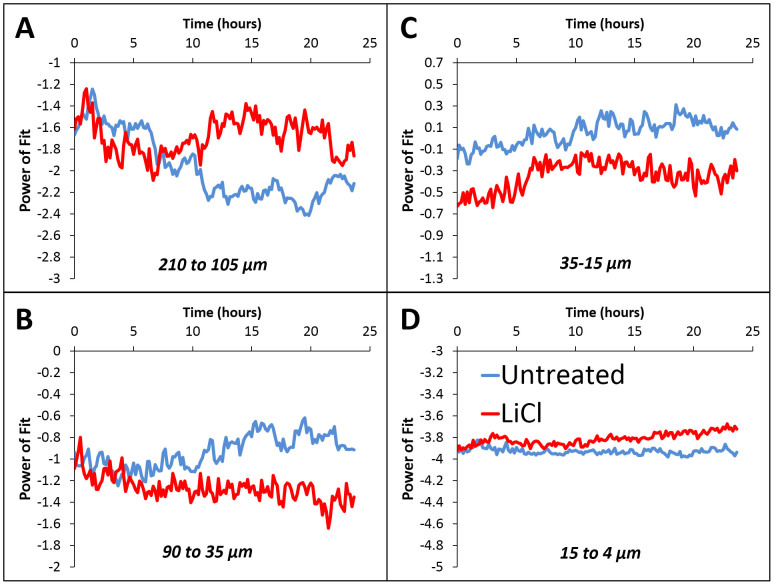
Figure 4.
Power law exponent of the angular scattering profiles at various spatial scales for both the untreated (blue) and LiCl treated (red) cultures (A) 210–105 μm, corresponding to clusters and inter-cluster distances (B) 90–35 μm, corresponding to small clusters (C) 35–15 μm, distances between cells (D) 15–4 μm, features comparable to and smaller than the soma. The power of the fit changes significantly for the untreated culture at scales corresponding to the cluster sizes and inter-cluster distances, whereas no such change is observed for the LiCl treated culture. This indicates that, although the level of spatial organization is increasing under normal conditions, it is inhibited by LiCl. Interestingly changes in organization of the untreated culture occur after 10 hours when the mass growth has stopped (see Fig. 1).
Dry mass growth
The total dry mass of the 0.87 mm × 1.16 mm area was calculated at each time point (at a rate of one frame every 10 minutes) as described in Materials and Methods. It can be seen in Fig. 1A that for the untreated culture the distribution of mass is drastically different between 0 hours and 24 hours, changing from compact isolated clusters to a highly connected network. By contrast, in the LiCl treated culture the clusters remain compact and isolated (See Supplementary Videos 1 and 2 for the complete 24-hour data). The dry mass measurements on these cultures (Fig. 1B) also show stark differences between the untreated and LiCL treated conditions. In the case of the untreated culture, after an initial period of growth that lasts 10 hours the mass growth plateaus. On the other hand, no significant increase in mass is observed for the LiCl culture over the entire 24-hour period. The images suggest that the mass increase in the untreated culture is largely due to the growth and extension of neurites from each cluster, which also explains why no increase is observed in the LiCl condition. Furthermore, once neurite growth slows down and the connections appear to be established (~10 hours), there is no significant increase in the total dry mass of the untreated culture despite the cells remaining viable and highly active. This indicates that there is a shift in the use of resources from growth to other functions. The measurements on changes in mass transport characteristics and spatial organization shown below provide insight on how the network switches priorities from growth to organization and mass transport and how these processes are disrupted when neurite growth is inhibited.
Mass transport
To quantify changes in the mass transport characteristics at the single cell and cluster level, DPS analysis was performed on 40×, 0.5 Hz data acquired at 0 (Supplementary Videos 3 and 5) and 24 hours (Supplementary Videos 4 and 6). The details of the analysis method can be found in the Materials and Methods. At least six data sets were acquired and analyzed at each time point for both the untreated and LiCl treated cultures. The average dispersion profiles calculated at 0 and 24 hours for both culture conditions are shown in Fig. 2A, see Supplementary Fig. 1 for data from each individual video.
The DPS analysis shows that for both the untreated and LiCl treated cultures the mass transport at 0 hours is primarily diffusive (Fig. 2A solid lines). In sharp contrast, the dispersion profile for the untreated (Fig. 2A, dotted blue line) culture shifts to a linear behavior after 24 hours when it is highly connected. This shift indicates that after 24 hours the mass transport is dominated by directed motion which can be explained by the fact that this bi-directional transport is occurring along defined tracks and is known to be facilitated by molecular motors such as dynein and kinesin49,50. As can be seen in Supplementary Video 4, there is a large amount of vesicular transport occurring in the processes that connect the clusters. Furthermore, after 24 hours, a statistically significant increase in the mean advection velocity was found for the untreated culture (Fig. 2B, blue boxes), confirming a shift toward deterministic transport of mass. On the other hand, the dispersion relation for the LiCl treated culture (Fig. 2A, red dotted line) shifts to a more quadratic behavior after 24 hours, indicating more diffusive transport. In fact, only one cluster measured after 24 hours showed signs of deterministic transport and thus no increase in the mean advection velocity was measured (Fig. 2B, red boxes).
Interestingly, there is no significant change in the mean diffusion coefficient after 24 hours (Fig. 2C, blue boxes). This result indicates that, at small scales, where diffusive transport is dominant, the overall mass transport is not affected by network formation. The spread of the diffusion coefficients is much larger at 24 hours suggesting that, with network formation, local transport becomes more inhomogeneous. Similar to the untreated culture, the mean diffusion coefficient for the LiCl culture does not change significantly, however, there is an increase in the range of measured values (Fig. 2C, red boxes).
These results support the hypothesis that, initially, the untreated network is primarily focused on extending processes towards other clusters and that once these processes successfully connect the focus shifts to transport of materials along them. Such measurements can serve as a way to quickly quantify and characterize the morphological and functional connectivity of a network and detect degeneracies in these processes.
Angular scattering
When measuring mostly transparent, optically thin samples, such as neurons, we can assume that the amplitude of the optical field is left unperturbed and that only the phase that is measured by SLIM is altered by the sample. In such a case, SLIM provides a measure of the complex optical field, U(r,t) = |U(r,t)|eiφ(r,t) at the sample plane. This field may then be numerically propagated to the far-field or scattering plane by simply calculating its spatial Fourier transform38 as 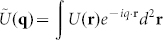 , where q is the scattering wave vector. The modulus square of this function,
, where q is the scattering wave vector. The modulus square of this function, 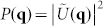 , is related to the spatial auto-correlation of the measured complex field through a Fourier transform as
, is related to the spatial auto-correlation of the measured complex field through a Fourier transform as 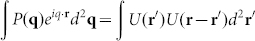 and thus describes the spatial correlation of the scattering particles in the sample. Since the signal is measured and reconstructed in the image plane, rather than in the far field as in traditional scattering experiments, all the scattering angles that are allowed by the numerical aperture of the microscope objective are measured simultaneously. This greatly enhances the sensitivity to scattering compared to the traditional approach of goniometric measurements38.
and thus describes the spatial correlation of the scattering particles in the sample. Since the signal is measured and reconstructed in the image plane, rather than in the far field as in traditional scattering experiments, all the scattering angles that are allowed by the numerical aperture of the microscope objective are measured simultaneously. This greatly enhances the sensitivity to scattering compared to the traditional approach of goniometric measurements38.
The scattered intensity is averaged over rings of constant scattering angle or scattering wave vectors, q = (4π/λ)sin(θ/2), where θ is the scattering angle. This radial average, calculated at each time point in the 24-hour data sets (at a rate of one frame per 10 minutes) is shown in Fig. 3 for both the untreated and treated cultures. The dashed lines indicate the spatial scales corresponding to various important structures in the cultures, as shown in Fig. 3A. As the untreated culture matures, the slope of the profiles in the log-log plots (power of exponential relationship) in different spatial ranges change consistently over time. (Fig. 3B), this monotonic narrowing of the scattering profiles indicates broadening in spatial correlations. In contrast, the scattering profiles from the LiCl treated culture do not show a consistent increase in slope in any region. In fact a decrease in the slope can be observed at the larger spatial scales and an overall broadening rather than narrowing of the power spectrum is observed indicating a decrease in spatial correlations. It is rather remarkable that our label-free imaging is able to monitor the temporal evolution of the neuron culture and quantify the trends of spatial correlations over time in such an uncomplicated manner. Note that the slopes in the spatial ranges corresponding to size of individual soma (15–4 μm) remain relatively constant, which suggest that local morphology remains essentially constant.
To quantitatively evaluate these changes in the power of the exponent over time (slopes in Fig. 3B), the profiles were analyzed at the various spatial scale ranges indicated by the dashed lines in Fig. 3B. The data in each of the regions were then fit to a power law function of the form P(q) = cqα. The power of the exponential relationship, α, for each of the spatial ranges vs. time resulting from these fits are shown in Fig. 4 for both the untreated (blue curves) and LiCl treated (red) cultures. It should be noted that trends towards more negative values in Fig. 4 relate to a steeper decay of the power spectrum Fig. 3, indicating broader spatial correlation. Stark differences at certain spatial scales can be observed between the two cultures in terms of the temporal evolution of the slopes. In the 210–105 μm range, corresponding to the inter-cluster distance (Fig. 4A), the slope of the untreated culture decreases for the first 10 hours and then stabilizes, while the slope of LiCl treated culture does not vary significantly. This suggests that in the untreated cells, there is an emerging spatial correlation that takes place predominantly in the first 10 hours, which does not occur in the treated samples. In the 90–35 μm range (Fig. 4B), the untreated culture exhibits a flatter slope, which corresponds to a reduction in the number of small clusters as they merge to larger clusters over time. For the LiCl treated culture, the power spectrum becomes steeper more gradually, which reflects the slow increase in the number of small clusters formed from the aggregation of individual cells found in close proximity of one another. In the 35–15 μm range, corresponding to inter-cellular distances, (Fig. 4C) both cultures exhibit an increase in the slope corresponding to a decrease in inter-cellular distances. Notice that the slope of the LiCl treated culture in this range stops increasing earlier than the untreated culture. This is because in the LiCl treated culture only the cells in close proximity (35–15 μm) are able connect to each other to form a cluster due to the suppressed neurite growth. Finally in the 15–4 μm range (Fig. 4D), which corresponds to the size of individual soma or very short neurites, the slope remains relatively stable for both cultures, suggesting that there is no change at these small scales. It is interesting to note that for the untreated culture, either the change in slope or the stabilization of slope, begin to occur at approximately the same time as the growth rate (Fig. 1) stabilizes. This suggests that in the untreated culture there is an active reorganization of the system once the neurons have already extended neurites to connect to neighboring clusters. Clearly, this mechanism is missing almost entirely at the large spatial scales in the LiCl treated cells, due to the neurite growth inhibition.
Discussion
In summary, we have shown that using quantitative phase imaging it is possible to characterize several fundamental properties of a forming human stem cell derived neuronal network. To our knowledge this is the first time that mass growth in a neural culture has been measured and linked to changes in transport dynamics and spatial organization. Under normal culture conditions, we found two distinct time periods with consistent growth trends, the first of which is marked by mass accumulation and the second by a relatively stable total mass. From the imaging data, we identified that the first time period corresponds to extension and growth of neurites and that the second time period is characterized by the aggregation of clusters and spatial reorganization of cells. By performing mass transport analysis we determined that there is a significant increase in deterministic transport after 24 hours, suggesting that the network has shifted from extending neurites to transport of materials along these extensions. Finally, by measuring the angular scattering from the culture we quantified the temporal evolution of spatial correlations in the system. Interestingly, the data shows that the increase in spatial correlations at the scales of cluster size and inter-cluster distances coincides with the shift in growth rate and transition from diffusive to deterministic intra-cellular transport. This observation suggests that, in this system, the majority of spatial organization occurs after clusters in the network are already well connected. In order to test this hypothesis, we performed the same measurements and analysis on a culture in which neurite outgrowth was chemically inhibited. In this case, as expected, we detected no increase in mass as there was no extension of neurites, no shift to deterministic transport after 24 hours, as there are no connections between clusters to transport material, and no increase in spatial correlations (self-organization).
The methods and analysis that have been developed and proven here can be applied to a variety of studies in neuroscience and beyond, with the potential to transform how basic behavior such as growth, spatial organization, and transport are measured and quantified. Since SLIM is easily interfaced with existing microscopes, it is also possible to incorporate commonly used tools to provide external perturbation such as microelectrode arrays, optical tweezers, microfluidics and micropipettes. Our analysis provides a practical way to characterize the spatial correlations in neural networks at all relevant spatial scales simultaneously. Combining external stimulation with the technology demonstrated here, it will be possible to quickly assess the effects of various forces on the natural evolution and maturation of a neural culture, opening the door to develop novel experimental strategies and potentially, new therapeutic methods. The correlation between the SLIM measurements and the in vivo outcomes of transplanted cells, such as level of sensory and motor function, will likely expedite efficacy and optimization of neural stem cell therapies.
Methods
Cell culture
Commercially available differentiated human neuronal cells (hN2 from ArunA Biomedical, Athens, GA) were used. Cells were grown in 35 mm Poly-l-lysine coated glass bottom dishes (20 mm well diameter, MatTek) which were first coated with Matrigel. AB2 Basal Neural Medium was supplemented with ANS supplement (both from ArunA Biomedical), Penicillin-streptomycin, GlutaMAX (both from Invitrogen) and Lukemia Inhibitory Factor (LIF, from Millipore). Cells were moved from the incubator to the imaging system 6 hours after plating. To retard neurite outgrowth the culture media was supplemented with 15 mM LiCl (Sigma Aldrich).
Cell imaging
Imaging was conducted using Spatial Light Interference Microscopy (SLIM) which has been described in detail previously29,30. In short, SLIM is a white light, common path, phase shifting interferometer which is designed as an add-on module for commercial phase contrast microscopes. SLIM operates by projecting the back focal plane of a phase contrast objective (which is a Fourier plane of the imaging system) onto a liquid crystal phase modulator (LCPM). The LCPM is then used to quantitatively control the phase shift between the scattered and un-scattered light. By recording 4 intensity images at phase shifts of 0, π/2, π and 3π/2 it is possible to uniquely calculate the phase shift between objects and the surrounding media at each pixel. Due to the white light illumination and common path geometry SLIM is remarkably sensitive and stable, with the capability to measure femtogram level changes in cell dry mass35.
During imaging the cells were kept at 37°C and in a 5% CO2 atmosphere with an incubator and heated stage insert (Zeiss). For the 24 hour dataset, the sample was scanned every 10 minutes in a 3 × 3 tile pattern with a Zeiss EC Plan-Neofluar 10×/0.3 PH1 objective providing a total field of view of 1.2 × 0.9 mm2. The exposure time was 15 ms for each image at full lamp power (3,200 K, or 10.7 V). Higher resolution datasets were recorded at 0 and 24 hours using a Zeiss EC Plan-Neofluar 40×/0.75 at a frame rate of 0.5 Hz for 5 minutes with a 50 ms exposure time.
Dry mass calculations
The dry mass density at each pixel is calculated as ρ = λφ/2πγ, where λ is the center wavelength of the illumination light, φ is the measured phase shift and γ = 0.2 ml/g is the refractive increment of protein33,35,51. The total dry mass or non-aqueous content of a region of interest is then calculated by integrating the dry mass density over the area. It is important to note that although the refractive increment may vary slightly depending on the cell type being considered, this variation will only affect the absolute value of the measurement, and not the rate, which is of most interest here.
Mass transport analysis
Mass transport activity is measured using Dispersion-relation Phase Spectroscopy (DPS), which has been described in detail previously36,37. In DPS the relationship between the decay rate and spatial mode, that is the dispersion relationship is used to estimate the mean diffusion coefficient and advection velocities in a field of view. Since the SLIM image is essentially a 2D dry mass density map, changes in density must satisfy an advection-diffusion equation. Taking a spatial Fourier transform, the temporal autocorrelation may be expressed in terms of the spatial frequency, q, as:
 |
thus relating the measured temporal autocorrelation function to diffusion coefficient, where D is the mean diffusion coefficient and v is the advection velocity. Taking into account the expected speeds of directed transport it can be calculated that the temporal autocorrelation decays exponentially:
 |
In practice, once Γ(q) is calculated from the time lapse data, the curve is fit in both the linear and quadratic regions to extract the mean diffusion coefficients and advection velocities.
Author Contributions
M.M., A.M., S.S., M.U.G. and G.P. designed research, M.M., T.K. and A.M. performed research; M.M., A.M., R.W. and S.S. contributed new reagents/analytic tools; M.M., T.K., A.M., M.X. and S.C.L. analyzed data; M.M., T.K., A.M., S.C.L., M.U.G., S.S. and G.P. wrote the paper.
Supplementary Material
Supplementary Info File #1
Supplementary Info File #2
Supplementary Info File #3
Supplementary Info File #4
Supplementary Info File #5
Supplementary Info File #6
Acknowledgments
This work was supported by the National Science Foundation (NSF) Grants: Science & Technology Center on Emergent Behaviors of Integrated Cellular Systems CBET 0939511 (to G.P., S.S., and M.U.G.), NSF CAREER 08-46660 (to G.P.), CBET-1040461 MRI (to G.P.), NeuroEngineering IGERT 0903622 (to S.C.L.) and National Institute of Mental Health R21MH 085220 (to M.U.G.). For more information, visit http://light.ece.illinois.edu/.
Footnotes
G.P. is the founder of Phi Optics, Inc., a company developing quantitative phase imaging technology for materials and life science applications, which, however, did not fund this research.
References
- Millet L. J. & Gillette M. U. New perspectives on neuronal development via microfluidic environments. Trends Neurosci. 35, 752–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. N. D. et al. Spatial organization of neuronal population responses in layer 2/3 of rat barrel cortex. J. Neurosci. 27, 13316–13328 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinvald A., Lieke E., Frostig R. D., Gilbert C. D. & Wiesel T. N. Functional Architecture of Cortex Revealed by Optical Imaging of Intrinsic Signals. Nature 324, 361–364 (1986). [DOI] [PubMed] [Google Scholar]
- Sato T. R., Gray N. W., Mainen Z. F. & Svoboda K. The functional microarchitecture of the mouse barrel cortex. PLoS. Biol. 5, 1440–1452 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet L. J., Bora A., Sweedler J. V. & Gillette M. U. Direct cellular peptidomics of supraoptic magnocellular and hippocampal neurons in low-density co-cultures. ACS Chem. Neurosci. 1, 36–48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch T., Lemmon V. & Banker G. Local presentation of substrate molecules directs axon specification by cultured hippocampal neurons. J. Neurosci. 19, 6417–6426 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozdar D. Y., Lee J. Y., Schmidt C. E. & Chen S. C. Selective axonal growth of embryonic hippocampal neurons according to topographic features of various sizes and shapes. Int. J. Nanomed. 6, 45–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton G. I. Dynamic neuronal-glial interactions: an overview 20 years later. Peptides 25, 403–411 (2004). [DOI] [PubMed] [Google Scholar]
- Kerlin A. M., Andermann M. L., Berezovskii V. K. & Reid R. C. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron 67, 858–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J. et al. Integrated microfluidics platforms for investigating injury and regeneration of CNS axons. Ann. Biomed. Eng. 40, 1268–1276 (2012). [DOI] [PubMed] [Google Scholar]
- Kleinfeld D., Kahler K. H. & Hockberger P. E. Controlled Outgrowth of Dissociated Neurons on Patterned Substrates. J. Neurosci. 8, 4098–4120 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar D., Gao Y. D., Li D. Y. & Webb D. J. Co-culture of neurons and glia in a novel microfluidic platform. J. Neurosci. Meth. 196, 38–44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelec S. et al. Concentration-dependent requirement for local protein synthesis in motor neuron subtype-specific response to axon guidance cues. J. Neurosci. 32, 1496–1506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobel W. & Helmchen F. In vivo calcium Imaging of neural network function. Physiology 22, 358–365 (2007). [DOI] [PubMed] [Google Scholar]
- Banker G. A. & Cowan W. M. Further Observations on Hippocampal-Neurons in Dispersed Cell-Culture. J. Comp. Neurol. 187, 469–493 (1979). [DOI] [PubMed] [Google Scholar]
- Valtorta F., Pennuto M., Bonanomi D. & Benfenati F. Synaptophysin: leading actor or walk-on role in synaptic vesicle exocytosis? Bioessays 26, 445–453 (2004). [DOI] [PubMed] [Google Scholar]
- Lundby A., Mutoh H., Dimitrov D., Akemann W. & Knopfel T. Engineering of a Genetically Encodable Fluorescent Voltage Sensor Exploiting Fast Ci-VSP Voltage-Sensing Movements. PLoS One 3, e2514 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. W. et al. Optically monitoring voltage in neurons by photo-induced electron transfer through molecular wires. Proc. Natl. Acad. Sci. USA 109, 2114–2119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J., Tooker A., Tai Y. C. & Pine J. Caged neuron MEA: A system for long-term investigation of cultured neural network connectivity. J. Neurosci. Meth. 175, 1–16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross G. W., Rieske E., Kreutzberg G. W. & Meyer A. A new fixed-array multi-microelectrode system designed for long-term monitoring of extracellular single unit neuronal activity in vitro. Neurosci. Lett. 6, 101–105 (1977). [DOI] [PubMed] [Google Scholar]
- Hardelauf H. et al. High fidelity neuronal networks formed by plasma masking with a bilayer membrane: analysis of neurodegenerative and neuroprotective processes. Lab Chip 11, 2763–2771 (2011). [DOI] [PubMed] [Google Scholar]
- Thery M. Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell. Sci. 123, 4201–4213 (2010). [DOI] [PubMed] [Google Scholar]
- Nelson C. M. & Tien J. Microstructured extracellular matrices in tissue engineering and development. Curr. Opin. Biotech. 17, 518–523 (2006). [DOI] [PubMed] [Google Scholar]
- Yang I. H., Siddique R., Hosmane S., Thakor N. & Hoke A. Compartmentalized microfluidic culture platform to study mechanism of paclitaxel-induced axonal degeneration. Exp. Neurol. 218, 124–128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Philipsborn A. C., Lang S., Jiang Z., Bonhoeffer F. & Bastmeyer M. Substrate-bound protein gradients for cell culture fabricated by microfluidic networks and microcontact printing. Sci. STKE 2007, pl6 (2007). [DOI] [PubMed] [Google Scholar]
- Wang J. Y. et al. Microfluidics: A new cosset for neurobiology. Lab Chip 9, 644–652 (2009). [DOI] [PubMed] [Google Scholar]
- Kunze A., Meissner R., Brando S. & Renaud P. Co-Pathological Connected Primary Neurons in a Microfluidic Device for Alzheimer Studies. Biotechnol. Bioeng. 108, 2241–2245 (2011). [DOI] [PubMed] [Google Scholar]
- Millet L. J., Collens M. B., Perry G. L. & Bashir R. Pattern analysis and spatial distribution of neurons in culture. Integr. Biol. (Camb) 3, 1167–1178 (2011). [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. Spatial light interference microscopy (SLIM). Opt Express 19, 1016–1026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. & Popescu G. Quantitative phase imaging with broadband fields. Appl. Phys. Lett. 96, 051117 (2010). [Google Scholar]
- Popescu G. Quantitative phase imaging of cells and tissues. (McGraw-Hill, New York, 2011). [Google Scholar]
- Mir M., Bhaduri B., Wang R., Zhu R. & Popescu G. Chapter 3 - Quantitative Phase Imaging. Progress in Optics, Wolf E. (ed.) 133–217 (Elsevier, Oxford, 2012). [Google Scholar]
- Barer R. Interference microscopy and mass determination. Nature 169, 366–367 (1952). [DOI] [PubMed] [Google Scholar]
- Popescu G. et al. Optical imaging of cell mass and growth dynamics. Am. J. Physiol. Cell. Physiol. 295, C538–C544 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M. et al. Optical measurement of cycle-dependent cell growth. Proc. Natl. Acad. Sci. USA 108, 13124–13129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. et al. One-dimensional deterministic transport in neurons measured by dispersion-relation phase spectroscopy. J. Phys-Condens. Mat. 23, 374107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. et al. Dispersion-relation phase spectroscopy of intracellular transport. Opt. Exp. 19, 20571–20579 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H., Wang Z., Nguyen F., Boppart S. A. & Popescu G. Fourier Transform Light Scattering of Inhomogeneous and Dynamic Structures. Phys. Rev. Lett. 101, 238102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H. F., Nguyen F., Boppart S. A. & Popescu G. Optical properties of tissues quantified by Fourier-transform light scattering. Opt. Lett. 34, 1372–1374 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H. F., Millet L. J., Gillette M. U. & Popescu G. Actin-driven cell dynamics probed by Fourier transform light scattering. Biomed. Opt. Exp. 1, 260–267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. et al. Born approximation model for light scattering by red blood cells. Biomed. Opt. Exp. 2, 2784–2791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M. et al. Tissue self-affinity and polarized light scattering in the born approximation: a new model for precancer detection. Phys. Rev. Lett. 97, 138102 (2006). [DOI] [PubMed] [Google Scholar]
- Dhara S. K. et al. Genetic Manipulation of Neural Progenitors Derived from Human Embryonic Stem Cells. Tissue. Eng. Pt. A 15, 3621–3634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhara S. K. et al. Human neural progenitor cells derived from embryonic stem cells in feeder-free cultures. Differentiation 76, 454–464 (2008). [DOI] [PubMed] [Google Scholar]
- Jin K. L. et al. Transplantation of human neural precursor cells in Matrigel scaffolding improves outcome from focal cerebral ischemia after delayed postischemic treatment in rats. J. Cereb. Blood Flow Metab. 30, 534–544 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder A. et al. Neurotrophic Effects of Leukemia Inhibitory Factor on Neural Cells Derived from Human Embryonic Stem Cells. Stem Cells 30, 2387–2399 (2012). [DOI] [PubMed] [Google Scholar]
- Young A. et al. Ion channels and ionotropic receptors in human embryonic stem cell derived neural progenitors. Neuroscience 192, 793–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill J. A., Freudenrich T. M., Robinette B. L. & Mundy W. R. Comparative sensitivity of human and rat neural cultures to chemical-induced inhibition of neurite outgrowth. Toxicol. Appl. Pharm. 256, 268–280 (2011). [DOI] [PubMed] [Google Scholar]
- Mackintosh F. C. & Schmidt C. F. Active cellular materials. Curr. Opin. Cell. Biol. 22, 29–35 (2010). [DOI] [PubMed] [Google Scholar]
- Brangwynne C. P., Koenderink G. H., MacKintosh F. C. & Weitz D. A. Intracellular transport by active diffusion. Trends. Cell. Biol. 19, 423–427 (2009). [DOI] [PubMed] [Google Scholar]
- Barer R. Determination of dry mass, thickness, solid and water concentration in living cells. Nature 172, 1097–1098 (1953). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Info File #1
Supplementary Info File #2
Supplementary Info File #3
Supplementary Info File #4
Supplementary Info File #5
Supplementary Info File #6