Abstract
Background
Insulin-like growth factor 1 receptor (IGF1R) represents a novel molecular target in non-small-cell-lung cancer (NSCLC). IGF1R and epidermal growth factor receptor (EGFR) activation are essential to mediate tumor cell survival, proliferation, and invasion. This study investigates the prognostic role of IGF1R and EGFR in surgically resected NSCLC.
Materials and methods
IGF1R and EGFR copy number gain (CNG) were tested by fluorescence in situ hybridization (FISH) and protein expression by immunohistochemistry (IHC) in 125 stage I–II–IIIA NSCLC patients.
Results
Fourty-six tumors (40.3 %) were IGF1R FISH-positive (FISH+), and 76 (67.2 %) were EGFR FISH+. Tumors with concomitant IGF1R/EGFR FISH+ were observed in 34 cases (30.1 %). IGF1R and EGFR FISH+ were associated with SCC histology (p = 0.01 and p = 0.04, respectively). IGF1R and EGFR protein over-expression (IHC+) were detected in 45 (36.0 %) and 69 (55.2 %) cases, respectively. Tumors with concomitant IGF1R/EGFR IHC+ were detected in 31 (24.8 %) patients. IGF1R/EGFR FISH+ and IGF1R/EGFR IHC+ were significantly associated (χ2 = 4.02, p = 0.04). Patients with IGF1R/EGFR FISH+ and IGF1R/EGFR IHC+ were associated with shorter disease-free survival (DFS) (p = 0.05 and p = 0.05, respectively). Patients with concomitant IGF1R/EGFR FISH+/IHC+ had a worse DFS and overall survival (p = 0.005 and p = 0.01, respectively). The multivariate model confirmed that IGF1R/EGFR FISH+/IHC+ (hazard ratio (HR), 4.08; p = 0.01) and tumor stage (II–III vs I) (HR, 4.77; p = 0.003) were significantly associated with worse DFS.
Conclusions
IGF1R/EGFR FISH+ correlates with IGF1R/EGFR IHC+. IGF1R/EGFR FISH+/IHC+ is an independent negative prognostic factor for DFS in early NSCLC. These features may have important implications for future anti-IGF1R therapeutic approaches.
Keywords: IGF1R, EGFR, Non-small-cell-lung cancer, Prognosis
Introduction
Lung cancer remains the most frequent cause of cancer death worldwide [1], and the 5-year survival remains below 15 % [2]. Variable survival outcome among patients within the same clinical stage suggests the existence of unknown biological factors affecting prognosis.
Insulin-like growth factor receptor 1 (IGF1R) is a transmembrane heterotetrameric protein encoded by the IGF1R gene located on chromosome 15q25–q26, which promotes oncogenic transformation, growth, and survival of cancer cells [3–6]. The binding of IGF-I and IGF-II to the extracellular subunity domain of IGF1R activates the tyrosine kinase activity of IGF1R and triggers a cascade of reactions involving signal transduction pathways: the Ras, Raf, mitogen-activated protein kinase, and the phosphoinositol-3-kinase (PI3K)/AKT/BAD (Bcl-xL/Bc12-associated death promoter) [7]. Currently several IGF1R inhibitors are undergoing clinical evaluation, including blocking antibodies and tyrosine kinase blockers. Phase I/II studies of these compounds indicated favorable toxicity profiles and promising activity [8–11]. However, phase III trials of anti-IGF1R agent in NSCLC were recently discontinued owing to futility. These results stress the need to identify patient subpopulations that may preferentially benefit from anti-IGF1R therapy.
Several lines of evidence suggest an association between epidermal growth factor receptor (EGFR) and IGF1R pathways. Results of preclinical studies indicate that both receptors may heterodimerize [12], are capable of transphosphorylation [13], and share the same adaptor proteins and downstream signaling pathways [14]. Data on cell lines suggest that IGF1R mediates resistance to anti-EGFR therapy through continued activation of the PI3K-Akt pathway [15, 16]. Numerous ongoing clinical trials explore the efficacy of combination of EGFR and IGF1R inhibitors in patients with lung cancer.
The prognostic role of IGF1R has been previously investigated in human malignancies including NSCLC, leading to conflicting results. In breast cancer, IGF1R expression and activation have been linked to disease progression, increased resistance to radiotherapy, and poor prognosis [17, 18]. Fidler et al. [19], in gefitinib-treated patients, showed counter-intuitive improved outcomes with the co-expression of IGF1R and EGFR in exploratory analyses. Cappuzzo et al. [20] observed that IGF1R expression was significantly associated with better survival in metastatic NSCLC patients treated with tyrosine kinase inhibitors (TKIs). The same authors, in another study [21], showed that IGF1R expression did not represent a prognostic factor in resected NSCLC patients. Instead, Nakagawa et al. [22] observed that IGF1R expression was associated with reduced disease-free survival (DFS) in NSCLC. Recently, Dziadziuszko et al. [23] reported that IGF1R protein and gene expression did not associate with survival, whereas high IGF1R gene copy number harbored positive prognostic value. In a previous study, we showed that IGF1R and EGFR protein expression levels were associated, and high-level co-expression was a significant prognostic factor for worse DFS in early-stage NSCLC patients [24]. The conflicting available data and the growing interest on anti-IGF1R agents support the present study aimed to evaluate the relationship between IGF1R and EGFR gene copy number by fluorescence in situ hybridization (FISH) and protein expression by immunohistochemistry (IHC), and to investigate the prognostic significance of these receptors in a cohort of patients with NSCLC who underwent pulmonary resection.
Materials and methods
Patients
Patients with a histological diagnosis of stage I–III NSCLC who had complete surgical resection of the primary tumor and were followed on a regular basis in a specified program were eligible for this study. Histological subtypes and grade of differentiation were determined according to the World Health Organization classification [25]. Patients were originally staged according to the 6th Edition of the IASLC TNM classification [26]. For this study, staging in all cases had been updated to reflect the 7th Edition [27]. Patients were classified as nonsmokers (never smoker), light smokers (<10 pack year and quit >10 years ago), and smokers. Neither chemotherapy nor radiotherapy had to be administered prior to surgery, and none of the patients had received EGFR target anticancer therapy during the follow-up period. Patients with disease recurrence or metastases were treated with platinum-based systemic chemotherapy. Follow-up including annual computed tomography (CT) scans of the chest and abdomen and chest X-rays in the 3-to 6-month intervals between the annual CT was planned for all patients in the first 3 years. Recurrences were detected by imaging techniques and when necessary confirmed by histological sampling.
The study was reviewed and approved by the institution's Ethics Committee, and the patients had to give their informed consent to surgical specimen analysis.
IGF1R and EGFR FISH analysis
Unstained 4 μm (+1 μm) sections of formalin-fixed paraffin-embedded (FFPE) tumor tissue were subjected to dual-color FISH assay using two probe sets: the laboratory-developed reagent IGF1R/CEP15 probe set combining the DNA insert encompassing IGF1R (BAC clone RP11 262P8) labeled with SpectrumRed d-UTPs and the SpectrumGreen CEP 15 probe and the commercial LSI EGFR Spectrum Orange/CEP7 Spectrum Green probe (Abbott Molecular). For both probe sets, assays were performed according to a previously described protocol [28]. Signals were enumerated in at least 100 tumor nuclei for each sample, using a fluorescence microscope (Zeiss AxioImager M1) equipped with single interference filters sets for green (FITC), red (Texas red), and blue (DAPI) as well as dual (red/green) and triple (blue, red, green) band pass filters. The copy number gain was evaluated utilizing a semi-quantitative grading system considering the increasing number of copies of the genes (disomy, low and high trisomy, low and high polysomy, and gene amplification) [29]. For documentation, images from representative fields were acquired by a charge-coupled device camera (Cool-Snap, Photometrics) and merged using dedicated software (CytoVision, Genetix, San Jose CA).
IHC staining of IGF1R and EGFR
The IHC staining of IGF1R and EGFR was performed according to a previously described protocol [24]. IGF1R and EGFR protein expression were evaluated using mouse antibodies (clones 24–31, 1:50 diluted, Lab Vision Neomarkers, and clone 3147, 1:100 diluted, Zymed, Laboratories Inc., respectively). There are at present no validated scoring systems for interpreting IHC staining for IGF1R and EGFR, thus we applied previous description for EGFR [30]. A cutoff value of 10 % positive cells was used in order to avoid inclusion of scattered positivity of the same intensity found in the normal bronchial tissue. A specimen was considered IHC-positive (IHC+) only when a distinct cell membrane staining was evident (Fig. 1e, f).
Fig. 1.
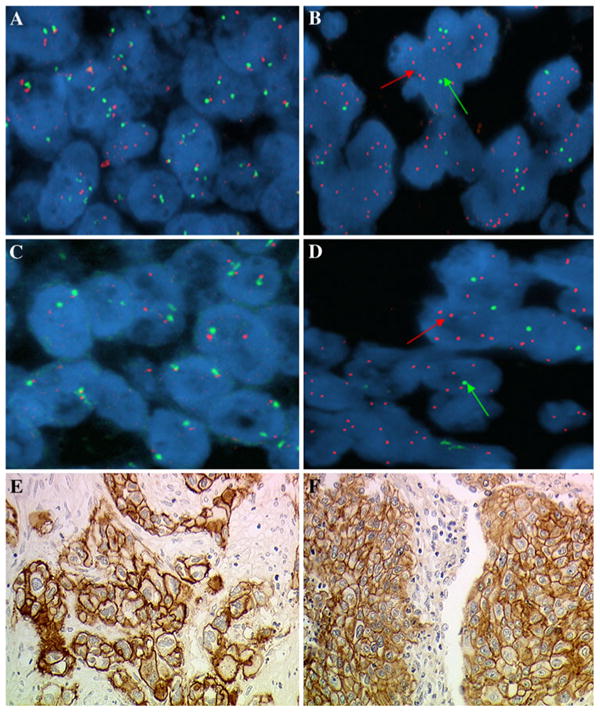
Dual color fluorescent in situ hybridization assays with probes for Insulin-like growth factor receptor 1(IGF1R; red signal indicated by the arrow) and chromosome 15 (CEP15; green signal indicated by the arrow). a IGF1R FISH−, b IGF1R FISH+. Dual color fluorescent in situ hybridization assays with probes for epidermal growth factor (EGFR; red signal indicated by the arrow) and chromosome 7 (CEP7; green signal indicated by the arrow). c EGFR FISH-, d EGFR FISH+. Examples of squamous-cell carcinoma with immunohistochemistry (IHC) positive for IGF1R (e) and for EGFR (f) (original magnification ×400)
Statistical analysis
The χ2-square or Fisher's exact test, where appropriate, was used to assess the association between clinical features and FISH analysis or the protein expression of biological factors. In a univariate exploratory analysis, maximum log-likelihood function was used to select the most discriminative cutoff values of IGF1R and EGFR FISH-positive (FISH+) for survival difference [31]. We found that the best discrimination was observed with a cutoff of ≥4.0 gene copies per cell displayed in 10 % of tumor cells to classify FISH+positive (FISH+) and FISH-negative (FISH-) patients. Overall survival (OS), disease-free survival (DFS), and the 95 % confidence intervals were evaluated by the Kaplan–Meier method comparing the different groups by log-rank test [32]. The Cox proportional hazards model was used to evaluate the prognostic role of each single studied parameter on OS and DFS, in univariate and multivariate analyses. Univariate survival analysis was conducted by exploring the interactions between the predictors, and the significant corrections were considered in the multivariate model (Cox, [33]). OS was defined as the time from diagnosis to the date of death from any cause; patients who were not reported as having died at the time of the analysis were censored at the date they were last known to be alive. DFS was defined as the time from diagnosis to first local, regional or distant recurrence, second primary malignancy or death from any cause, whichever came first. Patients who were alive and did not experience recurrence at the time of the analysis were censored at the last disease assessment date. Unless otherwise specified, all tests are with one degree of freedom (df). A probability value of <0.05 was considered as statistically significant. Statistical analysis was performed using Matlab software (The MathWorks ver. 7.2.0.232).
Results
Patient characteristics
From April 2002 to February 2006, we collected tissue samples of 125 consecutive early stage radically resected NSCLC patients, who were referred to the Thoracic Surgery Unit, Perugia University Hospital, Italy. Table 1 summarizes the demographic and clinical data. There were 96 males (84.8 %) and 19 females (15.2 %), with a median age of 66 years, ranging from 40 to 84. One hundred and sixteen patients (92.8 %) were current or former smokers. The histological types included 63 (50.4 %) squamous-cell carcinoma (SCC), 39 (31.2 %) adenocarcinoma, 3 (2.4 %) bronchioloalveolar, 12 (9.6 %) large cell and 8 (6.4 %) mixed histology. The majority of patients had poorly differentiated histology (57/125; 45.3 %) and surgery consisted of 102 lobectomies (81.6 %), 16 pneumonectomies (12.8 %), and 7 wedge resections (5.6 %), with hilar and mediastinal lymph node dissection. TNM staging consisted of 78 (62.4 %) Stage I, 20 (16.0 %) Stage II, 27 (21.6 %) Stage III. Following surgery, 3 patients (2.3 %) were treated with chemotherapy, 9 (6.9 %) with radiotherapy, 4 (3.1 %) with chemo-radiotherapy, and the remaining 109 (87.7 %) received no adjuvant treatment.
Table 1. Patient characteristics.
| Variable | Total patients | |
|---|---|---|
|
| ||
| No. | (%) | |
| Age (years) | ||
| Median (range) | 66 (40–84) | |
| Gender | ||
| Female | 19 | 15.2 |
| Male | 96 | 84.8 |
| Smoking history | ||
| Former/current smoker | 116 | 92.8 |
| Never smoker | 9 | 7.2 |
| Pathologic stage | ||
| I | 78 | 62.4 |
| II | 20 | 16.0 |
| III | 27 | 21.6 |
| Histology | ||
| Squamous-cell carcinoma | 63 | 50.4 |
| Adenocarcinoma | 39 | 31.2 |
| Bronchioloalveolar | 3 | 2.4 |
| Large cell carcinoma | 12 | 9.6 |
| Mixed histology | 8 | 6.4 |
| Grading | ||
| Well | 14 | 11.5 |
| Moderately | 54 | 43.2 |
| Poorly | 57 | 45.3 |
| Resection | ||
| Lobectomy | 102 | 81.6 |
| Pneumonectomy | 16 | 12.8 |
| Wedge | 7 | 5.6 |
At a median follow-up time of 48.9 months, 66 patients (52.8 %) had died: 64 (95.6 %) deaths were due to disease recurrence and two (4.4 %) to unrelated causes. Nine (15.2 %) of the 59 patients still on follow-up experienced recurrence. The median survival time was 61.4 months.
IGF1R and EGFR gene copy number
FISH analyses were successful in 114 patients for IGF1R and in 113 patients for EGFR due to yielded insufficient tumor tissue. Mean gene copy number was 2.48 copies per cell (range 1.47–7.2) for IGF1R and 2.80 copies per cell (range 1.56–15) for EGFR. Using a cutoff of ≥4.0 gene copies per cell displayed in 10 % of tumor cells, FISH+ was detected in 46 tumors (40 %) for IGF1R and 76 tumors (77 %) for EGFR. Both IGF1R and EGFR showed true gene amplification in 5 patients each (4.4 %). IGF1R and EGFR concurrent by FISH+ was observed in 34 cases (30 %). Examples of microscope fields from IGF1R and EGFR FISH- and IGF1R and EGFR FISH+ tumors are shown in Fig. 1a–d.
IGF1R and EGFR immunohistochemistry
IHC of IGF1R and EGFR was evaluated in 125 tumors. With a cutoff value of >10 % positive cells (2+, 3+), IGF1R protein overexpression was detected in 45 patients (36.0 %) and was associated with larger tumor size (p = 0.04), but not with other clinical or biological characteristics. IGF1R was more expressed in SCC histology than in non-SCC (57.8 vs 42.2 %, p = 0.59), although this difference was not statistically significance.
EGFR protein overexpression was observed in 69 patients (55.2 %), also more frequently in SCC than non-SCC (63.7 vs 36.3 %, p = 0.001). IGF1R protein expression was associated with EGFR protein expression (χ2 = 4.49, p = 0.03). Co-expression of both receptors was observed in 31 cases (24.8 %).
Correlation between IGF1R and EGFR FISH patterns and protein expression with clinical or demographic characteristics
Frequencies of IGF1R and EGFR FISH status, FISH status of both IGF1R and EGFR (indicated as IGF1R/EGFR), IHC status of IGF1R/EGFR and concomitant FISH and IHC status (indicated as FISH/IHC) of IGF1R/EGFR are shown in Table 2.
Table 2. Clinicopathologic characteristics of patients according to IGF1R, EGFR FISH status, IGF1R/EGFR FISH status, IGF1R/EGFR IHC status, and concomitant IGF1R/EGFR FISH/IHC status.
| IGF1R FISHa | EGFR FISHb | IGF1R/EGFR FISH | IGF1R/EGFR IHC | IGF1R/EGFR FISH-IHC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||
| Positive (%) |
Negative (%) |
Positive (%) |
Negative (%) |
Both positive (%) |
Other (%) |
Both positive (%) |
Other (%) |
Both positive (%) |
Other (%) |
|
| No. of Pts | 46 (40.3) | 68 (59.7) | 76 (67.2) | 37 (32.7) | 34 (30.1) | 79 (69.9) | 31 (24.8) | 94 (75.2) | 13 (10.4) | 112 (89.6) |
| Gender | ||||||||||
| Female | 6 (13.0) | 12 (17.6) | 14 (18.4) | 4 (10.8) | 4 (11.8) | 14 (17.7) | 3 (9.7) | 16 (17.1) | 1 (7.7) | 18 (16.1) |
| Male | 40 (87.0) | 56 (82.3) | 62 (81.6) | 33 (89.2) | 30 (88.2) | 65 (82.3) | 28 (90.3) | 78 (82.9) | 13 (92.3) | 94 (83.9) |
| P | 0.56 | 0.30 | 0.42 | 0.32 | 0.42 | |||||
| Smoking history | ||||||||||
| Smoker | 44 (94.6) | 61 (89.7) | 68 (89.5) | 36 (97.3) | 33 (97.0) | 71 (89.9) | 29 (93.5) | 87 (92.5) | 12 (7.7) | 104 (92.9) |
| Nonsmoker | 3 (3.4) | 7 (10.3) | 8 (10.5) | 1 (2.7) | 1 (3.0) | 8 (10.1) | 2 (6.5) | 7 (7.5) | 1 (92.3) | 8 (7.1) |
| P | 0.24 | 0.14 | 0.19 | 0.85 | 0.94 | |||||
| Histology | ||||||||||
| Squamous | 30 (65.2) | 29 (42.6) | 34 (44.8) | 24 (64.9) | 21 (61.3) | 37 (46.8) | 22 (71.0) | 41 (43.6) | 10 (76.9) | 53 (52.7) |
| Other | 16 (34.8) | 39 (57.4) | 42 (55.2) | 13 (35.1) | 13 (38.2) | 42 (53.2) | 9 (29.0) | 53 (56.4) | 3 (23.1) | 59 (47.3) |
| P | 0.01 | 0.04 | 0.14 | 0.008 | 0.04 | |||||
| pTNM stage | ||||||||||
| I | 28 (60.9) | 43 (63.2) | 48 (63.2) | 22 (59.4) | 21 (61.8) | 49 (62.0) | 18 (58.1) | 60 (63.8) | 5 (38.6) | 73 (65.2) |
| II | 7 (15.2) | 11 (16.2) | 10 (13.2) | 8 (21.6) | 4 (11.7) | 14 (17.7) | 2 (6.5) | 18 (19.2) | 1 (7.7) | 19 (16.7) |
| III | 11 (23.9) | 14 (20.6) | 18 (23.6) | 7 (19.0) | 9 (26.5) | 16 (20.3) | 11 (35.5) | 16 (17.0) | 7 (53.8) | 20 (17.9) |
| P | 0.17 | 0.48 | 0.62 | 0.04 | 0.01 | |||||
| Grading | ||||||||||
| Well | 5 (10.8) | 7 (10.3) | 9 (11.8) | 3 (8.11) | 5 (14.7) | 7 (8.9) | 4 (12.9) | 10 (10.6) | 2 (15.4) | 12 (10.7) |
| Moderately | 17 (36.9) | 35 (51.47) | 33 (43.4) | 18 (48.6) | 11 (32.3) | 40 (50.6) | 15 (48.4) | 39 (41.5) | 3 (23.1) | 51 (43.5) |
| Poorly | 24 (52.1) | 26 (38.2) | 34 (44.8) | 16 (43.2) | 18 (52.9) | 32 (40.5) | 12 (38.7) | 45 (47.9) | 8 (61.5) | 49 (43.8) |
| P | 0.28 | 0.78 | 0.18 | 0.67 | 0.25 | |||||
EGFR epidermal growth factor receptor, IGF1R insulin-like growth factor receptor 1, FISH fluorescence in situ hybridization, IHC immunohistochemistry
IGF1R FISH-positive = copy number gain (CNG), ≥ 4 gene copies in ≥ 10 % of tumor cells
EGFR FISH-positive = copy number gain (CNG), ≥ 4 gene copies in ≥ 10 % of tumor cells
IGF1R FISH+ was associated with EGFR FISH+ (p = 0.03). IGF1R and EGFR FISH+ were associated with SCC histology (p = 0.01 and p = 0.04, respectively) as well as IHC+ of IGF1R/EGFR (p = 0.008) and concomitant FISH +/IHC+ of IGF1R/EGFR (p = 0.04). IHC+ of IGF1R/EGFR and concomitant FISH +/IHC+ of IGF1R/EGFR were associated with advanced stage (p = 0.04 and p = 0.01, respectively). No other clinical or demographic association was detected with these biological markers.
IGF1R/EGFR FISH+ and IGF1R/EGFR IHC+ were significantly associated (χ2 = 4.02, p = 0.04). Thirteen patients (11.5 %) showed concomitant FISH+/IHC+ of both receptors.
Prognostic implication of IGF1R and EGFR gene copy number and protein expression
Table 3 presents the results of the univariate analysis for DFS and OS. No statistically significant difference in DFS and OS was observed between patients with negative or positive FISH pattern for IGF1R (HR 1.48; 95% CI 0.77–2.84, p = 0.10, and HR 1.55; 95 % CI 0.93–2.59, p = 0.08, respectively) or EGFR (HR 1.48; 95 % CI 0.77–2.84, p = 0.23, and HR 0.98; 95 % CI 0.56–1.71, p = 0.95, respectively). Similar results between patients with negative or positive IHC patterns for IGF1R (HR 1.18; 95 % CI 0.68–2.03, p = 0.53, and HR 1.42; 95 % CI 0.87–2.33, p = 0.15, respectively) or EGFR (HR 1.61; 95 % CI 0.93–2.77, p = 0.08, and HR 1.39; 95 % CI 0.85–2.27, p = 0.15, respectively) were found for DFS and OS (data not shown).
Table 3. Univariate analysis for variables considered (cox proportional hazard regression model).
| Feature | Disease-free survival | Overall survival | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95 % CI | P | HR | 95 % CI | P | |
| Gender | ||||||
| Female versus male | 0.82 | 0.38–1.73 | 0.60 | 0.77 | 0.38–1.57 | 0.48 |
| Age (years) | ||||||
| Continuous | 1.01 | 0.98–1.04 | 0.25 | 1.03 | 1.00–1.06 | 0.04 |
| Smoking history | ||||||
| Former/current smoker versus never smoker | 0.86 | 0.31–2.38 | 0.77 | 0.70 | 0.30–1.64 | 0.42 |
| Histology | ||||||
| Squamous versus other | 1.27 | 0.76–2.12 | 0.35 | 1.04 | 0.65–1.68 | 0.84 |
| Grading | ||||||
| G3 versus G1–G2 | 1.52 | 0.91–2.55 | 0.10 | 1.49 | 0.93–2.40 | 0.09 |
| TNM stage | ||||||
| II–III versus I | 2.86 | 1.64–4.97 | <0.001 | 2.47 | 1.48–4.12 | 0.001 |
| IGF1R FISHa | ||||||
| Positive versus negative | 1.59 | 0.91–2.77 | 0.10 | 1.55 | 0.93–2.59 | 0.08 |
| EGFR FISHb | ||||||
| Positive versus negative | 1.48 | 0.77–2.84 | 0.23 | 0.98 | 0.56–1.71 | 0.95 |
| IGF1R/EGFR FISH | ||||||
| Both positive versus other | 1.76 | 0.99–3.13 | 0.05 | 1.46 | 0.83–1.47 | 0.15 |
| IGF1R/EGFR IHC | ||||||
| Both positive versus other | 1.71 | 0.97–3.01 | 0.05 | 1.56 | 0.92–2.64 | 0.09 |
| IGF1R/EGFR FISH/IHC | ||||||
| Both positive versus other | 2.84 | 1.38–5.84 | 0.005 | 2.32 | 1.20–4.44 | 0.01 |
HR hazard ratio, CI confidence interval, EGFR epidermal growth factor receptor, IGF1R insulin-like growth factor receptor 1, FISH fluorescence in situ hybridization, IHC immunohistochemistry
IGF1R FISH-positive = copy number gain (CNG), ≥4 gene copies in ≥10 % of tumor cells
EGFR FISH-positive = copy number gain (CNG), ≥4 gene copies in ≥10 % of tumor cells
The different associations of IGF1R and EGFR gene copy number by FISH and protein expression by IHC were also compared. Patients were classified into three groups: group I with FISH+ pattern for both IGF1R and EGFR, group II with IHC+ pattern for both IGF1R and EGFR, and group III with FISH +/IHC+ patterns for both IGF1R and EGFR (Fig. 2). A statistically significant difference in DFS was observed between patients with FISH+ of both genes compared to other group (25 months vs not reached; HR 1.76; 95 % CI 0.99–3.13, p = 0.05, Fig. 2a). The subset of patients with IGF1R/EGFR IHC+ protein expression showed a worse DFS (25 months vs 90 months; HR 1.71, 95 % CI 0.97–3.01, p = 0.05, Fig. 2b) when compared with other group. Also, a very significant shorter DFS was observed in the group III (FISH +/IHC+ of both IGF1R and EGFR) (10 months vs 85 months; HR 2.84; 95 % CI 1.38–5.84, p = 0.005, Fig. 2c). The same group III were, also, significantly associated with a poorer OS (33 months vs 87 months; HR 2.32, 95 % CI 1.20–4.44, p = 0.01, Fig. 2f). The groups I and II were not associated with OS (p = 0.15, p = 0.09, respectively, Fig. 2d, e).
Fig. 2.

Kaplan-Meier curves of disease-free survival (DFS) and overall survival (OS) according to FISH status of IGF1R/EGFR (a, d), IHC status of IGF1R/EGFR (b, e), and FISH/IHC status of IGF1R/EGFR (c, f) in patients with non-small cell-lung cancer (NSCLC)
Among clinical variables, stage (III vs I–II) was significantly associated with worse DFS (HR 2.86, 95 % CI 1.64–4.97, p < 0.0001) and OS (HR 2.47, 95 % CI 1.48–4.12, p = 0.001), whereas age (as continuous variable) was significantly associated with worse OS (HR 1.03, 95 % CI 1.00–1.06, p = 0.04). The multivariate model confirmed that FISH +/IHC+ patterns for both IGF1R and EGFR (HR 4.08, 95 % CI 1.34–12.39, p = 0.01) and tumor stage (II–III vs I) (HR 4.77, 95 % CI 1.72–13.21, p = 0.003) were significantly associated with worse DFS (Table 4). Only stage (II–III vs I) was an independent prognostic factor for OS (HR 2.57, 95 % CI 1.38–4.79, p = 0.003) (data not shown).
Table 4. Prognostic effect of studied parameters on DFS—multivariate analysis.
| Variable | Disease-free survival | ||
|---|---|---|---|
|
| |||
| HR | 95 % CI | P value | |
| IGF1R/EGFR FISH | |||
| Both positive versus other groups | 1.44 | 0.61–3.40 | 0.40 |
| IGF1R/EGFR IHC | |||
| Both positive versus other groups | 1.36 | 0.49–3.75 | 0.54 |
| IGF1R/EGFR FISH/IHC | |||
| Both positive versus other | 4.08 | 1.34–12.39 | 0.01 |
| TNM stage | |||
| II–III versus I | 4.77 | 1.72–13.21 | 0.003 |
HR hazard ratio, CI confidence interval, EGFR epidermal growth factor receptor, IGF1R insulin-like growth factor receptor 1, FISH fluorescence in situ hybridization, IHC immunohistochemistry
Discussion
In this study, we evaluated the prognostic role of two of the most relevant biomarkers in NSCLC, and to our knowledge, report for the first time that FISH +/IHC+ of both IGF1R/EGFR receptors represents a negative prognostic factor in radically resected NSCLC. There is an increasing interest on IGF1R in NSCLC. Identification of biomarkers for selecting patients most likely to derive clinical benefit from IGF1R inhibitors is needed. In vitro studies showed that expression of IGF1R correlates with sensitivity to IGF1R inhibitors, such as BMS-536924 [34]. Preclinical models showed that IGF1R expression could be implicated in acquired resistance to anti-EGFR strategies [13, 15]. In breast and prostate cancer cells, Jones et al. [13] showed that increased signaling via the IGF1R pathway leads to acquired resistance to the EGFR tyrosine kinase inhibitor gefitinib. Chakravarti et al. [15] demonstrated that IGF1R can compensate for loss of EGFR function in primary glioma cell lines. Several new drugs, including mAbs and tyrosine kinase inhibitors, are currently under evaluation in patients with advanced NSCLC.
Gualberto et al. [35] reported prolonged clinical benefit to figitumumab (F) in tumors with intense membrane localization of IGF1R. They also saw that high IGF1R in the chemotherapy alone group was associated with rapid disease progression. However, these results should be considered preliminary due to small cohort of patients (N = 45) investigated.
A phase II study randomly assigned 150 chemonaive NSCLC patients to the standard combination of carboplatin (C) plus paclitaxel (P) versus the same regimen plus CP-751,871, an anti-IGF1R mAb [11]. The study showed a significant improvement in response rate favoring the experimental arm, with an impressive 78 % response rate in patients with squamous histology.
The higher expression of the IGF1R protein in patients with squamous-cell carcinoma compared with non-squamous histology observed in the Karp's study, as well as in our study, provides the biological background for the highest sensitivity to anti-IGF1R agents observed in clinical trials in this patient population. A recent phase III trial of F administered in combination with CP failed to demonstrate survival benefit in advanced NSCLC patients [36]. The study showed that the use of F with CP would be unlikely to improve overall survival compared to CP alone, mainly due to toxicity occurring in patients who randomly received F. However, in the subset of patients with high IGF serum levels, the addition of F appeared to offer benefit over CP. A second phase III trial investigating figitumumab with erlotinib versus erlotinib alone in the same population in the refractory setting (ADVIGO 1018; figitumumab plus erlotinib versus erlotinib in refractory advanced non-adenocarcinoma NSCLC) was also closed after an interim analysis in March 2010 because of the potential futility of the combination regimen. These results underscore the need to identify the patients most likely to benefit from anti–IGF1R therapy. The negative results in currently reported trials in unselected patients are disappointing, but they do not exclude the possibility that inhibition of the IGF-1R pathway could provide benefit in patients with specific molecular signatures. The preliminary results of presented trials suggest that high circulating levels of IGF-1 may be a potential, easy to test biomarker that may identify patients who may benefit from this class of agents.
A recent study has shown that the IGFIR and other molecules associated with the IGFIR pathway (e.g., IGF2R, IRS-1, -2) were overexpressed in NSCLC tumors undergoing epithelial-to-mesenchymal transition (EMT) [37]. The authors hypothesized that high circulating levels of free IGF-1(fIGF-1) could be associated with high IGF bioactivity in the tumor microenvironment, and this could favor EMT. Higher pre-treatment fIGF-1 levels were found to be predictive of the clinical benefit derived from the addition of Figitumumab to chemotherapy in NSCLC patients. In principle, the utilization of circulating factors as predictive biomarkers would appear to be more convenient than measurements requiring fresh or frozen neoplastic tissue. However, it should be noted that current assay methodologies, particularly those measuring IGF-1 bioactivity, are controversial, and it is necessary that assay methods be standardized [38]. In our study, IGF1R FISH+ pattern was detected in 40 % of NSCLC cases and EGFR FISH+ in 77 %, and there was a significant association between IGF1R and EGFR FISH+ and histologic subtypes, with the highest gene copy number in squamous-cell carcinoma. These data are consistent with a recent report [23].
The pattern of expression and prognostic value of IGF1R expression in NSCLC remains controversial. Prior studies have shown that IGF1R expression is associated with longer survival in patients treated with gefitinib [19, 20], poorer survival and higher expression in surgically treated lung adenocarcinomas versus squamous-cell carcinomas [39] or no association with survival in a similar population [40] and shorter DFS when co-expressed with EGFR in resected NSCLC [24].
Our study, conducted in early NSCLC, using FISH and IHC methods to understand the relationship between the gene and its protein levels, showed that tumors with IGF1R/EGFR FISH+/IHC+ were associated with a poorer DFS. The group of patients who had FISH+ patterns for both genes presents a DFS comparable to the group of patients with IHC+ patterns for both proteins (p = 0.05, p = 0.05, respectively, Fig. 2a, b). Finally, the group of patients who had FISH+/IHC+ of both genes had a significantly shorter DFS as shown in Fig. 2c, confirmed also in multivariate analysis. The FISH and IHC double positivity phenotype (IGF1R+/EGFR+) were found in about 10 % of the patients studied, and most of these patients were squamous-cell carcinoma. We could speculate that the cases FISH/IHC double positivity with this histological type (although the number is small) should be used to select patients for IGF1R or IGF1R + EGFR targeted therapy trials.
Furthermore, our results seem to suggest a correlation between high gene copy number and receptor activity, and they confirm the mathematical predictions of model developed by our group [41].
Interestingly, many of the genes regulated by IGF1R affect ribosome biogenesis and protein translation [3–5]. Thus, FISH+ of IGF1R and EGFR may cooperate in enhanced protein translation, either in a general way or for a specific subset of mRNAs. This finding generates the hypothesis that high gene copy number and overexpression of IGF1R with EGFR cooperatively shifts the repertoire of actively translated mRNAs to a set of genes that drive proliferation and growth, or inhibit apoptosis. Moreover, investigators have suggested that expression levels of IGF1R and EGFR are linked and that the IGF1R/EGFR ratio may serve as a more sensitive prognostic indicator of tumor phenotype and therapeutic response than expression of any single receptor [42, 43].
Given the possibility of cooperation and shared signaling pathways between the two systems, simultaneously targeting of the EGF and IGF systems may be more effective than targeting either system alone. Small molecule inhibitors and monoclonal antibodies that selectively inhibit either EGFR or IGF1R have to date predominated as effective treatments in co-targeting strategies. The EGFR inhibition may sensitize tumor cells to anti-IGF-IR treatment, particularly in tumors that are strongly driven by EGFR. Prior evidence has indicated that acquired resistance to EGFR inhibition in NSCLC may be associated with enhanced dependency on IGF-IR signaling [44].
Thus, co-targeting EGFR and IGF-IR could be expected to have additive or synergistic antitumor effects.
In conclusion, in the present study, we have shown that IGF1R and EGFR are frequently FISH+ and IHC+ in early NSCLC and more frequently present in squamous-cell carcinoma. The patients group with IGF1R/EGFR FISH +/IHC+ might represent a subpopulation that is able to develop a more aggressive behavior. Furthermore, our finding could be used as new biomarkers because tumors with IGF1R/EGFR FISH +/IHC+ showed a worse DFS; however, due to limited percentage of this subpopulation and the short follow-up of our study, these results must be interpreted cautiously and need to be confirmed in large prospective trials.
Acknowledgments
The authors acknowledge the technical assistance of the Cytogenetics Core of the University of Colorado Cancer Center for all FISH analyses. The authors thank the patients who participated in this study. This work was supported in part by a grant from the Italian Association for Cancer Research (AIRC) to A.F. and from Umbria Association Against Cancer (AUCC) for IHC reagents.
Footnotes
Conflict of interest: The author(s) indicated no potential conflicts of interest.
Contributor Information
V. Ludovini, Email: ludoviniv@hotmail.com, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy.
A. Flacco, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy
F. Bianconi, Department of Electronic and Information Engineering, Perugia University, Perugia, Italy
M. Ragusa, Department of Thoracic Surgery, Perugia University, Perugia, Italy
J. Vannucci, Department of Thoracic Surgery, Perugia University, Perugia, Italy
G. Bellezza, Institute of Pathological Anatomy and Histology, Division of Cancer Research, Perugia University, Perugia, Italy
R. Chiari, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy
V. Minotti, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy
L. Pistola, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy
F. R. Tofanetti, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy
A. Siggillino, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy
E. Baldelli, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy
A. Sidoni, Institute of Pathological Anatomy and Histology, Division of Cancer Research, Perugia University, Perugia, Italy
N. Daddi, Department of Thoracic Surgery, Perugia University, Perugia, Italy
F. Puma, Department of Thoracic Surgery, Perugia University, Perugia, Italy
M. Varella-Garcia, Department of Medicine/Medical Oncology, University of Colorado Cancer Center, Aurora, CO, USA
L. Crinò, Department of Medical Oncology, S. Maria Della Misericordia Hospital, 1, G. Dottori Street, 06132 Perugia, Italy
References
- 1.Maxwell PD. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Thomas A, Murray T, et al. Cancer statistics 2002. CA Cancer J Clin. 2002;2:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 3.Dufourny B, Alblas J, van Teeffelen HA, et al. Mitogenic signalling of insulin-like growth factor I in MCF-7 human breast cancer cells requires phosphatidylinositol 3-kinase and is independent of mitogen-activated protein kinase. J Biol Chem. 1997;272:31163–31171. doi: 10.1074/jbc.272.49.31163. [DOI] [PubMed] [Google Scholar]
- 4.Khandwala HM, McCutcheon IE, Flyvbjerg A, et al. The effects of insulin-like growth factors on tumorigenesis and neoplastic growth. Endocr Rev. 2000;21:215–244. doi: 10.1210/edrv.21.3.0399. [DOI] [PubMed] [Google Scholar]
- 5.Baserga R, Hongo A, Rubini M, et al. The IGF-I receptor in cell growth, transformation and apoptosis. Biochim Biophys Acta. 1997;1332:F105–F126. doi: 10.1016/s0304-419x(97)00007-3. [DOI] [PubMed] [Google Scholar]
- 6.Blakesley VA, Stannard BS, Kalebic T, et al. Role of the IGF-I receptor in mutagenesis and tumour promotion. J Endocrinol. 1997;152:339–344. doi: 10.1677/joe.0.1520339. [DOI] [PubMed] [Google Scholar]
- 7.LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195(2):127–137. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 8.Haluska P, Shaw HM, Batzel GN, et al. Phase I dose escalation study of the anti insulin-like growth factor-I receptor monoclonal antibody CP-751,871 in patients with refractory solid tumors. Clin Cancer Res. 2007;13:5834–5840. doi: 10.1158/1078-0432.CCR-07-1118. [DOI] [PubMed] [Google Scholar]
- 9.Kurzrock R, Patnaik A, Aisner J, et al. A phase I study of weekly R1507, a human monoclonal antibody insulin-like growth factor-I receptor antagonist, in patients with advanced solid tumors. Clin Cancer Res. 2010;16(8):2458–2465. doi: 10.1158/1078-0432.CCR-09-3220. [DOI] [PubMed] [Google Scholar]
- 10.Tolcher AW, Sarantopoulos J, Patnaik A, et al. Phase I, pharmacokinetic, and pharmacodynamic study of AMG 479, a fully human monoclonal antibody to insulin-like growth factor receptor 1. J Clin Oncol. 2009;27(34):5800–5807. doi: 10.1200/JCO.2009.23.6745. [DOI] [PubMed] [Google Scholar]
- 11.Karp DD, Paz-Ares LG, Novello S, et al. Phase II study of the anti-insulin-like growth factor type 1 receptor antibody CP-751,871 in combination with paclitaxel and carboplatin in previously untreated, locally advanced, or metastatic non-small-cell lung cancer. J Clin Oncol. 2009;27:2516–2522. doi: 10.1200/JCO.2008.19.9331. [DOI] [PubMed] [Google Scholar]
- 12.Morgillo F, Woo JK, Kim ES, et al. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer Res. 2006;66:10100–10111. doi: 10.1158/0008-5472.CAN-06-1684. [DOI] [PubMed] [Google Scholar]
- 13.Jones HE, Goddard L, Gee JM, et al. Insulin-like growth factor-I receptor signalling and acquired resistance to gefitinib (ZD1839; Iressa) in human breast and prostate cancer cells. Endocr Relat Cancer. 2004;11:793–814. doi: 10.1677/erc.1.00799. [DOI] [PubMed] [Google Scholar]
- 14.Knowlden JM, Jones HE, Barrow D, et al. Insulin receptor substrate-1 involvement in epidermal growth factor receptor and insulin-like growth factor receptor signaling: implication for gefitinib (‘Iressa’) response and resistance. Breast Cancer Res Treat. 2008;111:79–91. doi: 10.1007/s10549-007-9763-9. [DOI] [PubMed] [Google Scholar]
- 15.Chakravarti A, Loeffler JS, Dyson NJ. Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signalling. Cancer Res. 2002;62:200–207. [PubMed] [Google Scholar]
- 16.Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 17.Rocha RL, Hilsenbeck SG, Jackson, et al. Insulin-like growth factor binding protein-3 and insulin receptor substrate-1 in breast cancer: correlation with clinical parameters and disease-free survival. Clin Cancer Res. 1997;3:103–109. [PubMed] [Google Scholar]
- 18.Turner BC, Haffty BG, Narayanan L, et al. Insulin-like growth factor-I receptor overexpression mediates cellular radioresistance and local breast cancer recurrence after lumpectomy and radiation. Cancer Res. 1997;57:3079–3083. [PubMed] [Google Scholar]
- 19.Fidler MJ, Basu S, Buckingham L, et al. Utility of insulin-like growth factor receptor-1 expression in gefitinib-treated patients with non-small cell lung cancer. Anticancer Res. 2012;32(5):1705–1710. [PubMed] [Google Scholar]
- 20.Cappuzzo F, Toschi L, Tallini G, et al. Insulin-like growth factor receptor 1 (IGFR-1) is significantly associated with longer survival in non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2006;17:1120–1127. doi: 10.1093/annonc/mdl077. [DOI] [PubMed] [Google Scholar]
- 21.Cappuzzo F, Tallini G, Finocchiaro G, et al. Insulin-like growth factor receptor 1 (IGF1R) expression and survival in surgically resected non-small-cell lung cancer (NSCLC) patients. Ann Oncol. 2010;21:562–567. doi: 10.1093/annonc/mdp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makoto N, Hidetaka U, Soichi O, et al. Clinical significance of IGF1R expression in non-small-cell lung cancer. Clinical Lung cancer. 2011;13(2):136–142. doi: 10.1016/j.cllc.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Dziadziuszko R, Merrick DT, Witta SE, et al. Insulin-like growth factor receptor 1 (IGF1R) gene copy number is associated with survival in operable non–small-cell lung cancer: a comparison between IGF1R fluorescent in situ hybridization, protein expression, and mRNA expression. J Clin Oncol. 2010;28(13):2174–2180. doi: 10.1200/JCO.2009.24.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludovini V, Bellezza G, Pistola L, et al. High coexpression of both insulin-like growth factor receptor-1(IGFR-1) and epidermal growth factor receptor (EGFR) is associated with shorter disease-free survival in resected non-small-cell lung cancer patients. Ann Oncol. 2009;20:842–849. doi: 10.1093/annonc/mdn727. [DOI] [PubMed] [Google Scholar]
- 25.Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization. Classification of lung tumors. Semin Roentgenol. 2005;40(2):90–97. doi: 10.1053/j.ro.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Mountain CF. Revisions in the international system for staging lung cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 27.Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer stagging system. Chest. 2009;136:260–271. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 28.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 29.Varella-Garcia M, Diebold J, Eberhard DA, et al. EGFR fluorescence in situ hybridisation assay: guidelines for application to non-small-cell lung cancer. J Clin Pathol. 2009;62(11):970–977. doi: 10.1136/jcp.2009.066548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selvaggi G, Novello S, Torri V, et al. Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncol. 2004;15(1):28–32. doi: 10.1093/annonc/mdh011. [DOI] [PubMed] [Google Scholar]
- 31.Hosmer DW, Lemeshow S, May S. Applied survival analysis: regression modelling of time to event data. Wiley-Interscience; New York, NY: 2008. [Google Scholar]
- 32.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1985;53:457–481. [Google Scholar]
- 33.Cox DR. Regression models and life tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- 34.Huang F, Greer A, Hurlburt W, et al. The mechanisms of differential sensitivity to an insulin-like growth factor-1 receptor inhibitor (BMS-536924) and rationale for combining with EGFR/HER2 inhibitors. Cancer Res. 2009;69:161–170. doi: 10.1158/0008-5472.CAN-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gualberto A, Dolled-Filhart M, Gustavson M, et al. Molecular analysis of non–small cell lung cancer identifies subsets with different sensitivity to insulin-like growth factor I receptor inhibition. Clin Cancer Res. 2010;16(18):4654–4665. doi: 10.1158/1078-0432.CCR-10-0089. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Jassem J, Langer C, Karp D, Mok T, Benner R, et al. Randomized, open label, phase III trial of figitumumab in combination with paclitaxel and carboplatin versus paclitaxel and carboplatin in patients with non-small cell lung cancer (NSCLC) J Clin Oncol. 2010;28:539a. doi: 10.1200/JCO.2013.54.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gualberto A, Pollak M. Emerging role of insulin-like growth factor receptor inhibitors in oncology:early clinical trial results and future directions. Oncogene. 2009;28:3009–3021. doi: 10.1038/onc.2009.172. [DOI] [PubMed] [Google Scholar]
- 38.Frystyk J. Utility of free IGF-1 measurements. Pituitary. 2007;10:181–187. doi: 10.1007/s11102-007-0025-y. [DOI] [PubMed] [Google Scholar]
- 39.Merrick DT, Dziadziuszko R, Szostakiewicz B, et al. High insulin-like growth factor 1 receptor (IGF1R) expression is associated with poor survival in surgically treated non-small cell lung cancer (NSCLC) patients (pts) J Clin Oncol. 2007;25:18 s. Abstr 7550. [Google Scholar]
- 40.Lee YC, Jeon HJ, Kim JH, et al. Clinical significance of insulin-like growth factor-1 receptor expression in stage I non-small-cell lung cancer: immunohistochemical analysis. Korean J Intern Med. 2008;23:116–120. doi: 10.3904/kjim.2008.23.3.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bianconi F, Baldelli E, Ludovini V, Crinò L, Flacco A, Valigi P. Computational model of EGFR and IGF1R pathways in lung cancer: a systems biology approach for translational oncology. Biotechnol Adv. 2012;30(1):142–153. doi: 10.1016/j.biotechadv.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Van den Berg HW, Claffie D, Boylan M, McKillen J, Lynch M, McKibben B. Expression of receptors for epidermal growth factor and insulin-like growth factor I by ZR-75-1 human breast cancer cell variants is inversely related: the effect of steroid hormones on insulin-like growth factor I receptor expression. Br J Cancer. 1996;73:477–481. doi: 10.1038/bjc.1996.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cunningham MP, Essapen S, Thomas H, et al. Coexpression of the IGF-IR, EGFR and HER-2 is common in colorectal cancer patients. Int J Oncol. 2006;28:329–335. [PubMed] [Google Scholar]
- 44.Nguyen KS, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer. 2009;10:281–289. doi: 10.3816/CLC.2009.n.039. [DOI] [PMC free article] [PubMed] [Google Scholar]


